Abstract
NKT cells respond to a variety of CD1d-restricted glycolipid antigens that are structurally related to the prototypic antigen, α-galactosylceramide (α-GalCer). A modified analogue of α-GalCer with a carbon-based glycosidic linkage (α-C-GalCer) has generated great interest because of its apparent ability to promote prolonged, Th1-biased immune responses. Here we report the activation of spleen NKT cells to α-C-GalCer, and related C-glycoside ligands, is weaker than that of α-GalCer. Furthermore, the Vβ8.2 and Vβ7 NKT TCR affinity for CD1d-α-C-GalCer, and some related analogues, is approximately 10-fold lower than that for the NKT TCR-CD1d-α-GalCer interaction. Nevertheless, the crystal structure of the Vβ8.2 NKT TCR-CD1d-α-C-GalCer complex is similar to that of the corresponding NKT TCR-CD1d-α-GalCer complex, although subtle differences at the interface provide a basis for understanding the lower affinity of the NKT TCR-CD1d-α-C-GalCer interaction. Our findings support the concept that for CD1d-restricted NKT cells, altered glycolipid ligands can promote markedly different responses while adopting similar TCR docking topologies.
Introduction
Natural Killer T (NKT) cells express semi-invariant αβ T cell receptors (TCRs) that specifically recognise CD1d-restricted glycolipid antigens (Ag). Human NKT cells typically express an invariant Vα24-Jα18 TCR α-chain, and a Vβ11 TCR β-chain, whereas NKT cells from mice possess an invariant TCR α-chain (Vα14-Jα18) and commonly use up to three different TCR βchain V genes (Vβ8, Vβ7, Vβ2) (reviewed in (1, 2)). Upon activation from a variety of stimuli, NKT cells rapidly produce a variety of cytokines that enable them to influence immune outcomes in a broad range of immunological settings (3, 4)(reviewed in (5–7)). The most widely studied glycolipid antigen (Ag) for activating NKT cells is α-galactosylceramide (α-GalCer) (8), a potent NKT cell agonist and phase I therapeutic (reviewed in (9)).
Recently, the structures of NKT TCRs, unliganded (10), and in complex with CD1d-α-GalCer (11, 12), altered glycolipid ligands (AGLs) of α-GalCer (13, 14), α-galactosyldiacylglycerol (α-GalDAG) (15), the self-lipid phosphatidylinositol (PI) (16) and β-linked ligands (17) have been determined. Additionally, the structure of a Vα10 NKT TCR in complex with CD1d-α-glucosylceramide (α-GlcCer) has been reported (18). Collectively, these various NKT TCR-CD1d-Ag complexes exhibited a conserved docking strategy in which the NKT TCR was tilted and exhibited a parallel docking mode in relation to the Ag-binding cleft of CD1d, thereby contrasting that of TCR mediated recognition of pMHC (19). In each case, the NKT TCR α-chain dominated this interaction, with varied contributions from the Complementarity Determining Regions (CDRα) loops being observed among some of the Ags (reviewed in (20)). For example, while the CDR3α loop of the Vα14 NKT TCRs played a central role in interacting with α-GalCer, and AGLs thereof, it played a lesser role in interacting with PI (16). The role of the TCR-Vβ chain was restricted to interacting with CD1d, in which the CDR2β loop played a principal role (20–24). However, it has recently been shown that the hypervariable CDR3β loop can, in a sequence-dependent manner, mediate contacts with CD1d and determine the degree of NKT autoreactivity (16, 17, 25). Thus, within a conserved NKT TCR-CD1d-Ag docking topology, variations between the contributions between the CDR loops is observed in an Ag- and TCR β-dependent manner.
NKT TCRs can bind an array of different lipid-based antigens in complex with CD1d, including bacteria-derived lipid antigens and CD1d-presented mammalian lipid molecules. Furthermore, numerous agonist analogues of α-GalCer have been synthesised that exhibit profound effects on NKT cell function (26, 27). While some AGLs have the ability to promote Th2 biased responses (IL-4 > IFN-γ) downstream of NKT cell activation (28, 29), others promote Th1-biased responses (IFN-γ > IL-4) downstream of NKT cell activation. The AGL α-C-GalCer is considered the prototypical Th1-biasing Ag, also known as C-Glycoside, which possesses a CH2-based glycosidic linkage rather than the oxygen-based glycosidic linkage of α-GalCer and other natural glycosphingolipids (30). Further, a-1C-GalCer, a nonisosteric a-GalCer analogue in which the anomeric carbon of galactose is bonded directly to the sphingolipid backbone, was found to induce a higher IFN-γ/IL-13 ratio in human NKT cells than a-GalCer and a-C-GalCer (31). This C-glycoside modification is intended to provide resistance to enzymatic degradation at the glycosidic linkage by glycosidases, thus increasing the stability of the glycosphingolipid in vivo (30). NKT cell stimulation with α-C-glycoside resulted in a Th1-biased response in vivo, and this is thought to be attributable to more stable presentation by antigen presenting cells, more efficient activation of NKT cells, and prolonged downstream activation of IFN-γ production by NK cells (32). Consequently, α-C-GalCer provides superior protection against melanoma metastasis and malaria infection in mice (30, 33). It has also been suggested that the altered glycosidic linkage may change the way that this glycolipid sits in the CD1d Ag-binding cleft, resulting in structural differences and changes in NKT TCR interactions and affinity (30). This was supported by a recent study (34) that used a CD1d-α-C-GalCer tetramer dissociation assay, which indicated that CD1d-α-C-GalCer had significantly lower affinity for the NKT TCR than CD1d-α-GalCer.
Thus, studies using glycolipid AGLs demonstrate the feasibility of using these to manipulate the NKT cell response, which can translate to more tailored NKT cell based therapies (reviewed in (5)). However, to achieve more targeted NKT-based therapeutics requires a greater understanding of the molecular basis of antigenic modulation of the NKT cell response. Recently, we have provided insight into NKT TCR fine specificity against a range α-GalCer AGLs, including two Th2-biasing ligands (OCH and C20:2) (13). However, the molecular basis for TCR recognition of Th1-biasing ligands, such as the prototypical α-C-GalCer, and analogues thereof, was less clear. To address this, here we report the structural and functional correlates of NKT TCR recognition of CD1d-α-C-GalCer and analogues thereof.
Materials and Methods
Glycolipid Ags
The glycolipid analogues used in this study were synthesised as previously described, although α-1C-GalCer was prepared by a modification of a previously reported route (30, 31, 35). All of the synthesized compounds were characterized by 1H- and 13C-NMR spectroscopy and high-resolution mass spectrometry.
Mice
C57BL/6 mice were maintained in the animal facilities of the Department of Microbiology and Immunology, The University of Melbourne. All animal experiments were approved by, and carried out in accordance with the guidelines of, the University of Melbourne Animal Ethics Committee.
CFSE labelling and proliferation assay
Splenocytes from C57BL/6 mice were labelled with 2 μM 5,6-carboxyfluorescein diacetate succinimidyl ester (CFSE) (Molecular Probes) and incubated for 10 min at 37C. Splenocytes were pulsed with glycolipids at the indicated concentration and cultured overnight (5 × 105 cells per well). Culture media was replaced the next day without further addition of glycolipids. Cells and culture supernatants were harvested at 72 hours for proliferation and cytokine analysis. For proliferation analysis, cells were stained with α-GalCer-loaded CD1d tetramer, and the CFSE signal of gated NKT cells was measured.
In vivo stimulation of NKT cells
Mice were injected intraperitoneally with 1 μg of α-GalCer or α-C-GalCer dissolved in 200 μL PBS, and serum cytokines examined at 2 and 24 hr, using cytometric bead array as described below.
Cytometric bead array
Splenocyte culture supernatants were collected after 72 hr and analysed using cytometric bead array flex sets for mouse IFN-γ, IL-4, IL-10, IL-13, IL-17A, TNF and GM-CSF (BD Biosciences). Serum samples were collected at 2 and 24 hr and analysed for IFN-γ and IL-4.
Intracellular cytokine staining
Splenocytes from C57BL/6 mice were cultured (5 × 105 cells per well) with glycolipid for 8 hours, the final 4 hours in the presence of Golgistop (BD Biosciences). Four replicate wells were pooled for each determination and cells were surface stained with α-GalCer-loaded CD1d tetramer-PE and TCR-β-FITC, fixed, and permeabilized (Cytofix/Cytoperm Kit, BD Biosciences) prior to staining with anti-IFN-γ–allophycocyanin.
Protein expression, refolding, and purification of Vα14 NKT TCRs
The method for cloning, expression, and purification of the mouse Vα14 NKT TCRs has been previously described (11). After the initial refolding and dialysis, the NKT TCR was purified using gel filtration, ion exchange, and hydrophobic interaction chromatography. The refolded Vβ8.2 NKT TCR in 10 mM Tris, pH 8.0, and 150 mM NaCl was concentrated to 7–10 mg/ml for crystallography experiments. The Vβ8.2 and Vβ7 NKT TCRs were also used in the surface plasmon resonance experiments.
Protein expression, purification, and loading of mouse CD1d with glycolipid
Mouse CD1d cloned with a BirA and 6x His tag in a dual promoter baculovirus transfer vector, pBacp10pH was kindly provided by Dr. Mitchell Kronenberg (La Jolla Institute for Allergy and Immunology, CA, USA). Mouse CD1d expression, purification and lipid loading has been described previously (11). Loading of α-C-GalCer was improved using Triton X-100 (Sigma) at a final concentration of 0.05%.
Protein crystallization, structure determination, and refinement
The Vα14-Vβ8.2 NKT TCR-CD1d-α-C-GalCer complex (adjusted to 7 mg/ml in 10 mM Tris, pH 8.0, and 150 mM NaCl) crystallised in 15–17% PEG 10K, 0.1 M ammonium acetate, 0.1 M BisTris (pH 5.5) after 2–3 days. The crystal morphology was further improved by seeding in 22–25 % PEG 400. The crystals were flash frozen in mother liquor containing 25% PEG 400 as the cryoprotectant. The data were collected at the MX2 beamline at the Australian Synchrotron Facility in Melbourne, Australia. The crystals diffracted to 3.2 Å and belong to the space group P212121. The data were processed using Mosflm version 7.0.5 and scaled using SCALA from the CCP4 suite (36). The crystal structure was solved using the molecular replacement method with the PHASER program from the CCP4 suite. The Vα14-Vβ8.2 NKT TCR-CD1d-α-GalCer complex (Protein Data Bank ID code 3HE6 (11)) minus α-GalCer was used as the search model, and the structure was subsequently refined using REFMAC. To prevent model bias, the Rfree set of the Vα14-Vβ8.2 NKT TCR-CD1d-α-GalCer structure was used in the experimental intensities scaling using SCALA as well as the implementation of the simulated annealing protocol in Phenix (37). Model building was carried out using COOT, and at a later stage of refinement translation libration screw parameters were included. The quality of both structures was validated using the PDB validation website. All molecular graphics representations were created using PYMOL (38). The residues that could not be modelled in the structures were CD1d: residues 1–6, 300–302; β2m: residue 1; TCR α chain: residues 1, 134–135, 209–210; TCR β chain: residues 1–2, CDR3β loop 96–102. Electron density of atoms CAG to CAK in the acyl tail and of atoms C7, C8 and C13 to C18 in the sphingosine tail were not resolved and hence the occupancy was set to zero in the final model.
Surface plasmon resonance measurements and analysis
All surface plasmon resonance (SPR) experiments were conducted at 25°C on a Biacore 3000 instrument using HBS buffer (10 mM HEPES-HCl (pH 7.4), 150 mM NaCl, and 0.005% surfactant P20 supplied by the manufacturer). For kinetic experiments, loaded CD1d was coupled to research grade SA chips to a level of 300–500 resonance units (RU). Increasing concentrations of Vβ8.2 (0.078–10 μM) and Vβ7 TCR (0.19–25 μM) was injected over all flow cells at 50 μl/min for 60s. The final response was calculated by subtracting the response of the CD1d-endogenous complex from the Vβ8.2 or Vβ7-CD1d-Ag complex. BIAevaluation version 3.1 (Biacore AB) was used to fit the data to the 1:1 Langmuir binding model to calculate the kinetic constants. All measurements were taken in duplicate. For steady-state affinity measurements, loaded CD1d was coupled to research grade SA chips to a level of 2000 resonance units (RU). Increasing concentrations of Vβ8.2 and Vβ7 TCR (0.039–40 μM) was injected over all flow cells at 5 μl/min for 90s. The final response was calculated by subtracting the response of the CD1d-endogenous complex from the Vβ8.2 or Vβ7-CD1d-Ag complex. The equilibrium data were analyzed using GraphPad Prism.
Results
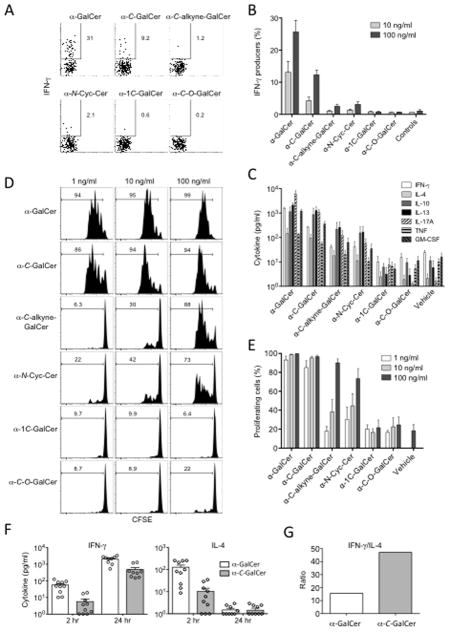
The proliferative and functional response of NKT cells to α-C-GalCer analogues
A series of α-GalCer analogues with modified α-glycosidic linkages were tested for their ability to stimulate splenic NKT cell proliferation and cytokine production. These α-glycoside analogues included α-C-GalCer (structurally identical to that used in previous publications) with a CH2 group in place of the glycosidic oxygen atom (30); a nonisosteric analogue that has one less CH2 group than α-C-GalCer in the link between the sugar and the sphingolipid (α-1C-GalCer) (31); a compound with a rigid triple bond in the link between the sugar and sphingolipid (α-C-alkyne-GalCer) (35); a variant bearing an ether oxygen atom in the linker (α-C-O-GalCer); and an aminocyclitol variant (α-N-Cyc-Cer)(39). The structures of these compounds and that of the O-glycoside standard (α-GalCer) are displayed in Figure 1. IFN-γ production was examined at 8 hours by intracellular cytokine staining (ICS) (Figure 2A and B) and several cytokines were examined at 72 hr spleen cell cultures by cytometric bead array (CBA) (Figure 2C). IFN-γ production at 8 hr showed that the strongest response was induced by α-GalCer with approximately 25% IFN-γ+ at the highest dose (100 ng/ml), whereas α-C-GalCer only triggered ~12% of the NKT cells to produce detectable levels of IFN-γ at this dose. Of the other AGLs, α-C-alkyne-GalCer and α-N-Cyc-Cer were ~10 fold lower again, and α-1C-GalCer and α-C-O-GalCer did not induce IFN-γ staining above background (Figure 2A and B). The CBA data showed a similar trend for other cytokines, with α-C-GalCer inducing 2–5 fold lower amounts compared to α-GalCer (Figure 2C) and the other AGLs inducing still lower amounts of the other cytokines assayed. There was little difference in the proliferative response to α-C-GalCer compared to α-GalCer, with nearly all cells proliferating at the two highest doses (100 and 10 ng/ml), and only a slight drop in proliferation at the lowest dose (1 ng/ml) (Figure 2D and 2E). Moderate proliferation was observed for α-C-alkyne-GalCer and α-N-Cyc-Cer especially at the highest dose, whereas no proliferation (above background) was detected for α-1C-GalCer and α-C-O-GalCer. The reduced IFN-γ response following α-C-GalCer compared to α-GalCer stimulation in vitro seemed inconsistent with α-C-GalCer’s apparent ability to promote Th1-biased responses in vivo (30). Therefore, we compared the serum cytokine response to in vivo challenge with these two glycolipids. Consistent with a recently published study (34) we found that α-C-GalCer induced lower amounts of both IL-4 and IFN-γ production in vivo compared to α-GalCer (Figure 2F), although the ratio of the peak IFN-γ response (at 24 hours) to the peak IL-4 response (at 2 hours) still revealed a Th1-like bias (Figure 2G), similar to that in earlier reports. This is thought to be due to greater in vivo stability of α-C-GalCer and prolonged production of IFN-γ by downstream NK cells in vivo, rather than more potent IFN-γ production by NKT cells (30, 34).
FIGURE 1. Structures of the glycolipid Ags used.
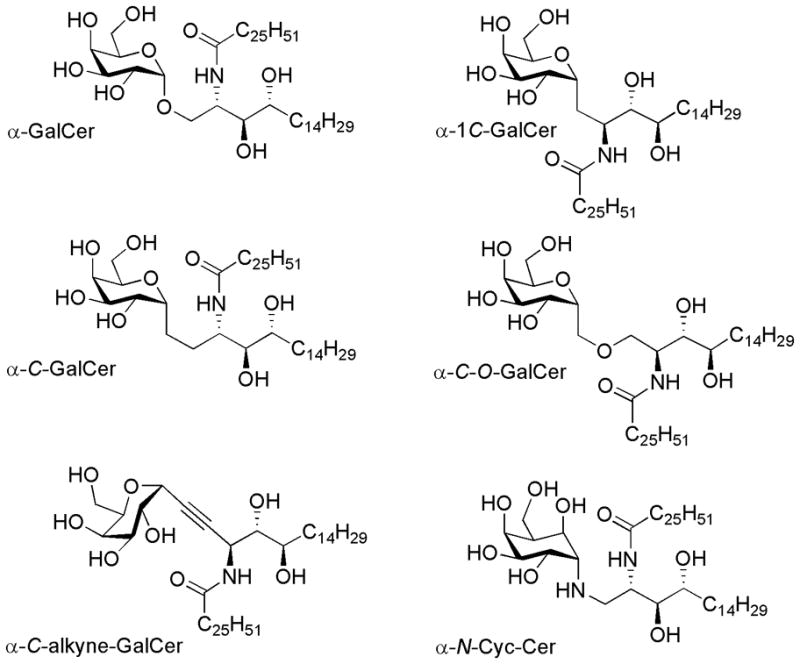
α-GalCer, α-C-GalCer, α-1C-GalCer, α-C-alkyne-GalCer, α-C-O-GalCer, α-N-Cyc-Cer.
FIGURE 2. In vitro stimulation of spleen NKT cells with α-C-GalCer and associated analogues.
Spleen cells were cultured with 100, 10, and 1 ng/ml of the glycolipids as indicated. A. After 8 hours, cell cultures were harvested and surface labelled with anti-TCR-β and α-GalCer-loaded CD1d tetramer then fixed and permeabilised for ICS for IFN-γ production. B. The mean ± SEM from three independent experiments testing IFN-γ production by ICS (as per A) are shown. C. Cytokine production at 72 hours from spleen cells cultured with the various analogues was determined by cytometric bead array. The mean ± SEM from three independent experiments are shown. The exception is IL-10, which was only measured in 2 experiments. D. Representative data showing the proliferation of NKT cells in response to the different ligands as determined by CFSE labelled spleen cells, gating on α-GalCer-CD1d tetramer + cells after 72 hr. E. The CFSE proliferation results for NKT cells (as depicted in C) from four independent experiments (mean ± SEM) are shown. F. Cytokines detected in the serum of mice determined by cytometric bead array following the i.p. administration of 1 μg of glycolipid. The combined results from three independent experiments (10 mice per group) are shown. G. The ratio of serum IFN-γ (24 hr) to serum IL-4 (2 hr). For B, C and E, the data readings for each individual experiment were the mean of duplicate cultures. Error bars represent SEM.
Taken together, these data are consistent with earlier studies suggesting that α-C-GalCer is a weaker NKT cell agonist than α-GalCer (34). Moreover, our data demonstrate that of the AGLs with modified glycosidic linkages tested in this study, α-C-GalCer is the most potent with respect to proliferative response and cytokine production, followed by α-C-alkyne-GalCer and α-N-Cyc-Cer, while the α-C-O-GalCer and α-1C-GalCer AGLs were greatly diminished in their potency.
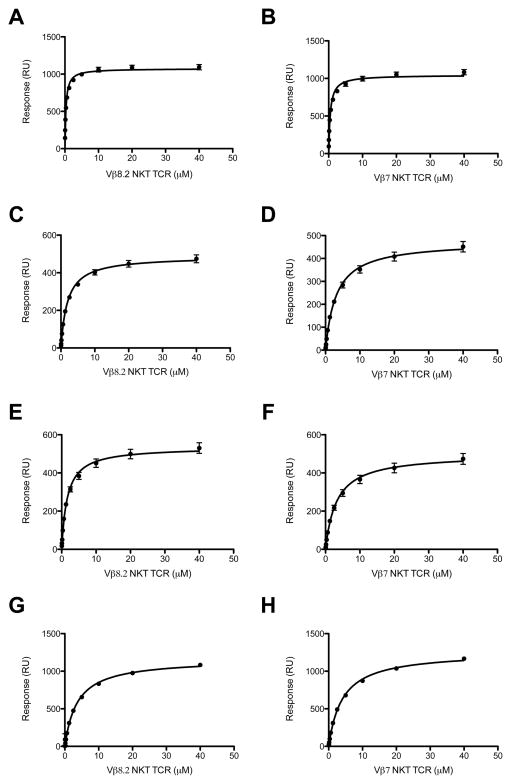
Affinity measurements
Previous studies using a tetramer-binding assay with Vα14+Vβ8.2+ NKT cell hybridomas showed that CD1d-α-C-GalCer tetramer bound to these cells with a much weaker avidity than the CD1d-α-GalCer complexes (34). We determined the affinity of the interaction between the Vβ8.2 and Vβ7 NKT TCRs and CD1d-α-C-GalCer complex (and analogues thereof) using surface plasmon resonance (SPR) and compared these values to the CD1d-α-GalCer interaction (Figure 3A-H). The affinity for the Vβ8.2 NKT TCR was ≈ 2 μM, which was determined by response at equilibrium (equilibrium dissociation constant, KD(eq)) (Figure 3C), and was significantly weaker than that of the corresponding NKT TCR-CD1d-α-GalCer interaction (300 nM) (Figure 3A)(Table I) (11). The affinity of the Vβ7 NKT TCR for CD1d-α-C-GalCer was ≈ 3 μM (Figure 3D) (and 470nM for the α-GalCer interaction, Figure 3B), indicating little difference between the affinity of Vβ8.2 and Vβ7 NKT TCRs for α-C-GalCer. We also determined the rate constants of the interaction between the Vβ8.2 NKT TCR and CD1d-α-C-GalCer. For the NKT TCR-CD1d-α-GalCer interaction, the on-rate was ka = 1.62 × 105 M−1s−1, the off-rate was kd = 0.02 s−1 and accordingly the half-life (t1/2) was long, approximately 35 s (13) (Supplementary Figure 2, Table 1). In contrast, for the NKT TCR-CD1d-α-C-GalCer interaction, the on-rate was approximately the same (ka = 2.25 × 105 M−1s−1), yet the off rate was faster (kd = 0.29 s−1), such that t1/2 of the interaction was only 2.4s (Supplementary Figure 2, Table 1). Recently, we have shown that for glycosyl-based modifications, the half-life on the interaction is linked to the potency of the ligand, which is consistent with α-C-GalCer being a less potent inducer of IFN-γ production from NKT cells compared with α-GalCer (Figure 2A) (13). We also measured the affinity of the interaction between the Vβ8.2 and Vβ7 NKT TCRs and the other AGLs depicted in Figure 1 (Table I, Figure 3E–H). For the five AGLs tested, the affinities between the Vβ8.2 and Vβ7 NKT TCRs were similar (ranging from ≈ 1.6 – 4μM), indicating no marked hierarchical recognition properties, whereas Vβ8.2 NKT TCRs trended towards preferential recognition of α-GalCer when compared to Vβ7 NKT TCRs (Table I and (24)). The binding affinity for the α-C-alkyne-GalCer and α-N-Cyc-GalCer analogues were similar to α-C-GalCer (Table I), indicating that the modifications did not appreciably affect the affinity of the interaction, despite the differing stimulatory activity to these Ags. This finding is consistent with previous studies, where other factors (eg Ag processing and presentation mechanisms and ligand stability) besides binding affinity of the NKT TCR can contribute to the biological responses of NKT cells (34, 40). Nevertheless, consistent with previous studies on α-GalCer analogues (13), the levels of cytokines produced by the NKT cells broadly reflects the NKT TCR affinity of these α-C-glycoside ligands.
FIGURE 3. Binding analysis of Vβ8.2 and Vβ7 to CD1d-Ag.
Equilibrium binding response curve for Vβ8.2 TCR to CD1d-α-GalCer (A), CD1d-α-C-GalCer (C), CD1d-α-C-alkyne-GalCer (E) and CD1d-α-N-Cyc-Cer (G) and Vβ7 TCR to CD1d-α-GalCer (B), CD1d-α-C-GalCer (D), CD1d-α-C-alkyne-GalCer (F) and CD1d-α-N-Cyc-Cer (H). The equilibrium dissociation constant (KDeq) by fitting to one-site binding model. All results are shown as one experiment performed in triplicate (representative of 2–3 experiments).
Table I.
Affinity measurements of C-glycoside analogues with Vβ8.2 TCR and Vβ7 TCR
| ka (M−1s−1) | kd (s−1) | KD (μM) | KD(eq)a (μM) | t1/2 (s) | |
|---|---|---|---|---|---|
| Vβ8.2 TCR | |||||
| α-GalCer | 1.62×105 ± 9.5×103 | 0.02 ± 4.5×10−4 | 0.13 ±0.01 | 0.31 ±0.02 | 34.65 |
| α-C-GalCer | 2.25×105 ± 4.1×104 | 0.29 ± 0.02 | 2.20 ± 0.73 | 1.95 ± 0.11 | 2.39 |
| α-N-Cyc-Cer | 5.67×104 ± 1.67×104 | 0.37 ± 0.01 | 7.10 ± 1.97 | 3.68 ± 0.27 | 1.87 |
| α-C-alkyne-GalCer | ND | ND | ND | 1.64 ± 0.13 | ND |
| α-1C-GalCer | NB | NB | NB | NB | NB |
| α-C-O-GalCer | NB | NB | NB | NB | NB |
| Vβ7 TCR | |||||
| α-GalCer | 2.31×105 ± 3.15×104 | 0.09 ± 0.003 | 0.38 ± 0.01 | 0.47 ± 0.03 | 7.70 |
| α-C-GalCer | ND | ND | ND | 3.11 ± 0.23 | ND |
| α-N-Cyc-Cer | ND | ND | ND | 4.06 ± 0.10 | ND |
| α-C-alkyne-GalCer | ND | ND | ND | 3.23 ± 0.29 | ND |
| α-1C-GalCer | NB | NB | NB | NB | NB |
| α-C-O-GalCer | NB | NB | NB | NB | NB |
steady-state affinity determined in separate experiment (see materials and methods)
NB= No binding; ND=not determined
No measurable binding affinity was observed for the α-1C-GalCer and the α-C-O-GalCer analogues (Table I), consistent with the lack of NKT cell stimulation in response to these ligands (Figure 2). Taken together, our findings suggest that these two analogues are not CD1d restricted antigens for mouse NKT cells.
The NKT TCR-CD1d-α-C-GalCer complex
Since the ceramide base and the α-galactosyl head group of α-C-GalCer is identical to α-GalCer, the only difference is the CH2 vs O-based glycosidic linkage; thus it is not obvious why there would be such a large difference in TCR-binding affinity to this glycolipid analogue. To determine how the NKT TCR binds to CD1d-α-C-GalCer, and to understand why the affinity of this interaction was much weaker than the NKT TCR-CD1d-α-GalCer interaction, we expressed and refolded the Vβ8.2 NKT TCR and ligated it to CD1d specifically loaded with α-C-GalCer. We subsequently determined the structure of Vα14Jα18-Vβ8.2 NKT TCR-CD1d-α-C-GalCer complex to 3.2 Å resolution to an Rfac and Rfree of 22.6 and 28.9%, respectively (Figure 4A and Supplemental Table I). The initial phases clearly showed unbiased electron density for α-C-GalCer head group (Supplemental Figure 1). The electron density at the NKT TCR-CD1d-α-C-GalCer interface was unambiguous, with the exception of a mobile CDR3β loop (residues 94 to 104) and side-chains of Leu99α and Arg103α of the CDR3α, which were not resolved in the final structure. Consistent with this, some previous Vβ8+ NKT TCR-CD1d-Ag structures have exhibited mobility in the CDR3β loop (11, 13). The structure of the NKT TCR-CD1d-α-C-GalCer complex enabled us to compare it to the closely related NKT TCR-CD1d-α-GalCer complex that had been solved to similar resolution (11) (Figure 4A,B). Further, the NKT TCR-CD1d-α-C-GalCer and NKT TCR-CD1d-α-GalCer complexes crystallized in identical space groups and isomorphous unit cell dimensions, and thus any structural differences observed between these two NKT TCR-CD1d-Ag complexes can be attributed to the impact of the specific modification of the α-C-GalCer.
FIGURE 4. Structure of Vα14-Vβ8.2 NKT TCRs in complex with CD1d-α-C-GalCer.
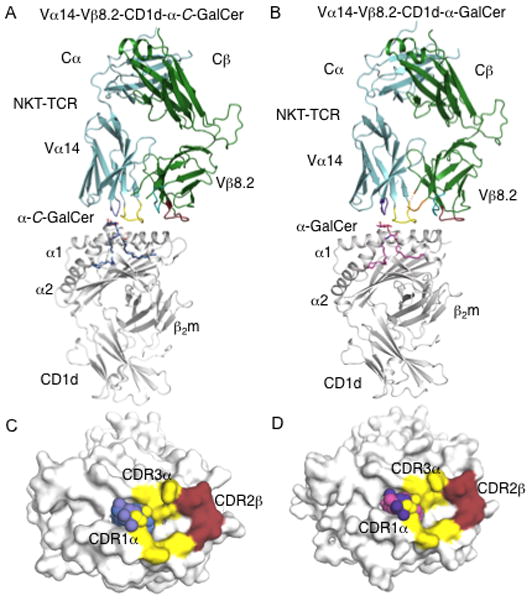
(A)Vα14-Vβ8.2 NKT TCR in complex with CD1d-α-C-GalCer. α-C-GalCer, blue; CD1d heterodimer, grey; Vα14, cyan; Vβ8.2, green. CDR1α, purple; CDR3α, yellow; CDR1β, teal; CDR2β, ruby; CDR3β, not modelled. B, Vα14-Vβ8.2 NKT TCR in complex with CD1d-α-GalCer. α-GalCer, magenta; CDR3β, orange; CD1d, Vα14, Vβ8.2, CDR1α, CDR3α, CDR1β, CDR2β colour coding as in A. C, Footprint of Vα14-Vβ8.2 on the surface of CD1d-α-C-GalCer. α-C-GalCer is shown in spheres. CD1d, α-C-GalCer and CDR loops colour coding as in A. D, footprint of Vα14-Vβ8.2 on the surface of CD1d-α-GalCer. CD1d, α-GalCer and CDR loops colour coding as in A and B.
The NKT TCR adopted the docking mode observed for the NKT TCR-CD1d-α-GalCer complex (11, 12), indicating that the α-C-GalCer did not cause a significant re-positioning of the NKT TCR. Namely, within the NKT TCR-CD1d-α-C-GalCer complex, the NKT TCR bound approximately parallel to, and above, the F′-pocket of the CD1d-Ag binding cleft (Figure 4C). The buried surface area (BSA) upon ligation is ≈ 690 Å2, which is lower than BSA ≈ 760 Å2 for the complex with α-GalCer (Figure 4C and D). The lower BSA is attributable to the increased mobility of the CDR3α loop within this complex, in which the Leu 99α and Arg 103α side chains were not resolved in the electron density, and thus were not included in the final refined model. In the NKT TCR-CD1d-α-GalCer complex, Leu 99α and Arg 103α make specificity-governing contacts with CD1d. Thus, the increased mobility of Leu 99α and Arg 103α in the NKT TCR-CD1d-α-C-GalCer complex would contribute to the lower affinity of this interaction. Similar observations were observed in the structure of the NKT TCR-CD1d-α-GlcCer complex, where increased mobility of the CDR3α loop was also attributed to a lower affinity interaction (13).
Within the NKT TCR-CD1d-α-C-GalCer interface, the TCR α-chain contributes 68% of the BSA, in which the CDR1α and CDR3α contributed 20% and 48% of the BSA, respectively. As observed previously in the Vα14-Vβ8.2 NKT TCR-CD1d-Ag structures, the Vβ8.2 interactions were mediated solely via the CDR2β (32% BSA). These values are very similar to those in the NKT TCR-CD1d-α-GalCer interaction, with the CDR1α, CDR3α, and CDR2β loops contributing 17%, 57%, and 26%, respectively (11). The slightly higher BSA of the CDR2β in the NKT TCR-CD1d-α-C-GalCer complex arises from small differences in the Vα-Vβ juxtapositioning between the two ternary complexes. The CDR2β interactions include the conserved interactions mediated by Tyr 48β, Tyr 50β, and Glu 56β, which have been observed in all Vβ8.2 NKT TCR-CD1d-Ag complexes solved to date (11, 13).
Regarding the TCR α-chain, most of the interactions observed in the NKT TCR-CD1d-α-GalCer complex were preserved in the NKT TCR-CD1d-α-C-GalCer complex (11), thereby collectively providing a focussed network of polar and salt-bridging interactions with CD1d (see Table II). However, in comparison to the NKT TCR-CD1d-α-GalCer complex, there was a loss of a H-bond between main-chain Gly96α and Asp153 of the α2 helix of CD1d in the NKT TCR-CD1d-α-C-GalCer complex (Figure 5A and B). The loss of this interaction was attributable to a 1 Å backbone shift at the Gly96α tip of the CDR3α loop, which was a result of a small shift in the head group position at the C3 carbon, whereby α-C-GalCer sat in a higher position at this carbon compared to that of α-GalCer (Figure 5C).
Table II.
Contacts at the NKT TCR-CD1d-α-C-GalCer interface
| CDR loop | Vβ8.2 in α-GalCer complex | CD1d | Bond | Vβ8.2 in α-C-GalCer complex | CD1d | Bond |
|---|---|---|---|---|---|---|
| CDR1α | - | - | - | Pro31 | Ser76 | VDW |
| CDR3α | Asp94Oδ1 | Arg79Nη1, Arg79Nη2 | Salt-bridge | Asp94Oδ1 | Arg79Nη2 | Salt-bridge |
| Asp94Oδ2 | Arg79Nη1, Arg79Nη2 | Salt-bridge | Asp94Oδ2 | Arg79Nη1, Arg79Nη2 | Salt-bridge | |
| Asp94 | Arg79 | VDW | Asp94 | Arg79 | VDW | |
| Arg95Nε | Asp80Oδ1, Asp80Oδ2 | Salt-bridge | Arg95Nε | Asp80Oδ1, Asp80Oδ2 | Salt-bridge | |
| Arg95Nη1 | Asp80Oδ1 Ser76Oγ |
Salt-bridge H-bond |
Arg95Nη1 | Asp80Oδ1 Arg79Nε |
Salt-bridge H-bond |
|
| Arg95 | Asp80, Arg79, Ser76 | VDW | Arg95 | Asp80, Ser76, Arg79 | VDW | |
| Gly96N | Asp153Oδ2 | H-bond | Gly96N | - | - | |
| Gly96 | Ala152, Asp153 | VDW | Gly96 | Ala152, Asp153 | VDW | |
| Ser97 | Val149 | VDW | Ser97 | Val149 | ||
| Leu99 | Arg79, Val149 | VDW | Leu99 | Arg79, Asp80 | VDW | |
| Leu99O | Arg79Nη2 | H-bond | Leu99O | Arg79Nη2 | H-bond | |
| Gly100 | Arg79 | VDW | Gly100 | Arg79 | VDW | |
| Arg103 | Arg79, Glu83 | VDW | Arg103 | Arg79 | VDW | |
| Arg103Nη1 | Glu83Oε2, Glu83Oε1 | Salt-bridge | ||||
| CDR2β | Tyr48Oη | Glu83Oε1, Glu83Oε2, Lys86Nζ | H-bond | Tyr48Oη | Glu83Oε1, Lys86Nζ | H-bond |
| Tyr48 | Glu83, Lys86 | VDW | Tyr48 | Glu83, Lys86 | VDW | |
| Tyr50Oη | Glu83Oε1 | H-bond | Tyr50Oη | Glu83Oε1 | H-bond | |
| Tyr50 | Glu83, Met87 | VDW | Tyr50 | Glu83 | VDW | |
| Glu56Oε1 | Lys86Nζ | Salt-bridge | Glu56Oε2 | Lys86Nζ | Salt-bridge | |
| Glu56 | Lys86 | VDW | Glu56 | Lys86 | VDW | |
| Vβ8.2 in α-GalCer complex | α-GalCer | Vβ8.2 in α-C-GalCer complex | α-C-GalCer | |||
| CDR1α | Pro28 | 6′-OH, 5′-O, C-1 | VDW | Pro28 | 6′-OH, 5′-O, C-1 | VDW |
| Asn30 | C-2, C-3, C-4 3′-OH, 4′-OH |
VDW | Asn30 | C-3, 3′-OH, 4′-OH | VDW | |
| Asn30Nδ2 | 3′-OH 4′-OH |
H-bond H-bond |
Asn30Nδ2 | 3′-OH | H-bond | |
| CDR3α | Asp94O | C-1 | VDW | Asp94O | C-1, C-2 | VDW |
| Arg95 | 2′-OH, C-2, 3-OHS | VDW | Arg95 | 2′-OH, 3-OHS | VDW | |
| Gly96N | 2′-OH | H-bond | Gly96N | 2′-OH | H-bond | |
| Gly96 | C-2, 3′-OH | VDW |
“-” interaction does not exist
FIGURE 5. Vα14-Vβ8.2 NKT TCR mediated interactions with mouse CD1d-glycolipid interface.
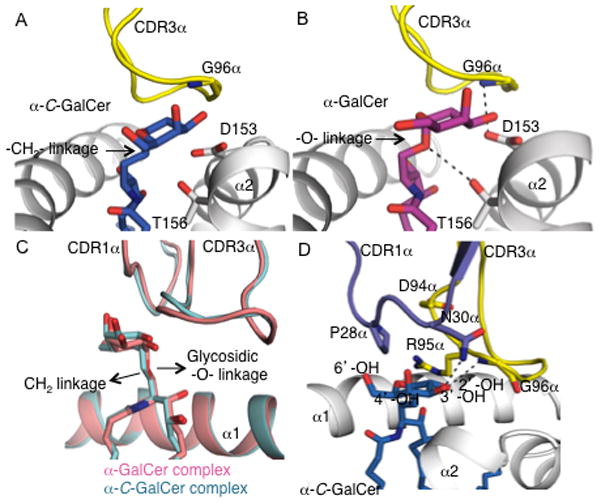
A, Vα14-Vβ8.2 NKT TCR CDR3α and mCD1d mediated contacts with α-C-GalCer. α-C-GalCer, blue; CDR3α, yellow; CD1d, grey. B, Vα14-Vβ8.2 NKT TCR CDR3α and CD1d mediated contacts with α-GalCer. α-GalCer, magenta; CDR3α, and mCD1d colour coding as in A. H-bonds are shown in black dashed lines. C, superposition of mouse Vα14-Vβ8.2 NKT TCR-CD1d-α-C-GalCer and Vα14-Vβ8.2 NKT TCR-CD1d-α-GalCer complexes. Vα14Vβ8.2 NKT TCR-CD1d-αC-GalCer is shown in cyan and Vα14-Vβ8.2 NKT TCR-CD1d-α-GalCer is shown in pink. The replacement of the glycosidic -O- linkage in α-GalCer to a hydrophobic -CH2- linkage in α-C-GalCer does not result in a major head group movement. D, Vα14-Vβ8.2 NKT TCR CDR1α and CDR3α mediated contacts with α-C-GalCer. α-C-GalCer, blue; CDR1α, purple; CDR3α, yellow; mCD1d, grey. H-bonds are shown in black dashed lines.
Interactions with the glycosyl head group
Similar to the CD1d-α-GalCer interactions observed in the NKT TCR-CD1d-α-GalCer complex, the 2′-OH and 3′-OH groups of the galactosyl moiety of α-C-GalCer H-bonded to Asp153, while the 3′-OH and 4′-OH of the sphingosine chain H-bonded to Asp80 (11). However due the presence of an –CH2- linkage in α-C-GalCer instead of a glycosidic –O-linkage in α-GalCer resulted in loss of H-bond between Thr156 and the lipid (Figure 5A and B). As previously observed with Vβ8.2 NKT TCRs, the CDR1α and CDR3α loops interact with the galactosyl head group of α-C-GalCer (11). Within the ternary complex, replacement of the glycosidic oxygen in α-GalCer with a CH2 group in α-C-GalCer caused no major re-orientation of the head group (Figure 5C). In comparison to NKT TCR-CD1d-α-GalCer, most of the interactions between α-C-GalCer and the TCR are maintained. Namely, the galactose ring sat underneath the CDR1α loop and abutted the CDR3α loop, forming van der Waals (VDW) contacts on one face of the sugar ring. Further, Gly96α H-bonded to the 2′-OH group, while Asn30α forms H-bonds to the 3′-OH group of the galactose ring. However, dissimilar to the NKT TCR-CD1d-α-GalCer complex, the H-bond between Asn30 and the 4′-OH group was absent, which was attributable to the slightly differing orientations of the respective galactosyl head groups (11) (Figure 5D). As we have recently shown that the 4′-OH group of α-GalCer is more critical than the 3′-OH group in mediating contacts with the Vβ8.2 NKT TCR, the observation of a loss of a H-bond between the 4′-OH group of α-C-GalCer and the NKT TCR is consistent with the lower affinity of the interaction compared with the α-GalCer interaction (Table I) (13).
Discussion
The structures of a number of NKT TCR-CD1d-Ag complexes determined to date have shown that a conserved docking topology underpins the interaction, regardless of the nature of the bound antigenic ligand, or indeed Vα-Jα and Vβ usage (13–18). Nevertheless some CD1d-restricted ligands can exert markedly differing biological effects, and the α-C-GalCer ligand is considered the prototypical Th1-biasing ligand. Our results show that the NKT TCR-CD1d-αGalCer and α-C-GalCer docking topologies are very similar. We provide a clear molecular basis for the lower affinity interaction observed with α-C-GalCer compared with α-GalCer, which was largely attributable to reduced contacts between the NKT TCR and the sugar head group of α-C-GalCer as well as the α-2 helix of CD1d. Similarly, in comparison to the Vβ8.2 NKT TCR-CD1d-α-GalCer complex, Aspeslagh et al reported a loss of an interaction between a Vβ8.2 NKT TCR and CD1d-C-GalCer, where a loss of H-bond between Arg95α to the 3-OH of phytosphingosine was observed (14) in comparison to the loss of H-bond between Asn30α of the Vβ8.2 NKT TCR to 4′-OH of galactose reported here. These differences could be attributable to the differing CDR3β sequences used in the respective studies, which is consistent with the much higher affinity (247nM) for α-C-GalCer reported by Aspeslagh et al (14) in contrast to the affinity measurements reported here (2μM). Regardless, the diminished contacts in the NKT TCR-CD1d-α-C-GalCer complex most likely explains the reduced effects on NKT cell activation and cytokine production against α-C-GalCer and its related analogues tested in this study. Moreover, these reduced contacts were consistent with the much shorter ½ life of the interaction when compared to the NKT TCR interaction with CD1d-α-GalCer. However, despite this, the proliferative response of NKT cells to α-C-GalCer was only slightly reduced compared to α-GalCer, which is consistent with our earlier report showing that TCR-affinity for glycolipid analogues does not directly correlate with proliferation. This may also reflect an earlier report demonstrating the sustained in vivo response to this analogue despite reduced short-term cytokine production (34). Taken together, our molecular and functional data for α-C-GalCer and analogues thereof, in combination with an earlier report (34), highlight the fact that even subtle changes in glycolipid structure can have profound effects on TCR-glycolipid-CD1d contacts and the affinity of the NKT TCR interaction. In turn, this can clearly affect the initial stages of NKT cell activation and cytokine production, whereas the duration and downstream effects of NKT cell activation appear to be less associated with the affinity of the ligand and more closely linked to the stability of its presentation.
Collectively, our findings provide a basis for understanding the fine specificity of the NKT cell mediated response that will be of enormous value in tailor making NKT cell AGLs for therapeutic benefit.
Supplementary Material
Atomic contacts determined using the CCP4i implementation of CONTACT and a cutoff of 4.5 A.
Van der Waals interactions defined as non-hydrogen bond contact distances of 4 Å or less.
Hydrogen bond interactions are defined as contact distances of 3.3 Å or less.
Salt bridge is defined as contact distance of 4.5 Å or less.
α-GalCer complex data is derived from (11) and shown for comparison.
Acknowledgments
We wish to thank the synchrotron staff at the MX2 beamline of the Australian synchrotron for assistance with data collection and David Taylor and the animal house staff at the University of Melbourne, Dept. of Microbiology and Immunology, for animal husbandry assistance. We thank the NIH tetramer facility for providing us with α-C-GalCer. We thank Maria Sandoval for technical assistance.
Footnotes
This work was supported by the Cancer Council of Victoria, the National Health and Medical Research Council of Australia (NHMRC), and the Australian Research Council. GC is supported by a Cancer Research Institute pre-doctoral scholarship. Financial support from NIH Grants AI45889 (to SAP) and HL083187 (to RB) is gratefully acknowledged. AL and RC are supported by MICINN and FEDER grant CTQ2008-01426. DIG is supported by an NHMRC Principal Research Fellowship; JR is supported by an ARC Federation Fellowship.
Accession codes: The coordinates for the NKT TCR-CD1d-α-C-GalCer complex have been deposition in the Protein Data Bank (3TN0) www.rcsb.org.
References
- 1.Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what’s in a name? Nat Rev Immunol. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 2.Bendelac A, Savage PB, Teyton L. The Biology of NKT Cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 3.Wingender G, Rogers P, Batzer G, Lee MS, Bai D, Pei B, Khurana A, Kronenberg M, Horner AA. Invariant NKT cells are required for airway inflammation induced by environmental antigens. The Journal of Experimental Medicine. 208:1151–1162. doi: 10.1084/jem.20102229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brigl M, Tatituri RVV, Watts GFM, Bhowruth V, Leadbetter EA, Barton N, Cohen NR, Hsu F-F, Besra GS, Brenner MB. Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. The Journal of Experimental Medicine. 208:1163–1177. doi: 10.1084/jem.20102555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerundolo V, Silk JD, Masri SH, Salio M. Harnessing invariant NKT cells in vaccination strategies. Nat Rev Immunol. 2009;9:28–38. doi: 10.1038/nri2451. [DOI] [PubMed] [Google Scholar]
- 6.Godfrey DI, Kronenberg M. Going both ways: immune regulation via CD1d-dependent NKT cells. J Clin Invest. 2004;114:1379–1388. doi: 10.1172/JCI23594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godfrey DI, Rossjohn J. New ways to turn on NKT cells. The Journal of Experimental Medicine. 2011;208:1121–1125. doi: 10.1084/jem.20110983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, Koseki H, Taniguchi M. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 9.Cerundolo V, Barral P, Batista FD. Synthetic iNKT cell-agonists as vaccine adjuvants†--†finding the balance. Current Opinion in Immunology. 2010;22:417–424. doi: 10.1016/j.coi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Kjer-Nielsen L, Borg NA, Pellicci DG, Beddoe T, Kostenko L, Clements CS, Williamson NA, Smyth MJ, Besra GS, Reid HH, Bharadwaj M, Godfrey DI, Rossjohn J, McCluskey J. A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition. J Exp Med. 2006;203:661–673. doi: 10.1084/jem.20051777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pellicci DG, Patel O, Kjer-Nielsen L, Pang SS, Sullivan LC, Kyparissoudis K, Brooks AG, Reid HH, Gras S, Lucet IS, Koh R, Smyth MJ, Mallevaey T, Matsuda JL, Gapin L, McCluskey J, Godfrey DI, Rossjohn J. Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity. 2009;31:47–59. doi: 10.1016/j.immuni.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, Rossjohn J. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 13.Wun KS, Cameron G, Patel O, Pang SS, Pellicci DG, Sullivan LC, Keshipeddy S, Young MH, Uldrich AP, Thakur MS, Richardson SK, Howell AR, Illarionov PA, Brooks AG, Besra GS, McCluskey J, Gapin L, Porcelli SA, Godfrey DI, Rossjohn J. A Molecular Basis for the Exquisite CD1d-Restricted Antigen Specificity and Functional Responses of Natural Killer T Cells. Immunity. 2011;34:327–339. doi: 10.1016/j.immuni.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aspeslagh S, Li Y, Yu ED, Pauwels N, Trappeniers M, Girardi E, Decruy T, Van Beneden K, Venken K, Drennan M, Leybaert L, Wang J, Franck RW, Van Calenbergh S, Zajonc DM, Elewaut D. Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis. Embo J. 2011;30:2294–2305. doi: 10.1038/emboj.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Girardi E, Wang J, Yu ED, Painter GF, Kronenberg M, Zajonc DM. The Va14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode. The Journal of Experimental Medicine. 2010;207:2383–2393. doi: 10.1084/jem.20101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mallevaey T, Clarke AJ, Scott-Browne JP, Young MH, Roisman LC, Pellicci DG, Patel O, Vivian JP, Matsuda JL, McCluskey J, Godfrey DI, Marrack P, Rossjohn J, Gapin L. A Molecular Basis for NKT Cell Recognition of CD1d-Self-Antigen. Immunity. 2011;34:315–326. doi: 10.1016/j.immuni.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pellicci DG, Clarke AJ, Patel O, Mallevaey T, Beddoe T, Le Nours J, Uldrich AP, McCluskey J, Besra GS, Porcelli SA, Gapin L, Godfrey DI, Rossjohn J. Recognition of [beta]-linked self glycolipids mediated by natural killer T cell antigen receptors. Nat Immunol. 2011 doi: 10.1038/ni.2076. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uldrich AP, Patel O, Cameron G, Pellicci DG, Day EB, Sullivan LC, Kyparissoudis K, Kjer-Nielsen L, Vivian JP, Cao B, Brooks AG, Williams SJ, Illarionov P, Besra GS, Turner SJ, Porcelli SA, McCluskey J, Smyth MJ, Rossjohn J, Godfrey DI. A semi-invariant V[alpha]10+ T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigen-recognition properties. Nat Immunol. 2011;12:616–623. doi: 10.1038/ni.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godfrey DI, Rossjohn J, McCluskey J. The Fidelity, Occasional Promiscuity, and Versatility of T Cell Receptor Recognition. Immunity. 2008;28:304V314. doi: 10.1016/j.immuni.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Godfrey DI, Pellicci DG, Patel O, Kjer-Nielsen L, McCluskey J, Rossjohn J. Antigen recognition by CD1d-restricted NKT T cell receptors. Semin Immunol. 2010;22:61–67. doi: 10.1016/j.smim.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Scott-Browne JP, Matsuda JL, Mallevaey T, White J, Borg NA, McCluskey J, Rossjohn J, Kappler J, Marrack P, Gapin L. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat Immunol. 2007;8:1105–1113. doi: 10.1038/ni1510. [DOI] [PubMed] [Google Scholar]
- 22.Florence WC, Xia C, Gordy LE, Chen W, Zhang Y, Scott-Browne J, Kinjo Y, Yu KO, Keshipeddy S, Pellicci DG, Patel O, Kjer-Nielsen L, McCluskey J, Godfrey DI, Rossjohn J, Richardson SK, Porcelli SA, Howell AR, Hayakawa K, Gapin L, Zajonc DM, Wang PG, Joyce S. Adaptability of the semi-invariant natural killer T-cell receptor towards structurally diverse CD1d-restricted ligands. EMBO J. 2009;28:3579–3590. doi: 10.1038/emboj.2009.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wun KS, Borg NA, Kjer-Nielsen L, Beddoe T, Koh R, Richardson SK, Thakur M, Howell AR, Scott-Browne JP, Gapin L, Godfrey DI, McCluskey J, Rossjohn J. A minimal binding footprint on CD1d-glycolipid is a basis for selection of the unique human NKT TCR. The Journal of Experimental Medicine. 2008;205:939–949. doi: 10.1084/jem.20072141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallevaey T, Scott-Browne JP, Matsuda JL, Young MH, Pellicci DG, Patel O, Thakur M, Kjer-Nielsen L, Richardson SK, Cerundolo V, Howell AR, McCluskey J, Godfrey DI, Rossjohn J, Marrack P, Gapin L. T cell receptor CDR2 beta and CDR3 beta loops collaborate functionally to shape the iNKT cell repertoire. Immunity. 2009;31:60–71. doi: 10.1016/j.immuni.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matulis G, Sanderson JP, Lissin NM, Asparuhova MB, Bommineni GR, Schumperli D, Schmidt RR, Villiger PM, Jakobsen BK, Gadola SD. Innate-like control of human iNKT cell autoreactivity via the hypervariable CDR3beta loop. PLoS Biol. 2010;8:e1000402. doi: 10.1371/journal.pbio.1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venkataswamy MM, Porcelli SA. Lipid and glycolipid antigens of CD1d-restricted natural killer T cells. Semin Immunol. 2009;22:68–78. doi: 10.1016/j.smim.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banchet-Cadeddu A, Henon E, Dauchez M, Renault J-H, Monneaux F, Haudrechy A. The stimulating adventure of KRN 7000. Organic & Biomolecular Chemistry. doi: 10.1039/c0ob00975j. [DOI] [PubMed] [Google Scholar]
- 28.Miyamoto K, Miyake S, Yamamura T. A synthetic glycolipid prevents autoimmune encephalomyelitis by inducing T(H)2 bias of natural killer T cells. Nature. 2001;413:531–534. doi: 10.1038/35097097. [DOI] [PubMed] [Google Scholar]
- 29.Yu KO, Im JS, Molano A, Dutronc Y, Illarionov PA, Forestier C, Fujiwara N, Arias I, Miyake S, Yamamura T, Chang YT, Besra GS, Porcelli SA. Modulation of CD1d-restricted NKT cell responses by using N-acyl variants of {alpha}-galactosylceramides. Proc Natl Acad Sci U S A. 2005;102:3383–3388. doi: 10.1073/pnas.0407488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmieg J, Yang G, Franck RW, Tsuji M. Superior protection against malaria and melanoma metastases by a C-glycoside analogue of the natural killer T cell ligand alpha-Galactosylceramide. J Exp Med. 2003;198:1631–1641. doi: 10.1084/jem.20031192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu X, Song L, Metelitsa LS, Bittman R. Synthesis and Evaluation of an α-C-Galactosylceramide Analogue that Induces Th1-biased Responses in Human Natural Killer T Cells. ChemBioChem. 2006;7:1750–1756. doi: 10.1002/cbic.200600197. [DOI] [PubMed] [Google Scholar]
- 32.Fujii S-i, Shimizu K, Hemmi H, Fukui M, Bonito AJ, Chen G, Franck RW, Tsuji M, Steinman RM. Glycolipid ±-C-galactosylceramide is a distinct inducer of dendritic cell function during innate and adaptive immune responses of mice. Proceedings of the National Academy of Sciences. 2006;103:11252–11257. doi: 10.1073/pnas.0604812103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teng MW, Westwood JA, Darcy PK, Sharkey J, Tsuji M, Franck RW, Porcelli SA, Besra GS, Takeda K, Yagita H, Kershaw MH, Smyth MJ. Combined natural killer T-cell based immunotherapy eradicates established tumors in mice. Cancer research. 2007;67:7495–7504. doi: 10.1158/0008-5472.CAN-07-0941. [DOI] [PubMed] [Google Scholar]
- 34.Sullivan BA, Nagarajan NA, Wingender G, Wang J, Scott I, Tsuji M, Franck RW, Porcelli SA, Zajonc DM, Kronenberg M. Mechanisms for glycolipid antigen-driven cytokine polarization by Valpha14i NKT cells. J Immunol. 2010;184:141–153. doi: 10.4049/jimmunol.0902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z, Byun H-S, Bittman R. Synthesis of Immunostimulatory ±-C-Galactosylceramide Glycolipids via Sonogashira Coupling, Asymmetric Epoxidation, and Trichloroacetimidate-Mediated Epoxide Opening. Organic Letters. 12:2974–2977. doi: 10.1021/ol1009976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 37.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC, Adams PD. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 38.DeLano WL. The PyMOL Molecular Graphics System. 2002 http://www.pymol.org.
- 39.Harrak Y, Barra CM, Delgado A, CastanÃÉo AR, Llebaria A. Galacto-Configured Aminocyclitol Phytoceramides Are Potent in Vivo Invariant Natural Killer T Cell Stimulators. Journal of the American Chemical Society. 2011;133:12079–12084. doi: 10.1021/ja202610x. [DOI] [PubMed] [Google Scholar]
- 40.Im JS, Arora P, Bricard G, Molano A, Venkataswamy MM, Baine I, Jerud ES, Goldberg MF, Baena A, Yu KO, Ndonye RM, Howell AR, Yuan W, Cresswell P, Chang YT, Illarionov PA, Besra GS, Porcelli SA. Kinetics and cellular site of glycolipid loading control the outcome of natural killer T cell activation. Immunity. 2009;30:888–898. doi: 10.1016/j.immuni.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.