Abstract
The production of mature and motile sperm is a detailed process that utilizes many molecular players to ensure the faithful execution of spermatogenesis. In most species that have been examined, spermatogenesis begins with a single cell that undergoes dramatic transformation, culminating with the hypercompaction of DNA into the sperm head by replacing histones with protamines. Precise execution of the stages of spermatogenesis results in the production of motile sperm. While comparative analyses have been used to identify similarities and differences in spermatogenesis between species, the focus has primarily been on vertebrate spermatogenesis, particularly mammals. To understand the evolutionary basis of spermatogenetic variation, however, a more comprehensive comparison is needed. In this review, we examine spermatogenesis and the final packaging of DNA into the sperm head in the insect Drosophila melanogaster and compare it to spermatogenesis in Homo sapiens.
Keywords: Homo sapiens, Drosophila, spermatogenesis, protamines, spermiogenesis, sperm, nebenkern
Introduction
Spermatogenesis is a highly orchestrated process that requires the correct interplay and timing of all molecular constituents to produce fully functional and motile sperm. Defects in spermatogenesis can impact a male’s overall fitness, which encompasses the ability to both survive and reproduce successfully.1-5 Aberrations during any stage within spermatogenesis can have profound effects on sperm quantity, motility, morphology and ability to fertilize an egg. In addition, poor packaging of chromatin within sperm nuclei can reduce the protection of DNA against chemical and physical damage, potentially leading to mutations and unfit offspring.
Spermatogenesis can be divided into three stages: pre-meiosis, meiosis and post-meiosis. During the course of the latter stages, two major transformations occur that drastically alter the cells produced from a single stem cell into mature sperm. First, primary spermatocytes developed in the pre-meiotic stage will transition during meiosis from a diploid genome to a haploid genome (reviewed in ref. 6). Second, modifications that occur post-meiotically act to condense chromatin to fit within the sperm nuclei.7,8 Modifications to sperm chromatin require the use of specialized DNA-binding proteins, referred to as protamines, which are capable of achieving the level of organization and compression necessary to fit the haploid genome into the compact sperm head.9,10
Much is known about protamine biochemistry, genetics, molecular structure and function in vertebrates, specifically mammals (reviewed in refs. 9 and 11). Recent work has also begun to identify the role of protamines in invertebrates, such as Drosophila.12-14 Although differences are apparent between Homo sapiens (human) spermatogenesis (reviewed in refs. 7 and 15) and Drosophila spermatogenesis (reviewed in ref. 16), the three main stages can be observed in both groups. Unfortunately, vertebrate and invertebrate spermatogenesis are rarely compared with each other, limiting our understanding of its evolutionary history and the different selection pressures that may exist. Here, we provide an overview of Drosophila spermatogenesis, with a particular emphasis on protamines and DNA packaging, and compare the process to what is observed in vertebrate spermatogenesis.
Drosophila Spermatogenesis: Cytological and Molecular Overview
Spermatogenesis in Drosophila has long been a model system for fertility, cell signaling, cytoskeletal/cytogenetic modifications, gene expression and evolution. The stages of spermatogenesis are readily identifiable through phase contrast microscopy, as they possess stereotypical morphology that appears to be well conserved among numerous species, and the genetic tools available in Drosophila melanogaster allow for relatively easy identification and characterization of the molecular underpinnings of spermatogenesis. Comparisons of spermatogenesis among species can provide some generalizations about the spermatogenic process. Spermatogenesis occurs within the testes, progressing distally from the initial stem cell division at the apical hub (Fig. 1A). Here, a group of eight to nine germline stem cells (GSCs), each surrounded by a pair of cyst progenitor cells (CPCs), physically associate with a somatic support hub, which allows for self-renewal as the GSC divides.17 The cell that maintains physical association has the Jak-STAT pathway activated through somatic secretions, which returns the cell to a GSC state, providing the template for the next round of division. The cell that is not in contact displaces from the hub and becomes the primary spermatogonium.17,18
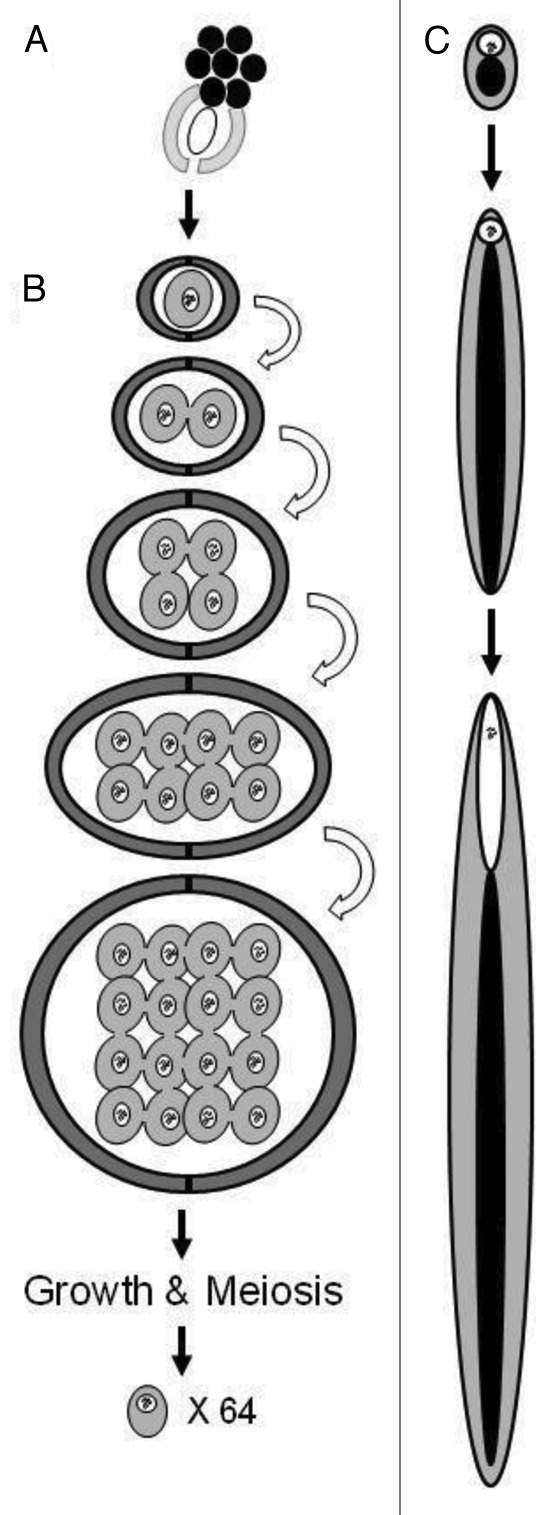
Figure 1. Spermatogenesis in Drosophila melanogaster. (A) Somatic hub cells (black) physically associate with GSCs (white oval) and CPCs (light gray). Both the GSCs and CPCs undergo self-renewal divisions, producing a (B) pair of daughter cyst cells (dark gray) surrounding a primary spermatogonium (gray). Black squiggles represent chromatin; note that chromatin varies in conformation depending on the stage in spermatogenesis, and this variation is not represented in the figure. The primary spermatogonium undergoes four mitotic divisions (arrows) resulting in a cyst of 16 primary spermatocyte cells. The spermatocytes will then undergo growth followed by meiosis to produce a cyst of 64 spermatids, each containing a nuclear (white circle) and mitochondrial (black circle) genome. (C) The mitochondria of each of the 64 spermatids aggregate to become nebenkern (black circle). The nucleus (white circle) maintains its size as the cell and nebenkern elongate to form the comet stage cell. As the cell elongates further, into the canoe stage cell, the nucleus begins to elongate and forms a shape that takes the appearance of a canoe with continuous extension of the former nebenkern.
The primary spermatogonium (Fig. 1B), also known as the gonioblast, enters the first mitotic stage of spermatogenesis to produce secondary spermatogonia. Further rounds of mitosis then occur, and the number of mitotic divisions is species-specific.19,20 Incomplete cytokinesis occurs after each round of mitosis, causing the secondary spermatogonia to remain interconnected by cytoplasmic bridges.21,22 Following meiosis, these cytoplasmic bridges may allow for the active or passive exchange of proteins that are post-meiotically transcribed,23,24 as seen in mammalian spermatogenesis.25-28 However, there is evidence in mice that some proteins are not shared equally (or at all) among the cells within a syncytium,29 which is potentially also true in Drosophila. As such, this can potentially produce an imbalance of protein product availability among different spermatids.
Following the final mitotic division, a syncytium of cells called primary spermatocytes (16 in D. melanogaster)16 is formed.30 Immature primary spermatocytes are morphologically indistinguishable from the preceding secondary spermatogonia. However, shortly after their formation, the immature primary spermatocytes begin drastic cellular and molecular changes as they enter interphase, where the cellular volume is estimated to experience a 20–25-fold increase. Upon completion of this growth phase, the primary spermatocyte will become the largest proliferating cell within the testes, and one of the largest cells in the organism.16,31
A second characteristic of the primary spermatocyte is its increase in transcription levels. Primary spermatocytes produce transcripts that are used at three different time points in spermatogenesis (reviewed in refs. 32 and 33). The first group of transcripts are produced to allow for the expansive growth that takes place during the immature primary spermatocyte stage. The second group of transcripts are those that are required for meiosis, where transcription effectively ceases. The third group includes transcripts that are used post-meiotically. These transcripts are translationally repressed until after meiosis.
The beginning of meiosis is marked by the simultaneous condensation and appearance of visible bivalents. In D. melanogaster, this appears as three condensed aggregates, each corresponding to a bivalent pair of homologous chromosomes.34,35 The bivalents at this stage typically occupy a region at the nuclear periphery.34,36 Here, they are maintained until migration toward the metaphase plate commences during the progression of meiosis. They congregate, appearing as a single large mass in the middle of the nucleus, where they are then pulled to distinct poles during anaphase I. As previously mentioned, cytokinesis occurs incompletely, and the cell quickly enters meiosis II.
After meiosis II is complete, the cells enter the “onion stage,” which is one of the most easily observed stages in Drosophila spermatogenesis. During this stage, all of the mitochondria within a single secondary spermatocyte fuse into two large mitochondrial derivatives. These two large derivatives interlock, forming a massive structure called the nebenkern (Fig. 1C).37 Due to the layered structure of the nebenkern, the appearance under electron microscopy looks like the layers of an onion, hence the name “onion stage.” The nebenkern becomes situated on one side of the secondary spermatocyte, while the nucleus lines up on the opposite side of the cell.37 The role of this stage is not clear, but its presence throughout insect spermatogenesis indicates that it likely has a vital role.
During the final stages of spermatogenesis, the nebenkern and nucleus of the developing spermatids undergo drastic remodeling (Fig. 1C).38 Initially, the nebenkern elongates down the axoneme,16 which will become the tail, producing a shape known as the “comet” stage. During this event, the chromatin condenses at the periphery of the nucleus. Subsequently, there is nuclear elongation resulting in the “canoe” stage, then the nucleus transitions from canoe-shaped to needle-shaped, and finally individualization of the sperm cell commences.39 Most of the cytoplasm is removed by individualizing machinery that traverse the bundle from the head of the sperm to the base. The last stage of sperm development within the bundles is coiling: following individualization, the entire cyst is coiled, after which the sperm are released into the testes.16 The sperm make their way to the seminal vesicle, through a currently undefined process, where they are stored until ejaculation.
While the overall morphology of Drosophila and human sperm are grossly similar—they both have a head and tail—they otherwise have divergent morphology resulting from differences in the spermatogenesis process.15,16 The sperm of Drosophila are long, strand-shaped structures, whose head and tail are almost indistinguishable by light microscopy; the head of the human sperm is a flattened pear shape that is easily distinguishable from the tail. During sperm elongation in Drosophila, each unit of the nebenkern extends along the length of the axoneme, ultimately forming the sperm tail. In humans, there is not a nebenkern, and instead the elongation process deposits individual mitochondria adjacent to the sperm head, occupying merely a third of the length of the tail.
The Role of Protamines in Spermatogenesis
During the post-meiotic stage of spermatogenesis, sperm undergo their final transformation and mature into fully functional and motile sperm. An integral process during the course of spermiogenesis is the reorganization of DNA into a highly condensed state that allows for proper packaging within the sperm head. In both vertebrates and invertebrates, the transition involves a series of steps that utilize transitional proteins that gradually replace the chromatin’s histones with protamines. In mammals, histones are removed after meiosis and are replaced by two major types of transition proteins (TP) that bind to DNA: TP1 and TP2. These transition proteins are known to condense DNA as they bind and replace histones, but do not condense the DNA to the degree seen in fully matured sperm.40-46 Although further investigation is needed to uncover the functional role of these transition proteins during sperm chromatin remodeling, TP1 appears to destabilize DNA to reduce the interaction between histones and DNA,47,48 whereas TP2 possesses two zinc fingers that may serve to condense DNA.49 Interestingly, neither transition protein is essential for fertility in mice.43,50 Thus, there is either functional redundancy among the transition proteins, or additional molecular players are present during the transition period.
Less is known about the transition stage in Drosophila. A testes-specific protein that is present during the transition period from nucleosome-based to protamine-based chromatin39 contains a high mobility group (HMG)-box motif, which has been proposed to play a role in remodeling DNA.51 Although this protein may be a strong candidate for a Drosophila transition protein, its sequence is not homologous to known mammalian transition proteins;39 as such, it has been named transition protein-like94D (tpl94D). The gene Mst77F also encodes a male-specific protein that has been proposed to act as a either a transition protein or a protamine-like protein.52 This gene has been demonstrated to be critical for both male fertility and nuclear shaping in D. melanogaster,12 and has high-sequence similarity to both the linker histone family histone H1/H512 and hHILS, which is a linker histone H1-like protein that is sperm-specific.53 However, Mst77F differs from hHILS in that it accumulates and remains in the sperm nucleus until after fertilization has occurred.12 Mst77F appears to play a role in sperm nuclei morphology, but does not affect condensation.14 Specifically, during the later stages of spermiogenesis, tubulin is weakly detectable resulting in unstable perinculear microtubules in Mst77F mutants.14 This presents an interesting aspect to sperm-specific proteins, wherein Mst77F plays a role in nuclear shaping by affecting microtubule stability.14 The gene also appears to have a dosage-dependent effect on male fertility: a single point missense mutation within the coding region results in fully sterile males with tiny nuclei, while a mutation within the promoter region greatly reduces the amount of transcript but does not appear to affect fertility.12
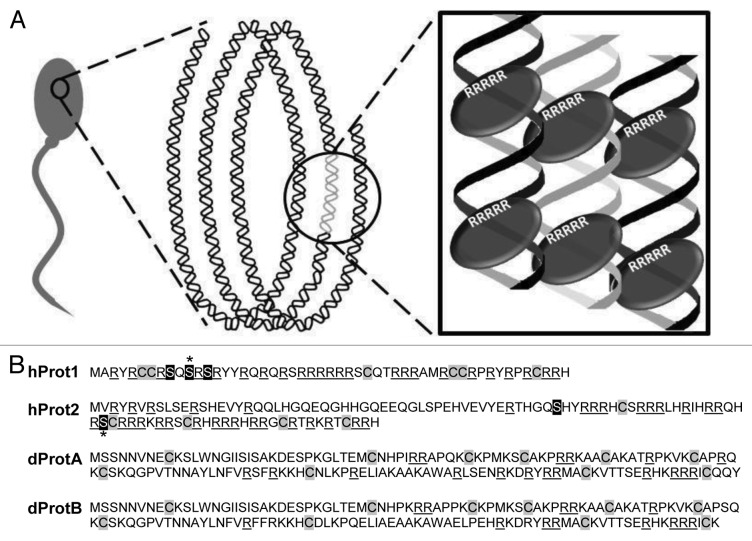
During the final stages of sperm transformation, transition proteins are replaced by protamines, which further condense DNA to the level of compaction needed to be properly packaged within the sperm heads,7,8 thus increasing sperm hydrodynamics and fertilization potential. Protamines are chromatin-binding proteins that serve to condense and organize the DNA within sperm heads, and as such, they are rich in amino acid residues that are known (human) or thought to be (Drosophila) involved in chromatin binding and linking neighboring protamines.9,54 In the past three decades, research in understanding the molecular interaction between protamines and DNA have generated a proposed model for the protamine-DNA complex and the conformation of chromatin within the sperm head (Fig. 2A). The DNA-binding domain wraps around one loop of DNA, fitting into the major groove.9,55-58 Protamines possess a high content of arginine and cysteine residues, a trend which is seen across a wide variety of species. The arginine residues reside within the DNA-binding domain, while the cysteine residues bind neighboring protamines, thus locking the DNA strands together.12,56,59-63 The protamine-based chromatin then coils into a toroid, or doughnut, conformation,44,64 wherein the DNA lies in side-by-side arrays. Protamines have a net positive charge that, when bound to DNA, neutralizes the negative charge from the DNA phosphate backbone.56,58
Figure 2. Proposed model for protamine-DNA complex and the conformation of DNA within a single sperm head (adapted from ref. 64). (A) Within the nucleus of a sperm, DNA is coiled into a doughnut shape conformation. In order to achieve this, arginine residues (white R’s) of protamines (gray ovals) bind to chromatin within the major groove of the DNA. Further interaction and binding between cysteine residues (not shown) of neighboring protamines lock adjacent DNA strands into place. (B) Amino acid sequences of human protamines 1 and 2 (hProt1 and hProt2, respectively) and Drosophila protamines A and B (dProtA and dProtB, respectively). Arginine residues are underlined; cysteine residues are highlighted in gray; serines that are phosphorylated in hProts are highlighted in black; the serine with the greatest phosphorylation is indicated with an asterisk.66
Extensive studies on the genes that encode protamines have mostly been performed in vertebrates, particularly in mammalian models.9 Human protamines (henceforth known as hProts) are often denoted as “true protamines” and their structure and amino acid composition differs greatly from Drosophila protamines (henceforth known as dProts), which are also referred to as protamine-like.65 In humans and other mammals, two main families of protamines have been identified: protamine P1 (hProt1) and protamine P2 (hProt2), encoded by the genes PRM1 and PRM2, respectively. hProt1 has been well characterized: the N terminus of hProt1 contains a high serine and threonine content, which upon phosphorylation, targets the DNA for protamine deposition (Fig. 2B).58,66 The binding patterns of hProt2, unlike hProt1, remain poorly understood; however, it is believed that they may be similar to hProt1.44 The precursor of hProt1 has an initial size that is much larger than that of hProt2, but hProtP1 undergoes multiple modification events until the two proteins are eventually equivalent in size.67-69 Like hProt1, hProt2 also possesses a string of arginine residues that act to bind the protamines to DNA,70,71 and is rich in cysteines, which are involved in the disulfide cross-linkage between neighboring protamines, either of the same or different family group.72-74 While hProt2 is also phosphorylated, this occurs at serine residues toward the center of the molecule rather than the N terminus, potentially indicating that hProt1 and hProt2 have different DNA binding patterns or different molecular interactions with other protamines (Fig. 2B).66
There are two transcripts in Drosophila whose sequences are similar to the protamine sequences found in humans: Mst35Ba and Mst35Bb.12 Further analysis determined that Mst35Ba and Mst35Bb encode chromatin proteins that serve to organize DNA to a level of compaction that would fit within the tiny volume of the sperm head and, thus, these proteins are denoted as Drosophila protamine A (dProtA) and Drosophila protamine B (dProtB), respectively.12 dProtA and dProtB are encoded by adjacent genes located on chromosome 2L and possess identical sequences for the 5`UTR, coding and 3`UTR regions, suggesting that they originated from a duplication event.12 When both protamine genes are deleted, sperm become more susceptible to mutation when exposed to X-ray irradiation, indicating that protamines, or the increased coiling they provide, serve to protect DNA from damage.14
Structurally, hProt1 and hProt2 differ in both their size and composition (Table 1).12,65 hProt2 contains twice as many amino acids as hProt1, but a smaller proportion of these are arginines and cysteines. In contrast, both dProtA and dProtB have almost identical amino acid composition and size, which indicates that they are likely functionally redundant. Amino acid composition differs greatly between human protamines and Drosophila protamines, particularly the lysine content, which is high in dProts and low in hProts. However, both the hProts and the dProts contain a relatively high content of arginine residues, which are likely involved in DNA binding, and cysteine residues, which likely bind protamines together through disulfide bonds (Fig. 2, Table 1).12,65 As such, the functional similarities between hProt and dProt may be high, and their differences in amino acid length and their amino acid content could be a simple reflection of their divergent evolutionary histories.
Table 1. Comparison of Drosophila and human protamines.
| Protamine | Total # aa | % Cysteine | % Lysine | % Arginine |
|---|---|---|---|---|
| hProt1 |
51 |
11.8 |
0 |
47.1 |
| hProt2 |
102 |
4.9 |
2.0 |
32.4 |
| dProtA |
146 |
6.8 |
14.4 |
12.3 |
| dProtB | 144 | 6.9 | 15.3 | 10.4 |
Protamine Expression During Spermatogenesis
There are two striking differences between human and Drosophila protamine expression. First, male fertility in humans requires that both alleles of each protamine gene are fully functional; both genes are haploinsufficient when mutated.75 In Drosophila, however, when both copies of dProtA and dProtB are simultaneously deleted, approximately 20% of sperm nuclei appear misshaped, but overall fertility is not affected. Thus, dProtA and dProtB are not essential for male fertility and may be functionally redundant with as-yet-unidentified nuclear proteins that serve to condense sperm chromatin.14 This contrasts with the previously discussed histone linker-like protein, Mst77F, which is essential for male sterility in a dosage-dependent manner.12
Second, the temporal expression of human and Drosophila protamine transcripts differ during spermatogenesis. In humans, both hProt1 and hProt2 are expressed post-meiotically, thus only one copy of each gene is expressed within the sperm head.11,76 Therefore, only one allele (from one parent) is expressed within each individual sperm derived in the offspring. However, during the course of spermatogenesis, sperm are interconnected via cytoplasmic bridges, allowing for the developing sperm to potentially share protamine transcripts.77 What is unknown is the extent that sharing occurs between maturing sperm, and whether subtle differences in protamine expression between two sibling sperm can cause differential fertilization success. In contrast to humans, in situ hybridization in D. melanogaster uncovered the presence of dProtA and dProtB transcripts in primary spermatocytes, which are diploid and have yet to undergo meiosis.12 Further research examining the allelic expression of protamines for two sibling species within the Drosophila genus demonstrated that both alleles for dProtB are present in each sperm cell.13 Therefore, in Drosophila, both parents contribute equally to the protamines found within a single sperm head.
The evolutionary forces acting on the temporal expression patterns of protamines, leading to differences between species, have yet to be determined. Variation in sperm functionality would have the greatest impact in species that experience sperm competition, either between individuals or within the ejaculate of a single male.78 This raises the question of whether there is a selective advantage of either haploid or diploid expression of protamines, as well as the number of species that use haploid vs. diploid protamine expression. When only one protamine allele is expressed, phenotypically different sperm could potentially be produced within one individual,11,76 which may increase a male’s fitness if this variation translates into a greater overall fertilization success. When both protamine alleles are expressed, however, there is functional duplication of the protamines, making it less likely that a defect in either gene would impact fertility; this could also increase a male’s overall fertilization success.79-82 Thus, the different allelic expression patterns could have resulted from different selective pressures in vertebrates vs. invertebrates. These selective pressures could have been historically influential but are no longer present, could be ongoing or could even be opposite in contemporary times to what they were when these divergent expression patterns initially arose. Thus, it will be difficult to assess the evolutionary basis of protamine expression patterns. Further research examining spermatogenesis in a variety of species will allow for a more thorough comparison between the stages of spermatogenesis and the packaging of DNA into the sperm head. This will enhance our understanding of how these processes have evolved, and how variations in these processes affect a male’s overall fitness.
Acknowledgments
We thank an anonymous reviewer for their very detailed feedback that significantly improved the manuscript. This work was supported by the Canada Research Chairs program and a Natural Sciences and Engineering Research Council Discovery Grant to A.J.M., and a Queen Elizabeth II Graduate Scholarship in Science and Technology to R.L K.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/spermatogenesis/article/24376
References
- 1.Agarwal A, Said TM. Role of sperm chromatin abnormalities and DNA damage in male infertility. Hum Reprod Update. 2003;9:331–45. doi: 10.1093/humupd/dmg027. [DOI] [PubMed] [Google Scholar]
- 2.Aoki VW, Liu L, Carrell DT. Identification and evaluation of a novel sperm protamine abnormality in a population of infertile males. Hum Reprod. 2005;20:1298–306. doi: 10.1093/humrep/deh798. [DOI] [PubMed] [Google Scholar]
- 3.Bianchi PG, Manicardi GC, Bizzaro D, Bianchi U, Sakkas D. Effect of deoxyribonucleic acid protamination on fluorochrome staining and in situ nick-translation of murine and human mature spermatozoa. Biol Reprod. 1993;49:1083–8. doi: 10.1095/biolreprod49.5.1083. [DOI] [PubMed] [Google Scholar]
- 4.Martin RH. Meiotic errors in human oogenesis and spermatogenesis. Reprod Biomed Online. 2008;16:523–31. doi: 10.1016/S1472-6483(10)60459-2. [DOI] [PubMed] [Google Scholar]
- 5.Oliva R. Protamines and male infertility. Hum Reprod Update. 2006;12:417–35. doi: 10.1093/humupd/dml009. [DOI] [PubMed] [Google Scholar]
- 6.Hecht NB. Molecular mechanisms of male germ cell differentiation. Bioessays. 1998;20:555–61. doi: 10.1002/(SICI)1521-1878(199807)20:7<555::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 7.de Kretser DM, Loveland KL, Meinhardt A, Simorangkir D, Wreford N. Spermatogenesis. Hum Reprod. 1998;13(Suppl 1):1–8. doi: 10.1093/humrep/13.suppl_1.1. [DOI] [PubMed] [Google Scholar]
- 8.Tokuyasu KT, Peacock WJ, Hardy RW. Dynamics of spermiogenesis in Drosophila melanogaster. I. Individualization process. Z Zellforsch Mikrosk Anat. 1972;124:479–506. doi: 10.1007/BF00335253. [DOI] [PubMed] [Google Scholar]
- 9.Balhorn R. The protamine family of sperm nuclear proteins. Genome Biol. 2007;8:227. doi: 10.1186/gb-2007-8-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward WS, Coffey DS. DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biol Reprod. 1991;44:569–74. doi: 10.1095/biolreprod44.4.569. [DOI] [PubMed] [Google Scholar]
- 11.Steger K, Pauls K, Klonisch T, Franke FE, Bergmann M. Expression of protamine-1 and -2 mRNA during human spermiogenesis. Mol Hum Reprod. 2000;6:219–25. doi: 10.1093/molehr/6.3.219. [DOI] [PubMed] [Google Scholar]
- 12.Jayaramaiah Raja S, Renkawitz-Pohl R. Replacement by Drosophila melanogaster protamines and Mst77F of histones during chromatin condensation in late spermatids and role of sesame in the removal of these proteins from the male pronucleus. Mol Cell Biol. 2005;25:6165–77. doi: 10.1128/MCB.25.14.6165-6177.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanippayoor RL, Moehring AJ. Allelic Expression of Drosophila Protamines during Spermatogenesis. Int J Evol Biol. 2012;2012:947381. doi: 10.1155/2012/947381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rathke C, Barckmann B, Burkhard S, Jayaramaiah-Raja S, Roote J, Renkawitz-Pohl R. Distinct functions of Mst77F and protamines in nuclear shaping and chromatin condensation during Drosophila spermiogenesis. Eur J Cell Biol. 2010;89:326–38. doi: 10.1016/j.ejcb.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Sharma R, Agarwal A. Spermatogenesis: An overview. In: Zini A, Agarwal A. eds. Sperm chromatin: Biological and clinical applications in male infertility and assisted reproduction. New York: Springer, 2011:19-44. [Google Scholar]
- 16.Fuller MT. Spermatogenesis. In: Development of Drosophila melanogaster Cold Spring Harbor, NY, 1993:71-147. [Google Scholar]
- 17.Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294:2542–5. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- 18.Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 2001;294:2546–9. doi: 10.1126/science.1066700. [DOI] [PubMed] [Google Scholar]
- 19.Pantazidis AC, Zouros E, Galanopoulos VK. Species-specific characteristics of spermatogenesis in Drosophila mojavensis (Patterson)(Diptera: Drosophilidae) Int J Insect Morphol Embryol. 1992;21:351–63. doi: 10.1016/0020-7322(92)90030-Q. [DOI] [Google Scholar]
- 20.Schärer L, Da Lage JL, Joly D. Evolution of testicular architecture in the Drosophilidae: a role for sperm length. BMC Evol Biol. 2008;8:143. doi: 10.1186/1471-2148-8-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hime GR, Brill JA, Fuller MT. Assembly of ring canals in the male germ line from structural components of the contractile ring. J Cell Sci. 1996;109:2779–88. doi: 10.1242/jcs.109.12.2779. [DOI] [PubMed] [Google Scholar]
- 22.Giansanti MG, Sechi S, Frappaolo A, Belloni G, Piergentili R. Cytokinesis in Drosophila male meiosis. Spermatogenesis. 2012;2:185–96. doi: 10.4161/spmg.21711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barreau C, Benson E, Gudmannsdottir E, Newton F, White-Cooper H. Post-meiotic transcription in Drosophila testes. Development. 2008;135:1897–902. doi: 10.1242/dev.021949. [DOI] [PubMed] [Google Scholar]
- 24.Vibranovski MD, Chalopin DS, Lopes HF, Long M, Karr TL. Direct evidence for postmeiotic transcription during Drosophila melanogaster spermatogenesis. Genetics. 2010;186:431–3. doi: 10.1534/genetics.110.118919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardy RW, Tokuyasu KT, Lindsley DL, Garavito M. The germinal proliferation center in the testis of Drosophila melanogaster. J Ultrastruct Res. 1979;69:180–90. doi: 10.1016/S0022-5320(79)90108-4. [DOI] [PubMed] [Google Scholar]
- 26.Burgos MH, Fawcett DW. Studies on the fine structure of the mammalian testis. I. Differentiation of the spermatids in the cat (Felis domestica) J Biophys Biochem Cytol. 1955;1:287–300. doi: 10.1083/jcb.1.4.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fawcett DW, Ito S, Slautterback D. The occurrence of intercellular bridges in groups of cells exhibiting synchronous differentiation. J Biophys Biochem Cytol. 1959;5:453–60. doi: 10.1083/jcb.5.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ventelä S, Toppari J, Parvinen M. Intercellular organelle traffic through cytoplasmic bridges in early spermatids of the rat: mechanisms of haploid gene product sharing. Mol Biol Cell. 2003;14:2768–80. doi: 10.1091/mbc.E02-10-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Y, Deng X, Martin-DeLeon PA. Lack of sharing of Spam1 (Ph-20) among mouse spermatids and transmission ratio distortion. Biol Reprod. 2001;64:1730–8. doi: 10.1095/biolreprod64.6.1730. [DOI] [PubMed] [Google Scholar]
- 30.Kurokawa H, Hihara F. Number of first spermatocytes in relation to phylogeny of Drosophila (Diptera: Drosophilidae) Int J Insect Morphol Embryol. 1976;5:51–63. doi: 10.1016/0020-7322(76)90021-0. [DOI] [Google Scholar]
- 31.Lin TY, Viswanathan S, Wood C, Wilson PG, Wolf N, Fuller MT. Coordinate developmental control of the meiotic cell cycle and spermatid differentiation in Drosophila males. Development. 1996;122:1331–41. doi: 10.1242/dev.122.4.1331. [DOI] [PubMed] [Google Scholar]
- 32.White-Cooper H. Molecular mechanisms of gene regulation during Drosophila spermatogenesis. Reproduction. 2010;139:11–21. doi: 10.1530/REP-09-0083. [DOI] [PubMed] [Google Scholar]
- 33.Lim C, Tarayrah L, Chen X. Transcriptional regulation during Drosophila spermatogenesis. Spermatogenesis. 2012;2:158–66. doi: 10.4161/spmg.21775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonaccorsi S, Pisano C, Puoti F, Gatti M. Y chromosome loops in Drosophila melanogaster. Genetics. 1988;120:1015–34. doi: 10.1093/genetics/120.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cenci G, Bonaccorsi S, Pisano C, Verni F, Gatti M. Chromatin and microtubule organization during premeiotic, meiotic and early postmeiotic stages of Drosophila melanogaster spermatogenesis. J Cell Sci. 1994;107:3521–34. doi: 10.1242/jcs.107.12.3521. [DOI] [PubMed] [Google Scholar]
- 36.Vazquez J, Belmont AS, Sedat JW. The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr Biol. 2002;12:1473–83. doi: 10.1016/S0960-9822(02)01090-4. [DOI] [PubMed] [Google Scholar]
- 37.Tokuyasu KT. Dynamics of spermiogenesis in Drosophila melanogaster. VI. Significance of “onion” nebenkern formation. J Ultrastruct Res. 1975;53:93–112. doi: 10.1016/S0022-5320(75)80089-X. [DOI] [PubMed] [Google Scholar]
- 38.Fabrizio JJ, Hime G, Lemmon SK, Bazinet C. Genetic dissection of sperm individualization in Drosophila melanogaster. Development. 1998;125:1833–43. doi: 10.1242/dev.125.10.1833. [DOI] [PubMed] [Google Scholar]
- 39.Rathke C, Baarends WM, Jayaramaiah-Raja S, Bartkuhn M, Renkawitz R, Renkawitz-Pohl R. Transition from a nucleosome-based to a protamine-based chromatin configuration during spermiogenesis in Drosophila. J Cell Sci. 2007;120:1689–700. doi: 10.1242/jcs.004663. [DOI] [PubMed] [Google Scholar]
- 40.Baskaran R, Rao MR. Interaction of spermatid-specific protein TP2 with nucleic acids, in vitro. A comparative study with TP1. J Biol Chem. 1990;265:21039–47. [PubMed] [Google Scholar]
- 41.Brewer L, Corzett M, Balhorn R. Condensation of DNA by spermatid basic nuclear proteins. J Biol Chem. 2002;277:38895–900. doi: 10.1074/jbc.M204755200. [DOI] [PubMed] [Google Scholar]
- 42.Lévesque D, Veilleux S, Caron N, Boissonneault G. Architectural DNA-binding properties of the spermatidal transition proteins 1 and 2. Biochem Biophys Res Commun. 1998;252:602–9. doi: 10.1006/bbrc.1998.9687. [DOI] [PubMed] [Google Scholar]
- 43.Meistrich ML, Mohapatra B, Shirley CR, Zhao M. Roles of transition nuclear proteins in spermiogenesis. Chromosoma. 2003;111:483–8. doi: 10.1007/s00412-002-0227-z. [DOI] [PubMed] [Google Scholar]
- 44.Balhorn R. Sperm chromatin: An overview. In: Zini A, Agarwal A. eds. Sperm chromatin: Biological and clinical applications in male infertility and assisted reproduction. New York: Springer, 2011:3-19. [Google Scholar]
- 45.Alfonso PJ, Kistler WS. Immunohistochemical localization of spermatid nuclear transition protein 2 in the testes of rats and mice. Biol Reprod. 1993;48:522–9. doi: 10.1095/biolreprod48.3.522. [DOI] [PubMed] [Google Scholar]
- 46.Oko RJ, Jando V, Wagner CL, Kistler WS, Hermo LS. Chromatin reorganization in rat spermatids during the disappearance of testis-specific histone, H1t, and the appearance of transition proteins TP1 and TP2. Biol Reprod. 1996;54:1141–57. doi: 10.1095/biolreprod54.5.1141. [DOI] [PubMed] [Google Scholar]
- 47.Pradeepa MM, Rao MR. Chromatin remodeling during mammalian spermatogenesis: role of testis specific histone variants and transition proteins. Soc Reprod Fertil Suppl. 2007;63:1–10. [PubMed] [Google Scholar]
- 48.Singh J, Rao MR. Interaction of rat testis protein, TP, with nucleosome core particle. Biochem Int. 1988;17:701–10. [PubMed] [Google Scholar]
- 49.Kundu TK, Rao MRS. Zinc dependent recognition of a human CpG island sequence by the mammalian spermatidal protein TP2. Biochemistry. 1996;35:15626–32. doi: 10.1021/bi961271i. [DOI] [PubMed] [Google Scholar]
- 50.Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176–8. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- 51.Travers AA. Priming the nucleosome: a role for HMGB proteins? EMBO Rep. 2003;4:131–6. doi: 10.1038/sj.embor.embor741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Russell SR, Kaiser K. Drosophila melanogaster male germ line-specific transcripts with autosomal and Y-linked genes. Genetics. 1993;134:293–308. doi: 10.1093/genetics/134.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan W, Ma L, Burns KH, Matzuk MM. HILS1 is a spermatid-specific linker histone H1-like protein implicated in chromatin remodeling during mammalian spermiogenesis. Proc Natl Acad Sci USA. 2003;100:10546–51. doi: 10.1073/pnas.1837812100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balhorn R. A model for the structure of chromatin in mammalian sperm. J Cell Biol. 1982;93:298–305. doi: 10.1083/jcb.93.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dadoune JP. Expression of mammalian spermatozoal nucleoproteins. Microsc Res Tech. 2003;61:56–75. doi: 10.1002/jemt.10317. [DOI] [PubMed] [Google Scholar]
- 56.Lewis JD, Song Y, de Jong ME, Bagha SM, Ausió J. A walk though vertebrate and invertebrate protamines. Chromosoma. 2003;111:473–82. doi: 10.1007/s00412-002-0226-0. [DOI] [PubMed] [Google Scholar]
- 57.Prieto MC, Maki AH, Balhorn R. Analysis of DNA-protamine interactions by optical detection of magnetic resonance. Biochemistry. 1997;36:11944–51. doi: 10.1021/bi971061l. [DOI] [PubMed] [Google Scholar]
- 58.Raukas E, Mikelsaar RH. Are there molecules of nucleoprotamine? Bioessays. 1999;21:440–8. doi: 10.1002/(SICI)1521-1878(199905)21:5<440::AID-BIES11>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 59.Ausió J. Histone H1 and evolution of sperm nuclear basic proteins. J Biol Chem. 1999;274:31115–8. doi: 10.1074/jbc.274.44.31115. [DOI] [PubMed] [Google Scholar]
- 60.Calvin HI. Comparative analysis of the nuclear basic proteins in rat, human, guinea pig, mouse and rabbit spermatozoa. Biochim Biophys Acta. 1976;434:377–89. doi: 10.1016/0005-2795(76)90229-4. [DOI] [PubMed] [Google Scholar]
- 61.Domenjoud L, Nussbaum G, Adham IM, Greeske G, Engel W. Genomic sequences of human protamines whose genes, PRM1 and PRM2, are clustered. Genomics. 1990;8:127–33. doi: 10.1016/0888-7543(90)90234-L. [DOI] [PubMed] [Google Scholar]
- 62.Gimenez-Bonafé P, Ribes E, Sautière P, Gonzalez A, Kasinsky H, Kouach M, et al. Chromatin condensation, cysteine-rich protamine, and establishment of disulphide interprotamine bonds during spermiogenesis of Eledone cirrhosa (Cephalopoda) Eur J Cell Biol. 2002;81:341–9. doi: 10.1078/0171-9335-00253. [DOI] [PubMed] [Google Scholar]
- 63.Biegeleisen K. The probable structure of the protamine-DNA complex. J Theor Biol. 2006;241:533–40. doi: 10.1016/j.jtbi.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 64.Braun RE. Packaging paternal chromosomes with protamine. Nat Genet. 2001;28:10–2. doi: 10.1038/ng0501-10. [DOI] [PubMed] [Google Scholar]
- 65.Alvi ZA, Chu TC, Schawaroch V, Klaus AV. Protamine-like proteins in 12 sequenced species of Drosophila. Protein Pept Lett. 2013;20:17–35. doi: 10.2174/092986613804096847. [DOI] [PubMed] [Google Scholar]
- 66.Pirhonen A, Linnala-Kankkunen A, Mäenpää PH. Identification of phosphoseryl residues in protamines from mature mammalian spermatozoa. Biol Reprod. 1994;50:981–6. doi: 10.1095/biolreprod50.5.981. [DOI] [PubMed] [Google Scholar]
- 67.Carré-Eusèbe D, Lederer F, Lê KH, Elsevier SM. Processing of the precursor of protamine P2 in mouse. Peptide mapping and N-terminal sequence analysis of intermediates. Biochem J. 1991;277:39–45. doi: 10.1042/bj2770039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chauvière M, Martinage A, Debarle M, Sautière P, Chevaillier P. Molecular characterization of six intermediate proteins in the processing of mouse protamine P2 precursor. Eur J Biochem. 1992;204:759–65. doi: 10.1111/j.1432-1033.1992.tb16691.x. [DOI] [PubMed] [Google Scholar]
- 69.Yelick PC, Balhorn R, Johnson PA, Corzett M, Mazrimas JA, Kleene KC, et al. Mouse protamine 2 is synthesized as a precursor whereas mouse protamine 1 is not. Mol Cell Biol. 1987;7:2173–9. doi: 10.1128/mcb.7.6.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bianchi F, Rousseaux-Prevost R, Bailly C, Rousseaux J. Interaction of human P1 and P2 protamines with DNA. Biochem Biophys Res Commun. 1994;201:1197–204. doi: 10.1006/bbrc.1994.1832. [DOI] [PubMed] [Google Scholar]
- 71.Brewer LR, Corzett M, Balhorn R. Protamine-induced condensation and decondensation of the same DNA molecule. Science. 1999;286:120–3. doi: 10.1126/science.286.5437.120. [DOI] [PubMed] [Google Scholar]
- 72.Balhorn R, Corzett M, Mazrimas J, Watkins B. Identification of bull protamine disulfides. Biochemistry. 1991;30:175–81. doi: 10.1021/bi00215a026. [DOI] [PubMed] [Google Scholar]
- 73.Bedford JM, Calvin HI. The occurrence and possible functional significance of -S-S- crosslinks in sperm heads, with particular reference to eutherian mammals. J Exp Zool. 1974;188:137–55. doi: 10.1002/jez.1401880203. [DOI] [PubMed] [Google Scholar]
- 74.Saowaros W, Panyim S. The formation of disulfide bonds in human protamines during sperm maturation. Experientia. 1979;35:191–2. doi: 10.1007/BF01920608. [DOI] [PubMed] [Google Scholar]
- 75.Cho C, Willis WD, Goulding EH, Jung-Ha H, Choi YC, Hecht NB, et al. Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nat Genet. 2001;28:82–6. doi: 10.1038/ng0501-82. [DOI] [PubMed] [Google Scholar]
- 76.Domenjoud L, Kremling H, Burfeind P, Maier WM, Engel W. On the expression of protamine genes in the testis of man and other mammals. Andrologia. 1991;23:333–7. doi: 10.1111/j.1439-0272.1991.tb02575.x. [DOI] [PubMed] [Google Scholar]
- 77.Caldwell KA, Handel MA. Protamine transcript sharing among postmeiotic spermatids. Proc Natl Acad Sci USA. 1991;88:2407–11. doi: 10.1073/pnas.88.6.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Immler S. Sperm competition and sperm cooperation: the potential role of diploid and haploid expression. Reproduction. 2008;135:275–83. doi: 10.1530/REP-07-0482. [DOI] [PubMed] [Google Scholar]
- 79.Parker GA. Sperm competition and its evolutionary consequences in the insects. Biol Rev Camb Philos Soc. 1970;45:525–67. doi: 10.1111/j.1469-185X.1970.tb01176.x. [DOI] [Google Scholar]
- 80.Parker GA. Sperm competition games: sperm size and sperm number under adult control. Proc Biol Sci. 1993;253:245–54. doi: 10.1098/rspb.1993.0110. [DOI] [PubMed] [Google Scholar]
- 81.Parker GA, Begon ME. Sperm competition games: sperm size and number under gametic control. Proc Biol Sci. 1993;253:255–62. doi: 10.1098/rspb.1993.0111. [DOI] [PubMed] [Google Scholar]
- 82.Snook RR. Sperm in competition: not playing by the numbers. Trends Ecol Evol. 2005;20:46–53. doi: 10.1016/j.tree.2004.10.011. [DOI] [PubMed] [Google Scholar]