Abstract
Transgenerational reprogramming of DNA methylation is important for transposon silencing and epigenetic inheritance. A stochastic regulation of methylation states in the germline may lead to epigenetic variation and the formation of epialleles that contribute to phenotypic variation. In Arabidopsis thaliana inbred lines, the frequency of single base variation of DNA methylation is much higher than genetic mutation and, interestingly, variable epialleles are pre-methylated in the male germline. However, these same alleles are targeted for demethylation in the pollen vegetative nucleus, by a mechanism that seems to contribute to the accumulation of small RNAs that reinforce transcriptional gene silencing in the gametes. These observations are paving the way toward understanding the extent of epigenetic reprogramming in higher plants, and the mechanisms regulating the stability of acquired epigenetic states across generations.
Keywords: DNA methylation, small RNAs, epiallele, transposable elements, germline, Arabidopsis
Introduction
Transposable elements (TEs) are epigenetically silenced by repressive chromatin marks such as DNA methylation and histone modifications,1 which in animals are extensively reprogrammed in primordial germ cells and early embryo development.2 DNA methylation in plants occurs in three different sequence contexts (CG, CHG and CHH, where H = A, C or T), which are established de novo by a mechanism known as RNA-directed DNA methylation (RdDM), guided by small interfering RNAs (siRNAs). CG and CHG methylation are strand symmetric and can be maintained by the DNA METHYLTRANSFERASE 1 (MET1, ortholog of mammalian Dnmt1) and CHROMOMETHYLASE 3 (CMT3), respectively. While maintenance of CG methylation seems to require uniquely hemimethylated DNA, CHG methylation relies on a self-reinforcing feedback loop mechanism that involves CMT3 and dimethylation of lysine 9 at histone 3 (H3K9me2).3 In contrast, CHH methylation is asymmetric and must be re-established de novo after each replication cycle, depending on RNA interference (RNAi) and the activity of the DNA methyltransferase DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2, ortholog of mammalian Dnmt3).4 The most accepted model for RdDM postulates that Pol IV transcripts are initially used as templates by RNA-DEPENDENT RNA POLYMERASE 2 (RDR2), while DICER-LIKE 3 (DCL3) cleaves the resulting dsRNAs into 24-nt siRNA products that are loaded into ARGONAUTE 4 (AGO4). Then, independent of this pathway of siRNA biogenesis, RNA Pol V produces nascent transcripts that serve as scaffolds for the binding of siRNA-loaded AGO4 complexes that recruit DRM2 in order to establish de novo DNA methylation. An important feature of epigenetic modifications is their reversibility, and in Arabidopsis cytosine methylation, can be removed by the DNA glycosylases DEMETER (DME), REPRESSOR OF SILENCING 1 (ROS1) AND DEMETER-LIKE 2 (DML2) and 3 (DML3) by a base excision repair mechanism.4
Over the past few years, the dynamics of DNA methylation in Arabidopsis have been studied in complex sporophytic tissues, but now with single cell-type resolution, we are starting to uncover intricate pathways of epigenetic regulation taking place in the gametophytes. In light of the genome-wide identification of hypervariable epialleles in Arabidopsis,5,6 and our recent methylome profiling of representative cell types throughout male gametogenesis,7 we will discuss here possible pathways toward epigenetic variation during genome reprogramming in the gametophytes and early embryogenesis, and possible implications for development and evolution.
Reprogramming of DNA Methylation in the Arabidopsis Germline
Most of our current understanding about epigenetic reprogramming of TEs in the plant germline comes from studies in the male gametophyte, as reliable methods to isolate the two differentiated cell types by flow cytometry are now available.8 Several lines of evidence suggested that reprogramming in the pollen vegetative cell might influence the epigenetic state of the neighboring sperm cells before fertilization. One of the most interesting ideas for a signaling mechanism involves small RNAs. Similarly to mutants with reactivated TEs, pollen accumulates epigenetically activated 21 nt siRNA from active retrotransposons in the vegetative cell. Interestingly, these siRNAs were detected at higher levels in sperm cells where TEs are transcriptionally silenced,9 thus suggesting that small RNAs are able to move between the two different cell lineages in pollen. The role of TE-derived 21 nt siRNAs during sperm cell specification and/or post-fertilization is still a mystery. However, it is interesting that at least some of these siRNAs can actively silence endogenous Arabidopsis genes by translational repression or transcript cleavage depending on the tissue type,10 perhaps reflecting the activity of different Argonaute proteins. As such, it is possible that TE-derived siRNAs play a critical role modulating the sperm and early seed transcriptomes.11
Recently, using genome-wide DNA methylation analysis, we observed that gains and losses of DNA methylation occur after meiosis in the developing pollen grain. Unexpectedly, most CHH methylation is lost in the reduced haploid genome and re-established only in the vegetative cell, while the resulting sperm cells remain largely hypomethylated at CHH.7,12 In contrast, CG methylation is widely maintained throughout plant development and in the differentiating germline. The absence of CHH methylation in the germline is particularly striking in pericentromeric regions where LTR retrotransposons are more abundant, suggesting that re-establishment of CHH methylation in the paternal genome occurs only after fertilization, possibly guided by maternal siRNAs.7 These results indicated that in the Arabidopsis male germline transcriptional gene silencing is globally impaired, possibly explaining why 21 nt siRNAs matching to retrotransposons are important to control TE silencing at the post-transcriptional level. However, particular loci retain CHH methylation in the microspore and sperm cells, especially at TEs neighboring imprinted genes that are maternally expressed in the endosperm,7 thus indicating that RdDM activity is not completely lost in sperm.
In the female gametophyte, Jullien et al.13 have recently shown that DRM2 is weakly expressed in the central cell, and completely absent in the developing endosperm. This indicates that the classical pathway for de novo DNA methylation is switched off during endosperm proliferation, but alternative pathways may exist considering the relatively high levels of CHH methylation that were still detected in wild-type endosperm.14 In contrast, DRM2 is highly abundant in the egg cell, but curiously CHH methylation at some imprinted genes seems to be only gradually restored in the embryo after fertilization when the other methyltransferases MET1 and CMT3 are also expressed.13 In summary, these results strongly suggest that DNA methylation and small RNA dynamics in the Arabidopsis gametophytes might dictate the extent of epigenetic reprogramming during early embryo development. Therefore, a stochastic regulation of genome reprogramming in the germline might contribute to the epigenetic variation observed across generations in Arabidopsis.5,6
Epigenetic Variation by DNA Demethylation
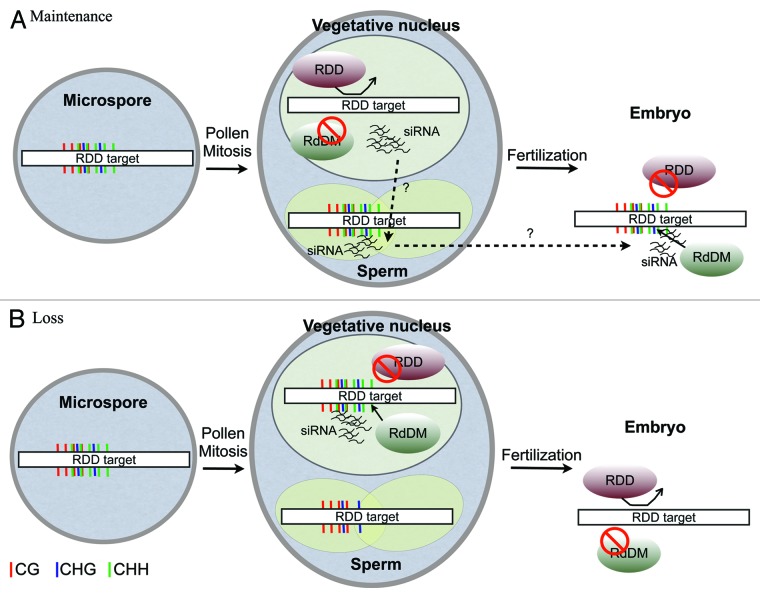
Hypervariable epialleles were detected by spontaneous variation of DNA methylation at specific loci in inbred Arabidopsis wild-type lines.5,6 Epiallele formation represents a major source of phenotypic variation observed in natural populations,15 as it often leads to changes in the transcriptional status of protein-coding genes and regulatory RNAs. Interestingly, most of these variable epialleles are targets of the DNA glycosylases ROS1, DML2 and DML3 (RDD) that are expressed throughout the sporophyte and in the vegetative cell of pollen, but not in the germline.16 This expression pattern explains the higher levels of DNA methylation observed at RDD targets in sperm cells, compared with the vegetative cell nucleus,7 allowing speculation about a possible mechanism to generate epigenetic variation of heritable DNA methylation in Arabidopsis. For example, if 24 nt siRNAs matching to these loci are able to accumulate in the gametes, they might be able to trigger RdDM and prevent targeted demethylation by RDD proteins after fertilization (Fig. 1). This idea goes in agreement with the recently proposed activity for DME in the vegetative cell that is important to reinforce DNA methylation in the germline, as dme/+ sperm cells showed reduced levels of CHH in comparison with wild-type.12 In fact, methylome profiling throughout pollen development supports this idea, as DNA demethylation at TEs neighboring maternally expressed imprinted genes occurs exclusively in the vegetative cell after the first pollen mitosis, and corresponding 24 nt siRNAs accumulate at higher levels in sperm cells.7 As DME is expressed preferentially in the gametophytes and endosperm, we can rule out at this point any sporophytic contribution for the observed variations. Taken together, is still difficult to explain how targeted TE demethylation in the vegetative cell nucleus leads to reinforcement of RdDM activity in sperm, as active demethylation by RDD proteins usually leads to loss of 24 nt siRNA accumulation from targeted loci.17 One hypothesis, considering that in the vegetative nucleus both DNA demethylation and RdDM pathways compete for the same loci, is that demethylation is able to prevent RdDM activity, so that 24 nt siRNAs initially produced from these loci can be subsequently routed to the sperm cells to induce silencing (Fig. 1). In contrast, if DNA demethylation is not sufficient to counteract RdDM activity, the corresponding siRNAs will remain in the vegetative cell nucleus within the RdDM loop (Fig. 1). Based on this model, we are proposing an important role for RDD proteins and RdDM in the male gametophyte, to uncover loci that should remain transcriptionally silenced in the germline and following generations.
Figure 1. Hypothetical mechanism regulating variable epialleles by DNA demethylation and RdDM in Arabidopsis pollen. Epigenetic variation in plants might occur with additional rounds of cell division in post-meiotic gametophytes. In Arabidopsis pollen, two mitotic divisions originate a larger vegetative cell embedding two sperm cells. One sperm cell will fuse with the egg cell giving rise to the diploid embryo. (A) Transgenerational maintenance of acquired DNA methylation might require the activity of the DNA glycosylases ROS1, DML2 and DML3 (RDD) in the vegetative cell nucleus, which may this way direct accumulated siRNAs to the neighboring sperm cells to reinforce transcriptional gene silencing in the gametes and following generations. (B) The failure to demethylate RDD targets might retain siRNAs in the vegetative nucleus within the RNA-directed DNA methylation (RdDM) pathway, resulting in reduced CHH methylation in sperm, and active demethylation in the embryo when the activity of RDD proteins is restored. CG, red lines; CHG, blue lines; CHH, green lines.
Triggering Epiallele Formation with RNAi
The formation of pure epialleles occurs independently of a new transposition event or genetic variation, but the mechanisms responsible for triggering de novo DNA methylation are still poorly understood. With the sperm cell methylome we observed a pre-methylated state at the CG context for some epialleles that were found hypomethylated in leaf tissue of the parental lines analyzed in two recent studies.5,6 One possible explanation is that CG hypermethylation at particular loci represents a default state at undifferentiated cell lineages that will later give rise to gametophytes, depending exclusively on MET1 activity for its maintenance. This way, this methylation pattern is passed on to the germline, but its stability requires RdDM and accumulation of corresponding 24 nt siRNA when the expression of DNA glycosylases is restored in the embryo. The initial accumulation of 24 nt siRNA from targeted loci is not yet well understood, and remains an important question in the field. One possible mechanism was previously described as a stepwise pathway involving primary RdDM activity that is then able to recruit the machinery for secondary siRNA biogenesis.18 This is an interesting possibility as pathways triggering siRNA production exist in Arabidopsis, by transcript cleavage with 22 nt siRNAs that are used as template for RDR6-dependent double-stranded RNA synthesis resembling the trans-acting siRNA (tasiRNA) pathway.19 However, this involves the activity of DCL4 producing mainly 21 nt siRNAs that are not able to trigger primary RdDM through the classical Pol V/AGO4 pathway.18 As such, future studies should focus on finding novel pathways or specific conditions that could lead to an initial accumulation of 24 nt siRNA through the RDR2/DCL3 pathway, or alternative ways to recruit Pol IV and Pol V to specific loci. Several studies have started to uncover the intricate control of Pol IV and Pol V recruitment;4 however, recent results have shown that even in the absence Pol IV and Pol V, half of the CHH methylation was still detected at wild-type levels, while the remainder was essentially relocated to pericentromeric regions.20 These unexpected results indicated that the pathways controlling de novo DNA methylation in Arabidopsis might be more complex than what was previously proposed, or that alternative pathways may exist. For example, it was recently proposed based on genome-wide DNA methylation analysis that the histone methyltransferases KRYPTONITE (KYP/SUVH4), SUVH5 and SUVH6 in association with CMT3, might be involved in establishing and maintaining CHH methylation in a siRNA-independent manner.21 The spontaneous formation of pure epialleles will therefore represent a good working model to elaborate future hypotheses.
Future Perspectives
Epigenomic analysis of complex tissues constitutes a substantial problem to unravel the many overlapping epigenetic mechanisms that evolved in eukaryotic genomes, as it includes many differentiated cell types with different epigenetic states. For this reason, future analysis with single cell-type resolution will certainly expand our understanding on the dynamics of epigenetic marks in different cell lineages and natural epigenetic variation. In this respect, mutants or environmental conditions that lead to reactivation of TEs will remain an important line of research: for example, one hypervariable inbred line had acquired a point mutation in MEE57 that encodes a homolog of the maintenance methyltransferase MET1.6 Mutagenic epigenetic variation was originally demonstrated in inbred mutants such as for the chromatin remodeler DDM1 (DECREASE IN DNA METHYLATION 1) that results in a genome-wide decrease of DNA methylation and upregulation of TE expression.22 Transposition and retrotransposition occurs in ddm1 mutant plants originating cumulative morphological phenotypes and sterility after a few rounds of inbreeding.23 However, the frequency and mechanisms controlling the transmission of new insertions to the next generation via the germline remain unclear, as well as the role of RNAi in this process. As the molecular characterization of the gametophytes with single-cell resolution is now possible,8,24-26 further studies will allow addressing these important questions by characterizing the pre- and post-meiotic transpositional landscape in higher plants, thus providing new insight into the mechanisms by which TEs propagate across generations as a driving force toward biological diversity and evolution.
Acknowledgments
We thank Yannick Jacob for critically reading the manuscript, and all the members of the Martienssen lab for insightful discussions. Research in the Martienssen laboratory is supported by the National Institute of Health (NIH) grant R01 GM067014.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/24085
References
- 1.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–85. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 2.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–7. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, Caro E, et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell. 2012;151:167–80. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–20. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitz RJ, Schultz MD, Lewsey MG, O’Malley RC, Urich MA, Libiger O, et al. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–73. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker C, Hagmann J, Müller J, Koenig D, Stegle O, Borgwardt K, et al. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature. 2011;480:245–9. doi: 10.1038/nature10555. [DOI] [PubMed] [Google Scholar]
- 7.Calarco JP, Borges F, Donoghue MTA, Van Ex F, Jullien PE, Lopes T, et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell. 2012;151:194–205. doi: 10.1016/j.cell.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borges F, Gardner R, Lopes T, Calarco JP, Boavida LC, Slotkin RK, et al. FACS-based purification of Arabidopsis microspores, sperm cells and vegetative nuclei. Plant Methods. 2012;8:44. doi: 10.1186/1746-4811-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slotkin RK, Vaughn M, Borges F, Tanurdzić M, Becker JD, Feijó JA, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–72. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCue AD, Nuthikattu S, Reeder SH, Slotkin RK. Gene expression and stress response mediated by the epigenetic regulation of a transposable element small RNA. PLoS Genet. 2012;8:e1002474. doi: 10.1371/journal.pgen.1002474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCue AD, Slotkin RK. Transposable element small RNAs as regulators of gene expression. Trends Genet. 2012;28:616–23. doi: 10.1016/j.tig.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Ibarra CA, Feng X, Schoft VK, Hsieh TF, Uzawa R, Rodrigues JA, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337:1360–4. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jullien PE, Susaki D, Yelagandula R, Higashiyama T, Berger F. DNA methylation dynamics during sexual reproduction in Arabidopsis thaliana. Curr Biol. 2012;22:1825–30. doi: 10.1016/j.cub.2012.07.061. [DOI] [PubMed] [Google Scholar]
- 14.Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, et al. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324:1451–4. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaughn MW, Tanurdzić M, Lippman Z, Jiang H, Carrasquillo R, Rabinowicz PD, et al. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 2007;5:e174. doi: 10.1371/journal.pbio.0050174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schoft VK, Chumak N, Choi Y, Hannon M, Garcia-Aguilar M, Machlicova A, et al. Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc Natl Acad Sci USA. 2011;108:8042–7. doi: 10.1073/pnas.1105117108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–36. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daxinger L, Kanno T, Bucher E, van der Winden J, Naumann U, Matzke AJ, et al. A stepwise pathway for biogenesis of 24-nt secondary siRNAs and spreading of DNA methylation. EMBO J. 2009;28:48–57. doi: 10.1038/emboj.2008.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molnar A, Melnyk CW, Bassett A, Hardcastle TJ, Dunn R, Baulcombe DC. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science. 2010;328:872–5. doi: 10.1126/science.1187959. [DOI] [PubMed] [Google Scholar]
- 20.Wierzbicki AT, Cocklin R, Mayampurath A, Lister R, Rowley MJ, Gregory BD, et al. Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev. 2012;26:1825–36. doi: 10.1101/gad.197772.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stroud H, Greenberg MVC, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–64. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lippman Z, Gendrel A-V, Black M, Vaughn MW, Dedhia N, McCombie WR, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–6. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 23.Kakutani T, Jeddeloh JA, Flowers SK, Munakata K, Richards EJ. Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc Natl Acad Sci USA. 1996;93:12406–11. doi: 10.1073/pnas.93.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikeda Y, Kinoshita Y, Susaki D, Ikeda Y, Iwano M, Takayama S, et al. HMG domain containing SSRP1 is required for DNA demethylation and genomic imprinting in Arabidopsis. Dev Cell. 2011;21:589–96. doi: 10.1016/j.devcel.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 25.Wuest SE, Vijverberg K, Schmidt A, Weiss M, Gheyselinck J, Lohr M, et al. Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr Biol. 2010;20:506–12. doi: 10.1016/j.cub.2010.01.051. [DOI] [PubMed] [Google Scholar]
- 26.Gebert M, Dresselhaus T, Sprunck S. F-actin organization and pollen tube tip growth in Arabidopsis are dependent on the gametophyte-specific Armadillo repeat protein ARO1. Plant Cell. 2008;20:2798–814. doi: 10.1105/tpc.108.061028. [DOI] [PMC free article] [PubMed] [Google Scholar]