Abstract
Myogenic differentiation plays an important role in muscle regeneration and is regulated by two transcription factor families, MRFs and MEF2, which induce differentiation of myoblasts through expression of the muscle-specific gene, myogenin. In addition, many intracellular signaling pathways are also involved in myogenic differentiation, including p38 MAPK, ERK/MAPK and PI3K/AKT. The JAK-STAT pathway is activated by various cytokines and positively or negatively regulates the differentiation of myoblasts. JAK1 plays a notable role in proliferation; whereas, JAK2 and JAK3 function mainly in differentiation. The STATs, molecules downstream of JAK, regulate myogenesis. With JAK1, STAT1 promotes proliferation, while STAT3 has a dual effect on proliferation and differentiation. The JAK-STAT negative regulator, SOCS, is also associated with myogenesis; although, its role is controversial. In this review, we will discuss the role of the JAK-STAT pathway on myogenic differentiation.
Keywords: JAK1, JAK2, JAK3, STAT1, STAT2, STAT3, SOCS, myogenic differentiation
Introduction
Multiple intracellular signaling pathways play roles in myogenic differentiation via many hormones and growth factors. These include the extracellular signal-regulated kinase (ERK)/MAPK, p38 MAPK and phosphatidylinositol 3-kinase (PI3K)/AKT pathways. First, the ERK/MAPK pathway elicits differentiation signals to promote or inhibit differentiation and fusion in the presence of mitogens.1-5 The p38 MAPK pathway is required for myogenic differentiation and also promotes skeletal muscle differentiation, at least in part via activation of myocyte enhancer-binding factor 2 (MEF2)C.4,6-8 In addition, PI3K/AKT signaling positively regulates myogenic differentiation when insulin-like growth factor (IGF) is stimulated.6,9,10 The Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathway also has an important role in the regulatory mechanism of myogenic differentiation; however, each factor affects myogenic differentiation in an opposite manner.11-13 This review provides an overview of current research on the role of the JAK-STAT pathway and its regulatory mechanism in myogenic differentiation.
Myogenic Differentiation
Myogenic differentiation is a highly orchestrated sequential program to generate mature skeletal muscle. This event is initiated from muscle precursors called myoblasts, which arise from the somatic mesoderm.14 Most myoblasts are mitotically quiescent. However, a muscle injury, exercise, or a pathological state may lead to activation, causing myoblasts to re-enter the cell cycle and proliferate. After proliferation, cells irreversibly withdraw from the cell cycle, differentiate and fuse with existing myofibrils.15 Myoblasts differentiate into mononucleated myocytes in the early differentiation stage, and they fuse into multinucleated myotubes in the late stage of differentiation. The multinucleated myotubes express many muscle structural proteins, such as myosin heavy chain (MHC), muscle creatine kinase (MCK) and α-actin.16 Muscle fibers form multinucleated myotubes during terminal differentiation. The steps of myogenic differentiation are illustrated in Figure 1.
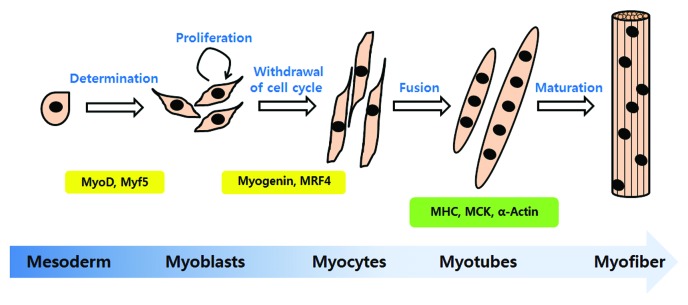
Figure 1. The steps of myogenic differentiation. Myoblasts originate from the mesoderm and are converted to skeletal muscle lineage myoblasts after MyoD and Myf5 expression. First, myoblasts enter the cell cycle and proliferate. When the growth factor or mitogen from myoblast cultures is withdrawn, proliferating cells exit from the cell cycle and initiate differentiation. Myogenin and MRF4 are involved in the initiation of differentiation. In this step, myoblasts changed to an elongated shape and are called myocytes. Myocytes fuse with neighboring cells into multinucleated myotubes. The multinucleated myotubes express the muscle specific proteins, MHC (myosin heavy chain), muscle creatine kinase (MCK) and α-actin. The mature form of myotubes is turned to myofiber.
Two transcription factor families, MRFs (myogenic regulatory factors) and MEF2, are known to be critical for regulating myogenic differentiation. At the onset of myogenesis in the embryo, Myf5 and then MyoD are directly activated in the dorsal somite by signals from adjacent tissues.17 Myf5 and MyoD are members of the MRF family, which belongs to the basic helix-loop-helix (bHLH) protein family18 that consists of MyoD, Myf5, MRF4 and myogenin; these proteins are able to initiate the myogenic program and convert non-muscle cells to the skeletal muscle lineage.19 Myf5 and MyoD act as muscle lineage components and also play important roles in the induction of the myogenin gene.20 Furthermore, Myf5 deficiency, leading to a lack of amplification of myoblasts and loss of MyoD, reduced myogenic differentiation.21-24 This suggests that Myf5 regulates the proliferation rate of myoblasts; whereas MyoD is required for the differentiation potential of skeletal myoblasts.25 The appearance of myogenin reflects that cells have irreversibly withdrawn from the cell cycle and entered differentiation.26,27 MRFs normally form heterodimers with E2A gene products (E12/E47), and bind to the E box consensus sequences (CANNTG) in the promoters of muscle-specific genes and then induce transcription of target genes.25 The other family is the myocyte enhancer-binding factor 2 (MEF2) proteins; this group includes MEF2A, 2B, 2C and 2D.15,28 The MEF2 family functions as accessory regulators of muscle gene expression and differentiation. In addition, they can physically bind to and cooperate with MRFs to synergically induce many muscle-specific genes.29 However, silencing of Mef2 at early stages of adult myogenesis impairs myoblast fusion and muscle structural protein gene expression, suggesting MEF2 is critical at the early stages of adult myoblast fusion.30
The transcription factors regulate myogenic differentiation through several signaling pathways, mainly, the IRS-PI3K-AKT and MEK/ERK. The IRS-PI3K-AKT pathway promotes myogenic differentiation through myogenin expression by MEF2.31 In contrast, phosphorylated MEK1 directly associates with MyoD, which prevents MyoD binding to the myogenin promoter, resulting in inhibition of muscle differentiation.32 The C2C12 cells are murine myoblast cells derived from mouse satellite cells, which differentiate into multinucleated myotubes upon withdrawal of serum or mitogens from myoblast cultures. The morphology of C2C12 cells change to elongated, multinucleated myotubes during differentiation.33 Therefore, C2C12 cells are very useful for studying myoblast differentiation and proliferation in vitro. Moreover, myotoxins such as bupivacaine (Marcaine), cardiotoxin (CTX) and notexin (NTX), venoms isolated from snakes, are the easiest and most reproducible way to induce regeneration in vivo.34 Injection of CTX in the adult mouse tibialis anterior muscle is a widely used method to study myogenic differentiation for muscle regeneration.
JAKs-STATs Pathway in Myogenic Differentiation
JAKs are a family of non-receptor tyrosine kinases, comprised of JAK1, JAK2, JAK3 and TYK2 in mammals. The STATs family are located downstream of JAKs and seven proteins are identified in mammals: STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B and STAT6.35 The JAK-STAT pathway is activated by ligands (such as cytokines or growth factors) binding to their cognate receptor. The binding of ligand to the receptor activates receptor-associated JAKs by phosphorylation of specific tyrosine residues. Subsequently, activated JAKs lead to phosphorylation of their selective STATs. Activated STATs dissociate from the receptor and, in turn, dimerize and translocate into the nucleus where they function as transcription factors to regulate expression of target genes.35,36 The JAK-STAT pathway is important in hematopoiesis, inflammation, immunity and immune-related diseases. The JAK-STAT pathway is also linked to cell growth, proliferation and differentiation in many cell types, including immune cells, hematopoietic stem cells, osteoblasts, neuronal precursor cells and myoblasts.37-41
Guillet-Deniau et al. suggested the possibility that activation of the JAK-STAT pathway is correlated with myogenic differentiation.42 In that report, skeletal muscle serotonin increased the expression of myogenin and triggered activation of the JAK-STAT pathway in primary myoblasts. Serotonin also induced translocation of STAT3 into the nucleus. They elucidated that the nuclear translocation of STAT3 by serotonin is related to the upregulation of myogenin, a transcription factor involved in myogenic differentiation.42 These findings provided the basis for further understanding of the role of the JAK-STAT pathway in myogenic differentiation. In addition, among transcriptional profile of GTP (guanosine 5′-triphosphate)-mediated differentiation of C2C12 myoblasts, genes such as interleukin 6, Pim-1 and IL1Ra (interleukin 1 receptor antagonist homolog 1) are upregulated, and it is suggested that JAK-STAT pathway can play a crucial role in myogenic differentiation.43 In next section, we will focus on the role of the JAK-STAT pathway in the differentiation and proliferation of skeletal muscle myoblasts.
First, JAK1-STAT1-STAT3 promotes proliferation and prevents premature differentiation of myoblasts.11 JAK1 knockdown studies showed that myogenic differentiation was accelerated and proliferation was inhibited in both C2C12 cells and primary myoblast cells. JAK1-siRNA induced the accelerated expression of MyoD and MEF2 and enhanced expression of MEF2-dependent genes. JAK1-siRNA also upregulated the prominent cyclin-dependent kinase 2 inhibitors, p21Cip1 and p27Kip1, in differentiating cells as well as proliferating cells and downregulated the inhibitor of differentiation, Id1. The ERK pathway contributed to the repressive effect of JAK1 on myogenic differentiation. In addition, JAK1-overexpressed cells had decreased expression of MHC and MGN (myogenin), which was rescued by STAT1-siRNA transfection. This result implied that STAT1 mediates the anti-differentiation effect of JAK1. Leukemia inhibitory factor (LIF) induced myogenic proliferation through the formation of a STAT3 and STAT1 complex. In an in vivo study, JAK1-STAT1-STAT3 was upregulated and activated when CTX was injected into mouse muscle. This result revealed JAK1-STAT1-STAT3 is involved in myogenic regeneration, required for myoblast proliferation and serves as a key check point to prevent myoblasts from premature differentiation.11 The repressive role of JAK1-STAT1-STAT3 on myogenic differentiation was supported by another published paper from the same group. A member of the cytokine interleukin (IL)-6 family, oncostatin M (OSM), inhibited differentiation of myoblasts through activation the JAK1-STAT1-STAT3 pathway.44 OSM decreased expression of MEF2A and the transcriptional activity of MEF2 and MyoD; whereas, expression of Id1 and Id2 increased. Interestingly, MEF2 co-precipitated with STAT1, but an N-terminal deletion mutant of MEF2 did not. This result revealed that MEF2 interacts with STAT1 through the N-terminus of MEF2. In addition, a pull down assay of MEF2 showed the C-terminal portion of STAT1 is involved in interacting with MEF2.44 These results implied that the STAT1 interaction with MEF2 is able to inhibit myogenic differentiation through repression of MEF2 transcriptional activity. In CTX-injected mice, OSM mRNA expression was reduced when mRNA expression of myogenin peaked. In addition, a tibialis anterior muscle transfected with an OSM-expressing vector, still had large unrepaired regions induced by CTX.44 Taken together, these data suggested that JAK1-STAT1-STAT3 activation inhibited myoblast differentiation and muscle regeneration, which resulted from the formation of the MEF2 and STAT1 complex and that MEF2 and STAT1 are the main effectors for suppression of myogenic differentiation.
Cardiotrophin-1 (CT-1), another member of the IL-6 family, is also a potent inhibitor of skeletal muscle differentiation.45 CT-1 activated both the STAT3 signaling pathway and MEK/ERK pathway during differentiation. However, the inhibitory effect of CT-1 on differentiation is not due to STAT3 signaling, but to the MEK/ERK pathway.45 Even though CT-1 activated STAT3, inhibition of the transcriptional activity of MyoD through MEK activation induces suppression of myogenic differentiation.3,45 Therefore, the activation of STAT3 may not be essential to repress myogenic differentiation.
Second, JAK2-STAT2-STAT3 promotes differentiation.12 Expression of MHC, MGN, MEF2 and MyoD decreased after treatment with the JAK2 inhibitor AG490 and JAK2-siRNA transfection. JAK2-siRNA also significantly reduced the expression of myogenin-, MRF-, MEF2- and MyoD-dependent luciferase reporter genes. STAT2 or STAT3 siRNA-transfection led to the same results.12 These findings indicated that the pro-differentiation effect of the JAK2-STAT2-STAT3 signaling pathway is partially mediated by MyoD and MEF2. In general, MyoD directly binds to MEF2 and enhances MEF2-dependent transcription through its own transcriptional activities, which results in cooperative enhancement of MyoD- and MEF-2-dependent myogenic differentiation.46 In addition, the HGF-ERK and IGF-AKT pathways were regulated by the JAK2-STAT2-STAT3 pathway.12 Therefore, the study’s results suggested that JAK2-STAT2-STAT3 signaling pathway is indispensable for myogenic differentiation. The expression profiles of JAK1, JAK2, STAT1 and STAT3 were confirmed in primary human skeletal muscle cells.13
In contrast, activation of JAK2-STAT3 also regulates proliferation of muscle cells.47 LIF increased the number of C2C12 cells and primary rat satellite cells, which were decreased by incubation with AG490. The phosphorylation of JAK2 and STAT3 and STAT3-activated luciferase activity increased in the presence of LIF, which indicated that LIF induces proliferation of muscle satellite cells and that activation of the JAK2 and STAT3 signaling pathway at least partially contributes to the increased satellite cell proliferation.47
Third, our group reported on the negative role of JAK3 in myogenic differentiation.48 We provided the first evidence that JAK3 is expressed in myoblast cells, and JAK3 inhibition promotes precocious myogenic differentiation and plays important roles in terminal differentiation. We demonstrated an inhibitor of JAK3, WHIp154, inhibited the formation of multinucleated myotubes by cell fusion and also induced precocious myogenic differentiation. JAK3-siRNA increased expression of MGN at the mRNA and protein levels and endogenous IGF-II mRNA expression during differentiation. Interestingly, when undergoing differentiation, WHIp154 not only decreased STAT1 phosphorylation but STAT3 phosphorylation also increased. JAK3 activation was gradually inhibited by WHIp154 at all incubation times in differentiation media. These data showed that the precocious myogenic differentiation by JAK3 inhibition is prompted via downregulation of STAT1 and upregulation of STAT3; however, unfortunately it is not clear how the activation of STAT1 and STAT3 is regulated in opposite directions. The AKT and ERK pathway also contributed to myogenic differentiation by JAK3 inhibition. JAK3 inhibition increases expression of a marker of early differentiation, MGN, and a marker of late differentiation, MHC. In contrast with a previous study, the JAK2 inhibitor AG490 did not have any change on myogenic differentiation. We explained that this effect is due to the dosage of AG490 and also concluded that JAK2 may be more important for survival of myoblasts with differentiation potential.48
The role of STATs in myoblast differentiation or proliferation was verified in many studies. STAT1 activation inhibits differentiation of myoblasts, which probably results from physical association with MEF2.44 However, STAT3 seems to have dual effects. STAT3 positively regulates both myogenic differentiation and proliferation. STAT3 activity increased in proliferation conditions, and knockdown of STAT3 reduced LIF-induced myoblast proliferation.11,47 When the expression of STAT3 was induced by 4-HT (hydroxytamoxifen), myogenin expression was reduced and the suppression of differentiation-induced growth (the S-G2/M phase in the cell cycle) was restored.49 These data are consistent with the role of STAT3 in proliferation. On the other hand, STAT3 activity gradually increased during differentiation and knockdown of STAT3 inhibited differentiation, suggesting that STAT3 promotes myoblast differentiation.12,13,39,48 The dual role of STAT3 in proliferation and differentiation is regulated by interaction with the transcription factor, MyoD. The direct interaction of STAT3-MyoD inhibits differentiation through reciprocal inhibition of their DNA binding activities, which are proved through transfection of MyoD or STAT3 construct.49 On the contrary to this, STAT3 bound with MyoD contributed to the stage of differentiation, suggesting that activation of STAT3 induced myogenic differentiation through binding with MyoD.50 Therefore, the interaction between STAT3 and MyoD is considered an important mechanism for the role of STAT3 during proliferation and differentiation of myoblasts.
Fer is an intracellular tyrosine kinase that associates with STAT3 in mammalian cells and is thought to be a mediator for regulation of STAT3 activity in myogenic differentiation.51 A study demonstrated that Fer is a mediator for STAT3 activation in insulin-PI3K pathway-eliciting myogenic differentiation. Fer and JAK1 co-immunoprecipitated in proliferating cells. However, insulin caused phosphorylation of Fer that led to dissociation from JAK1. In addition, insulin phosphorylated STAT3 and active Fer associated. This interaction between Fer and STAT3 was enhanced and sustained until 48 h after insulin treatment when myotubes appeared in the treated culture. A relevant explanation for this is that Fer mediates activation of STAT3 to enhance myogenic differentiation by insulin stimulation in myogenic cells.51 However, further studies are needed to identify the definite role of Fer in differentiation.
Finally, the SOCS (suppressors of cytokine signaling) family affects myogenic differentiation.52-54 SOCS is one of the transcripts of STAT, which acts as a negative regulator in the positive autoregulatory loop of the JAK-STAT pathway to suppress further signaling. The SOCS, PIAS (protein inhibitors of activated stats) and PTPs (protein tyrosine phosphatases) are major classes of negative regulators.55 The SOCS family of proteins consists of SOCS1 to SOCS7 and CIS.56 SOCS3 positively regulates myoblast differentiation.52 IGF-I was produced endogenously from myotubes during differentiation. This study showed that IGF-I increased SOCS3 transcription through STAT3 activation during differentiation. Moreover, overexpression of SOCS3 enhanced SRF (serum response factor)-dependent α-actin promoter activity during differentiation. SRF is a transcription factor that is associated with the differentiation and fusion of myoblasts. Consequently, activation of STAT3 by IGF-I upregulated SOCS3 stimulates SRF transcription factor-mediated myogenic differentiation. Therefore, it is likely that SOCS3 leads to endogenous IGF-I-mediated myoblast differentiation.52
SOCS1 also promotes myogenic differentiation and its role was tested with SOCS2 and PIAS.53 PIAS is another negative regulator of the JAK-STAT pathway. Individual knockdown of SOCS1, SOCS3 and PIAS inhibited myogenic differentiation of both primary myoblasts and C2C12 cells; whereas, overexpression promoted myogenic differentiation. This result revealed that SOCS1, SOCS3 and PIAS are necessary for myogenic differentiation. Importantly, this report proved strongly that SOCS1 and SOCS3 promote differentiation through inhibition of JAK1 activation and phosphorylation of the upstream receptor of JAK, glycoprotein of 130 (gp130), respectively. Also, PIAS prevents STAT1 or STAT3 binding to DNA, which contributes to the promotion of myogenic differentiation.53 These findings suggested how to repress JAK1-STAT1-STAT3 activation in proliferation and to induce differentiation.
Another view is that SOCS1 suppresses muscle differentiation.54 According to Inaba and colleagues, modulation of IGF-I receptor signal transduction is a suppressive mechanism of SOCS1 on myogenic differentiation.54 Generally, IGF-I promotes myogenic differentiation via the IRS-PI3K-AKT pathway and inhibits myogenic differentiation via MEK/MAPK. However, SOCS1 induced imbalance between the MEK/MAPK and IRS-PI3K-AKT pathways by enhancement of MEK and suppression of the IRS-1/AKT signaling pathway.54 The IRS-PI3K-AKT pathway promotes myogenic differentiation through myogenin expression by MEF2;31 while, phosphorylated MEK1 directly associates with MyoD, which prevents MyoD binding to the myogenin promoter and results in inhibition of muscle differentiation.32 Although the effect of JAK or STAT on the negative role of SOCS1 was not examined, SOCS may promote myogenic differentiation through regulation of STAT activity.
Conclusion and Perspectives
We have described the role of the JAKs-STATs pathways in the proliferation and differentiation of myoblasts. The findings are summarized in Table 1. JAK1-STAT1-STAT3 activation plays a crucial role in proliferation; whereas, activation of JAK2 and inhibition of JAK3 are responsible for myogenic differentiation. JAK2 activation results in phosphorylation of both STAT2 and STAT3 for differentiation. Interestingly, unlike JAK2, JAK3 regulates myogenic differentiation negatively, through STAT1 downregulation and STAT3 upregulation. This implies that JAK2 and JAK3 have contradictory effects on differentiation. STAT3 positively affects both proliferation and differentiation, depending on the type of JAK. The specificity of JAK type is likely able to determine differentiation or proliferation in myoblasts. It is also important which transcription factor interacts with STAT3 to determine proliferation or differentiation.
Table 1. The JAKs-STATs family and associated components in regulation of myogenic proliferation and differentiation.
| Proliferation | Differentiation | |
|---|---|---|
| JAKs family |
JAK1 |
JAK2, JAK3 |
| STATs family |
STAT1, STAT3 |
STAT2, STAT3, STAT1 |
| Transcription factor |
MEF2-STAT1 MyoD-STAT3* |
MyoD-MEF2, MyoD-STAT3*(Fer?only in IGF stimulation) |
| Signaling pathway |
MEK/ERK |
PI3K/AKT |
| Negative regulator of STATs |
SOCS1 |
SOCS1, SOCS3, PIAS1 |
| Representative inducer | LIF | IGF |
JAK1 activates STAT1 and STAT3, and activated STAT1 associates with transcription factor MEF2 in proliferation. Activated STAT3 interacts with MyoD, which inhibits differentiation and increases myoblast growth.* The MEK/ERK pathway is involved in this process. During proliferation, SOCS1, a negative regulator of STATs, is expressed, which inhibits differentiation of myoblasts. LIF induces proliferation and represses differentiation. In contrast, activation of JAK2 and inhibition of JAK3 promote differentiation. The activation of JAK2 phosphorylates STAT2 and STAT3 for differentiation. However, activation of JAK3 phosphorylates STAT1 and suppresses STAT3 activation, which inhibits differentiation. Activated STAT3 forms a complex with MyoD and the MyoD-MEF2 interaction, which stimulates transcription of MyoD- and MEF2-dependent muscle specific genes.* It is a mystery whether Fer, a mediator of STAT3 activity, is associated with this process. However, it is certain that the PI3K/AKT pathway contributes to differentiation, especially in IGF-induced myogenic differentiation. SOCS1, SOCS3 and PIAS inhibit activation of JAK1, gp130 and STAT1/3 binding to DNA, respectively. *Explains the dual effect of STAT3 on myoblast proliferation and differentiation.
Although the role of JAK1, 2 and 3 in myogenic differentiation was elucidated, the role of TYK2 is not determined yet. It is also unclear how STAT3 regulates proliferation and differentiation. Moreover, most previous studies have verified the effect of the JAK-STAT pathway on muscle differentiation in in vitro culture systems. Therefore, we need more evidence from in vivo animal experiments to better understand muscle regeneration. Future studies are expected to define more detailed roles of the JAK-STAT pathway and other regulatory mechanisms on myogenic differentiation.
Acknowledgments
This work was supported NRF funded by the MEST (2012011417), (20110002759) and KOSEF through Chronic Inflammatory Disease Research Center (2012051428).
Glossary
Abbreviations:
- bHLH
basic helix-loop-helix
- CTX
cardiotoxin
- ERK
extracellular signal-regulated kinase
- HGF
hepatocyte growth factor
- HT
hydroxytamoxifen
- IGF
insulin-like growth factor
- IL-6
interleukin-6
- JAK
Janus kinase
- LIF
leukemia inhibitory factor
- MCK
muscle creatine kinase
- MEF2
myocyte enhancer-binding factor 2
- MGN
myogenin
- MHC
myosin heavy chain
- MRFs
myogenic regulatory factors
- GTP
guanosine 5′-triphosphate
- IL1Ra
interleukin 1 receptor antagonist homolog 1
- NTX
notexin
- OSM
oncostatin M
- PI3K
phosphatidylinositol 3-kinase
- PIAS
protein inhibitors of activated STATs
- PTPs
protein tyrosine phosphatases
- SOCS
suppressor of cytokine signaling
- SRF
serum response factor
- STAT
signal transducer and activator of transcription
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/jak-stat/article/23282
References
- 1.Yang W, Chen Y, Zhang Y, Wang X, Yang N, Zhu D. Extracellular signal-regulated kinase 1/2 mitogen-activated protein kinase pathway is involved in myostatin-regulated differentiation repression. Cancer Res. 2006;66:1320–6. doi: 10.1158/0008-5472.CAN-05-3060. [DOI] [PubMed] [Google Scholar]
- 2.Tortorella LL, Milasincic DJ, Pilch PF. Critical proliferation-independent window for basic fibroblast growth factor repression of myogenesis via the p42/p44 MAPK signaling pathway. J Biol Chem. 2001;276:13709–17. doi: 10.1074/jbc.M100091200. [DOI] [PubMed] [Google Scholar]
- 3.Jo C, Kim H, Jo I, Choi I, Jung SC, Kim J, et al. Leukemia inhibitory factor blocks early differentiation of skeletal muscle cells by activating ERK. Biochim Biophys Acta. 2005;1743:187–97. doi: 10.1016/j.bbamcr.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Wu Z, Woodring PJ, Bhakta KS, Tamura K, Wen F, Feramisco JR, et al. p38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Mol Cell Biol. 2000;20:3951–64. doi: 10.1128/MCB.20.11.3951-3964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Johnson SE. ERK2 is required for efficient terminal differentiation of skeletal myoblasts. Biochem Biophys Res Commun. 2006;345:1425–33. doi: 10.1016/j.bbrc.2006.05.051. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Jiang B, Ensign WY, Vogt PK, Han J. Myogenic differentiation requires signalling through both phosphatidylinositol 3-kinase and p38 MAP kinase. Cell Signal. 2000;12:751–7. doi: 10.1016/S0898-6568(00)00120-0. [DOI] [PubMed] [Google Scholar]
- 7.Zetser A, Gredinger E, Bengal E. p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation. Participation of the Mef2c transcription factor. J Biol Chem. 1999;274:5193–200. doi: 10.1074/jbc.274.8.5193. [DOI] [PubMed] [Google Scholar]
- 8.Keren A, Tamir Y, Bengal E. The p38 MAPK signaling pathway: a major regulator of skeletal muscle development. Mol Cell Endocrinol. 2006;252:224–30. doi: 10.1016/j.mce.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 9.Coolican SA, Samuel DS, Ewton DZ, McWade FJ, Florini JR. The mitogenic and myogenic actions of insulin-like growth factors utilize distinct signaling pathways. J Biol Chem. 1997;272:6653–62. doi: 10.1074/jbc.272.10.6653. [DOI] [PubMed] [Google Scholar]
- 10.Tureckova J, Wilson EM, Cappalonga JL, Rotwein P. Insulin-like growth factor-mediated muscle differentiation: collaboration between phosphatidylinositol 3-kinase-Akt-signaling pathways and myogenin. J Biol Chem. 2001;276:39264–70. doi: 10.1074/jbc.M104991200. [DOI] [PubMed] [Google Scholar]
- 11.Sun L, Ma K, Wang H, Xiao F, Gao Y, Zhang W, et al. JAK1-STAT1-STAT3, a key pathway promoting proliferation and preventing premature differentiation of myoblasts. J Cell Biol. 2007;179:129–38. doi: 10.1083/jcb.200703184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K, Wang C, Xiao F, Wang H, Wu Z. JAK2/STAT2/STAT3 are required for myogenic differentiation. J Biol Chem. 2008;283:34029–36. doi: 10.1074/jbc.M803012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trenerry MK, Della Gatta PA, Cameron-Smith D. JAK/STAT signaling and human in vitro myogenesis. BMC Physiol. 2011;11:6. doi: 10.1186/1472-6793-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes SM. Muscle development: reversal of the differentiated state. Curr Biol. 2001;11:R237–9. doi: 10.1016/S0960-9822(01)00114-2. [DOI] [PubMed] [Google Scholar]
- 15.Sabourin LA, Rudnicki MA. The molecular regulation of myogenesis. Clin Genet. 2000;57:16–25. doi: 10.1034/j.1399-0004.2000.570103.x. [DOI] [PubMed] [Google Scholar]
- 16.Lluís F, Perdiguero E, Nebreda AR, Muñoz-Cánoves P. Regulation of skeletal muscle gene expression by p38 MAP kinases. Trends Cell Biol. 2006;16:36–44. doi: 10.1016/j.tcb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Buckingham M, Montarras D. Skeletal muscle stem cells. Curr Opin Genet Dev. 2008;18:330–6. doi: 10.1016/j.gde.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–95. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 19.Choi J, Costa ML, Mermelstein CS, Chagas C, Holtzer S, Holtzer H. MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proc Natl Acad Sci U S A. 1990;87:7988–92. doi: 10.1073/pnas.87.20.7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, et al. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol. 2005;25:3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabourin LA, Girgis-Gabardo A, Seale P, Asakura A, Rudnicki MA. Reduced differentiation potential of primary MyoD-/- myogenic cells derived from adult skeletal muscle. J Cell Biol. 1999;144:631–43. doi: 10.1083/jcb.144.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cornelison DD, Olwin BB, Rudnicki MA, Wold BJ. MyoD(-/-) satellite cells in single-fiber culture are differentiation defective and MRF4 deficient. Dev Biol. 2000;224:122–37. doi: 10.1006/dbio.2000.9682. [DOI] [PubMed] [Google Scholar]
- 23.Ustanina S, Carvajal J, Rigby P, Braun T. The myogenic factor Myf5 supports efficient skeletal muscle regeneration by enabling transient myoblast amplification. Stem Cells. 2007;25:2006–16. doi: 10.1634/stemcells.2006-0736. [DOI] [PubMed] [Google Scholar]
- 24.Gayraud-Morel B, Chrétien F, Flamant P, Gomès D, Zammit PS, Tajbakhsh S. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Dev Biol. 2007;312:13–28. doi: 10.1016/j.ydbio.2007.08.059. [DOI] [PubMed] [Google Scholar]
- 25.Le Grand F, Rudnicki MA. Skeletal muscle satellite cells and adult myogenesis. Curr Opin Cell Biol. 2007;19:628–33. doi: 10.1016/j.ceb.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang P, Wong C, Liu D, Finegold M, Harper JW, Elledge SJ. p21(CIP1) and p57(KIP2) control muscle differentiation at the myogenin step. Genes Dev. 1999;13:213–24. doi: 10.1101/gad.13.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walsh K, Perlman H. Cell cycle exit upon myogenic differentiation. Curr Opin Genet Dev. 1997;7:597–602. doi: 10.1016/S0959-437X(97)80005-6. [DOI] [PubMed] [Google Scholar]
- 28.Molkentin JD, Olson EN. Defining the regulatory networks for muscle development. Curr Opin Genet Dev. 1996;6:445–53. doi: 10.1016/S0959-437X(96)80066-9. [DOI] [PubMed] [Google Scholar]
- 29.Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–36. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 30.Bryantsev AL, Baker PW, Lovato TL, Jaramillo MS, Cripps RM. Differential requirements for Myocyte Enhancer Factor-2 during adult myogenesis in Drosophila. Dev Biol. 2012;361:191–207. doi: 10.1016/j.ydbio.2011.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamir Y, Bengal E. Phosphoinositide 3-kinase induces the transcriptional activity of MEF2 proteins during muscle differentiation. J Biol Chem. 2000;275:34424–32. doi: 10.1074/jbc.M005815200. [DOI] [PubMed] [Google Scholar]
- 32.Perry RLS, Parker MH, Rudnicki MA. Activated MEK1 binds the nuclear MyoD transcriptional complex to repress transactivation. Mol Cell. 2001;8:291–301. doi: 10.1016/S1097-2765(01)00302-1. [DOI] [PubMed] [Google Scholar]
- 33.Burattini S, Ferri P, Battistelli M, Curci R, Luchetti F, Falcieri E. C2C12 murine myoblasts as a model of skeletal muscle development: morpho-functional characterization. Eur J Histochem. 2004;48:223–33. [PubMed] [Google Scholar]
- 34.Chargé SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–38. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 35.Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene. 2002;285:1–24. doi: 10.1016/S0378-1119(02)00398-0. [DOI] [PubMed] [Google Scholar]
- 36.O’Sullivan LA, Liongue C, Lewis RS, Stephenson SE, Ward AC. Cytokine receptor signaling through the Jak-Stat-Socs pathway in disease. Mol Immunol. 2007;44:2497–506. doi: 10.1016/j.molimm.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 37.Hankey PA. Regulation of hematopoietic cell development and function by Stat3. Front Biosci. 2009;14:5273–90. doi: 10.2741/3597. [DOI] [PubMed] [Google Scholar]
- 38.Egwuagu CE. STAT3 in CD4+ T helper cell differentiation and inflammatory diseases. Cytokine. 2009;47:149–56. doi: 10.1016/j.cyto.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bellido T, Borba VZ, Roberson P, Manolagas SC. Activation of the Janus kinase/STAT (signal transducer and activator of transcription) signal transduction pathway by interleukin-6-type cytokines promotes osteoblast differentiation. Endocrinology. 1997;138:3666–76. doi: 10.1210/en.138.9.3666. [DOI] [PubMed] [Google Scholar]
- 40.Kim YH, Chung JI, Woo HG, Jung YS, Lee SH, Moon CH, et al. Differential regulation of proliferation and differentiation in neural precursor cells by the Jak pathway. Stem Cells. 2010;28:1816–28. doi: 10.1002/stem.511. [DOI] [PubMed] [Google Scholar]
- 41.Weber-Nordt RM, Mertelsmann R, Finke J. The JAK-STAT pathway: signal transduction involved in proliferation, differentiation and transformation. Leuk Lymphoma. 1998;28:459–67. doi: 10.3109/10428199809058353. [DOI] [PubMed] [Google Scholar]
- 42.Guillet-Deniau I, Burnol A-F, Girard J. Identification and localization of a skeletal muscle secrotonin 5-HT2A receptor coupled to the Jak/STAT pathway. J Biol Chem. 1997;272:14825–9. doi: 10.1074/jbc.272.23.14825. [DOI] [PubMed] [Google Scholar]
- 43.Mancinelli R, Pietrangelo T, Burnstock G, Fanò G, Fulle S. Transcriptional profile of GTP-mediated differentiation of C2C12 skeletal muscle cells. Purinergic Signal. 2012;8:207–21. doi: 10.1007/s11302-011-9266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao F, Wang H, Fu X, Li Y, Ma K, Sun L, et al. Oncostatin M inhibits myoblast differentiation and regulates muscle regeneration. Cell Res. 2011;21:350–64. doi: 10.1038/cr.2010.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyake T, Alli NS, Aziz A, Knudson J, Fernando P, Megeney LA, et al. Cardiotrophin-1 maintains the undifferentiated state in skeletal myoblasts. J Biol Chem. 2009;284:19679–93. doi: 10.1074/jbc.M109.017319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–36. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 47.Spangenburg EE, Booth FW. Multiple signaling pathways mediate LIF-induced skeletal muscle satellite cell proliferation. Am J Physiol Cell Physiol. 2002;283:C204–11. doi: 10.1152/ajpcell.00574.2001. [DOI] [PubMed] [Google Scholar]
- 48.Jang YN, Lee IJ, Park MC, Baik EJ. Role of JAK3 in myogenic differentiation. Cell Signal. 2012;24:742–9. doi: 10.1016/j.cellsig.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 49.Kataoka Y, Matsumura I, Ezoe S, Nakata S, Takigawa E, Sato Y, et al. Reciprocal inhibition between MyoD and STAT3 in the regulation of growth and differentiation of myoblasts. J Biol Chem. 2003;278:44178–87. doi: 10.1074/jbc.M304884200. [DOI] [PubMed] [Google Scholar]
- 50.Yang Y, Xu Y, Li W, Wang G, Song Y, Yang G, et al. STAT3 induces muscle stem cell differentiation by interaction with myoD. Cytokine. 2009;46:137–41. doi: 10.1016/j.cyto.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 51.Taler M, Shpungin S, Salem Y, Malovani H, Pasder O, Nir U. Fer is a downstream effector of insulin and mediates the activation of signal transducer and activator of transcription 3 in myogenic cells. Mol Endocrinol. 2003;17:1580–92. doi: 10.1210/me.2002-0328. [DOI] [PubMed] [Google Scholar]
- 52.Spangenburg EE. SOCS-3 induces myoblast differentiation. J Biol Chem. 2005;280:10749–58. doi: 10.1074/jbc.M410604200. [DOI] [PubMed] [Google Scholar]
- 53.Diao Y, Wang X, Wu Z. SOCS1, SOCS3, and PIAS1 promote myogenic differentiation by inhibiting the leukemia inhibitory factor-induced JAK1/STAT1/STAT3 pathway. Mol Cell Biol. 2009;29:5084–93. doi: 10.1128/MCB.00267-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inaba M, Saito H, Fujimoto M, Sumitani S, Ohkawara T, Tanaka T, et al. Suppressor of cytokine signaling 1 suppresses muscle differentiation through modulation of IGF-I receptor signal transduction. Biochem Biophys Res Commun. 2005;328:953–61. doi: 10.1016/j.bbrc.2005.01.046. [DOI] [PubMed] [Google Scholar]
- 55.Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci. 2004;117:1281–3. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- 56.Alexander WS. Suppressors of cytokine signalling (SOCS) in the immune system. Nat Rev Immunol. 2002;2:410–6. doi: 10.1038/nri818. [DOI] [PubMed] [Google Scholar]


