Abstract
Longevity is influenced by genetic and environmental factors. The brain's dopamine system may be particularly relevant, since it modulates traits (e.g., sensitivity to reward, incentive motivation, sustained effort) that impact behavioral responses to the environment. In particular, the dopamine D4 receptor (DRD4) has been shown to moderate the impact of environments on behavior and health. We tested the hypothesis that the DRD4 gene influences longevity and that its impact is mediated through environmental effects. Surviving participants of a 30-year-old population-based health survey (N = 310; age range, 90–109 years; the 90+ Study) were genotyped/resequenced at the DRD4 gene and compared with a European ancestry-matched younger population (N = 2902; age range, 7–45 years). We found that the oldest-old population had a 66% increase in individuals carrying the DRD4 7R allele relative to the younger sample (p = 3.5 × 10−9), and that this genotype was strongly correlated with increased levels of physical activity. Consistent with these results, DRD4 knock-out mice, when compared with wild-type and heterozygous mice, displayed a 7–9.7% decrease in lifespan, reduced spontaneous locomotor activity, and no lifespan increase when reared in an enriched environment. These results support the hypothesis that DRD4 gene variants contribute to longevity in humans and in mice, and suggest that this effect is mediated by shaping behavioral responses to the environment.
Introduction
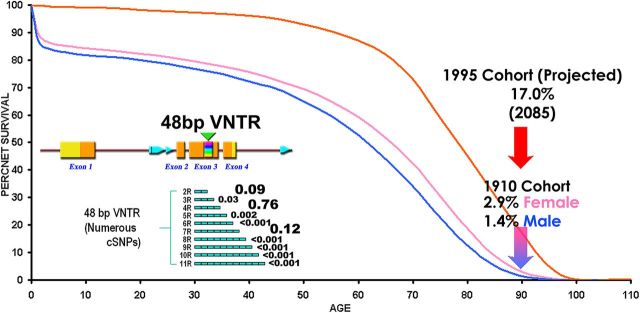
Individuals ≥90 years of age constitute the fastest growing segment of our population (Fig. 1) (Day, 1996), yet little is known about the factors that contribute to their extreme longevity. Addressing this gap would help develop smarter strategies to increase the number of people who reach old age in good health.
Figure 1.
Survival curves of the oldest-old cohort. The known male (blue) and female (pink) survival curves of European ancestry individuals born in 1910 (the average birth year of participants) are shown (source: Berkeley Mortality Database, http://www.demog.berkeley.edu/∼bmd/; UK and USA data). Individuals older than 90 years of age represent 1.4% (male) to 2.9% (female) of their birth cohort. The projected curve of the 1995 birth cohort (orange) suggests that 17% of this cohort (males and females) will reach 90+ years of age (Social Security Administration, Alternative II Forecast, 1998, http://www.demog.berkeley.edu/∼bmd/). Inset, Diagrammatic representation of the human DRD4 gene region. Exon positions are indicated by blocks (yellow = noncoding; orange = coding), and the positions of alu repetitive sequences are represented by pointed blue blocks. The position of a coding 48 bp VNTR in exon 3 is indicated by a green triangle. The 2R to 11R variants of this repeat are indicated below exon 3, along with their frequencies in European ancestry populations (Ding et al., 2002; Grady et al., 2003; Wang et al., 2004; Grady et al., 2005; this study).
Undoubtedly, genetic factors contribute to both longevity (Sebastiani et al., 2012) and health among the oldest-old (Martin et al., 2007). Surprisingly, little attention has been directed toward genes that influence personality as “longevity genes” (Eaton et al., 2012). This is puzzling not only because the impact of life-style choices on longevity is self-evident (e.g., smoking and early death from pulmonary disease or cancer), but also because of the strong genetic contributions to human personality (Bouchard and Loehlin, 2001).
A good example is the human dopamine receptor D4 (DRD4) gene, whose associations with the personality trait of novelty seeking (MIM 601696) (Kluger et al., 2002; Gillespie et al., 2003) and with attention deficit hyperactivity disorder (ADHD) (MIM 126452) (Faraone et al., 2001; Grady et al., 2003, 2005; Li et al., 2006) have been replicated numerous times. The DRD4 gene exhibits an unusually high amount of expressed polymorphism (Fig. 1, inset). Most of this variation is the result of a 48 bp variable number tandem repeat (VNTR), located in exon 3 that encodes the third intracellular loop of the receptor. Alleles containing from 2 to 11 repeats (2R to 11R) have been identified, with the three most common variants (2R, 4R, and 7R) accounting for >90% of the allelic diversity (Ding et al., 2002; Wang et al., 2004).
The functional significance of these length/sequence changes in the DRD4 protein, in a region that couples to G-proteins, has been studied extensively. The 7R variant shows a blunted response to dopamine when compared with the 4R variant (Asghari et al., 1995; Jovanovic et al., 1999; Wang et al., 2004) and does not form heteromers with the short isoform of the presynaptic dopamine D2 receptor (Borroto-Escuela et al., 2011; González et al., 2012). The DRD4 protein is expressed in several brain regions, with high levels in prefrontal cortex (Oak et al., 2000). DRD4 knock-out mice display better performance on complex motor tasks and reduced exploration of novel stimuli (Rubinstein et al., 1997; Dulawa et al., 1999). ADHD individuals with a 7R allele have faster reaction times than non-7R individuals (Swanson et al., 2000; Langley et al., 2004).
Given that the DRD4 7R allele is associated with increased activity/novelty-seeking in children and young adults (Faraone et al., 2001; Kluger et al., 2002; Grady et al., 2003, 2005; Li et al., 2006), we hypothesized that this allele would be associated with increased physical activity in the elderly. In addition, since physical activity is associated with increased lifespan and health (Adlard et al., 2005; Larson et al., 2006; Willcox et al., 2006; Paganini-Hill et al., 2011), we hypothesized that the DRD4 7R allele would be over-represented in the oldest old.
Materials and Methods
Study participants.
The 90+ Study cohort (N = 1151) consists of all participants aged ≥90 years who were alive on January 1, 2003, and were part of the initial Leisure World Cohort Study (Corrada et al., 2008; Paganini-Hill et al., 2011). The Leisure World Cohort Study was established in 1981, when a health survey was mailed to all residents who at that time owned homes in the Leisure World retirement community, Laguna Woods, California. A total of 13,979 residents (61%) returned the questionnaire and constitute this cohort. The median age of subjects when they joined the cohort was 73 years (range, 44–107 years). Reflecting the characteristics of the community, the cohort was primarily of European ancestry (98%) and highly educated (67% with some college to advanced degree). An additional questionnaire on current (2003 to present) activity levels was obtained from 233 participants, using questions appropriate for this age group. The longitudinal data in this large cohort have been used to conclude that spending more time in physical activities is significantly associated with lower mortality risks (15–35%), and is not confounded by smoking, alcohol intake, caffeine consumption, body mass index (BMI), or a history of hypertension, angina, heart attack, stroke, diabetes, rheumatoid arthritis, or cancer (Paganini-Hill et al., 2011).
Cell lines/DNA and genotyping/resequencing.
Genotypes were obtained from DNA isolated from blood cell pellets from a subset of 310 subjects (91 males and 219 females, ∼60% of individuals still alive) representative of the entire 90+ cohort with respect to gender, education, BMI, exercise, drinking, or medical histories. Two hundred seventy participants had complete information available from the Leisure World Cohort Study (Paganini-Hill et al., 2011). Lymphoblastoid cell lines were established for all 310 participants, using a modified protocol involving up to 5 months of cell culture (Grady et al., 2003). Detailed ancestry information was collected from the 90+ Study participants to confirm self-declared ancestry for matching to European ancestry population-based control sample DRD4 genotypes (N = 2902) from ongoing studies (Ding et al., 2002; Grady et al., 2003, 2005; Wang et al., 2004). Controls ranged in age from 7 to 45 years of age (mostly adolescents and college students). Previously established methods to genotype and resequence the DRD4 VNTR variant were used (Ding et al., 2002; Grady et al., 2003, 2005; Wang et al., 2004). Single-nucleotide polymorphisms in other genes used as controls were genotyped by DNA sequencing. The Institutional Review Board of the University of California, Irvine, approved all procedures.
Genotypes obtained for the 90+ sample were as follows: 6 2R/2R, 27 2R/4R, 4 2R/7R, 4 3R/3R, 25 3R/4R, 7 3R/7R, 135 4R/4R, 1 4R/5R, 3 4R/6R, 82 4R/7R, 1 4R/11R, 1 5R/6R, 1 5R/7R, 1 6R/6R, 11 7R/7R, and 1 7R/8R. The genotypes of the control individuals were as follows: 60 2R/2R, 16 2R/3R, 345 2R/4R, 1 2R/5R, 58 2R/7R, 1 2R/8R, 4 3R/3R, 127 3R/4R, 17 3R/7R, 1715 4R/4R, 5 4R/5R, 1 4R/6R, 493 4R/7R, 1 4R/8R, 1 4R/9R, 2 5R/5R, 2 5R/7R, 2 6R/7R, and 51 7R/7R. There were no significant deviations from the expected Hardy–Weinberg equilibrium. Observed allele frequencies for the oldest-old sample were 2R = 0.07, 3R = 0.06, 4R = 0.66, 5R = 0.005, 6R = 0.009, 7R = 0.189, and 8R = 0.0016. Observed allele frequencies for the population and ancestry-matched controls were 2R = 0.09, 3R = 0.03, 4R = 0.76, 5R = 0.002, 6R < 0.001, 7R = 0.12, 8R = <0.001, and 9R < 0.001. The DRD4 7R/x category contains individuals with at least one 7R (or derived 7R, i.e., 5R, 6R, 8R, 9R, 10R, and 11R) allele, and the non-7R/x category the remaining individuals (Ding et al., 2002; Wang et al., 2004). A representative sample of 140 oldest-old individuals was resequenced at the DRD4 exon 3 polymorphism to determine the haplotype (273 alleles). The common 7R(1-2-6-5-2-5-4) haplotype or 7R derivatives were the predominant 7R sequences, with the fraction of common to rare haplotypes similar to that observed in prior population studies (Ding et al., 2002; Wang et al., 2004).
Mouse longevity studies.
DRD4 wild-type (WT), heterozygous (HT), and homozygous knock-out (KO) mice (Rubinstein et al., 1997) were assigned to either an enriched environment (EE; housed with other mice and with access to toys and running wheels) or a deprived environment (DE; housed in isolation in standard cages) and maintained under these conditions for their lifespan. Mice were divided into six groups distinguished by their genotype (WT, HT, and KO) and environmental exposure (EE and DE) with the following sample sizes: WT (DE = 66, EE = 51), HT (DE = 31, EE = 26), and KO (DE = 34, EE = 34). Age at time of death was recorded for all animals following guidelines previously outlined (Ullman-Culleré and Foltz, 1999). Locomotor activity was tested bimonthly for 90 min using an activity monitoring system (Mini Mitter). Differences in longevity and locomotor activity between groups were assessed using SAS 9.1 and STRATA 7.0. Experiments followed the Guide for the Care and Use of Laboratory Animals (Quimby et al., 2010).
Results
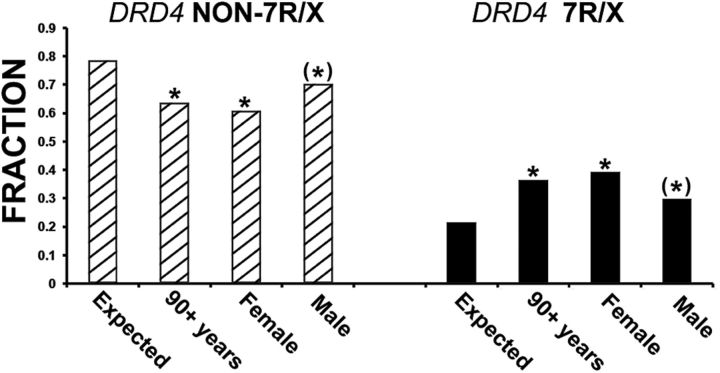
To test our hypothesis, we determined DRD4 exon 3 VNTR genotypes/sequences for 310 90+ study individuals (the “oldest-old”; mean age, 95.2 years; age range, 90–109 years; 91 males) (Fig. 1). These individuals represent ∼1.4–2.9% of their initial birth cohort and, by definition, should have altered allele frequencies of genes that impact health and longevity. When compared with matched European ancestry controls of individuals 50–90 years younger (N = 2902; age range, 7–45 years), the oldest-old had a 66% increase in observed 7R/x genotype (Fig. 2) (0.364 vs 0.219, χ2 = 33.28, p = 3.5 × 10−9; φ2 = 0.026). This extremely small p value would be significant at a genome-wide level following Bonferroni correction. Comparing allele frequency rather than genotype, the frequency of the 7R allele is greater in the oldest-old population by 72% (0.205 vs 0.119, χ2 = 38.27, p = 6.15 × 10−10). This increase was preferentially observed in females (0.393 vs 0.297 males, χ2 = 9.65, p = 0.002), among whom the difference was significant at a genome-wide level (0.393 vs 0.219, χ2 = 34.66, p = 1.7 × 10−9). In contrast, the difference in males, while significant, did not reach genome-wide level significance (0.297 vs 0.219, χ2 = 3.11, p = 0.034) (Fig. 2). This gender difference could reflect potentially negative (and often risky) behaviors associated with DRD4 7R alleles earlier in life, such as ADHD and drug abuse, which have a significant male bias (Swanson et al., 2000; Grady et al., 2003; Olsson et al., 2011). Allele frequencies at 20 randomly chosen polymorphisms in nine unrelated genomic regions (CFTR, ERCC8, ERBB4, FANCC, HTERT, NRF1, RTN1, SLC6A3, and UCP3) did not differ significantly among the oldest-old individuals, younger control individuals, or HapMap European ancestry populations (International HapMap Consortium et al., 2007).
Figure 2.
Comparison of expected to observed DRD4 genotypes. Oldest-old individuals (N = 310) were genotyped at the exon 3 DRD4 polymorphism (Fig. 1, inset) and compared with population-based European ancestry control sample DRD4 genotypes (Expected, N = 2902). The DRD4 7R/x category (black bars) contains individuals with at least one 7R (or derived 7R, i.e., 5R, 6R, 8R, 9R, 10R, and 11R) allele, and the non-7R/x category (striped bars) contains the remaining individuals. The expected and observed fraction for each category is shown. Expected 7R/x (N = 635), Non 7R/x (N = 2267); 90+ observed 7R/x (N = 113), Non 7R/x (N = 197); Female 7R/x (N = 86), Non 7R/x (N = 133); Male 7R/x (N = 27), Non 7R/x (N = 64). Asterisks denote observed differences significant at p = 3.5 × 10−9 (90+ years), p = 1.7 × 10−9 (Female), and p = 0.034 (Male) (see text).
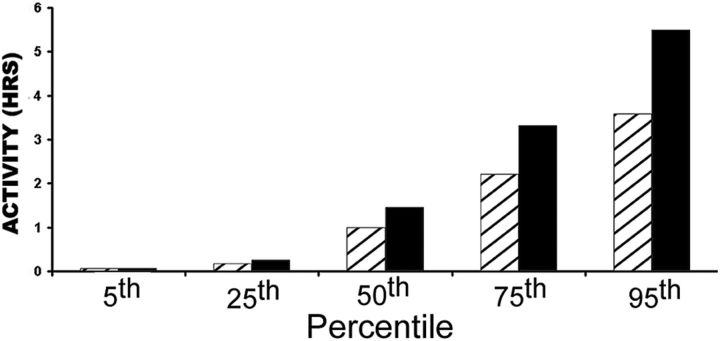
In the oldest-old, we also observed a significant association between the 7R/x genotype and higher reported activity levels in both males and females (Fig. 3). This association was observed not only for measures obtained in 1981, when the mean self-reported activity level differed by 45% (1.42 vs 0.98 h; χ2 = 5.07, p = 0.024 when measured as a categorical variable; and t = 2.40, p = 0.018 when measured as a continuous variable), but also when the measured variable was current self-reported activity (χ2 = 4.43, p = 0.033). Oldest-old participants with the DRD4 7R/x genotype were twice as likely to report exercising ≥2 h/d in 1981 (top 20%; χ2 = 17.33, p = 1.64 × 10−5; Fig. 3). The correlation of higher activity with DRD4 7R/x genotype was not confounded by age, gender, alcohol intake, BMI, or medical history, similar to results obtained in the larger Leisure World cohort (Paganini-Hill et al., 2011). The proportion of the variance in physical activity explained by DRD4 7R in this oldest-old population was r2 = 0.023.
Figure 3.
DRD4 genotype and self-reported activity. The number of hours per day of activity reported in 1981 is plotted versus participants with a DRD4 7R/x genotype (black bars) or non-DRD4 7R/x genotype (striped bars). Comparisons are shown at the 5th, 25th, 50th, 75th, and 95th percentiles. Including DRD4 2R/x and 3R/x individuals with the DRD4 7R/x individuals instead of the non-7R/x individuals (Ding et al., 2002; Grady et al., 2003; Wang et al., 2004; Grady et al., 2005) gave comparable activity level differences (i.e., an ancestral 4R/4R vs non-4R/4R division).
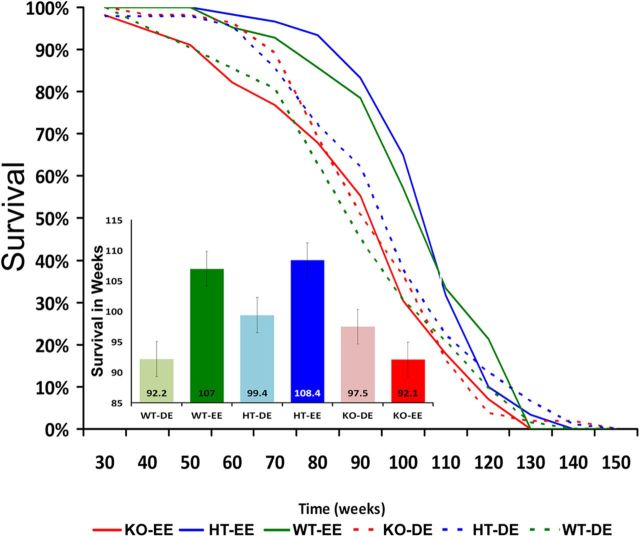
To test the association of the DRD4 gene with longevity and responses to the environment directly, we compared the lifespans of mice that expressed DRD4 (HT and WT) versus those of KO mice (Rubinstein et al., 1997) that lacked the DRD4 gene, when reared in either a DE (standard laboratory cage) or EE (group cages with multiple options for physical activity and exploration) environment (Ullman-Culleré and Foltz, 1999; Quimby et al., 2010). DRD4 genotype significantly affected the lifespan (F = 5.74, p = 0.004, η2 = 0.03; Fig. 4). DRD4 KO mice (N = 68) had a 9.7% shorter lifespan (mean and SEs: 94.8 ± 2.08 weeks; 95% CI, 91–99) than HT mice (N = 57; 104 ± 1.82 weeks; 95% CI, 100–108; p < 0.05), and 7% shorter than WT mice (N = 117; 101.4 ± 2.38 weeks; 95% CI, 97–106; p < 0.05). The rearing environment also influenced longevity (F = 10.58, p = 0.001, η2 = 0.03; Fig. 4). Mice in EE (N = 111; 102.2 ± 1.76 weeks; 95% CI, 98–106) lived 5.7% longer than mice in DE (N = 131; 96.7 ± 1.54 weeks; 95% CI, 94–100; p < 0.05), but these effects were moderated by the DRD4 genotype, as shown by a significant environment × genotype interaction (F = 8.05, p < 0.001, η2 = 0.04; Fig. 4). Specifically, whereas the lifespan was increased in an EE for HT (9% increase) and WT (16% increase) mice, the longevity of DRD4 KO mice did not differ between DE and EE rearing conditions (p = 0.15; Fig. 4, inset).
Figure 4.
Kaplan–Meier survival curves of mice as a function of genotype and exposure to an EE or DE. Insert, Lifespan per group (mean and SE). WT and HT mice reared in an EE lived significantly longer than when reared in a DE. In contrast, survival of DRD4 KO mice was not increased when reared in an EE compared with a DE and was shorter than that of WT and HT mice.
Discussion
We have shown that a human DRD4 variant associated with activity in younger individuals is at higher observed frequency in the oldest-old (Figs. 1, 2, 3), and that the DRD4 gene in mice moderated the beneficial effects of an enriched environment on lifespan (Figs. 4, 5). These results are consistent with human studies showing that sensitivity to environmental factors, both positive and negative, is modulated by the DRD4 7R variant (Sheese et al., 2007; Belsky et al., 2009; Das et al., 2011; Olsson et al., 2011). The reduced spontaneous locomotor activity observed in DRD4 KO mice compared with WT and HT mice (Fig. 5) supports this interpretation, as well as the higher reported physical activity levels in oldest-old individuals with a DRD4 7R variant (Fig. 3). However, from our experimental design it cannot be determined whether the beneficial effects of an enriched environment on longevity in WT and HT mice is moderated through social factors rather than increased locomotor activity per se.
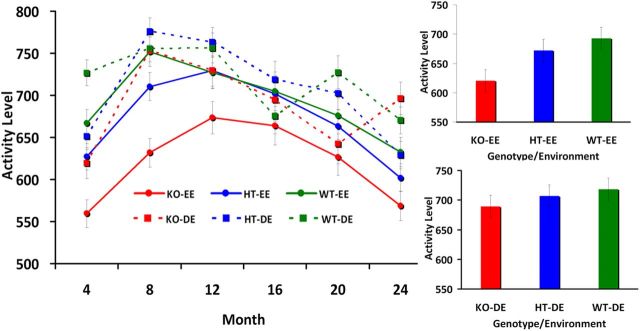
Figure 5.
Spontaneous locomotor activity for different ages in WT, HT, and KO mice reared in a DE and an EE. Values correspond to means (±SE) and correspond to average activity over 90 min. The genotype-by-age interaction on locomotor activity was significant (F = 3.61, p < 0.001, ηp2 = 0.034) with overall significantly lower activity levels for KO mice than for WT or HT mice (p < 0.05). The interaction of environment by age with locomotor activity was also significant (F = 2.77, p = 0.017, ηp2 = 0.013); overall, DE mice were significantly more active than EE mice (p < 0.05). However, since animals in an EE decrease in spontaneous locomotor activity (Garland et al., 2011) and we did not record 24 h activity levels, we cannot compare activity levels across the two environmental conditions.
The mechanism by which the DRD4 variants would differentially influence environmental responses is unclear. The variant DRD4 7R protein has a blunted response for cAMP reduction compared with the 4R protein (Asghari et al., 1995; Jovanovic et al., 1999; Swanson et al., 2000; Wang et al., 2004). Also, the DRD4 7R protein, unlike the DRD4 4R protein, does not form functional heteromers with the short presynaptic isoform of the dopamine D2 receptor in the striatum (Borroto-Escuela et al., 2011; González et al., 2012). Dysfunctional D2-D4 7R heteromers may impair dopaminergic control of corticostriatal glutamatergic neurotransmission. Thus, it is plausible that either a dopamine signaling deficit or enhanced glutamate release, mediated by DRD4 7R, could enhance reactivity to environmental stimuli.
While the biochemical/physiological significance of DRD4 variants is under continuing investigation, there is solid molecular evolutionary evidence that functional differences exist at this locus. Extensive linkage disequilibrium (LD) surrounds the DRD4 7R allele in individuals sampled worldwide (compared with the random LD at the ancestral 4R allele), as determined by direct DNA resequencing of the entire gene and genotyping of polymorphisms extending 100 kb in both directions (Ding et al., 2002; Wang et al., 2004, 2006). The high frequency of an allele coupled with extensive LD is a powerful indicator of recent adaptive selection (Wang et al., 2006; Hawks et al., 2007) (LDD test). The pattern of LD decay surrounding the DRD4 7R allele, together with the highest LD near the VNTR decaying in both directions, suggests that the DRD4 7R VNTR is the “target” of this Darwinian selection, with a recent evolutionary origin (∼40,000–50,000 years ago). Consistent with this recent evolutionary origin and the known biochemical differences noted above, we have speculated that a “response-ready” adaptation influenced by the DRD4 7R allele might have played a role in the Out-of-Africa human exodus (Ding et al., 2002; Wang et al., 2004).
In further discussing our findings, it is important to note their limitations. The ideal control for the genotype/haplotype frequencies of DRD4 alleles would have been the nonliving birth cohorts of our oldest-old population, which is clearly not possible. However, human gene frequencies cannot change radically in a few generations, except following extreme population bottlenecks, which have not occurred in the last 100 years in European ancestry populations. A possible selective force that could have introduced intergenerational variation is the high mortality due to infectious disease experienced early in life by this oldest-old cohort, which has been largely eliminated through the use of immunization and antibiotics in the younger cohort (Fig. 1). Thus, any hypothetical “resistance” allele might exhibit higher frequencies in the oldest-old population than currently observed, due to this environmental intervention. However, it is unlikely that the DRD4 gene (or any of the three closest genes in modest LD with it: DEAF1, SCT, and MUPCDH) (International Human Genome Sequencing Consortium, 2004) would confer such resistance. Moreover, allele frequencies at 20 randomly chosen polymorphisms in nine unrelated genomic regions (CFTR, ERCC8, ERBB4, FANCC, HTERT, NRF1, RTN1, SLC6A3, and UCP3) did not differ significantly among the oldest-old individuals, younger control individuals, or HapMap European ancestry populations (International HapMap Consortium et al, 2007); nor was the observed DRD4 7R male/female allele frequency bias (Fig. 2) found at any of these randomly chosen sites. It is difficult to conceive of a mechanism that explains the variations in allele frequencies between males and females except for one that involves the functional significance of the underlying variant locus. We cannot, however, rule out alternative behavioral possibilities to explain why 7R/x individuals are disproportionately found in our specific cohort, such as the ability/desire to live in a planned retirement community with good access to leisure activities [see the discussion of the potential impact of socioeconomic status (SES)]. Clearly, further studies of the allele frequencies of DRD4 throughout the lifespan, rather than just at the extremes (Fig. 1) are warranted.
A strength and a weakness of the current study is that the oldest-old population studied is of predominantly European ancestry. We specifically chose this population to reduce the genetic diversity and allow for better case-control matching, essential for genetic studies. It should be cautioned, therefore, that it is unclear how broadly such results can be inferred beyond the ancestry group examined. The ubiquitous presence of DRD4 7R (and its derived 2R allele) in most human populations (Ding et al., 2002; Wang et al., 2004), however, make it likely that this study can be generalized (and potentially replicated) in individuals whose ancestry is from different geographic origins. While a prospective design would allow for much stronger inferences, the logistics and cost of a prospective study to assess the effects of genotype on longevity are enormous, and data would not be available for analysis until the cohorts died.
The current study was conducted on a population with high educational achievement and socioeconomic status (SES), again, to minimize the effect of these variables on our genetic studies. SES is known to influence both longevity and research participation (Seeman et al., 2004). While this homogeneity is an advantage, making it less likely that individuals with different genotypes would be differentially recruited, replication studies in other SES populations should be pursed. Indeed, one could hypothesize that the “enriched environment” found in the currently investigated 90+ population (Paganini-Hill et al., 2011) would not be available to individuals with lower SES, and hence DRD4 7R frequency might not be elevated in such populations.
Finally, it would be useful to obtain measures of physical activity and DRD4 genotype in younger populations. DRD4 variation is not associated with higher BMI (Guo et al., 2007; Fuemmeler et al., 2008), except in African American populations, but few studies on DRD4/activity have been conducted except for individuals diagnosed with ADHD (Swanson et al., 2000). Measurements of the heritability of physical activity vary widely, with a value of ∼0.30 often reported (Pérusse et al., 1989; Horimoto et al., 2011). The proportion of the variance in physical activity explained by DRD4 7R in the oldest-old population (Fig. 3) is r2 = 0.023, or ∼7.7% of the variance attributable to genes measured in heritability studies. However, heritability studies assume that multiple additive genes contribute to the phenotype, but an interacting gene model, as proposed for the DRD4 7R/ADHD association (Grady et al., 2003), would make the contribution of DRD4 7R to the variance in activity level significantly greater (Zuk et al., 2012). Clearly, further studies of DRD4 7R contributions to the variance of activity level in other populations should be pursed. Unfortunately, there are no genetic markers on commercially available genotyping arrays that are in strong LD with the DRD4 7R VNTR (Saccone et al., 2009), making direct genotyping/sequencing essential.
For genetic studies of complex phenotypes, most associated variants are observed to have relatively small effect sizes (<1%), as expected for polygenic traits (Manolio et al., 2009). Studies of longevity in European ancestry populations similar to the oldest-old population studied here have indicated that longevity has modest heritability (∼25%; Herskind et al., 1996; Mitchell et al., 2001). Given the estimated effect size for the DRD4 7R/longevity association (φ2 = 0.026; Fig. 2), we can estimate that ∼10.2% of the variance attributable to genes, measured in heritability studies, is associated with DRD4 7R in our oldest-old population. The effect sizes of DRD4 variants on longevity, estimated in our mouse studies, are comparable (η2 = 0.03–0.04; Fig. 4). These values are quite large for a phenotype expected to be associated with many genes, and suggests that variants of DRD4 make a significant impact on longevity, again making direct genotyping/sequencing essential.
In conclusion, we observed a striking increase in DRD4 7R allele frequency in the oldest-old population and a strong association of this allele with increased activity levels. These findings are consistent with our animal model results showing that the DRD4 gene moderates the beneficial effects of an enriched environment in increasing the rodent's lifespan. We propose that DRD4 gene variants moderate longevity by altering behavioral responses to environmental factors. It is likely that such gene–environmental interactions underlie much human variability and many common disorders afflicting humankind (Wang et al., 2006; Manolio et al., 2009).
Footnotes
This work was supported by grants from the U.S. Department of Energy and National Institute of Aging (to R.K.M.), a National Library of Medicine National Research Service Award (to E.W.), the National Institute of Aging (to C.H.K.), the 111 Gene/Brain/Behavior Project from the Ministry of Education of China (to Q.D.), and the National Institute on Alcohol Abuse and Alcoholism Intramural Program (to N.D.V.). We dedicate this paper to Hubert H. M. Van Tol, whose pioneering work on the DRD4 protein will not be forgotten. We thank the participants, especially those of the 90+ Study, for their generous involvement in this project; Hao-Yuan Cheng and Simin Hakim for technical assistance; and Ruben Baler for editorial assistance.
The authors declare no competing financial interests.
References
- Adlard PA, Perreau VM, Pop V, Cotman CW. Voluntary exercise decreases amyloid load in a transgenic model of Alzheimer's disease. J Neurosci. 2005;25:4217–4221. doi: 10.1523/JNEUROSCI.0496-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghari V, Sanyal S, Buchwaldt S, Paterson A, Jovanovic V, Van Tol HH. Modulation of intracellular cyclic AMP levels by different human dopamine D4 receptor variants. J Neurochem. 1995;65:1157–1165. doi: 10.1046/j.1471-4159.1995.65031157.x. [DOI] [PubMed] [Google Scholar]
- Belsky J, Jonassaint C, Pluess M, Stanton M, Brummett B, Williams R. Vulnerability genes or plasticity genes? Mol Psychiatry. 2009;14:746–754. doi: 10.1038/mp.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borroto-Escuela DO, Van Craenenbroeck K, Romero-Fernandez W, Guidolin D, Woods AS, Rivera A, Haegeman G, Agnati LF, Tarakanov AO, Fuxe K. Dopamine D2 and D4 receptor heteromerization and its allosteric receptor-receptor interactions. Biochem Biophys Res Commun. 2011;404:928–934. doi: 10.1016/j.bbrc.2010.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard TJ, Jr, Loehlin JC. Genes, evolution, and personality. Behav Genet. 2001;31:243–273. doi: 10.1023/a:1012294324713. [DOI] [PubMed] [Google Scholar]
- Corrada MM, Brookmeyer R, Berlau D, Paganini-Hill A, Kawas CH. Prevalence of dementia after age 90: results from the 90+ Study. Neurology. 2008;71:337–343. doi: 10.1212/01.wnl.0000310773.65918.cd. [DOI] [PubMed] [Google Scholar]
- Das D, Cherbuin N, Tan X, Anstey KJ, Easteal S. DRD4-exonIII-VNTR moderates the effect of childhood adversities on emotional resilience in young-adults. PLoS One. 2011;6:e20177. doi: 10.1371/journal.pone.0020177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day J. Population projections of the United States by age, sex, race, and hispanic origin: 1995 to 2050, Series P25–P1130. Washington, DC: U.S. Bureau of the Census, Government Printing Office; 1996. [Google Scholar]
- Ding YC, Chi HC, Grady DL, Morishima A, Kidd JR, Kidd KK, Flodman P, Spence MA, Schuck S, Swanson JM, Zhang YP, Moyzis RK. Evidence of positive selection acting at the human dopamine receptor D4 gene locus. Proc Natl Acad Sci U S A. 2002;99:309–314. doi: 10.1073/pnas.012464099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulawa SC, Grandy DK, Low MJ, Paulus MP, Geyer MA. Dopamine D4 receptor-knock-out mice exhibit reduced exploration of novel stimuli. J Neurosci. 1999;19:9550–9556. doi: 10.1523/JNEUROSCI.19-21-09550.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton NR, Krueger RF, South SC, Gruenewald TL, Seeman TE, Roberts BW. Genes, environments, personality, and successful aging: toward a comprehensive developmental model in later life. J Gerontol A Biol Sci Med Sci. 2012;67:480–488. doi: 10.1093/gerona/gls090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraone SV, Doyle AE, Mick E, Biederman J. Meta-analysis of the association between the 7-repeat allele of the dopamine D(4) receptor gene and attention deficit hyperactivity disorder. Am J Psychiatry. 2001;158:1052–1057. doi: 10.1176/appi.ajp.158.7.1052. [DOI] [PubMed] [Google Scholar]
- Fuemmeler BF, Agura-Collins T, McClernon FJ, Kollins SH, Kail M, Bergen AW, Ashley-Koch AE. Genes implicated in serotonergic and dopaminergic functioning predict BMI categories. Obesity (Silver Spring) 2008;16:348–356. doi: 10.1038/oby.2007.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T, Jr, Schutz H, Chappell MA, Keeney BK, Meek TH, Copes LE, Acosta W, Drenowatz C, Maciel RC, van Dijk G, Kotz CM, Eisenmann JC. The biological control of voluntary exercise, spontaneous physical activity and daily energy expenditure in relation to obesity: human and rodent perspectives. J Exp Biol. 2011;214:206–229. doi: 10.1242/jeb.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie N, Cloninger C, Heath A, Martin N. The genetic and environmental relationship between Cloninger's dimensions of temperament and character. Pers Individ Diff. 2003;35:1931–1946. doi: 10.1016/S0191-8869(03)00042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González S, Rangel-Barajas C, Peper M, Lorenzo R, Moreno E, Ciruela F, Borycz J, Ortiz J, Lluís C, Franco R, McCormick PJ, Volkow ND, Rubinstein M, Floran B, Ferré S. Dopamine D4 receptor, but not the ADHD-associated D4.7 variant, forms functional heteromers with the dopamine D2S receptor in the brain. Mol Psychiatry. 2012;17:650–662. doi: 10.1038/mp.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady DL, Chi HC, Ding YC, Smith M, Wang E, Schuck S, Flodman P, Spence MA, Swanson JM, Moyzis RK. High prevalence of rare dopamine receptor D4 alleles in children diagnosed with attention-deficit hyperactivity disorder. Mol Psychiatry. 2003;8:536–545. doi: 10.1038/sj.mp.4001350. [DOI] [PubMed] [Google Scholar]
- Grady DL, Harxhi A, Smith M, Flodman P, Spence MA, Swanson JM, Moyzis RK. Sequence variants of the DRD4 gene in autism: further evidence that rare DRD4 7R haplotypes are ADHD specific. Am J Med Genet B Neuropsychiatr Genet. 2005;136B:33–35. doi: 10.1002/ajmg.b.30182. [DOI] [PubMed] [Google Scholar]
- Guo G, North KE, Gorden-Larsen P, Bulik CM, Choi S. Body mass, DRD4, physical activity, sedentary behavior, and family socioeconomic status: the add health study. Obesity. 2007;15:1199–1206. doi: 10.1038/oby.2007.640. [DOI] [PubMed] [Google Scholar]
- Hawks J, Wang ET, Cochran GM, Harpending HC, Moyzis RK. Recent acceleration of human adaptive evolution. Proc Natl Acad Sci U S A. 2007;104:20753–20758. doi: 10.1073/pnas.0707650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskind AM, McGue M, Holm NV, Sørensen TI, Harvald B, Vaupel JW. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum Genet. 1996;97:319–323. doi: 10.1007/BF02185763. [DOI] [PubMed] [Google Scholar]
- Horimoto AR, Giolo SR, Oliveira CM, Alvim RO, Soler JP, de Andrade M, Krieger JE, Pereira AC. Heritability of physical activity traits in Brazilian families: the Baependi Heart Study. BMC Medical Genetics. 2011;12:155–163. doi: 10.1186/1471-2350-12-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Jovanovic V, Guan HC, Von Tol HH. Comparative pharmacological and functional analysis of the human dopamine D4.2 and D4.10 receptor variants. Pharmacogenetics. 1999;9:561–568. [PubMed] [Google Scholar]
- Kluger AN, Siegfried Z, Ebstein RP. A meta-analysis of the association between DRD4 polymorphism and novelty seeking. Mol Psychiatry. 2002;7:712–717. doi: 10.1038/sj.mp.4001082. [DOI] [PubMed] [Google Scholar]
- Langley K, Marshall L, van den Bree M, Thomas H, Owen M, O'Donovan M, Thapar A. Association of the dopamine D4 receptor gene 7-repeat allele with neuropsychological test performance of children with ADHD. Am J Psychiatry. 2004;161:133–138. doi: 10.1176/appi.ajp.161.1.133. [DOI] [PubMed] [Google Scholar]
- Larson EB, Wang L, Bowen JD, McCormick WC, Teri L, Crane P, Kukull W. Exercise is associated with reduced risk for incident dementia among persons 65 years of age and older. Ann Intern Med. 2006;144:73–81. doi: 10.7326/0003-4819-144-2-200601170-00004. [DOI] [PubMed] [Google Scholar]
- Li D, Sham PC, Owen MJ, He L. Meta-analysis shows significant association between dopamine system genes and attention deficit hyperactivity disorder (ADHD) Hum Mol Gen. 2006;15:2276–2284. doi: 10.1093/hmg/ddl152. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GM, Bergman A, Barzilai N. Genetic determinants of human health span and life span: progress and new opportunities. PLoS Genet. 2007;3:e125. doi: 10.1371/journal.pgen.0030125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell BD, Hsueh WC, King TM, Pollin TI, Sorkin J, Agarwala R, Schäffer AA, Shuldiner AR. Heritability of life span in the Old Order Amish. Am J Med Genet. 2001;102:346–352. doi: 10.1002/ajmg.1483. [DOI] [PubMed] [Google Scholar]
- Oak JN, Oldenhof J, Van Tol HH. The dopamine D4 receptor: one decade of research. Eur J Pharmacol. 2000;405:303–327. doi: 10.1016/s0014-2999(00)00562-8. [DOI] [PubMed] [Google Scholar]
- Olsson CA, Moyzis RK, Williamson E, Ellis JA, Parkinson-Bates M, Patton GC, Dwyer T, Romaniuk H, Moore EE. Gene-environment interaction in problematic substance use: interaction between DRD4 and insecure attachments. Addict Biol. 2011 doi: 10.1111/j.1369-1600.2011.00413.x. Advance online publication. Retrieved November 28, 2012. [DOI] [PubMed] [Google Scholar]
- Paganini-Hill A, Kawas CH, Corrada MM. Activities and mortality in the elderly: the Leisure World cohort study. J Gerontol A Biol Sci Med Sci. 2011;66:559–567. doi: 10.1093/gerona/glq237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérusse L, Tremblay A, Leblanc C, Bouchard C. Genetic and environmental influences on level of habitual physical activity and exercise participation. Am J Epidemiol. 1989;129:1012–1022. doi: 10.1093/oxfordjournals.aje.a115205. [DOI] [PubMed] [Google Scholar]
- Quimby F, Turner P, Wood G, Würbel H. Guide for the care and use of laboratory animals. Ed 8. Washington, DC: The National Academies Press; 2010. [Google Scholar]
- Rubinstein M, Phillips TJ, Bunzow JR, Falzone TL, Dziewczapolski G, Zhang G, Fang Y, Larson JL, McDougall JA, Chester JA, Saez C, Pugsley TA, Gershanik O, Low MJ, Grandy DK. Mice lacking dopamine D4 receptors are supersensitive to ethanol, cocaine, and methamphetamine. Cell. 1997;90:991–1001. doi: 10.1016/s0092-8674(00)80365-7. [DOI] [PubMed] [Google Scholar]
- Saccone SF, Bierut LJ, Chesler EJ, Kalivas PW, Lerman C, Saccone NL, Uhl GR, Li CY, Philip VM, Edenberg HJ, Sherry ST, Feolo M, Moyzis RK, Rutter JL. Supplementing high-density SNP microarrays for additional coverage of disease-related genes: addiction as a paradigm. PLoS One. 2009;4:e5225. doi: 10.1371/journal.pone.0005225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani P, Solovieff N, Dewan AT, Walsh KM, Puca A, Hartley SW, Melista E, Andersen S, Dworkis DA, Wilk JB, Myers RH, Steinberg MH, Montano M, Baldwin CT, Hoh J, Perls TT. Genetic signatures of exceptional longevity in humans. PLoS One. 2012;7:e29848. doi: 10.1371/journal.pone.0029848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman TE, Crimmins E, Huang MH, Singer B, Bucur A, Gruenewald T, Berkman LF, Reuben DB. Cumulative biological risk and socioeconomic differences in mortality: MacArthur studies of successful aging. Soc Sci Med. 2004;58:1985–1997. doi: 10.1016/S0277-9536(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Sheese BE, Voelker PM, Rothbart MK, Posner MI. Parenting quality interacts with genetic variation in dopamine receptor D4 to influence temperament in early childhood. Dev Psychopathol. 2007;19:1039–1046. doi: 10.1017/S0954579407000521. [DOI] [PubMed] [Google Scholar]
- Swanson J, Oosterlaan J, Murias M, Schuck S, Flodman P, Spence MA, Wasdell M, Ding Y, Chi HC, Smith M, Mann M, Carlson C, Kennedy JL, Sergeant JA, Leung P, Zhang YP, Sadeh A, Chen C, Whalen CK, Babb KA, et al. Attention deficit/hyperactivity disorder children with a 7-repeat allele of the dopamine receptor D4 gene have extreme behavior but normal performance on critical neuropsychological tests of attention. Proc Natl Acad Sci U S A. 2000;97:4754–4759. doi: 10.1073/pnas.080070897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman-Culleré MH, Foltz CJ. Body condition scoring: a rapid and accurate method for assessing health status of mice. Lab Anim Sci. 1999;49:319–323. [PubMed] [Google Scholar]
- Wang ET, Kodama G, Baldi P, Moyzis RK. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc Natl Acad Sci U S A. 2006;103:135–140. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Ding YC, Flodman P, Kidd JR, Kidd KK, Grady DL, Ryder OA, Spence MA, Swanson JM, Moyzis RK. The genetic architecture of selection at the human dopamine receptor D4 (DRD4) gene locus. Am J Hum Genet. 2004;74:931–944. doi: 10.1086/420854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox BJ, He Q, Chen R, Yano K, Masaki KH, Grove JS, Donlon TA, Willcox DC, Curb JD. Midlife risk factors and healthy survival in men. JAMA. 2006;296:2343–2350. doi: 10.1001/jama.296.19.2343. [DOI] [PubMed] [Google Scholar]
- Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability:Genetic interactions create phantom heritability. Proc Natl Acad Sci U S A. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]