Abstract
Initiation of reverse transcription of genomic RNA is a key early step in replication of the human immunodeficiency virus (HIV) upon infection of a host cell. Viral reverse transcriptase (RT) initiates from a specific RNA-RNA complex formed between a host transfer RNA (tRNALys3) and region at the 5'end of genomic RNA; the 3'end of the tRNA acts as a primer for reverse transcription of genomic RNA. We report here the secondary structure of the HIV genomic RNA-human tRNALys3 initiation complex using heteronuclear nuclear magnetic resonance methods. We show that both RNAs undergo large-scale conformational changes upon complex formation. Formation of the 18 base pair primer helix with the 3' end of tRNALys3 drives large conformational rearrangements of the tRNA at the 5' end, while maintaining the anticodon loop for potential loop-loop interactions. HIV RNA forms an intramolecular helix adjacent to the intermolecular primer helix. This helix, which must be broken by reverse transcription, likely acts as a kinetic block to reverse transcription.
Keywords: HIV, reverse transcriptase, initiation complex, RNA, NMR
Introduction
RNA structure guides key functions in HIV infection and replication. The viral genome is RNA, and RNA-RNA and RNA-protein interactions pervade the viral life cycle. The first step in viral replication after entry into an infected cell is reverse transcription, which is catalyzed by a viral enzyme, reverse transcriptase (RT)1. RT must initiate from a specific RNA assembly and then navigate the complex secondary structure of genomic RNA while copying RNA to DNA2. RT is a major target of therapeutic intervention in treatment of Acquired Immunodeficiency (AIDS).
HIV RT is a virally encoded RNA/DNA directed DNA polymerase, which consists of two subunits, p66 and p51. RT has both polymerase activity, as well as an RNase H domain, which cleaves RNA-DNA hybrids. The general mechanism of reverse transcription has been determined3. RT initiates reverse transcription from a specific RNA complex formed between a host tRNALys34; 5 and a specific region (primer binding site or PBS) of the HIV genome near the 5'-end of HIV RNA6; 7; 8 (shown schematically in Fig. 1a). This initiation complex is preassembled in the virion, and initiation occurs by addition of deoxynucleotides to the free 3'OH of the tRNALy33 (nts)9; 10. RT copies the 5'-end of viral RNA into DNA, using its RNAase H activity to digest the RNA template. The DNA product is complementary to the 3' long terminal repeat (LTR) region of genomic RNA; interaction of the two strands allows reverse transcription to continue through the viral RNA 3' to 5'. Short stretches of genomic RNA remain that are resistant to RNase H cleavage, which then prime synthesis by RT of positive-strand DNA. Complementary regions of the single-stranded DNA hybridize and lead to eventual synthesis of fully double-stranded DNA that is competent for integration into the host genome.
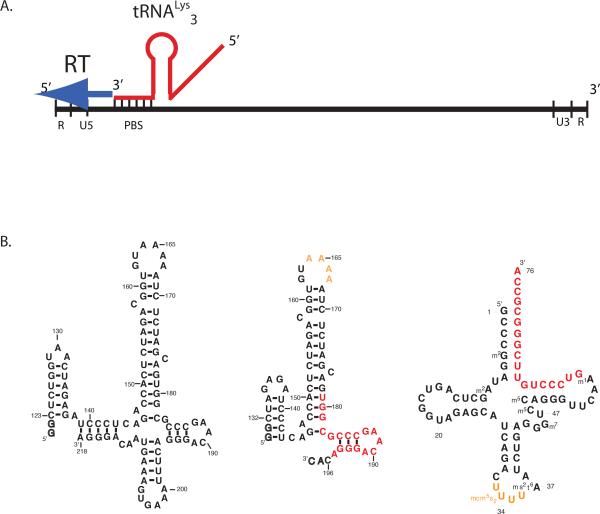
Figure 1.
(A.) Schematic of the reverse transcription initiation complex within the HIV genome. The RNA genome of HIV RNA has the standard structure for retroviral genomes, with long terminal repeat regions (U5, U3, R), and the primer binding site (PBS) near the 5'end. Host tRNALys3 interacts with the PBS through an 18 base-pair interaction, providing the 3'-OH group to initiate reverse transcription by viral reverse transcriptase (RT).
(B.) RNA oligonucleotides corresponding to HIV-1 genomic RNAs used in the current study (left) a 99 nt and (right) 69 nt RNA with numbering according to the Mal isolate. Additional nucleotides added at the 5'end for transcription with T7 RNA polymerase are indicated as outlines. The primer binding site region of 18bp of complementarity with tRNA is highlighted in red, whereas the A-rich loop is in yellow. (c) Secondary structure of human tRNALys3 with modified nucleotides; Coloring as in (b).
Initiation of reverse transcription in HIV is directed from a specific RNA-RNA complex formed between host tRNALys3 and HIV genomic RNA11. The PBS forms an 18 bp duplex with the 3' end of tRNALys3 but sequences outside this region of tRNA-HIV RNA complementarity are also critical for viral replication. Attempts to force HIV to use other tRNAs by making the PBS sequence complementary to the 3'-most 18 nts of the tRNA lead to rapid reversion of the virus back to using tRNALys3 by mutation of the PBS12; 13; 14; 15; 16. Thus, sequences outside the simple PBS-tRNA base-pairing region are required for efficient initiation. Chemical probing of the viral RNA focused on a ca. 100 nt region whose reactivity to chemical probes changed upon binding of the viral RNA6; 7. Mutations within this region can change both the efficiency of tRNA-viral RNA complex formation and the rate of initiation of reverse transcription in vitro8; 17 and in vivo18; 19. Different viral isolates have sequence changes in this region20; 21; 22.
Biochemical experiments have defined the general RNA-RNA interactions that direct formation of the initiation complex. The RNA-RNA complex that forms between host tRNALys3 and HIV PBS is >40 kDa in size, and global features of its secondary structure have been determined by chemical and enzymatic probing7; 8; 23. Modified nucleotides in tRNALys3 facilitate proper initiation6, and a specific loop-loop interaction between an A-rich loop region of the viral RNA and the U-rich anticodon of tRNALys3 has been proposed to be critical for initiation complex formation and activity, with a modified 2-thio-uridine at position 34 in the anticodon stabilizing this interaction6; 24; 25. A secondary structure for the tRNA-HIV RNA complex has been proposed, but no structural data beyond the experiments discussed above are available.
NMR spectroscopy allows determination of RNA secondary and tertiary structures26; 27; 28; 29. Base-pair formation can be monitored through the exchangeable imino proton resonances; these protons are protected from solvent exchange upon base-pair formation, and appear in a distinct 1H frequency in an NMR spectrum. Isotopic labeling of RNAs with 13C and 15N facilitate assignment and analysis of RNA NMR spectra26; 27; 30. High-resolution structures of RNAs up to 100–120nts have been painstakingly achieved by using a combination of distance constraints and residual dipolar couplings obtained through heteronuclear NMR31; 32; 33; 34. Large RNA complexes suffer from line broadening caused by slow molecular tumbling, although TROSY methods greatly improve linewidths for RNA base 1H resonances27. Secondary structure and overall folds for large RNAs (>25 kDa) can be achieved by analysis of experiments on imino 1H resonances35; 36.
Here we use heteronuclear NMR methods to probe the structure of the tRNALys3-HIV RNA complex. Biochemical analyses, coupled with prior experiments, defined the region of HIV RNA required for interaction with human tRNALys3. We show that that large scale conformational changes occur in both the HIV genomic RNA and host tRNALys3 upon formation of a binary RNA complex; the expected 18 base pair duplex that forms the central feature of the PBS is observed, as is the predicted downstream duplex within HIV genomic RNA. These results support prior models of the initiation complex and are a starting point for further mechanistic and structural studies of HIV reverse transcription.
Results
We first defined a system to probe structurally the initiation complex in vitro. Prior groundbreaking work by Marquet and co-workers defined the essential features of the HIV RNA-RNA initiation complex using biochemical and chemical probing (discussed above). Their results defined a 300 nt fragment of the HIV-1 RNA genome, spanning the PBS region that was required for complex formation6; 7; 8; 23; 37. These experiments more specifically defined a ca. 100 nt region (120–220 in the Mal isolate) that formed a specific complex with modified native tRNALys3. This was our starting point for biophysical experiments. We initially explored the conformation of a 99nt HIV fragment of PBS (Fig. 1b), spanning this region, by NMR. Size-exclusion chromatography demonstrated a monomeric conformation for this RNA. However, the imino proton NMR spectrum (not shown) of this RNA was broad and showed evidence for multiple conformers, and no evidence for the formation of the A-rich loop, which has distinct chemical shifts determined by our prior structure determination38. We concluded that the 99mer RNA has a misfolded conformation in solution and in addition undergoes hydrolysis at a UpA step between positions 199 and 200. We thus abandoned this construct for further biophysical study.
The segment of HIV viral RNA between nts 130 to 200 directs tRNALys3 hybridization. A 69mer RNA (Fig. 1b) that corresponds to this region did not hydrolyze under standard conditions, and NMR spectroscopy suggested a single conformation for this RNA. We prepared unlabeled and 13C, 15N-labeled versions of the 69mer, and confirmed a general secondary structure as proposed from chemical probing experiments. The NMR data showed that the A-rich loop, whose structure was previously determined by NMR on the isolated domain oligonucleotide38, was formed in this construct.
Likewise, we have examined previously the secondary and tertiary structure of human tRNALys3 (Fig. 1b) using NMR spectroscopy35. Using 15N, 13C labeled tRNA transcripts (described in Materials & Methods), we showed that the correct tRNA secondary and tertiary, L-shaped fold is formed in the presence of 5–10mM MgCl2. We assigned the imino 1H resonances for the four helical stems of the tRNA, confirming their formation. These data also confirmed the formation of the key base triple interactions: A9--U12-A23 and C13-G22-G46 as well as the U8-A14 tertiary pair. Thus, modified nucleotides are not required for correct folding of this tRNA.
To observe whether the HIV RNA constructs could form a specific initiation complex with tRNALys3, we performed chemical probing footprinting experiments. Natural, fully modified tRNALys3 was purified from bovine liver as described previously39, and was used as a control in comparison to transcribed tRNALys3. RNAs were hybridized at 1:1 stoichiometry at 90°C for 3 minutes in low salt (10mM Na phosphate or cacodylate, pH 7), followed by cooling to 60°C, addition of 100mM NaCl, and slow cooling to room temperature. MgCl2 was only added at room temperature, to avoid hydrolysis of the RNAs. Hybridization of tRNA and HIV RNA was readily confirmed by native gel electrophoresis.
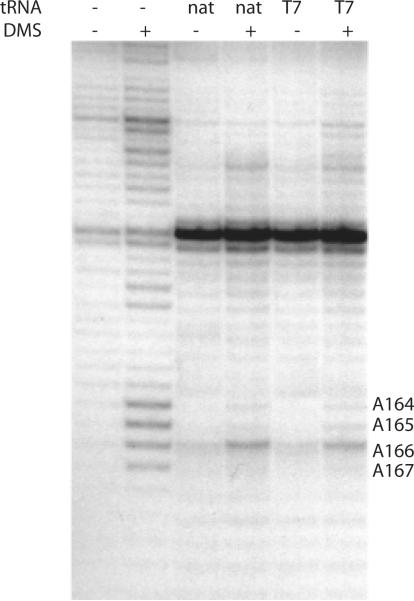
RNA structure in the free and complexed HIV 69mer was probed using chemical reactivity with dimethyl sulfate, which reacts with the N1 position of A and N3 position of C40. In the absence of tRNA, the 69nt RNA shows chemical reactivity within the A-rich loop region between A164–167 (Fig. 2). Upon addition of 1 molar equivalent of either natural tRNALys3 or transcribed tRNALys3, similar protections are observed in the A-rich loop region, indicating formation of RNA structure in this region. These results confirmed that tRNA transcripts or native tRNAs form similar complexes with the 69nt HIV genomic RNA construct. We thus used the transcribed tRNA, which is readily labeled with 15N and 13C, for subsequent NMR experiments on the RNA-RNA complex.
Figure 2. Chemical probing experiments on HIV initiation complexes with 69nt model HIV1 genomic RNA.
Accessibility of adenosine N1 and cytosine N3 positions to reaction with dimethyl sulfate (DMS) was detected in the presence or absence of 1:1 stoichiometry of either pure bovine native tRNALys3 (nat) or T7 RNA polymerase transcript (T7). Reactivity with DMS was detected using primer extension with reverse transcription using a DNA primer in the absence (−) and presence (+) of dimethyl sulfate reaction. Reactions were performed at room temperature as described.
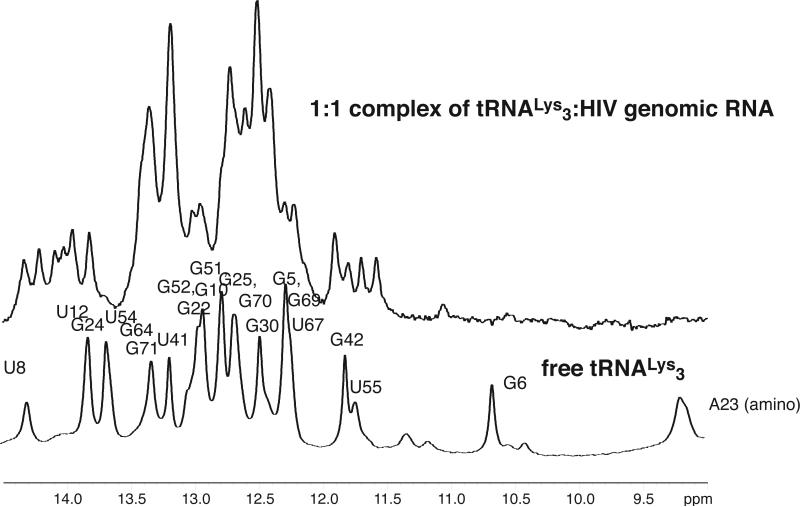
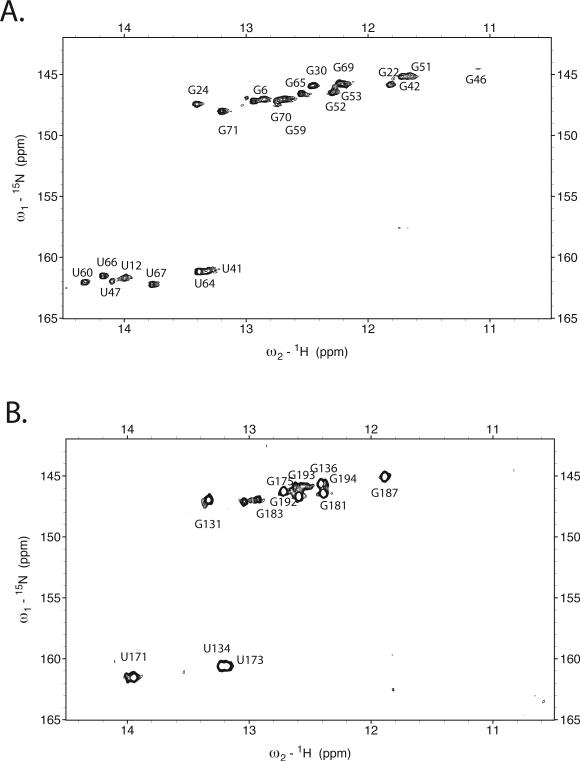
Despite the large molecule weight of the 1:1 tRNALys3-HIV69 RNA complex (MW 47,000), the imino 1H NMR spectrum was well resolved and interpretable. Large changes in the imino spectrum of tRNA (and HIV 69 mer) is observed upon 1:1 complex formation (Fig. 3), indicating large scale structural rearrangements in the tRNA. Strikingly, resonances that originate from the tRNA acceptor stem (e.g. G6), D stem, and tRNA tertiary structure (amino resonances, U8, U55)35 are lost or shifted in the RNA-RNA complex. A large number of new resonances that originate from imino protons within the tRNA are observed. These qualitative observations are suggestive of large changes in the tRNA structure upon complex formation.
Figure 3. Imino 1H NMR spectra of folded human tRNA Lys3 transcript (bottom) compared to that of 1:1 complex of human tRNALys3 with 69nt HIV1 genomic RNA (top).
Spectra were acquired at 800 MHz in 10mM MgCl2, 100mM NaCl, 10mM Na phosphate, pH 6.5 at 25°C. Assignments of resonances in folded tRNALys3 were reported previously.
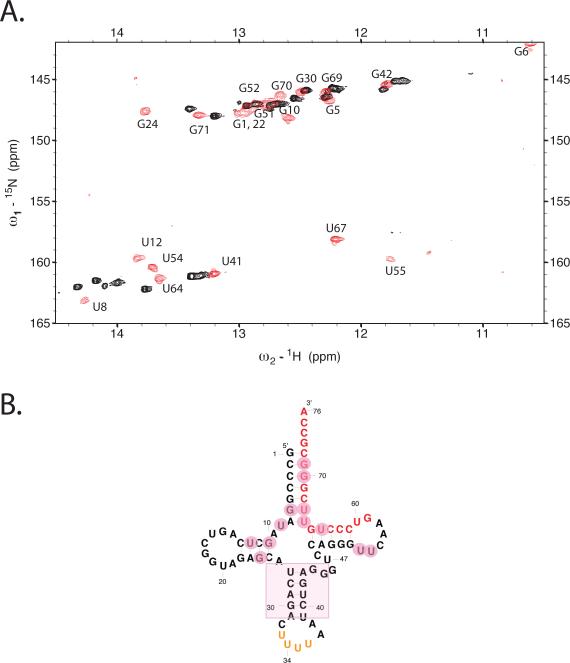
To confirm this observation and focus on changes in only tRNALys3, we prepared 100% 13C, 15N-labeled tRNALys3 and formed a 1:1 complex with unlabeled HIV genomic RNA (69mer). We have previously assigned the exchangeable 1H spectrum for folded tRNALys335. Within the 1:1 RNA-RNA complex, drastic changes in the spectrum for tRNALys3 are observed (Fig. 4a). The D-stem, with its set of tertiary interactions involving G10-C25, C11-G24, U12-A23, and U8, undergoes large chemical shift changes in the complex. Likewise, shifts are observed in the T- and acceptor stem resonances G6-U67, C2-G71, U55 in the T loop (Fig. 4b). These changes are consistent with the proposed hybridization of 18 nts at the 3'end of tRNALys3 (located in the T- and acceptor stems) with the complementary PBS sequence in HIV RNA. Only minor shifts were observed in the anticodon stem loop resonances. The results suggest changes in the D, T and acceptor stems of the tRNA upon binding to HIV genomic RNA.
Figure 4. Changes in tRNALys3 conformation upon initiation complex formation.
(A.) 15N-1H TROSY spectrum of uniformly 13C, 15N labeled human tRNALys3 either alone (red) or in 1:1 complex with unlabeled HIV-1 69nt RNA (black). The comparison of the two spectra highlights the large changes in tRNA structure that occurs upon complex formation. (B.) Secondary structure of human tRNALys3; major perturbations (pink circles) of the folded tRNA spectrum upon complex formation with HIV RNA occur in the acceptor, T and D-stems, whereas resonances for the anticodon stem are relatively unperturbed. The spectra were acquired at 800 MHz in 10mM MgCl2 50mM NaCl, 10mM Na phosphate, pH 6.5 at 25°C using a T1-relaxation optimized TROSY pulse sequence as described in the text for improved signal-to-noise.
To assign in detail the secondary structure of the tRNA-HIV complex, the following general strategy was taken. First, imino proton resonances were assigned to either tRNA or HIV RNA by preparing two individual samples with one component uniformly labeled with 13C, 15N nucleotides, and the partner unlabeled (Fig. 5a, b). An unlabeled complex sample was prepared for homonuclear NOESY experiments to map imino-imino NOEs and allow assignment of helical elements. Finally, intramolecular (tRNA-tRNA or HIV-HIV) helical elements were assigned using HNN COSY approaches. We explore each step in detail below.
Figure 5. 1H-15N TROSY experiments for the HIV initiation complex at 1:1 tRNALys3-HIV RNA stoichiometry.
(A.) Spectrum for initiation complex formed with uniformly labeled 13C, 15N-tRNALys3 and unlabeled HIV-1 69nt RNA. (B.) Spectrum for initiation complex formed with uniformly labeled 13C, 15N-HIV-1 69nt RNA and unlabeled tRNALys3. Spectra were acquired at 800 MHz 1H frequency T1-relaxation optimized TROSY at 25°C. Spectral assignments of the imino resonances as discussed in the text are indicated.
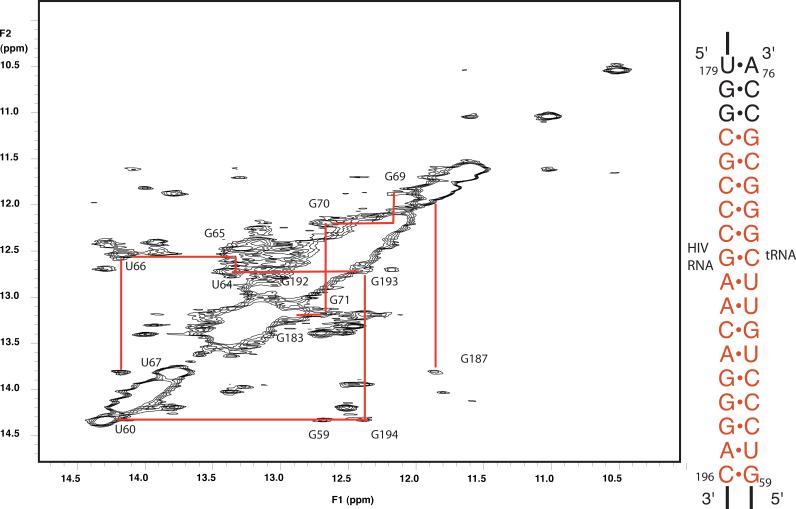
Based on the changes in the tRNA data, we assumed that the 18bp PBS helix was formed. We first assigned the imino 1H resonances for base pairs in the 18 bp PBS helix. We performed homonuclear NOESYs at several mixing times (50, 100, 200ms) to observe imino-imino 1H NOES characteristic of A-form RNA helices. The NOESY experiments confirm that 18 bp helix is indeed formed in our tRNA-HIV complex (Fig. 6). Connectivity of imino resonances in the NOESY was observed for 16 continuous base pairs from CH196-GT59 until GH181-CT74. These assignments were confirmed by synthesis of an oligonucleotide containing the 18 base pair helix in isolation, capped at one end by a UUCG tetraloop (Supplementary Fig. 1). Assignment of the base pairing imino 1H spectrum of this RNA agreed with those of the same helix in the HIV-tRNA complex. This further suggests that the PBS helix forms without extensive tertiary contacts to the rest of the RNA in the binary complex.
Figure 6. NOESY spectrum of unlabeled 1:1 complex of human tRNALys3 and HIV1 69nt RNA.
The region of imino 1H-1H NOES is shown; data were acquired at 800 MHz with 100ms mixing time at 25°C. NOE connectivities are observed that allow assignment of 16 of the 18 base pairs formed between the tRNA and HIV RNA.
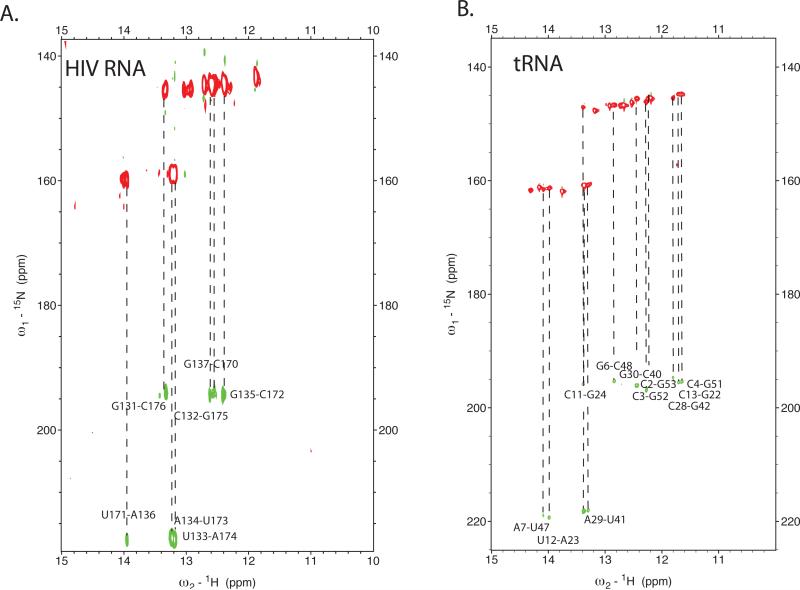
To assign remaining features of the complex structure, we used the heteronuclear NMR experiments27; 41 on complexes with one component of the complex isotopically labeled and the other unlabeled (see Materials and Methods). First, assignment of the 18 bp helix was confirmed by the sorting of imino protons from either tRNA or HIV RNA, as determined by the 15N-TROSY experiments on complexes with either uniformly 15N-labeled tRNA or HIV RNA. Intramolecular base pairing within an individual RNA component was identified by direct measurement of 15N-15N scalar couplings across base pairs using HNN-COSY experiments42.
We next confirmed formation of an intramolecular RNA-RNA helix within HIV RNA in the initiation complex. The helical connectivity extends from base pairs C132-G175 until U140-A168. The existence of this helix is supported by imino 1H NOESY crosspeaks, and the observed correlation peaks in the HNN NOESY experiment using an initiation complex formed with 13C,15N-labeled HIV RNA and unlabeled tRNALys3 (Fig. 7a). Several imino resonances arise from the HIV RNA that we have not assigned, indicating possible additional pairing within the large hairpin loop formed by the HIV RNA.
Figure 7. H-NN COSY experiments showing through-space 15N-15N scalarcouplings across base-pairing hydrogen bonds.
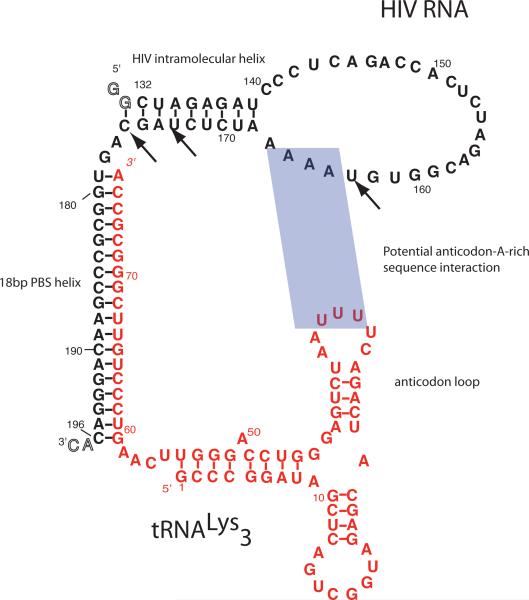
(A.) H-NN COSY spectrum of the initiation complex formed with uniformly labeled 13C, 15N-tRNALys3 and unlabeled HIV-1 69nt RNA. (B.) H-NN COSY spectrum for initiation complex formed with uniformly labeled 13C, 15N-HIV-1 69nt RNA and unlabeled tRNALys3. Both experiments only show correlations for intramolecular base pairing for the 15N-labeled component of the complex. Data were acquired at 25°C at 800 MHz using a T1-relaxation optimized (BEST-HNN COSY) version that uses band selective pulses centered on the imino proton resonances as described in the materials and methods. (C). Secondary Structure of the HIV initiation complex determined here by NMR spectroscopy. HIV 1 genomic RNA (Mal-1 isolate) is in black, tRNALys3 in red. The key regions of the structure, including 18 bp PBS helix, HIV genomic RNA intramolecular helix, and the conservation of the tRNA anticodon stem-loop are highlighted. Potential U-rich anticodon loop interaction with A-rich sequence in the HIV RNA is shaded. Arrows indicate pausing sites observed in kinetic investigations of the HIV-1 initiation complex with reverse transcriptase. These strong pauses are at positions +3, +6 and +16.
We determined rearrangements in the tRNALys3 secondary structure within the initiation complex. tRNALys3 was uniformly 13C, 13N labeled and HIV RNA was unlabeled. HNN COSY experiments confirmed the presence of extensive intratRNA base pairing within the initiation complex (Fig. 7b). As shown above, the 3' portion of the acceptor stems and T-stems have rearranged to form the 18 bp helix. This explains the large changes in chemical shift observed for imino resonances for nucleotides G59-A76. In addition, significant changes are also observed for resonances from nucleotides in the D stem and 5' side of the acceptor stem; this suggests rearrangements driven by loss of secondary and tertiary structure in the tRNA upon binding. Interestingly, resonances from the anticodon stem did not undergo significant changes upon complex formation, suggesting that this structural feature, which is independent of the surrounding secondary and tertiary structure, is maintained in the initiation complex. This observation is confirmed by the more detailed analysis presented below.
The secondary structure of the tRNALys3 in the initiation complex was confirmed by NOESY experiments. The helices of the anticodon stem (U27-A43 to G30-C40) and D-stem (G10-C25 to C13-G22) are formed in the complex; the chemical shifts of the anticodon stem residues are similar to those in the tRNA, whereas significant shifts are observed in D-stem resonances, reflecting the disruption of tertiary interactions in free tRNALys3 upon initiation complex formation. The 5'-end of the acceptor stem now pairs with a region formerly in the T-loop. A helix interrupted by a single-nucleotide bulge is formed from nucleotides G1 to A9 pairing with nucleotides G45 to U54. This pairing is confirmed by both HNN-COSY crosspeaks and imino-imino proton NOEs.
Discussion
The NMR results presented here allowed definition of the secondary structure for the initiation complex (Fig. 7c). We first defined a system for NMR using biochemical approaches, building on the pioneering studies of Marquet and co-workers7; 39. We focused on the PBS region of HIV-1-RNA, initially defined as a 300 nt region of the HIV genome. The initiation complex between HIV RNA and tRNALys3 was formed biochemically by hybridization of the two RNAs using heat annealing. Longer constructs (>90nt) of HIV RNA gave poor biophysical behavior on their own, likely due to multiple RNA conformations and potential aggregation; efficient complex formation with these longer RNAs requires fully modified natural tRNALys36. However, a 69 nt fragment of the HIV 1 Mal isolate primer binding site is well-behaved biophysically and allows efficient hybridization of either naturally modified or unmodified transcript human tRNALys3. Correct complex formation, including interaction of the HIV A-rich loop, was observed with 1:1 stoichiometry of 69nt HIV RNA and unmodified tRNALys3 using chemical probing. These experiments defined a tractable, biochemically relevant system for subsequent NMR studies of the initiation complex.
The 50 kDa tRNALys3-HIV RNA complex was a challenging target for NMR study. Here, we used both homonuclear and heteronuclear NMR to define the secondary structure of the initiation complex (Fig. 7c). Imino proton resonances were assigned and intermolecular hydrogen bonding and NOEs used to define base pairing interactions. NMR spectra were simplified and imino 1H resonances were assigned to either the HIV RNA or tRNA component of the complex through selective 15N, 13C- labeling of either the HIV RNA or tRNA. Imino resonances were then assigned using a combination of sorting of imino proton resonances to the two component, HNN COSY data to define base pairing and imino-imino 1H NOEs to define patterns of secondary structure. Where possible, assignments were confirmed by synthesis and NMR analysis of shorter oligonucleotides corresponding to domains of the initiation complex. A total of 38 of 42 observed imino proton resonances were assigned using this approach.
The secondary structure of the initiation complex determined by NMR shows multiple features that may modulate its function (Fig. 7c). As expected, nucleotides 179–196 of HIV genomic RNA pairs with nts 69–76 at the 3'end of tRNA. This stable intermolecular 18 base pair primer helix must bind within the active site cleft of reverse transcriptase, positioning HIV nucleotide G178 as the template for the first round of DNA synthesis. Downstream from the 18bp primer helix, there is a 3bp stretch of single stranded nucleotides. Then, HIV RNA forms an intramolecular helix as proposed and demonstrated by Marquet et al.7 with base pairs from nts C132-G175 to G137-C170. We could not assign any additional significant secondary structure for the HIV RNA after this region.
tRNALys3 undergoes significant remodeling of its secondary structure upon initiation complex formation. The 3'end of tRNA forms the 18 bp primer helix, which deprives the 5'end of the acceptor stems and T-stems of their pairing partners. These regions refold to pair with each other in the initiation complex; nts G1 to A9 form an imperfect duplex with nucleotide 45–54. The anticodon and D-stems remained base paired as in the tRNA secondary structure, although chemical shift and NOE data support that tertiary interactions that occur in folded tRNA within the D-stem do not occur in the initiation complex.
The secondary structure that we observe by NMR confirms prior predictions made by a variety of biochemical and modeling experiments23. The 18 base pair helix between the tRNA and HIV is a central feature of the secondary structure, as demonstrated by many biochemical and viral replication experiments. Formation of this thermodynamically stable intermolecular helix drives structural rearrangements in both the HIV genomic RNA and tRNALys3. These conformational rearrangements in vivo are likely facilitated by the nucleocapsid protein and other chaperones43; 44.
The tRNA rearrangement that occurs upon formation of the initiation complex further explains the choice of tRNALys3 as a primer. The stable pairing between the T and acceptor stems leads to a stable refolding of the tRNA sequence upon initiation complex formation. The refolded tRNA structure maintains the U-rich anticodon stem loop to make potential contacts with the A-rich loop within the HIV RNA. We do not observe imino proton resonances consistent with anticodon-A-loop interaction, but chemical probing data, coupled with recent NMR experiments by Agris and co-workers25, suggests that such a pairing occurs; the absence of modified nucleotides in our tRNA construct likely increases the solvent exchange rate for these potential base pairs. The A-loop region in the complex structure is presented as single-stranded nucleotides available for additional pairing. The stability of this loop-loop interaction is likely such that the imino resonances from the anticodon loop are broadened by solvent exchange.
Upon initiation complex formation, the HIV genomic RNA rearranges to form a large hairpin loop structure with an intramolecular helix starting just downstream from the 18bp PBS helix. This secondary structure element, first proposed by Marquet and coworkers7, represents a block to reverse transcriptase20; 45, which must melt this helix during the early rounds of transcription. This helix is site of two reverse transcription pauses during minus strand strong stop DNA synthesis at positions +3 and +620. (see Fig. 7c); rates of reverse transcription are 3400-fold slower for the first 15 nts compared to subsequent nucleotides, with limited pausing also after the potential A-rich sequence-anticodon loop interactions. The RNA interactions observed in our structure present a plausible explanation for the slow elongation rates during the early phase of reverse transcription, and may explain the limited extension observed in the preformed initiation complex within the HIV virion.
The initiation complex must be recognized by HIV reverse transcriptase. The 18bp helix would fit within the cleft of HIV RT, where other nucleic acid helical structures (DNA-DNA and DNA-RNA duplexes) have been observed to bind1; 46. This would place the downstream HIV helix adjacent to the active site. How HIV RT recognizes the initiation complex and perhaps reorganizes the tRNA-HIV RNA complex structure observed here awaits more detailed structural investigation of the initiation complex.
Recent single-molecule fluorescence experiments on HIV RT bound to the initiation complex have underscored the role of RNA structure in modulating protein-RNA dynamics45. Using fluorescence resonance energy transfer (FRET), Legrice, Zhuang and co-workers found dynamic switching between correct, active initiation complex orientation within the binding cleft of RT, and an inactive orientation in which the +1 position is flipped 180° and located near the RNAase H domain. The presence of the intramolecular HIV helix that we observe here by NMR stabilizes RT in the incorrect orientation, explaining the poor kinetics of elongation through this helix; once RT traverses the +6 position, the active orientation of RT is greatly favored.
The results presented here represent a first level of understanding of the initiation complex. Our data highlight the power of modern NMR experiments to map global RNA folds, and demonstrate how RNA structure may control the activity of reverse transcription. More detailed structural data on the RNA initiation complex using NMR and crystallography are required to provide a tertiary structure of both the RNA-RNA complex and its interaction with RT.
Materials and Methods
RNA synthesis
RNA samples were prepared by in vitro transcription using T7 polymerase as described47; 48. HIV RNA was purified using Superdex 200 (26/60) gel filtration as described48; 49. Natural tRNALys3 was purified from 10 kg of bovine livers using modifications of published protocols6; 39. Unmodified human tRNALys3 was prepared by in vitro transcription and purified using preparative polyacrylamide gel electrophoresis as described previously50. Uniformly 13C, 5N-labeled RNAs were transcribed using 13C, 15N-labeled ribonucleoside triphosphates prepared in house51; 52. All RNA samples were concentrated by Vivaspin (cutoff 5,000Da) and kept in 10 mM phosphate buffer, pH 6.5. Molar extinction coefficients were calculated from the A260 values at 20□C by extrapolation from the values at 90□C. All RNAs were characterized for the oligomerization state using analytical gel filtration chromatography and native gel electrophoresis to confirm monomeric species, or bimolecular RNA-RNA complexes.
Chemical Probing
Chemical probing was performed using dimethyl sulfate reaction53 with 20 nM RNA complexes at 25°C in 100mM NaCl, 10mM MgCl2 10mM Na phosphate pH 6.5; the 1:1 binary complexes of HIV 69 nt RNA with either native bovine tRNALys3 or transcribed human tRNALys3 were incubated with a 1:600 dilution of DMS for 7 minutes at 25°C and quenched by the addition of 4 volumes of 94% EtOH, 63 mM βME, 94 mM NaOAc (pH 5.5), 54 mM Tris-HOAc (pH 7.5), 30 μg/mL carrier tRNA and the reactions were precipitated by spinning at 16,000 × g for 15 minutes, washed with 70% EtOH and re-suspended in water. A 18nt DNA primer was hybridized to the 5'-end of HIV genomic RNA complementary to the PBS. The DNA primers are diluted to 1 μM in 225 mM HEPES-KOH (pH 7.0) and 450 mM KCl and added to an equal volume of resuspended RNA, mixed and then heated to 90°C for 1 min and then slow cooled to 42°C. The primed RNA was then incubated with AMV-RT in 130 mM Tris-HCl (pH 8.5), 10 mM MgCl2, 10 mM DTT, 6 μCi α-32P-dTTP, 110 μM dATP, dCTP, dGTP, and 6 μM dTTP at 50°C for 30 minutes. The reaction was stopped by the addition of 1/10th volume of 3 M NaOAc (pH 5.2) and then phenol:chloroform extracted before being EtOH precipitated. The cDNA pellet was then suspended in a small volume (10 μL) of denaturing urea loading buffer, heated to 90°C for 5 minutes and immediately loaded onto a pre-run 15% denaturing polyacrylamide sequencing gel. Subsequent to electrophoresis, the gel was transferred to Whatman filter paper, dried and exposed to film or phosphorimager to detect the radioactive bands.
NMR Spectroscopy
All NMR spectra were collected at 25°C on a Varian INOVA 800 NMR spectrometer equipped with triple resonance x,y,z-axis gradient probes within the Stanford Magnetic Resonance Laboratory (SMRL). 1H and 15N assignments were obtained using standard homonuclear and heteronuclear methods optimized for RNA structure determination (RNAPack, http://www.varianinc.com)27; 41 following detailed published procedures, except where outlined below. NMR data were processed with VNMR (http://www.varianinc.com) and spectra were analyzed with SPARKY54.
Several RNA samples were prepared: unlabeled HIV1 99mer genomic RNA, unlabeled HIV1 69mer genomic RNA, unlabeled human tRNALys3, uniformly 13C, 15N-labeled 69mer HIV1 genomic RNA and uniformly 13C, 15N-labeled tRNALys3. 1:1 stoichiometric complexes of tRNALys3-69mer HIV genomic RNA samples were formed by heat annealing the two RNAs at 95°C for 3 min in 10mM Na Phosphate, pH 6.5, incubation at 60°C for 5 min with adjustment of the monovalent ion concentration to 100mM NaCl, and slow cooling to room temperature; where indicated, MgCl2 was added at room temperature to avoid hydrolysis. Samples were 0.5–0.8 mM for unlabeled RNAs and 0.25–0.4mM for labeled RNAs in 250μL in Shigemi NMR tubes.
To assign exchangeable resonances of individual RNAs and RNA-RNA complexes, 2D NOESY (30, 50, 75, 100 and 200 ms mixing times; 2.0 s delay time) were obtained using shifted laminar pulses for solvent suppression (SSNOESY)55. 1H-15N TROSY, 3D 1H −15N NOESY-HSQC, and 2D HNN-COSY were collected in 90% H2O/10% 2H2O at 298K or 303K. For the large (50 kDa) RNA-RNA complexes, T1-optimized spectroscopy was used to obtain improved signal-to-noise for 1H-15N TROSY and HNN COSY experiments56; these experiments use selective pulses centered on the imino proton region of the spectrum to enhance population differences with neighboring amino proton resonances, thus facilitating T1 relaxation through dipolar mechanisms. For these experiments, recycle delay was set to 0.5 s for optimal signal, and good-quality HNN-COSY spectra were obtained in 2–8 hrs.
Supplementary Material
The secondary structure and sequence of the oligonucleotide is shown; the first base pair was changed to allow transcription. The NOESY spectrum shows imino-imino 1H NOEs obtained using shifted laminar pulse solvent suppression and a mixing time of 200ms at 800 MHz and 25°C. Assignment of imino-imino NOES is shown, and the region assigned is highlighted in red in the oligonucleotide. The numbering uses that of the equivalent positions in the tRNA and HIV RNA (see Fig. 6 & 7c).
Acknowledgements
The authors thank Dr. Insil Kim, Joren Retel and Dr. Corey Liu for assistance in sample preparation and NMR spectroscopy, and Dr. Marat Yusupov and Dr. Gulnara Yusupova for advice. Supported by GM69314. The Stanford Magnetic Resonance Laboratory is partially supported by the Stanford University School of Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J Mol Biol. 2009;385:693–713. doi: 10.1016/j.jmb.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barraud P, Gaudin C, Dardel F, Tisne C. New insights into the formation of HIV-1 reverse transcription initiation complex. Biochimie. 2007 doi: 10.1016/j.biochi.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 3.Hu W-S, Temin HM. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 4.Mak J, Kleiman L. Primer tRNAs for reverse transcription. J Virol. 1997;71:8087–95. doi: 10.1128/jvi.71.11.8087-8095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu M, Wainberg MA. Interactions between human immunodeficiency virus type 1 reverse transcriptase, tRNA primer, and nucleocapsid protein during reverse transcription. J Hum Virol. 2000;3:16–26. [PubMed] [Google Scholar]
- 6.Isel C, Marquet R, Keith G, Ehresmann C, Ehresmann B. Modified nucleotides of tRNALys3 modulate primer/template loop-loop interaction in the initiation complex of HIV-1 reverse transcription. J. Biol. Chem. 1993;268:25269–25272. [PubMed] [Google Scholar]
- 7.Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R. Initiation of reverse transcription of HIV-1: Secondary structure of the HIV-1 RNA/tRNALys (template/primer) complex. J. Mol. Biol. 1995;247:236–250. doi: 10.1006/jmbi.1994.0136. [DOI] [PubMed] [Google Scholar]
- 8.Isel C, Keith G, Ehresmann B, Ehresmann C, Marquet R. Mutational analyses of the tRNA3Lys/HIV-1 RNA (primer/template) complex. Nucl.Acids. Res. 1998;26:1198–1204. doi: 10.1093/nar/26.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Zhang Y, Spicer T, Henrard D, Poiesz BJ. Nascent human immunodeficiency virus type 1 reverse transcription occurs within an enveloped particle. J Virol. 1995;69:3675–82. doi: 10.1128/jvi.69.6.3675-3682.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Zhang Y, Spicer TP, Abbott LZ, Abbott M, Poiesz BJ. Reverse transcription takes place within extracellular HIV-1 virions: potential biological significance. AIDS Res Hum Retroviruses. 1993;9:1287–96. doi: 10.1089/aid.1993.9.1287. [DOI] [PubMed] [Google Scholar]
- 11.Mougel M, Houzet L, Darlix JL. When is it time for reverse transcription to start and go? Retrovirology. 2009;6:24. doi: 10.1186/1742-4690-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wakefield JK, Rhim H, Morrow CD. Minimal sequence requirements of a functional human immunodeficiency virus type 1 primer binding site. J Virol. 1994;68:1605–14. doi: 10.1128/jvi.68.3.1605-1614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wakefield JK, Wolf AG, Morrow CD. Human immunodeficiency virus type 1 can use different tRNAs as primers for reverse transcription but selectively maintains a primer binding site complementary to tRNA(3Lys) J Virol. 1995;69:6021–9. doi: 10.1128/jvi.69.10.6021-6029.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang SM, Wakefield JK, Morrow CD. Mutations in both the U5 region and the primer-binding site influence the selection of the tRNA used for the initiation of HIV-1 reverse transcription. Virology. 1996;222:401–14. doi: 10.1006/viro.1996.0437. [DOI] [PubMed] [Google Scholar]
- 15.Wakefield JK, Kang SM, Morrow CD. Construction of a type 1 human immunodeficiency virus that maintains a primer binding site complementary to tRNA(His) J Virol. 1996;70:966–75. doi: 10.1128/jvi.70.2.966-975.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wakefield JK, Morrow CD. Mutations within the primer binding site of the human immunodeficiency virus type 1 define sequence requirements essential for reverse transcription. Virology. 1996;220:290–8. doi: 10.1006/viro.1996.0317. [DOI] [PubMed] [Google Scholar]
- 17.Isel C, Lanchy JM, Le Grice SF, Ehresmann C, Ehresmann B, Marquet R. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-transcriptional modifications of primer tRNA3Lys. Embo J. 1996;15:917–24. [PMC free article] [PubMed] [Google Scholar]
- 18.Liang C, Rong L, Gotte M, Li X, Quan Y, Kleiman L, Wainberg MA. Mechanistic studies of early pausing events during initiation of HIV-1 reverse transcription. J Biol Chem. 1998;273:21309–15. doi: 10.1074/jbc.273.33.21309. [DOI] [PubMed] [Google Scholar]
- 19.Liang C, Rong L, Gotte M, Li X, Quan Y, Kleiman L, Wainberg MA. Mechanistic studies of early pausing events during initiation of HIV-1 reverse transcription. Journal of Biological Chemistry. 1998;273:21309–15. doi: 10.1074/jbc.273.33.21309. [DOI] [PubMed] [Google Scholar]
- 20.Goldschmidt V, Rigourd M, Ehresmann C, Le Grice SF, Ehresmann B, Marquet R. Direct and indirect contributions of RNA secondary structure elements to the initiation of HIV-1 reverse transcription. J Biol Chem. 2002;277:43233–42. doi: 10.1074/jbc.M205295200. [DOI] [PubMed] [Google Scholar]
- 21.Goldschmidt V, Ehresmann C, Ehresmann B, Marquet R. Does the HIV-1 primer activation signal interact with tRNA3(Lys) during the initiation of reverse transcription? Nucleic Acids Res. 2003;31:850–9. doi: 10.1093/nar/gkg187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldschmidt V, Paillart JC, Rigourd M, Ehresmann B, Aubertin AM, Ehresmann C, Marquet R. Structural variability of the initiation complex of HIV-1 reverse transcription. J Biol Chem. 2004;279:35923–31. doi: 10.1074/jbc.M404473200. [DOI] [PubMed] [Google Scholar]
- 23.Isel C, Westhof E, Massire C, Le Grice SF, Ehresmann B, Ehresmann C, Marquet R. Structural basis for the specificity of the initiation of HIV-1 reverse transcription. Embo J. 1999;18:1038–48. doi: 10.1093/emboj/18.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bajji AC, Sundaram M, Myszka DG, Davis DR. An RNA complex of the HIV-1 A-loop and tRNA(Lys,3) is stabilized by nucleoside modifications. J Am Chem Soc. 2002;124:14302–3. doi: 10.1021/ja028015f. [DOI] [PubMed] [Google Scholar]
- 25.Bilbille Y, Vendeix FA, Guenther R, Malkiewicz A, Ariza X, Vilarrasa J, Agris PF. The structure of the human tRNALys3 anticodon bound to the HIV genome is stabilized by modified nucleosides and adjacent mismatch base pairs. Nucleic Acids Res. 2009;37:3342–53. doi: 10.1093/nar/gkp187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tzakos AG, Grace CR, Lukavsky PJ, Riek R. NMR techniques for very large proteins and RNAs in solution. Annu Rev Biophys Biomol Struct. 2006;35:319–42. doi: 10.1146/annurev.biophys.35.040405.102034. [DOI] [PubMed] [Google Scholar]
- 27.Lukavsky PJ, Puglisi JD. Structure determination of large biological RNAs. Methods Enzymol. 2005;394:399–416. doi: 10.1016/S0076-6879(05)94016-0. [DOI] [PubMed] [Google Scholar]
- 28.Davis JH, Tonelli M, Scott LG, Jaeger L, Williamson JR, Butcher SE. RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex. J Mol Biol. 2005;351:371–82. doi: 10.1016/j.jmb.2005.05.069. [DOI] [PubMed] [Google Scholar]
- 29.Dominguez C, Schubert M, Duss O, Ravindranathan S, Allain FH. Structure determination and dynamics of protein-RNA complexes by NMR spectroscopy. Prog Nucl Magn Reson Spectrosc. 2011;58:1–61. doi: 10.1016/j.pnmrs.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 30.Lu K, Miyazaki Y, Summers MF. Isotope labeling strategies for NMR studies of RNA. J Biomol NMR. 2010;46:113–25. doi: 10.1007/s10858-009-9375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nature Structural Biology. 2003;10:1033–8. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 32.D'Souza V, Summers MF. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature. 2004;431:586–90. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]
- 33.D'Souza V, Dey A, Habib D, Summers MF. NMR structure of the 101-nucleotide core encapsidation signal of the Moloney murine leukemia virus. J Mol Biol. 2004;337:427–42. doi: 10.1016/j.jmb.2004.01.037. [DOI] [PubMed] [Google Scholar]
- 34.Miyazaki Y, Irobalieva RN, Tolbert BS, Smalls-Mantey A, Iyalla K, Loeliger K, D'Souza V, Khant H, Schmid MF, Garcia EL, Telesnitsky A, Chiu W, Summers MF. Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography. J Mol Biol. 2010;404:751–72. doi: 10.1016/j.jmb.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puglisi EV, Puglisi JD. Probing the conformation of human tRNA(3)(Lys) in solution by NMR. FEBS Lett. 2007;581:5307–14. doi: 10.1016/j.febslet.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 36.Hart JM, Kennedy SD, Mathews DH, Turner DH. NMR-assisted prediction of RNA secondary structure: identification of a probable pseudoknot in the coding region of an R2 retrotransposon. J Am Chem Soc. 2008;130:10233–9. doi: 10.1021/ja8026696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baudin F, Marquet R, Isel C, Darlix JL, Ehresmann B, Ehresmann C. Functional sites in the 5' region of human immunodeficiency virus type 1 RNA form defined structural domains. J. Mol. Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 38.Puglisi EV, Puglisi JD. HIV-1 A-rich RNA loop mimics the tRNA anticodon structure. Nat Struct Biol. 1998;5:1033–6. doi: 10.1038/4141. [DOI] [PubMed] [Google Scholar]
- 39.Yusupova G, Lanchy JM, Yusupov M, Keith G, Le Grice SF, Ehresmann C, Ehresmann B, Marquet R. Primer selection by HIV-1 reverse transcriptase on RNA-tRNA(3Lys) and DNA-tRNA(3Lys) hybrids. J Mol Biol. 1996;261:315–21. doi: 10.1006/jmbi.1996.0463. [DOI] [PubMed] [Google Scholar]
- 40.Moazed D, Stern S, Noller HF. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- 41.Lukavsky PJ, Puglisi JD. RNAPack - An integrated NMR approach to RNA structure determination. Methods. 2001;25:316–332. doi: 10.1006/meth.2001.1244. [DOI] [PubMed] [Google Scholar]
- 42.Dingley AJ, Grzesiek S. Direct Observation of Hydrogen Bonds in Nucleic Acid Base Pairs by Internucleotide 2JNN Couplings. J. Am. Chem. Soc. 1998;120:8293–8297. [Google Scholar]
- 43.Zeng Y, Liu HW, Landes CF, Kim YJ, Ma X, Zhu Y, Musier-Forsyth K, Barbara PF. Probing nucleation, reverse annealing, and chaperone function along the reaction path of HIV-1 single-strand transfer. Proc Natl Acad Sci U S A. 2007;104:12651–6. doi: 10.1073/pnas.0700350104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 2010;7 doi: 10.4161/rna.7.6.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu S, Harada BT, Miller JT, Le Grice SF, Zhuang X. Initiation complex dynamics direct the transitions between distinct phases of early HIV reverse transcription. Nat Struct Mol Biol. 2010;17:1453–60. doi: 10.1038/nsmb.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–75. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 47.Lukavsky PJ, Puglisi JD. Large-scale preparation and purification of polyacrylamide-free RNA oligonucleotides. RNA. 2004;10:889–893. doi: 10.1261/rna.5264804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKenna SA, Kim I, Puglisi EV, Lindhout DA, Aitken CE, Marshall RA, Puglisi JD. Purification and characterization of transcribed RNAs using gel filtration chromatography. Nat Protoc. 2007;2:3270–7. doi: 10.1038/nprot.2007.480. [DOI] [PubMed] [Google Scholar]
- 49.Kim I, McKenna SA, Viani Puglisi E, Puglisi JD. Rapid purification of RNAs using fast performance liquid chromatography (FPLC) RNA. 2007;13:289–94. doi: 10.1261/rna.342607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Puglisi JD, Wyatt JR. Biochemical and NMR studies of RNA conformation with an emphasis on RNA pseudoknots. Methods Enzymol. 1995;261:323–50. doi: 10.1016/s0076-6879(95)61016-2. [DOI] [PubMed] [Google Scholar]
- 51.Batey RT, Inada M, Kujawinski E, Puglisi JD, Williamson JR. Preparation of isotopically labeled ribonucleotides for multidimensional NMR spectroscopy of RNA. Nucleic Acids Research. 1992;20:4515–23. doi: 10.1093/nar/20.17.4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Batey RT, Battiste JL, Williamson JR. Preparation of isotopically enriched RNAs for heteronuclear NMR. Methods Enzymology. 1995;261:300–323. doi: 10.1016/s0076-6879(95)61015-4. [DOI] [PubMed] [Google Scholar]
- 53.Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nature Structural Biology. 2000;7:1105–10. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 54.Goddard TG, Kneller DG. Sparky 3. University of California; San Francisco: 2007. [Google Scholar]
- 55.Smallcombe S. Solvent suppression with symmetrically-shifted pulses. J Am Chem Soc. 1993;115 [Google Scholar]
- 56.Farjon J, Boisbouvier J, Schanda P, Pardi A, Simorre JP, Brutscher B. Longitudinal-relaxation-enhanced NMR experiments for the study of nucleic acids in solution. J Am Chem Soc. 2009;131:8571–7. doi: 10.1021/ja901633y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The secondary structure and sequence of the oligonucleotide is shown; the first base pair was changed to allow transcription. The NOESY spectrum shows imino-imino 1H NOEs obtained using shifted laminar pulse solvent suppression and a mixing time of 200ms at 800 MHz and 25°C. Assignment of imino-imino NOES is shown, and the region assigned is highlighted in red in the oligonucleotide. The numbering uses that of the equivalent positions in the tRNA and HIV RNA (see Fig. 6 & 7c).