Abstract
Long RNAs often exist as multiple conformers in equilibrium. For the genomes of single-stranded RNA viruses, one of these conformers must include a compacted state allowing the RNA to be confined within the virion. We have used single molecule fluorescence correlation spectroscopy to monitor the conformations of viral genomes and sub-fragments in the absence and presence of coat proteins. Cognate RNA-coat protein interactions in two model viruses cause a rapid collapse in the hydrodynamic radii of their respective RNAs. This is caused by protein binding at multiple sites on the RNA that facilitate additional protein-protein contacts. The collapsed species recruit further coat proteins to complete capsid assembly with great efficiency and fidelity. The specificity in RNA-coat protein interactions seen at single-molecule concentrations reflects the packaging selectivity seen for such viruses in vivo. This contrasts with many in vitro reassembly measurements performed at much higher concentrations. RNA compaction by coat protein or polycation binding are distinct processes, implying that defined RNA-coat protein contacts are required for assembly.
Keywords: RNA folding, viral genomes, single molecule fluorescence
Introduction
Single-stranded (ss) RNA viruses are major pathogens in every kingdom of life. They have mono- or multipartite genomes ranging in length up to > 10 kb, encoding multiple open reading frames. These genomes often serve multiple functions acting as both mRNAs for viral protein expression and templates for RNA-dependent RNA polymerase during replication. During these processes, the RNAs are not in compact conformations. Formation of progeny virions, however, requires that the RNAs be confined in the restricted space formed by assembly of a protective protein shell. The precise mechanism that controls both the conformational changes accompanying virion assembly and the selective packaging of cognate genomes, rather than potential competitor cellular RNAs, has remained vague. The current paradigm, extrapolating from the fact that many viral coat proteins contain positively charged domains and will self-assemble in vitro around non-viral RNAs, assumes that genome packaging is non-sequence specific.1-3 Our recent results with two simple viruses that infect bacteria and plants overturn this view.4 Using single molecule fluorescence assembly assays that avoid artifacts due to high protein concentration, we show that packaging is both sequence-specific and two-stage. The first stage is a rapid compaction of the RNA that is required to allow it to fit into the capsid, driven by multiple coat protein-RNA and coat protein-coat protein interactions. The second stage is recruitment of the remaining complement of coat proteins to these partially formed compact complexes. This mechanism mirrors aspects of other RNA folding reactions, such as ribosome assembly, and provides novel insights into the biology of RNA viruses that could be exploited therapeutically.
RNA compaction
Folding into a compact state plays an important role in the function of many RNAs. This has been demonstrated for both short RNAs, such as ribozymes or riboswitches (< 500 nt long), and also for longer rRNA (16S bacterial rRNA, 1,530 nt). Ribozymes, riboswitches and shorter fragments of rRNAs (e.g., the 5′ domain of 16S rRNA)5 fold in the presence of multivalent cations. Folding of these RNAs usually proceeds through rapid formation of compact structures, which may contain non-native tertiary contacts that are resolved during a subsequent slower phase.6
Longer rRNAs also undergo rapid formation of secondary structure and collapse into a compact ensemble but require ribosomal proteins for stabilization of the native fold under physiological salt concentrations.7 Ribosomal protein assembly onto the largely pre-folded RNA core is a co-operative, step-wise process,8,9 characterized by a gradual decrease in RNA flexibility upon addition of proteins.10 Some ribosomal proteins, such as S4 and S7, are largely disordered prior to interaction with the rRNA and may also fold upon assembly. Such co-folding further increases the co-operativity and specificity of assembly.7,11 Formation of secondary structure and initial compaction of long rRNAs is thought to occur co-transcriptionally in cells and in the later stages is assisted by assembly of ribosomal proteins.7 Parallel assembly pathways may be utilized12 to bypass potential roadblocks due to mutations or protein deficiency.13
Considerably less is known about the structures of other long RNAs, such as mRNAs, long non-coding (lnc) RNAs and viral genomic RNAs. While local secondary and tertiary structures are important in mRNAs for control of gene expression, processing and stability,14-16 their overall structures are less well defined. Due to their limited lifetimes and coding functions, they are not required to adopt a uniquely folded structure.
Likewise, viral RNAs were thought not to harbor extensive regions of tertiary structure, especially those which also act as mRNAs (e.g., viruses with positive sense ssRNA genomes). Medium resolution cryo-EM reconstructions have demonstrated, however, that substantial portions of genomic RNA are ordered inside the capsids of many viruses.17-21 This order includes regions in contact with the coat protein layer as well as regions free of protein. Specific packaging signals (PSs), short conserved sequences/motifs that are recognized by viral coat proteins during assembly, have been identified in many viral RNAs.22-29 These sequences are thought to facilitate selective packaging of viral genomic RNA. However, many viruses do not exhibit such clearly defined packaging signals and most of those which do can also package non-specific RNA in vitro or cellular mRNA when their proteins are overexpressed from a DNA vector in cells, or even during normal infection cycles.30
Since RNA is a polyanion, buffer salt concentration and composition are crucial for folding and condensation, e.g., for ribosomal and other structured RNAs. Many viral coat proteins possess positively charged polypeptide arms or domains which are in direct contact with RNA in the virion and help to neutralize RNA charge. Some viruses also encapsidate polyvalent cations, such as spermidine, to aid charge neutralization.31-33 As a result of these features, charge neutralization has been thought to be the essential step in viral genome packaging,1,3,34-36 although such a mechanism cannot easily explain the observed preference for encapsidation of cognate genomes in vivo.
How can a virus selectively package its own RNA inside an infected cell cytoplasm full of heterologous RNAs? One strategy, adopted by dsRNA viruses and some ssRNA viruses, is to sequester the sites of replication and assembly away from the cytoplasm into “virus factories,” inclusion bodies or onto membrane surfaces.37-39 This process requires a level of sophistication and specialized gene products that organize these assemblies, leading to co-localization and direct coupling between RNA replication and packaging.40 Simpler viruses, with limited coding capacity, including major pathogens, do not have such luxury and replicate in the cytoplasm and, therefore, have to select their own RNAs based on coat protein affinity. Given the apparent paucity of high affinity packaging signals and the relatively high concentration of cellular competitors, affinity alone may be insufficient to achieve specificity. Nature's solution to this problem seems to be the evolution of co-operativity in the packaging process, based on multiple weak RNA-coat protein interactions rather than on recognition of a single high affinity site.4 A key to observing such co-operativity during co-assembly of RNA and viral coat proteins was to mimic early stages in virus assembly when the coat protein concentration is naturally low. This was achieved using single molecule detection of assembling intermediates, based on fluorescence correlation spectroscopy (FCS).
RNA virus assembly
Coat proteins of RNA viruses, such as bacteriophage MS241 and the small plant virus satellite tobacco necrosis virus (STNV) (Fig. 1),42,43 interact with genomic RNA primarily via two mechanisms. The MS2 coat protein recognizes a high affinity, RNA stem-loop within the cognate genome (TR),44-47 which interacts with both subunits of a coat protein dimer.41,48-50 This complex initially represses translation of the phage replicase and is thought to nucleate assembly (Fig. 1). STNV is an example of a large group of viruses in which the coat proteins interact with RNA, at least in part, using positively charged extended polypeptide arms. In different viral proteins, these can be at the N or C terminus and have been thought to neutralize RNA charge with no sequence specificity. However, in vitro SELEX against the STNV coat protein allowed us to identify aptamers with sequence/structure matches to multiple degenerate potential stem-loop structures positioned throughout the known STNV genomes, suggesting sequence selectivity.22 The latter has been demonstrated in in vitro reassembly assays.51 Multiple RNA-coat protein contacts are effective at promoting efficient assembly.52 Similar mechanisms seem to be adopted by other RNA viruses, including some plant (TCV), animal and human viruses.23-29
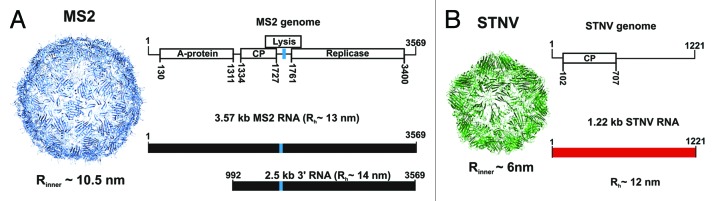
Figure 1. The bacteriophage MS2 and satellite tobacco necrosis virus (STNV) components. (A) Structure of the MS2 phage capsid (inner capsid radius, Rinner~10.5 nm), its genomic map and a cartoon showing the RNA fragments used for the smFCS experiments described here (below). The location of TR is indicated by a blue stripe. (B) Structure of the STNV capsid (inner capsid radius, Rinner~6.0 nm), together with its genomic map and corresponding transcript employed in assembly experiments (1.22 kb, shown in red below). Average hydrodynamic radii of the protein-free RNAs in a polyvalent ion-free buffer (see text) are also shown.
Many viruses can, however, package non-cognate RNAs in vitro53-55 or assemble without any RNA,56 leading to the dominance of a protein-centric assembly model in which coat protein binding neutralizes RNA charge, gradually condensing it to fit into the capsid. This concept cannot account for the highly specific packaging observed for RNA viruses in vivo, perhaps reflecting the difference from in vitro conditions, which usually employ high protein and nucleic acid concentrations. In vivo, protein and RNA concentrations build up from scratch during viral infection. In many cases, replication is completed during the early stages of infection and precedes coat protein production. A plausible scenario in the infected cell is that assembly is initiated on viral RNA at coat protein concentrations that are much lower than those used in vitro. Consequently, most of the newly synthesized coat would be incorporated into the growing capsids leaving little free coat protein for non-specific interactions. This scenario begs a number of questions: Can assembly be triggered at low concentrations? How is the viral RNA compacted at such low protein concentrations? Is there any difference between the packaging of cognate vs. non-cognate RNA? Answers to these came from single-molecule assays of RNA packaging during capsid assembly, which are described in the following section.
Single molecule techniques in RNA folding
FCS is a correlation-based method which exploits spontaneous fluctuations in fluorescent signals in order to obtain characteristic time scales (e.g., relaxation times or rates) for molecular processes.57,58 The fluctuations are due to changes in the number density of fluorescent species within the measurement volume and are related to rates of chemical (e.g., the conversion of a fluorescent to non-fluorescent substrate by quenching) or photo-physical reactions (e.g., the dynamics of triplet state formation, on the ~1 µs time scale). When performed at the single molecule level using a small confocal volume (~1 fL, the number density changes due to molecules diffusing in and out of the detection volume (Fig. 2). The resulting fluctuations are then transformed in real time, usually by a specialized hardware correlator, to an autocorrelation function (CF) (Fig. 2), and the characteristic times (τd) are obtained from fitting the correlation function to a model.57 At single molecule concentrations (~1 nM), the diffusion represents the longest correlated process and the corresponding diffusion time, which is related to the average time the molecule takes to transverse the confocal volume, is readily identifiable from the CF profile (Fig. 2). A small molecule (e.g., fluorescent dye), which has a large diffusion coefficient, will spend a relatively short time in the measured volume and, thus, the fluorescent signal will only be correlated during this short period (~80 μs for an AlexaFluor 488 dye molecule, Fig. 2). Viral coat proteins, assembly intermediates, genomic RNAs and capsids diffuse progressively more slowly producing considerably longer correlations in the fluorescent signal (~0.4−1 ms, Fig. 2). This provides a way to size selectively labeled molecules in the presence of unlabeled species. Their hydrodynamic radii (Rh) are then computed using calibration of the confocal volume with free dye and Stokes-Einstein equation.57 For the viral systems described here, these Rh values agree very well with separate estimations based on X-ray structures or mass spectrometry.41,43,59-61
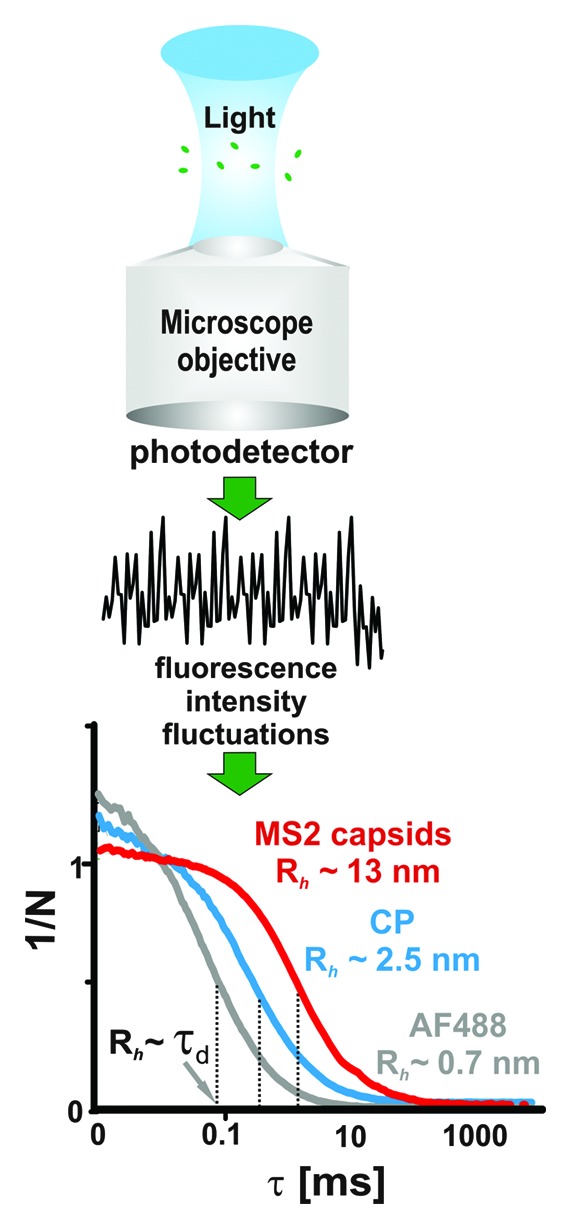
Figure 2. Single-molecule fluorescence correlation spectroscopy. The fluorescent signal is collected from single molecules passing through the confocal volume of the microscope objective. Rapid fluctuations in the fluorescent signal (black trace) are collected by the detector and correlated to obtain an autocorrelation function (CF). Typical CF values for Alexa Fluor 488 dye (gray), MS2 bacteriophage coat protein dimer (blue) and capsid (red) are shown below. The τd values are indicated by dotted vertical lines and the corresponding hydrodynamic radii (Rh) are shown. The CF amplitude scales inversely with the average number of molecules in the confocal volume (N) as indicated on the Y axis.
FCS assays of viral assembly rely on selective and efficient labeling of RNA and their cognate interacting proteins. Many strategies have been developed for covalent labeling of proteins, ranging from relatively non-specific lysine-reactive dye conjugates to more selective cysteine-reactive maleimide derivatives and CLICK chemistry.62-66 Specific covalent labeling of RNA can be achieved at either 5′ or 3′ termini.67-71 Two 3′-end strategies were employed in the work described here: (1) end ligation of a dye labeled dA10 oligomer; (2) incorporation of 3′-amino adenosine by poly-A-polymerase followed by amine-specific labeling.4 The advantage of the former is the elimination of chemical labeling steps, while the latter is more effective in situations where the 3′ end of the RNA is partially occluded, preventing the incorporation of the bulky oligomer.
Other single molecule techniques, such as Förster resonance energy transfer (smFRET), provide a more detailed view of conformational changes, as elegantly demonstrated for the folding pathway of the S-adenosyl methionine regulated riboswitch72,73 and for protein-assisted folding of telomerase RNA.74 Unlike FCS, smFRET interrogates local, specific structural details and employs detailed structural information to position the donor and acceptor dyes. In principle, such an approach is also applicable to large RNAs but positioning of the dyes needs to be carefully guided (e.g., by secondary structure probing) in order to avoid interfering with RNA folding and function (e.g., virus assembly).
The single molecule methods discussed here may be useful in many other situations involving large RNA molecules. For instance, it would be an excellent way to investigate the structures and interactions of long non-coding RNAs (e.g., Xist, HOTAIR, NRON).75-78
Sizes of protein-free viral RNAs
Given the need for compaction during assembly, it is important to consider the structure of the protein-free viral RNA. All long ssRNAs, with the exception of synthetic homopolymers, will form short stretches of secondary structure by intramolecular base pairing.79 Raman spectra of MS2 and other viral RNAs suggest that at least 85% of their nucleotides are base-paired,80-83 and are consistent with structure probing studies.84-86 This contrasts with early electron microscopy (EM) images of bacteriophage RNAs, which revealed extended conformations with few loops and low secondary structure content. It is now presumed that this appearance was due to disruption of the native structure during sample preparation.87 Estimation of RNA size and shape in solution has also proved difficult. Analysis of MS2 RNA by SAXS was complicated by self-association due to the high concentrations required.88,89 Sedimentation velocity measurements of MS2 genomic and sub-genomic RNAs under native conditions, however, indicates that these viral RNAs have compact conformations.60 Attempts have been made to estimate the overall shape and compactness of long RNA molecules.90 The extensive branching of viral RNAs and the requirement that they fit into the cramped space of a capsid led to the proposal that they are more compact than RNAs with the same base composition but randomized sequences. A recent SAXS study and direct visualization of a few examples of long RNA molecules in frozen solution by cryoEM demonstrated that they are indeed folded into highly branched, relatively compact but elongated structures.91 However, the limited number of RNA types examined by this method thus far precludes a definitive conclusion on the overall compactness of viral vs. non-viral RNA.
SmFCS, coupled with non-disruptive end-labeling with dye, is an important development. It provides a convenient tool for sizing RNA molecules in extremely dilute solutions in which intermolecular RNA interactions would be expected to be negligible. Hydrodynamic radii estimated from such measurements demonstrate that the solution sizes of STNV and MS2 genomic RNAs, as well as several sub-genomic MS2 RNA fragments (Fig. 1), are larger than the volume available inside their cognate capsids ((Rh values of the genome and a fragment lacking roughly a third of the 5′ end−3′ RNA, are ~13 and ~14 nm, respectively, compared with Rinner ~10.5 nm, for the RNA volume in a capsid). This confirms the need for compaction during assembly. Similar assays with a range of non-viral RNAs have demonstrated that they may be as compact as viral RNA.4 Hence, the initial size of viral RNA cannot be the sole factor by which the coat protein selects the genome. Are there other properties which might distinguish the cognate RNA from others during packaging?
Cooperative collapse
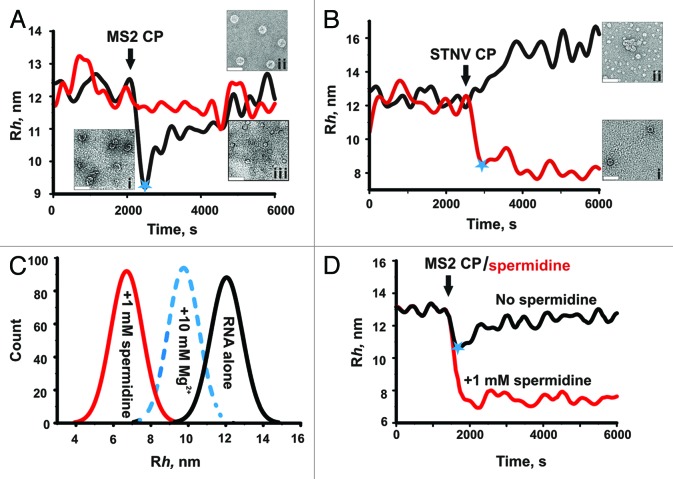
The answer to this question came from following viral assembly reactions by FCS in real time.4 The results of the most revealing experiment are summarized in Figure 3. When the hydrodynamic radius of labeled MS2 genomic RNA is measured before and after addition of sufficient MS2 coat protein to complete capsid assembly on every RNA (Fig. 3A, black line), a large (~30%) and rapid (faster than the experimental dead time, 60 s) collapse in the Rh value is seen. This is followed by an increase in Rh in a second stage of assembly that appears to reflect the completion of capsid assembly from pre-compacted, cognate coat protein-RNA complexes. In the EM, these have the appearance of partially formed shells. Under the same conditions, no collapse occurs with STNV genomic RNA mixed with MS2 coat protein (Fig. 3A, red line). In contrast, when STNV coat protein was used, STNV RNA undergoes a collapse (Fig. 3B, red line). Hydrodynamic collapse is therefore specific to viral RNAs making cognate interactions with their respective coat proteins. For MS2, a mutant coat protein that binds its RNA normally but is deficient in protein-protein interactions fails to elicit the collapse, showing that both interactions are required for this effect and that the collapsed state is not simply a random complex of protein and RNA. This result can only be explained if there are multiple, correctly placed protein binding sites, i.e., packaging signals (PSs), along the viral RNAs. No collapse was seen with non-viral RNAs, or for non-cognate viral RNAs.4 Interestingly, sub-genomic RNA fragments retain the property of CP-induced collapse, indicating that the co-operativity extends throughout the RNA. Perhaps surprisingly, given the above, all the RNAs tested in these assays trigger assembly of capsid-like aggregates. However, only cognate interactions yield capsids of the correct size and symmetry (T = 1 for STNV and T = 3 for MS2), whereas non-cognate assembly reactions are relatively low yield and produce a high proportion of misassembled species.
Figure 3. Two-stage assembly of cognate viral RNAs. Time-resolved changes in the apparent Rh of MS2 (black) and STNV (red) genomic RNAs are shown before and after addition (black arrow) of stoichiometric amounts of MS2 (A) or STNV (B) coat proteins, i.e., sufficient protein to allow each RNA to form a complete capsid. Blue stars denote the end of the respective compaction stages for interactions between cognate molecules. For MS2, this is followed by a slower increase in Rh correlated with formation of the capsids with high yield and fidelity. Capsid formation may already be complete for STNV following stage 1. Non-cognate interactions do not lead to collapse but do lead to inefficient formation of mostly aberrant aggregates. Electron micrographs of negatively stained assembly reactions at defined points in the pathway are shown. ( (A) i, assembly intermediates observed ~1 min after addition of MS2 CP to MS2 RNA; ii, T = 3 MS2 capsids present at the end of stage 2, assembled with MS2 RNA; iii, aberrant assembly products and aggregates formed by co-assembly of MS2 CP and STNV RNAs (scale bars 50 nm). (B) i, T = 1 STNV capsids at the end of the assembly reaction and ii, aberrant aggregates formed by co-assembly of STNV CP and MS2 genomic RNA. (C) Effects of multivalent cations on the apparent Rh of the MS2 3′ RNA sub-genomic fragment (Fig. 1) shown as size (Rh) distributions. RNA in a monovalent buffer is shown in black, followed by separate reactions in which divalent (Mg2+, 10 mM, blue) or trivalent (spermidine, 1mM, red) ions were added. (D) Condensation by counterions or coat protein subunits are not equivalent processes. The MS2 3′ RNA undergoes condensation upon addition of 1 mM spermidine. Addition of 200 nM MS2 CP2 to this sample (red) or one in the monovalent buffer (black) has no effect or results in compaction and then recovery, respectively. The latter sample produces capsids, whereas no assembly occurs under these conditions in the presence of spermidine unless the coat protein concentration is raised significantly (not shown).
Given the roles of multivalent cations in RNA folding, and the paradigm of electrostatically controlled assembly, a question arises of whether simple charge neutralization, e.g., by magnesium ions or spermidine, could also trigger RNA collapse? The data shown in Figure 3C suggest that these cations do indeed collapse RNA conformations to approximately the same (Mg2+) or even greater (spermidine) degrees than binding of coat proteins. Interestingly, once the RNA is compacted by spermidine, it is no longer a good substrate for capsid assembly, although increasing the CP concentrations significantly can reverse this effect (Fig. 3D). Thus, it seems that while both coat protein binding and electrostatic neutralization are sufficient to compact the viral RNA, they are not equivalent processes on the pathway to capsid assembly. The role(s) of electrostatics in virus assembly may therefore be less prominent than previously thought.1,34
The requirement for protein-protein interactions to drive the RNA collapse implies that assembly is in fact a cooperative process. Sub-stoichiometric (with respect to capsid) amounts of coat protein can also cause the full collapse, implying a nucleated effect governed by coat protein-RNA packaging signal affinities. Putative packaging signals for both MS2 and STNV have now been identified and consist of stem-loops dispersed throughout their genomes. These have only a minimal consensus recognition motif (ref. 22; Dykeman, submitted), explaining why they have not been identified previously. The collapse is rapid, occurring within the dead time of the FCS instrument (60 s per CF). This is consistent with previous solution structure probing of the MS2 genome that suggests that many of the predicted PSs should be present in the RNA and, thus, able to bind coat proteins immediately.92,93 The first stage of assembly resembles the millisecond compaction of smaller RNAs by Mg2+, previously observed using SAXS.94 Genomic RNA compaction by a sub-stoichiometric amount of cowpea chlorotic mottle virus (CCMV) coat protein has also been observed.95 However, that process was slow and indiscriminate, compacting both viral as well as non-viral RNA, perhaps because the assays were at higher concentrations than those reported here.
A two-stage cooperative mechanism of selective RNA packaging
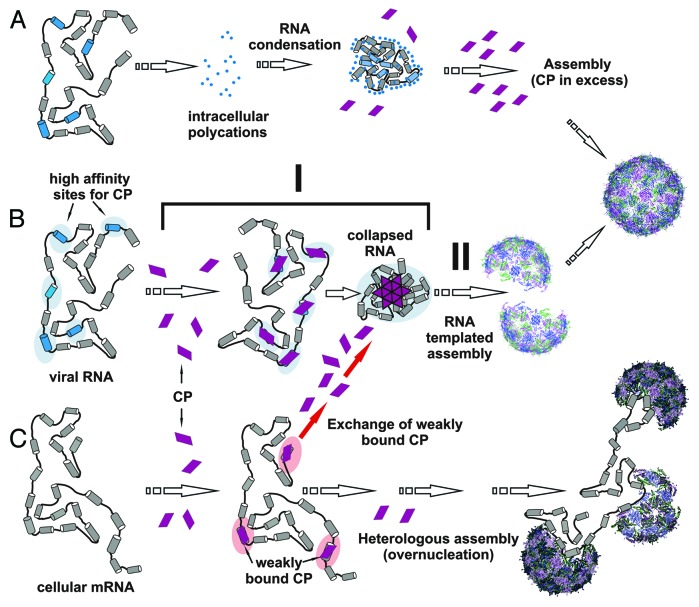
The new findings challenge the prevailing view that RNA is gradually condensed during capsid polymerization. While non-cognate (e.g., cellular) RNA may be gradually condensed, the cognate viral RNA is rapidly and cooperatively collapsed (c.f. pathways B and C in Fig. 4). The collapsed state is relatively stable since it is formed by multiple RNA-CP and CP-CP interactions. The affinities of individual PS-CP complexes within the collapsed state could be very low but are augmented by their relative locations and the protein-protein interactions that they promote. The stability of this nucleation complex facilitates recruitment of the additional coat proteins required to complete the capsid during a second stage of assembly that is protein concentration dependent.
Figure 4. The two-stage assembly mechanism for viral RNA. Our results suggest that there are three forms of RNA condensation/compaction in the context of virus assembly. (A) Non-specific condensation of viral RNA by multivalent ions (Mg2+, spermidine) inhibits assembly by coat proteins (middle). Only at very high CP concentrations (> 5 μM) is this block overcome (right), showing that simple electrostatic condensation is not on pathway to capsid formation. (B) A two-stage mechanism of assembly in which CPs first bind to cognate RNAs displaying multiple packaging sites (blue segments), distributed throughout the viral genome to facilitate protein-protein interactions, thus mediating a rapid RNA collapse (stage I). The collapse is followed by cooperative recruitment of additional CP subunits (stage II), even at low concentration (< 1 μM), to complete capsid assembly. (C) Assembly with non-cognate (cellular) RNAs leads to weak interactions without observable initial RNA collapse. Dissociation of coat proteins from these complexes allows them to be captured by the cognate assembly pathway (red arrows, middle). A low yield alternative pathway occurs when non-specifically bound coat proteins nucleate assembly on cellular RNA. Since the coat protein binding sites are not correctly positioned, these RNAs do not collapse and there can be multiple nucleation events leading to misassembled and multishell structures. Such pathways explain the assembly of non-cognate RNAs in vitro at relatively high coat protein concentrations.
In contrast, coat proteins will bind to non-cognate RNAs at individual stem-loops that resemble cognate packaging sites, but these will be randomly distributed and, thus, unable to support co-operative collapse. Individual coat protein-RNA complexes are therefore likely to dissociate without nucleating assembly. This provides an explanation for the effective discrimination between the cognate and non-cognate RNAs in vivo. Coat proteins binding and dissociating from sites on cellular RNAs will eventually be captured by the stable nucleation complexes and used to complete capsids (Fig. 4, red arrows). The energetic cost of genome confinement in these viruses is paid by the free energy of coat protein-packaging signal interactions.
Simple charge neutralization and condensation by multivalent cations produces collapsed RNAs that assemble poorly at low coat protein concentrations (nanomolar). Since this block can be overcome at high (micromolar) coat protein concentrations (not shown), the cations may simply bind to the packaging sites and compete with coat protein or act indirectly by folding the RNA into structures in which the packaging sites are obscured (Fig. 4A). Only high coat protein concentration can overcome this effect and complete assembly. This again highlights the role of multiple packaging sites which are accessible throughout the viral genome and distributed in a way to promote cooperative assembly of the nucleation complex.
Conclusions
The RNAs of ssRNA viruses seem to have evolved multiple packaging signals that are dispersed throughout their genomes facilitating cooperative assembly of an extended nucleation complex that simultaneously compacts the RNA during the first stage of assembly. In the second stage, the collapsed nucleation complex is efficiently encapsidated, effectively outcompeting protein binding to cellular RNA. An analogy of this process is the idea of packing a suitcase. In this instance, the suitcase (capsid) folds its own contents (the viral RNA). Since we see the same phenomenon in two viruses drawn from widely different families, one of which has an extremely widespread coat protein fold,96 it is highly likely that similar conserved assembly mechanisms exist in many viruses in this class. Note the molecular details driving the hydrodynamic collapse can vary widely in the different cases. Some viruses, especially animal viruses, appear only to package nascent genomes into progeny virions. This behavior could be due to the need for kinetic folding of the PSs in those cases. Even though the genome would not collapse in the same way, virion assembly needs to compensate for the entropic cost of confining its RNA by making the favorable CP-PS contacts. Interrupting the interactions between coat proteins and their cognate packaging signals would inhibit these processes and may provide a novel target for anti-virals. Given the co-operative nature of the assembly mechanism described here, such drugs may be more difficult to escape by mutation.
Acknowledgements
A.B. thanks the Wellcome Trust for support of his postgraduate studentship (089310/09/Z) and support of the facilities used (062164) (090932/Z/09/Z). Single molecule work within the Astbury Centre has been supported by The University of Leeds.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/23838
References
- 1.Belyi VA, Muthukumar M. Electrostatic origin of the genome packing in viruses. Proc Natl Acad Sci USA. 2006;103:17174–8. doi: 10.1073/pnas.0608311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forrey C, Muthukumar M. Electrostatics of capsid-induced viral RNA organization. J Chem Phys. 2009;131:105101–9. doi: 10.1063/1.3216550. [DOI] [Google Scholar]
- 3.Devkota B, Petrov AS, Lemieux S, Boz MB, Tang L, Schneemann A, et al. Structural and electrostatic characterization of pariacoto virus: implications for viral assembly. Biopolymers. 2009;91:530–8. doi: 10.1002/bip.21168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borodavka A, Tuma R, Stockley PG. Evidence that viral RNAs have evolved for efficient, two-stage packaging. Proc Natl Acad Sci USA. 2012;109:15769–74. doi: 10.1073/pnas.1204357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adilakshmi T, Ramaswamy P, Woodson SA. Protein-independent folding pathway of the 16S rRNA 5′ domain. J Mol Biol. 2005;351:508–19. doi: 10.1016/j.jmb.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 6.Russell R, Millett IS, Tate MW, Kwok LW, Nakatani B, Gruner SM, et al. Rapid compaction during RNA folding. Proc Natl Acad Sci USA. 2002;99:4266–71. doi: 10.1073/pnas.072589599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woodson SA. RNA folding pathways and the self-assembly of ribosomes. Acc Chem Res. 2011;44:1312–9. doi: 10.1021/ar2000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Traub P, Nomura M. Structure and function of Escherichia coli ribosomes. VI. Mechanism of assembly of 30 s ribosomes studied in vitro. J Mol Biol. 1969;40:391–413. doi: 10.1016/0022-2836(69)90161-2. [DOI] [PubMed] [Google Scholar]
- 9.Traub P, Nomura M. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc Natl Acad Sci USA. 1968;59:777–84. doi: 10.1073/pnas.59.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stagg SM, Mears JA, Harvey SC. A structural model for the assembly of the 30S subunit of the ribosome. J Mol Biol. 2003;328:49–61. doi: 10.1016/S0022-2836(03)00174-8. [DOI] [PubMed] [Google Scholar]
- 11.Williamson JR. Proteins that bind RNA and the labs who love them. Nat Struct Biol. 2001;8:390–1. doi: 10.1038/87540. [DOI] [PubMed] [Google Scholar]
- 12.Talkington MW, Siuzdak G, Williamson JR. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–32. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bubunenko M, Korepanov A, Court DL, Jagannathan I, Dickinson D, Chaudhuri BR, et al. 30S ribosomal subunits can be assembled in vivo without primary binding ribosomal protein S15. RNA. 2006;12:1229–39. doi: 10.1261/rna.2262106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bag J, Bhattacharjee RB. Multiple levels of post-transcriptional control of expression of the poly (A)-binding protein. RNA Biol. 2010;7:5–12. doi: 10.4161/rna.7.1.10256. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Kiledjian M. Regulation of mRNA decapping. Wiley Interdiscip Rev RNA. 2010;1:253–65. doi: 10.1002/wrna.15. [DOI] [PubMed] [Google Scholar]
- 16.Tuerk C, Gauss P, Thermes C, Groebe DR, Gayle M, Guild N, et al. CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci USA. 1988;85:1364–8. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toropova K, Basnak G, Twarock R, Stockley PG, Ranson NA. The three-dimensional structure of genomic RNA in bacteriophage MS2: implications for assembly. J Mol Biol. 2008;375:824–36. doi: 10.1016/j.jmb.2007.08.067. [DOI] [PubMed] [Google Scholar]
- 18.Toropova K, Stockley PG, Ranson NA. Visualising a viral RNA genome poised for release from its receptor complex. J Mol Biol. 2011;408:408–19. doi: 10.1016/j.jmb.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 19.Bakker SE, Ford RJ, Barker AM, Robottom J, Saunders K, Pearson AR, et al. Isolation of an asymmetric RNA uncoating intermediate for a single-stranded RNA plant virus. J Mol Biol. 2012;417:65–78. doi: 10.1016/j.jmb.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang L, Johnson KN, Ball LA, Lin T, Yeager M, Johnson JE. The structure of pariacoto virus reveals a dodecahedral cage of duplex RNA. Nat Struct Biol. 2001;8:77–83. doi: 10.1038/83089. [DOI] [PubMed] [Google Scholar]
- 21.Larson SB, Koszelak S, Day J, Greenwood A, Dodds JA, McPherson A. Double-helical RNA in satellite tobacco mosaic virus. Nature. 1993;361:179–82. doi: 10.1038/361179a0. [DOI] [PubMed] [Google Scholar]
- 22.Bunka DHJ, Lane SW, Lane CL, Dykeman EC, Ford RJ, Barker AM, et al. Degenerate RNA packaging signals in the genome of Satellite Tobacco Necrosis Virus: implications for the assembly of a T=1 capsid. J Mol Biol. 2011;413:51–65. doi: 10.1016/j.jmb.2011.07.063. [DOI] [PubMed] [Google Scholar]
- 23.D’Souza V, Summers MF. How retroviruses select their genomes. Nat Rev Microbiol. 2005;3:643–55. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 24.Hutchinson EC, von Kirchbach JC, Gog JR, Digard P. Genome packaging in influenza A virus. J Gen Virol. 2010;91:313–28. doi: 10.1099/vir.0.017608-0. [DOI] [PubMed] [Google Scholar]
- 25.Kim DY, Firth AE, Atasheva S, Frolova EI, Frolov I. Conservation of a packaging signal and the viral genome RNA packaging mechanism in alphavirus evolution. J Virol. 2011;85:8022–36. doi: 10.1128/JVI.00644-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lever A, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–7. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu F, Morris TJ. Encapsidation of turnip crinkle virus is defined by a specific packaging signal and RNA size. J Virol. 1997;71:1428–35. doi: 10.1128/jvi.71.2.1428-1435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao AL. Genome packaging by spherical plant RNA viruses. Annu Rev Phytopathol. 2006;44:61–87. doi: 10.1146/annurev.phyto.44.070505.143334. [DOI] [PubMed] [Google Scholar]
- 29.Terasaki K, Murakami S, Lokugamage KG, Makino S. Mechanism of tripartite RNA genome packaging in Rift Valley fever virus. Proc Natl Acad Sci USA. 2011;108:804–9. doi: 10.1073/pnas.1013155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Routh A, Domitrovic T, Johnson JE. Host RNAs, including transposons, are encapsidated by a eukaryotic single-stranded RNA virus. Proc Natl Acad Sci USA. 2012;109:1907–12. doi: 10.1073/pnas.1116168109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen SS, Greenberg ML. Spermidine, an intrinsic component of turnip yellow mosaic virus. Proc Natl Acad Sci USA. 1981;78:5470–4. doi: 10.1073/pnas.78.9.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raina A, Tuomi K, Mäntyjärvi R. Roles of polyamines in the replication of animal viruses. Med Biol. 1981;59:428–32. [PubMed] [Google Scholar]
- 33.Nickerson KW, Lane LC. Polyamine content of several RNA plant viruses. Virology. 1977;81:455–9. doi: 10.1016/0042-6822(77)90160-X. [DOI] [PubMed] [Google Scholar]
- 34.Hagan MF. A theory for viral capsid assembly around electrostatic cores. J Chem Phys. 2009;130:114902. doi: 10.1063/1.3086041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cadena-Nava RD, Hu Y, Garmann RF, Ng B, Zelikin AN, Knobler CM, et al. Exploiting fluorescent polymers to probe the self-assembly of virus-like particles. J Phys Chem B. 2011;115:2386–91. doi: 10.1021/jp1094118. [DOI] [PubMed] [Google Scholar]
- 36.van der Schoot P, Bruinsma R. Electrostatics and the assembly of an RNA virus. Phys Rev E Stat Nonlin Soft Matter Phys. 2005;71:061928. doi: 10.1103/PhysRevE.71.061928. [DOI] [PubMed] [Google Scholar]
- 37.Wang RY, Nagy PD. Tomato bushy stunt virus co-opts the RNA-binding function of a host metabolic enzyme for viral genomic RNA synthesis. Cell Host Microbe. 2008;3:178–87. doi: 10.1016/j.chom.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 38.Novoa RR, Calderita G, Arranz R, Fontana J, Granzow H, Risco C. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol Cell. 2005;97:147–72. doi: 10.1042/BC20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nugent CI, Johnson KL, Sarnow P, Kirkegaard K. Functional coupling between replication and packaging of poliovirus replicon RNA. J Virol. 1999;73:427–35. doi: 10.1128/jvi.73.1.427-435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seo JK, Kwon SJ, Rao ALN. A physical interaction between viral replicase and capsid protein is required for genome-packaging specificity in an RNA virus. J Virol. 2012;86:6210–21. doi: 10.1128/JVI.07184-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valegård K, Liljas L, Fridborg K, Unge T. The three-dimensional structure of the bacterial virus MS2. Nature. 1990;345:36–41. doi: 10.1038/345036a0. [DOI] [PubMed] [Google Scholar]
- 42.Montelius I, Liljas L, Unge T. Structure of EDTA-treated satellite tobacco necrosis virus at pH 6.5. J Mol Biol. 1988;201:353–63. doi: 10.1016/0022-2836(88)90143-X. [DOI] [PubMed] [Google Scholar]
- 43.Lane SW, Dennis CA, Lane CL, Trinh CH, Rizkallah PJ, Stockley PG, et al. Construction and crystal structure of recombinant STNV capsids. J Mol Biol. 2011;413:41–50. doi: 10.1016/j.jmb.2011.07.062. [DOI] [PubMed] [Google Scholar]
- 44.Stockley PG, Stonehouse NJ, Murray JB, Goodman STS, Talbot SJ, Adams CJ, et al. Probing sequence-specific RNA recognition by the bacteriophage MS2 coat protein. Nucleic Acids Res. 1995;23:2512–8. doi: 10.1093/nar/23.13.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.LeCuyer KA, Behlen LS, Uhlenbeck OC. Mutants of the bacteriophage MS2 coat protein that alter its cooperative binding to RNA. Biochemistry. 1995;34:10600–6. doi: 10.1021/bi00033a035. [DOI] [PubMed] [Google Scholar]
- 46.Beckett D, Wu HN, Uhlenbeck OC. Roles of operator and non-operator RNA sequences in bacteriophage R17 capsid assembly. J Mol Biol. 1988;204:939–47. doi: 10.1016/0022-2836(88)90053-8. [DOI] [PubMed] [Google Scholar]
- 47.Johansson HE, Liljas L, Uhlenbeck OC. RNA recognition by the MS2 phage coat protein. Semin Virol. 1997;8:176–85. doi: 10.1006/smvy.1997.0120. [DOI] [Google Scholar]
- 48.Convery MA, Rowsell S, Stonehouse NJ, Ellington AD, Hirao I, Murray JB, et al. Crystal structure of an RNA aptamer-protein complex at 2.8 A resolution. Nat Struct Biol. 1998;5:133–9. doi: 10.1038/nsb0298-133. [DOI] [PubMed] [Google Scholar]
- 49.Gell C, Sabir T, Westwood J, Rashid A, Smith DAM, Harris SA, et al. Single-molecule fluorescence resonance energy transfer assays reveal heterogeneous folding ensembles in a simple RNA stem-loop. J Mol Biol. 2008;384:264–78. doi: 10.1016/j.jmb.2008.08.088. [DOI] [PubMed] [Google Scholar]
- 50.Horn WT, Tars K, Grahn E, Helgstrand C, Baron AJ, Lago H, et al. Structural basis of RNA binding discrimination between bacteriophages Qbeta and MS2. Structure. 2006;14:487–95. doi: 10.1016/j.str.2005.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ford RJ, Barker AM, Bakker SE, Coutts RH, Ranson NA, Phillips SEV, et al. Sequence-specific, RNA-protein interactions overcome electrostatic barriers preventing assembly of Satellite Tobacco Necrosis Virus coat protein. J Mol Biol. 2013 doi: 10.1016/j.jmb.2013.01.004. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elsawy KM, Caves LS, Twarock R. The impact of viral RNA on the association rates of capsid protein assembly: bacteriophage MS2 as a case study. J Mol Biol. 2010;400:935–47. doi: 10.1016/j.jmb.2010.05.037. [DOI] [PubMed] [Google Scholar]
- 53.Comas-Garcia M, Cadena-Nava RD, Rao AL, Knobler CM, Gelbart WM. In vitro quantification of the relative packaging efficiencies of single-stranded RNA molecules by viral capsid protein. J Virol. 2012;86:12271–82. doi: 10.1128/JVI.01695-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Annamalai P, Apte S, Wilkens S, Rao AL. Deletion of highly conserved arginine-rich RNA binding motif in cowpea chlorotic mottle virus capsid protein results in virion structural alterations and RNA packaging constraints. J Virol. 2005;79:3277–88. doi: 10.1128/JVI.79.6.3277-3288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Annamalai P, Rao AL. Dispensability of 3′ tRNA-like sequence for packaging cowpea chlorotic mottle virus genomic RNAs. Virology. 2005;332:650–8. doi: 10.1016/j.virol.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 56.Adolph KW, Butler PJ. Studies on the assembly of a spherical plant virus. I. States of aggregation of the isolated protein. J Mol Biol. 1974;88:327–41. doi: 10.1016/0022-2836(74)90485-9. [DOI] [PubMed] [Google Scholar]
- 57.Gell C, Brockwell D, Smith A. Handbook of Single Molecule Fluorescence Spectroscopy. Oxford: Oxford University Press, 2006. [Google Scholar]
- 58.Rigler R, Elson ES, eds. Fluorescence Correlation Spectroscopy. Berlin: Springer, 2001. [Google Scholar]
- 59.Rolfsson O. The roles of MS2 RNA in MS2 capsid assembly. Faculty of Biological Sciences. University of Leeds, 2009:176. [Google Scholar]
- 60.Rolfsson O, Toropova K, Ranson NA, Stockley PG. Mutually-induced conformational switching of RNA and coat protein underpins efficient assembly of a viral capsid. J Mol Biol. 2010;401:309–22. doi: 10.1016/j.jmb.2010.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stockley PG, Rolfsson O, Thompson GS, Basnak G, Francese S, Stonehouse NJ, et al. A simple, RNA-mediated allosteric switch controls the pathway to formation of a T=3 viral capsid. J Mol Biol. 2007;369:541–52. doi: 10.1016/j.jmb.2007.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hermanson GT, Hermanson GT. Bioconjugate techniques. Bioconjugate techniques. San Diego: Academic Press, 1996:785. [Google Scholar]
- 63.Fauster K, Hartl M, Santner T, Aigner M, Kreutz C, Bister K, et al. 2′-Azido RNA, a versatile tool for chemical biology: synthesis, X-ray structure, siRNA applications, click labeling. ACS Chem Biol. 2012;7:581–9. doi: 10.1021/cb200510k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ishizuka T, Kimoto M, Sato A, Hirao I. Site-specific functionalization of RNA molecules by an unnatural base pair transcription system via click chemistry. Chem Commun (Camb) 2012;48:10835–7. doi: 10.1039/c2cc36293g. [DOI] [PubMed] [Google Scholar]
- 65.Rao H, Sawant AA, Tanpure AA, Srivatsan SG. Posttranscriptional chemical functionalization of azide-modified oligoribonucleotides by bioorthogonal click and Staudinger reactions. Chem Commun (Camb) 2012;48:498–500. doi: 10.1039/c1cc15659d. [DOI] [PubMed] [Google Scholar]
- 66.Winz ML, Samanta A, Benzinger D, Jäschke A. Site-specific terminal and internal labeling of RNA by poly(A) polymerase tailing and copper-catalyzed or copper-free strain-promoted click chemistry. Nucleic Acids Res. 2012;40:e78. doi: 10.1093/nar/gks062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vauléon S, Ivanov SA, Gwiazda S, Müller S. Site-specific fluorescent and affinity labelling of RNA by using a small engineered twin ribozyme. Chembiochem. 2005;6:2158–62. doi: 10.1002/cbic.200500215. [DOI] [PubMed] [Google Scholar]
- 68.Vilfan ID, Kamping W, van den Hout M, Candelli A, Hage S, Dekker NH. An RNA toolbox for single-molecule force spectroscopy studies. Nucleic Acids Res. 2007;35:6625–39. doi: 10.1093/nar/gkm585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Logsdon N, Lee CG, Harper JW. Selective 5′ modification of T7 RNA polymerase transcripts. Anal Biochem. 1992;205:36–41. doi: 10.1016/0003-2697(92)90575-R. [DOI] [PubMed] [Google Scholar]
- 70.Kinoshita Y, Nishigaki K, Husimi Y. Fluorescence-, isotope- or biotin-labeling of the 5 ’-end of single-stranded DNA/RNA using T4 RNA ligase. Nucleic Acids Res. 1997;25:3747–8. doi: 10.1093/nar/25.18.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smith GJ, Sosnick TR, Scherer NF, Pan T. Efficient fluorescence labeling of a large RNA through oligonucleotide hybridization. RNA. 2005;11:234–9. doi: 10.1261/rna.7180305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heppell B, Blouin S, Dussault A-M, Mulhbacher J, Ennifar E, Penedo JC, et al. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat Chem Biol. 2011;7:384–92. doi: 10.1038/nchembio.563. [DOI] [PubMed] [Google Scholar]
- 73.Eschbach SH, St-Pierre P, Penedo JC, Lafontaine DA. Folding of the SAM-I riboswitch: a tale with a twist. RNA Biol. 2012;9:535–41. doi: 10.4161/rna.19648. [DOI] [PubMed] [Google Scholar]
- 74.Stone MD, Mihalusova M, O’connor CM, Prathapam R, Collins K, Zhuang X. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446:458–61. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12:136–49. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gralla J, Steitz JA, Crothers DM. Direct physical evidence for secondary structure in an isolated fragment of R17 bacteriophage mRNA. Nature. 1974;248:204–8. doi: 10.1038/248204a0. [DOI] [PubMed] [Google Scholar]
- 80.Thomas GJ, Jr., Prescott B, McDonald-Ordzie PE, Hartman KA. Studies of virus structure by laser-Raman spectroscopy. II. MS2 phage, MS2 capsids and MS2 RNA in aqueous solutions. J Mol Biol. 1976;102:103–24. doi: 10.1016/0022-2836(76)90076-0. [DOI] [PubMed] [Google Scholar]
- 81.Thomas GJ, Hartman KA. Laser-Excited Raman-Spectra of R17 Phage and R17 RNA. Fed Proc. 1974;33:1372–3. [Google Scholar]
- 82.Thomas GJ, Jr., Hartman KA. Raman studies of nucleic acids. 8. Estimation of RNA secondary structure from Raman scattering by phosphate-group vibrations. Biochim Biophys Acta. 1973;312:311–32. doi: 10.1016/0005-2787(73)90376-6. [DOI] [PubMed] [Google Scholar]
- 83.Hartman KA, Clayton N, Thomas GJ., Jr. Studies of viral structure by Raman spectroscopy. I. R17 virus and R17 RNA. Biochem Biophys Res Commun. 1973;50:942–9. doi: 10.1016/0006-291X(73)91336-3. [DOI] [PubMed] [Google Scholar]
- 84.Beekwilder J, Nieuwenhuizen R, Poot R, van Duin J. Secondary structure model for the first three domains of Q beta RNA. Control of A-protein synthesis. J Mol Biol. 1996;256:8–19. doi: 10.1006/jmbi.1996.0064. [DOI] [PubMed] [Google Scholar]
- 85.Skripkin EA, Adhin MR, de Smit MH, van Duin J. Secondary structure of the central region of bacteriophage MS2 RNA. Conservation and biological significance. J Mol Biol. 1990;211:447–63. doi: 10.1016/0022-2836(90)90364-R. [DOI] [PubMed] [Google Scholar]
- 86.Schmidt BF, Berkhout B, Overbeek GP, van Strien A, van Duin J. Determination of the RNA secondary structure that regulates lysis gene expression in bacteriophage MS2. J Mol Biol. 1987;195:505–16. doi: 10.1016/0022-2836(87)90179-3. [DOI] [PubMed] [Google Scholar]
- 87.Jacobson AB. Studies on secondary structure of single-stranded RNA from bacteriophage MS2 by electron microscopy. Proc Natl Acad Sci USA. 1976;73:307–11. doi: 10.1073/pnas.73.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ribitsch G, De Clercq R, Folkhard W, Zipper P, Schurz J, Clauwaert J. Small-angle X-ray and light scattering studies on the influence of Mg2+ ions on the structure of the RNA from bacteriophage MS2. Z Naturforsch C. 1985;40:234–41. doi: 10.1515/znc-1985-3-417. [DOI] [PubMed] [Google Scholar]
- 89.Zipper P, Folkhard W. A small-angle x-ray scattering investigation on the structure of the RNA from bacteriophage MS2. FEBS Lett. 1975;56:283–7. doi: 10.1016/0014-5793(75)81110-0. [DOI] [PubMed] [Google Scholar]
- 90.Yoffe AM, Prinsen P, Gopal A, Knobler CM, Gelbart WM, Ben-Shaul A. Predicting the sizes of large RNA molecules. Proc Natl Acad Sci USA. 2008;105:16153–8. doi: 10.1073/pnas.0808089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gopal A, Zhou ZH, Knobler CM, Gelbart WM. Visualizing large RNA molecules in solution. RNA. 2012;18:284–99. doi: 10.1261/rna.027557.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Olsthoorn RCL. Structure and Evolution of RNA Phages. Leiden: Leiden University, 1996. [Google Scholar]
- 93.Groeneveld H. Secondary Structure of Bacteriophage MS2 RNA: Translational Control by Kinetics of RNA folding. Leiden: Leiden University, 1997. [Google Scholar]
- 94.Fang X, Littrell K, Yang XJ, Henderson SJ, Siefert S, Thiyagarajan P, et al. Mg2+-dependent compaction and folding of yeast tRNAPhe and the catalytic domain of the B. subtilis RNase P RNA determined by small-angle X-ray scattering. Biochemistry. 2000;39:11107–13. doi: 10.1021/bi000724n. [DOI] [PubMed] [Google Scholar]
- 95.Johnson JM, Willits DA, Young MJ, Zlotnick A. Interaction with capsid protein alters RNA structure and the pathway for in vitro assembly of cowpea chlorotic mottle virus. J Mol Biol. 2004;335:455–64. doi: 10.1016/j.jmb.2003.10.059. [DOI] [PubMed] [Google Scholar]
- 96.Abrescia NGA, Bamford DH, Grimes JM, Stuart DI. Structure unifies the viral universe. Annu Rev Biochem. 2012;81:795–822. doi: 10.1146/annurev-biochem-060910-095130. [DOI] [PubMed] [Google Scholar]