Abstract
Accurate 3′end processing depends on the correct recognition of polyadenylation regulatory elements by specific protein complexes. In addition to the well-known hexanucleotide motif and downstream sequence element (DSE), less-defined auxiliary elements are usually found to modulate cleavage and polyadenylation. They are generally located in close proximity to the core polyadenylation elements but, in most of the cases, the molecular mechanisms involved are not well defined. We concentrated our studies on the regulation of the mouse β adducin (Add2) pre-mRNA cleavage and polyadenylation. It contains two proximal erythroid-specific (PAS1 and PAS2-3) and one distal brain-specific (PAS4) polyadenylation sites along the 3′UTR. Using an in vivo approach based in the transfection of minigenes containing the Add2 polyadenylation signals, we previously identified the core regulatory elements responsible for PAS4 activity. Here, we have identified two novel non-canonical cis-acting elements regulating 3′end processing at PAS4, which show long-distance activities. The first of these elements, which spans for 257 nucleotides and is located at more than 5 kb upstream the PAS4, was essential to enable processing at the Add2 PAS4. The second element, located at about 4.5 kb upstream of the PAS4, reduces PAS4 processing. Both elements display long-distance activities and, to our knowledge, long-distance upstream polyadenylation regulatory elements have not been previously described in non-viral eukaryotic transcripts. These results highlight the complexity of the regulatory mechanisms directing Add2 pre-mRNA 3′end processing, and suggests that pre-mRNA 3′ end processing of other genes may also be unexpectedly regulated by non-canonical auxiliary elements.
Keywords: 3′ end processing, long-distance, mouse beta adducin (Add2), non-canonical, upstream sequence element (USE)
Introduction
Eukaryotic pre-mRNA 3′end processing includes endonucleolytic cleavage of the nascent transcript, generally followed by the non-templated addition of adenosines at the remaining 3′end.1,2 Inefficiently cleaved and polyadenylated transcripts are retained and degraded in the nucleus or inefficiently transported to the cytoplasm.3,4 Correct recognition and efficient processing at the cleavage and polyadenylation site (PAS) depend on the accurate assembly of the 3′end processing factors onto adjacent regulatory elements. The hexanucleotide motif, located 20–30 bases upstream of the PAS, and an adjacent downstream sequence element (DSE), represent the core polyadenylation elements. They are recognized by the basal polyadenylation factors, cleavage and polyadenylation specificity factor (CPSF) and cleavage stimulation factor (Cstf), respectively. For more than 50% of human and mouse PASs, the hexanucleotide motif is represented by the highly conserved AAUAAA sequence, whereas AAUAAA variants or no evident hexamer motifs are present in the rest of PAS.5 The DSE is usually represented by a variable number of U-rich and/or G/U-rich elements, although a precise consensus motif is not well-defined.6 The efficiency of 3′end processing can be further enhanced by the presence of auxiliary DSE (auxDSE), as well as upstream sequence elements (USE). These elements are of particular importance when the hexanucleotide motif is represented by one of its less efficient variants and/or when the DSE is weak, such as those having low GU/U context. AuxDSEs are characterized by a G-rich sequence (GRS) positioned just downstream of the DSE,6 although a GRS enhancer has been reported to be positioned 440 nt downstream of the PAS.7 Variable number of U-rich sequences placed immediately upstream of the hexanucleotide motif generally represents an USE. As an exception, a long-distance USE has been found for the Rous sarcoma virus (RSV) polyadenylation signal.8,9 The USEs are well characterized for some viral and cellular polyadenylation signals and are known to modulate 3′end processing.10-15
We have previously identified and functionally mapped the core elements (the AGUAAA hexanucleotide motif and GU-rich DSE) which were necessary for the correct definition and precise processing of the mouse β adducin (Add2) pre-mRNA at the distal brain-specific PAS (PAS4).16 Add2 gene expression is mainly restricted to hematopoietic and neuronal tissues, which is correlated with the use of alternative promoters and PASs.17-19 In mice, the use of proximal erythroid-specific PASs (PAS1 and PAS2-3) results in shorter 3.1–3.7 kb mRNA forms, with a 3′ UTR of about 800 bases. Instead, usage of a distal brain-specific PAS (PAS4) generates an 8.3 kb mRNA, having an unusually long 3′UTR of about 5.7 kb. Both short and long mRNA forms encode for the same protein. The presence and use of the different alternative PASs in the Add2 gene are very well conserved among species,17 suggesting an important and specific role for each of the generated 3′UTRs. The long Add2 mRNA isoform is expressed at high levels in the hippocampus, cerebellum and neocortex, showing dendritic localization.20 Beta adducin is involved in the stabilization of the synapses21 and its absence in mice leads to motor coordination, behavioral and learning deficits associated with impairment of long-term potentiation (LTP) and complete abolishment of long-term depression (LTD).20
In the present study, we determined that, besides the presence of the core elements, additional non-canonical long-distance cis-acting elements regulate 3′end processing at the Add2 PAS4. We identified long-range polyadenylation enhancer and silencer elements that are located, in the minigene context, more than 2 Kb and about 0.8 Kb upstream from the PAS4, and more than 5 Kb and about 4 Kb in the genomic context, respectively. Deletion of the silencer element resulted in a 2- to 3-fold increase of the levels of the mRNA cleaved and polyadenylated at the PAS4. The enhancer, which flanks the stop codon of the Add2 ORF, was necessary to obtain 3′ end processing at the brain-specific Add2 PAS4. To our knowledge, this is the first report of the presence of long-distance upstream polyadenylation regulatory elements in non-viral eukaryotic pre-mRNAs.
Results
Pre-mRNA cleavage and polyadenylation at the brain-specific distal PAS4 of Add2 is regulated by long-range elements
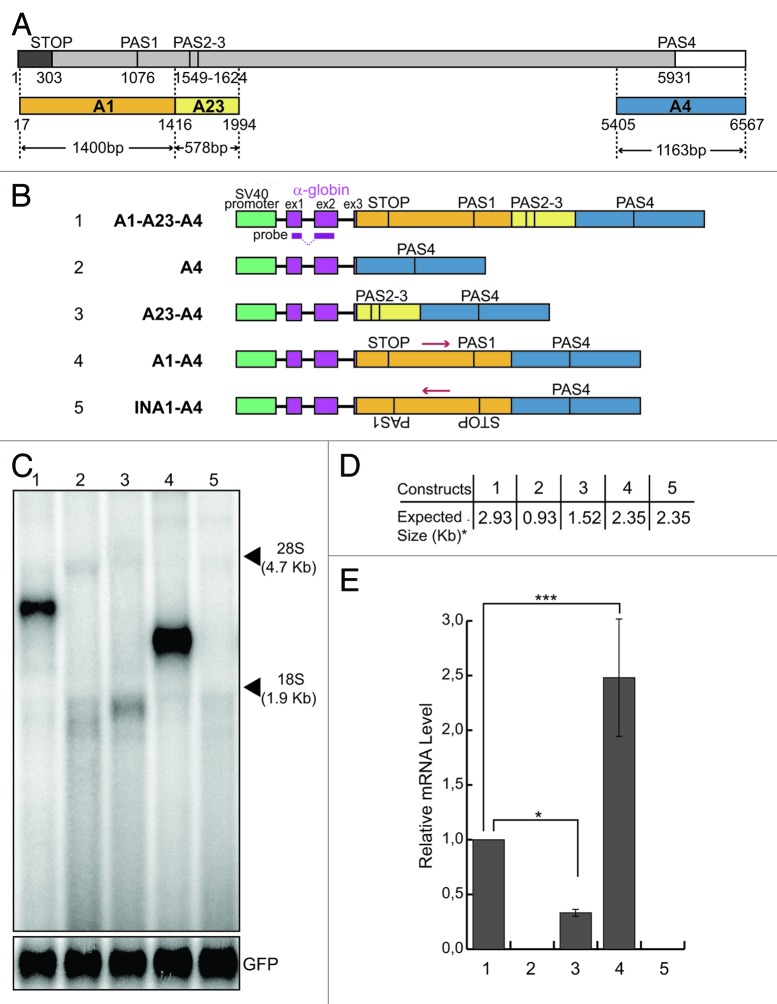
We have previously shown that we accurately reproduced the use of the brain-specific PAS4 in transcripts derived from a chimeric minigene construct containing all Add2 polyadenylation regions, after transient transfection in HeLa cells (Fig. 1, construct A1-A23-A4, lane 1).16 These transcripts were cleaved and polyadenylated at the same position normally used in the endogenous Add2 gene in neuronal tissues.16,17 In the same study, using constructs containing all three PASs of Add2, we have identified the core cis-acting elements necessary for the correct definition and 3′end processing of the Add2 pre-mRNA at the PAS4.
Figure 1. The presence of the A1 region is necessary to get mRNA expression. (A) Schematic representation of the Add2 last exon; numbers indicate nucleotide positions from the first base of the Add2 last exon; a dark gray box indicates the coding region; a light gray box indicates the 3′UTR; a white box indicates 3′genomic flanking region; STOP, stop codon; PAS1, the first polyadenylation site; PAS23, the second polyadenylation site; PAS4, the fourth polyadenylation site; different regions (A1, A23 and A4) containing Add2 polyadenylation sites (PAS1, PAS2-3 and PAS4) are represented as orange, yellow and blue boxes, respectively; arrowheads indicate the size of each region; (B) schematic representation of the Add2 chimeric minigene constructs A1-A23-A4 (1), A4 (2), A23-A4 (3), A1-A4 (4) and INA1-A4 (5); the green box indicates the SV40 promoter; violet boxes indicate exons of the α globin gene; the α globin fragment used as a probe for the northern blots is indicated; (C) northern blot analysis of total RNA prepared from HeLa cells previously transfected with the constructs shown in (B); the p-EGFP-C2 plasmid was used for the normalization of the transfection efficiency, the positions of the 28S and 18S rRNAs are indicated on the right; (D) the expected sizes of the mRNAs of the constructs shown in Panel B, up to the predicted PAS4 (i.e., the asterisk indicates without the pA tail). (E) The relative mRNA levels were calculated as the ratio between the analyzed mRNAs and GFP mRNA represented in (C); quantification of the northern blot signals was performed using the Cyclone Phosphorimager and the OptiQuant software (Packard); the mean +/− SD of three independent experiments is shown; data were analyzed using one-way ANOVA and Tukey's multiple comparison test; *** p < 0.0001, * p < 0.05.
Unexpectedly, we observed that after transfection into HeLa cells, no mRNA product was detected from a construct lacking the A1 and A23 regions (A4 construct, Fig. 1C, lane 2). This result suggested that the presence of the proximal A1 and A23 regions may be important for the usage of the distal PAS4.
In order to map more precisely the sequences participating in PAS4 polyadenylation activity, we added back the A1 and A23 regions to the A4 construct to generate the A1-A4 and A23-A4 constructs (Fig. 1B). The addition of the A23 region (Fig. 1B, construct A23-A4) did not restore polyadenylation activity at the PAS4 (Fig. 1C−E, compare lane 3 to lane 1). Interestingly, adding back only the A1 region (Fig. 1B, construct A1-A4) restored the activity of the PAS4, showing an increase of more than 2-folds in the amount of mRNA when compared with the product of the original A1-A23-A4 construct (Fig. 1C and E, compare lane 4 to 1), suggesting the presence of a silencer element in the A23 region (compare lane 1 to lane 4), and the presence of an enhancer element in the A1 region (compare lane 2 to lane 4). To rule out the possibility that the increase in mRNA levels produced by the A1-A4 construct could be related to the distance of the PAS4 from the start of the exon, we generated a construct containing the A1 region in the antisense orientation (Fig. 1A, construct INA1-A4). This construct showed no activity (Fig. 1C, lane 5), supporting the hypothesis of the presence of a long-distance enhancer element in the A1 region.
The observed differences in A1-A23-A4, A23-A4 and A1-A4 mRNA levels are related to changes in cleavage efficiency
We reasoned that the absence of expression of the A4 and INA1-A4 constructs could be related to different mechanisms: (1) degradation of the transcribed mRNA, potentially triggered by the mRNA surveillance pathway activated during translation;22 (2) differences in stability of the different mRNAs, or (3) inefficient 3′end processing of the pre-mRNA at the PAS4 leading to pre-mRNA degradation.
Therefore, we tested the first of these three possibilities by blocking mRNA translation with cycloheximide. We expect that if mRNAs produced from the A4 and INA1-A4 constructs were degraded after the first round of translation, blocking translation of mRNAs should lead to an increase in the levels of A4- and INA1-A4-derived mRNAs, which should become evident by northern blot analysis. However, no recovery of the signal was observed in the northern blot analysis after cycloheximide treatment (Fig. S1A). Therefore, we concluded that the absence of expression of the A4 and INA1-A4 constructs was not due to mRNA degradation by the mRNA surveillance pathways activated during translation.
To determine differences in mRNA stability, we used actinomycin D to block new synthesis of mRNAs and to monitor the decay of the already-synthesized ones. mRNA products from the A4 and INA1-A4 constructs were not detected preventing the determination of the stability of these specific mRNAs. Therefore, we determined whether differences in mRNA levels generated by the A1-A23-A4 and A1-A4 constructs were associated to changes in mRNA stability. Thus, HeLa cells were transfected with these constructs, treated with actinomycin D and total RNA collected at different time points (Fig. S2). As expected, the detected signal decreased with the actinomycin D treatment, but the rate of decay was similar for all the tested mRNAs, suggesting that there was no link between mRNA levels and stability for the constructs analyzed.
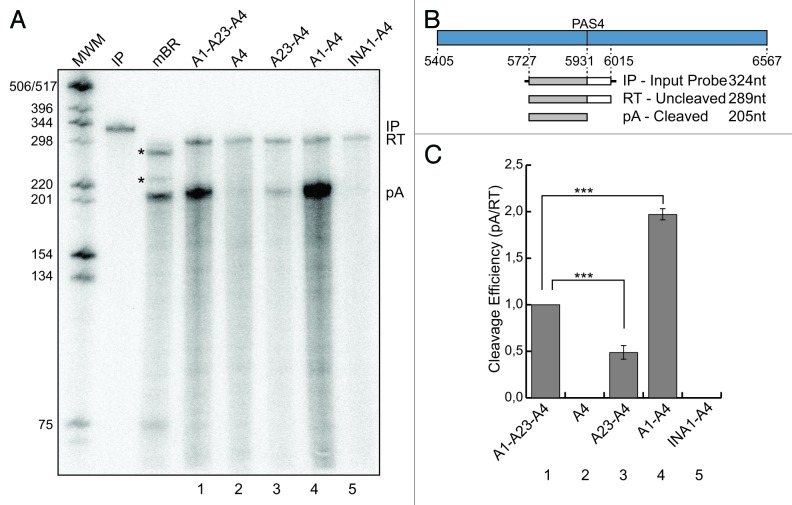
To test whether the absence of expression of the A4 and INA1-A4 constructs could be related to inefficient 3′end processing of the pre-mRNAs at the PAS4 leading to degradation, we performed an RNase protection assay. To this end, we utilized a 324 nt-long input probe (IP) having 289 nt complementary to the pre-mRNA transcript (205 nt upstream and 84 nt downstream of the PAS4) (Fig. 2B). The cleaved protected product (pA) was clearly detected for the A1-A23-A4, A23-A4 and A1-A4 constructs (Fig. 2A, lanes 1, 3 and 4), but not for A4 and INA1-A4 ones (Fig. 2A, lanes 2 and 5), as already observed in the northern blot analysis (Fig. 1C). The uncleaved protected products (RT, read-through) were detected in all tested constructs at roughly the same levels, suggesting that all constructs were transcriptionally active. Additionally, when we analyzed pre-mRNA levels by performing RT-qPCR with primers located in the introns, the A4, A23-A4, A1-A4 and INA1-A4 constructs had similar levels, which were roughly half of that of the A1-A23-A4 pre-mRNA (Fig. S3). Cleavage efficiency was calculated as the ratio between the signal obtained for the protected product (pA) and that of the unprocessed (RT) band. The A1-A4 construct showed a 2-fold increase in cleavage efficiency in comparison with the A1-A23-A4 one (Fig. 2C, lane 4 compared with lane 1). The A23-A4 construct showed a decrease of more than 2-fold in its cleavage efficiency when compared with the A1-A23-A4 construct (Fig. 2C, lane 3 compared with lane 1). Interestingly, there is a positive correlation between the observed differences in the cleavage efficiency and the amount of mRNAs detected by the northern blot analysis (compare Fig. 2C with Fig. 1E). These results strongly support the hypothesis that the major reason for the observed differences in expression of the described constructs is associated to differences in 3′end processing.
Figure 2. Cleavage efficiency of the pre-mRNA is enhanced by the presence of the A1 region. (A) RNase protection analysis of total RNA prepared from mouse brain (mBR) and HeLa cells previously transfected with the constructs shown in Figure 1B (1–5); IP, input probe; RT, uncleaved protected product; pA, cleaved protected product; MWM, molecular weight marker; the asterisks indicate unidentified products in the mBR sample. (B) Schematic representation of the A4 region containing PAS4 and the probe used in the RNase protection experiment shown below; the expected protected fragments, with their expected sizes are shown; nucleotide positions are numbered respective to the first base of the Add2 last exon. (C) The cleavage efficiency was calculated as the ratio between pA and RT represented in (A, 1–5). The quantification of the protected signals has been performed using the Cyclone Phosphorimager and the OptiQuant software (Packard); the mean +/− SD of three independent experiments is shown; data were analyzed using one-way ANOVA and Tukey's multiple comparison test,;*** p < 0.0001.
Summarizing the above-mentioned results, we concluded that the A1 and A23 regions probably contain cis-acting elements, which participate in 3′end processing of the Add2 PAS4. The A1 region contains a positive element(s), whereas the A23 region contains a negative cis-element(s). The next series of studies were directed to the fine mapping of the enhancer and silencer elements present in the A1 and A23 regions, respectively.
A silencer element downregulates pre-mRNA cleavage and polyadenylation at the brain-specific distal PAS4 of Add2
To map more in detail the negative cis-acting element, we introduced deleted segments of the A23 region at the place of the “full-length” A23 region in the A1-A23-A4 construct, generating the A1-A23(1,416-1,781)-A4 and A1-A23(1,416-1,581)-A4 constructs (Fig. 3A). Removal of the negative cis-acting element should result in an increase in mRNA levels, similar to that one observed after deletion of the whole A23 region (Figs. 1C and 2A, compare lane 1 to lane 4). We observed such an increase for the A1-A23(1,416-1,581)-A4 construct (Fig. 3B and D, compare lanes 1 and 3), which was even higher to that seen when the whole A23 region was absent (Fig. 1, construct A1-A4, lanes 1 and 4). This result suggests the location of the silencer element within the 200 bp region that spans from 1,581−1,781 bp relative to the first base of the last exon of the Add2 gene.
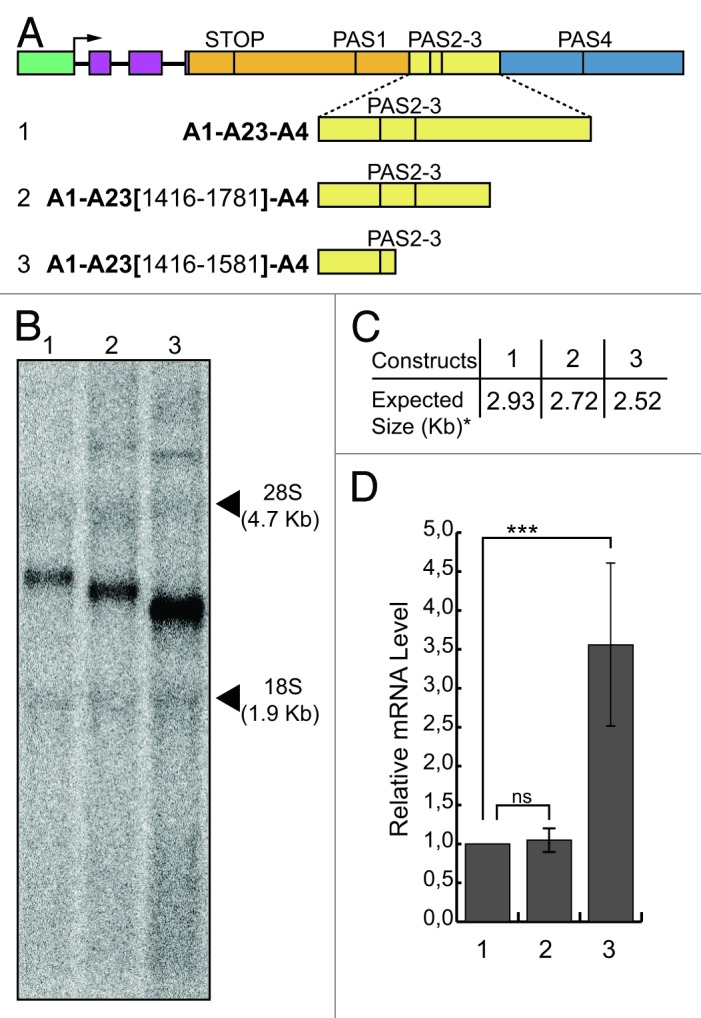
Figure 3. Mapping of the silencer element. (A) Schematic representation of the chimeric minigene constructs: A1-A23-A4 (1), A1-A23(1,416-1,781)-A4 (2) and A1-A23(1,416-1,581)-A4 (3). (B) Northern blot analysis of total RNA prepared from HeLa cells previously transfected with the constructs shown in (A); the α globin probe was used to detect the mRNAs of interest; phRG-TK (Renilla) plasmid was used for the normalization of the transfection efficiency; the positions of the 28S and 18S rRNAs are indicated on the right; (C) the expected sizes of the mRNAs of the constructs shown in (A), up to the predicted PAS4 (i.e., the asterisk indicates without the pA tail). (D) RT-qPCR analysis of the relative mRNA levels calculated as the ratio between analyzed mRNAs and Renilla control mRNA; the mean +/− SD of three independent experiments is shown; experiments were performed in duplicates; data were analyzed using one-way ANOVA and Tukey's multiple comparison test,; *** indicates p < 0.0001.
The RNase protection analysis showed that the differences between the A1-A23-A4 and A1-A4 mRNA levels were related to the decrease in cleavage efficiency at the PAS4 in the presence of the A23 region (see above, Fig. 2C). The possibility that the A23 region could contain destabilizing elements was ruled out, as the mRNAs derived from the A1-A23-A4 and the A1-A4 constructs showed similar stability after actinomycin D treatment (Fig. S2). These results strongly suggested that these effects are due to the presence of a negative cis-acting element within the A23 region affecting cleavage efficiency.
A long-range polyadenylation enhancer is necessary to activate pre-mRNA cleavage and polyadenylation at the brain-specific distal PAS4 of Add2
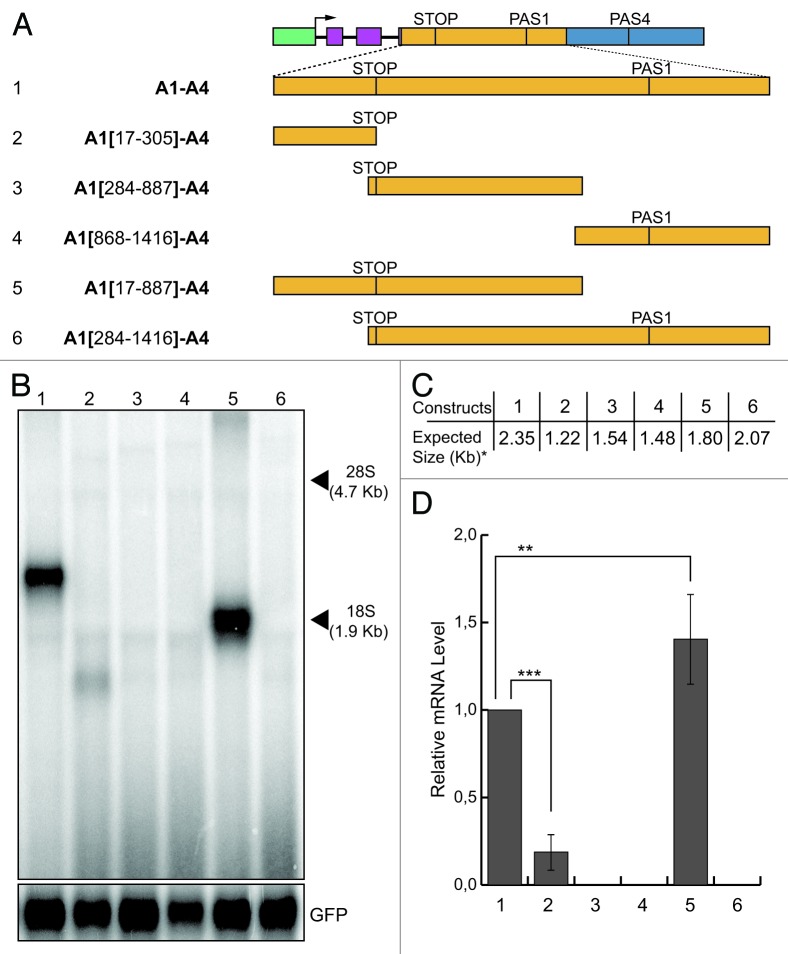
The A1 region used in the original A1-A23-A4 construct is 1,400 bp long. It contains coding and non-coding sequences of the last Add2 exon (Fig. 1A). To map more precisely the enhancer elements present in the A1 region, we initially divided this region into three partially overlapping segments (Fig. 4A): the coding part of the A1 region, ending at the stop codon (TGA at position 303 as indicated in Fig. 1A), the second segment corresponded to the region spanning for 604 bases, initiating just upstream of the stop codon and the last segment comprised the last 549 bases of the A1 region, including the PAS1 and its flanking sequences. The full A1 region present in the A1-A4 construct was replaced by each one of these three sub-regions [Fig. 4A, constructs A1(17-305)-A4, A1(284-887)-A4 and A1(868-1,416)-A4]. The expression pattern of the new constructs was determined after transfection of HeLa cells followed by northern blot analysis. Only the A1(17-305)-A4 construct (containing the most proximal sub-region) produced a signal visible by northern blot analysis, but the intensity of this signal was very faint (Fig. 4B, lane 2). Complete absence of the signal was observed for the other two constructs (Fig. 4B, lanes 3 and 4). These results suggest that the enhancer element may be disrupted. Therefore, to identify those elements necessary to recover full function of the PAS4, we joined the first to the second sub-region, and the second to the third one [Fig. 4A, constructs A1(17-887)-A4 and A1(284-1,416)-A4]. We observed that the presence of the first two segments was enough to completely restore the signal obtained with the full A1 region, whereas the A1(284-1,416)-A4 construct, which contained the second and third A1 sub-segments, produced no signal (Fig. 4B, lanes 5 and 6). Consequently, we can conclude that the third sub-segment containing the PAS1 had no influence on the processing at the PAS4. This result strongly suggested that regions flanking the Add2 stop codon contain elements that enhance the activity of the PAS4. Next, we started with a more detailed analysis of the enhancer element present in the A1 region.
Figure 4. Mapping of the enhancer element present in the A1 region. (A) Schematic representation of the chimeric minigene constructs: A1-A4 (1), A1(17-305)-A4 (2), A1(284-887)-A4 (3), A1(868-1,416)-A4 (4), A1(17-887)-A4 (5) and A1(284-1,416)-A4 (6). (B) Northern blot analysis of total RNA prepared from HeLa cells previously transfected with the constructs shown in panel A; the α globin fragment used as a probe for the northern blots is indicated in Figure 1; the p-EGFP-C2 plasmid was used for the normalization of the transfection efficiency; the positions of the 28S and 18S rRNAs are indicated on the right; (C) the expected sizes of the mRNAs of the constructs shown in (A), up to the predicted PAS4 (i.e., the asterisk indicates without the pA tail). D: the relative mRNA levels were determined as described for Figure 1; the mean +/− SD of three independent experiments is shown; data were analyzed using one-way ANOVA and Tukey's multiple comparison test,; *** and ** indicate p < 0.0001 and p < 0.01, respectively.
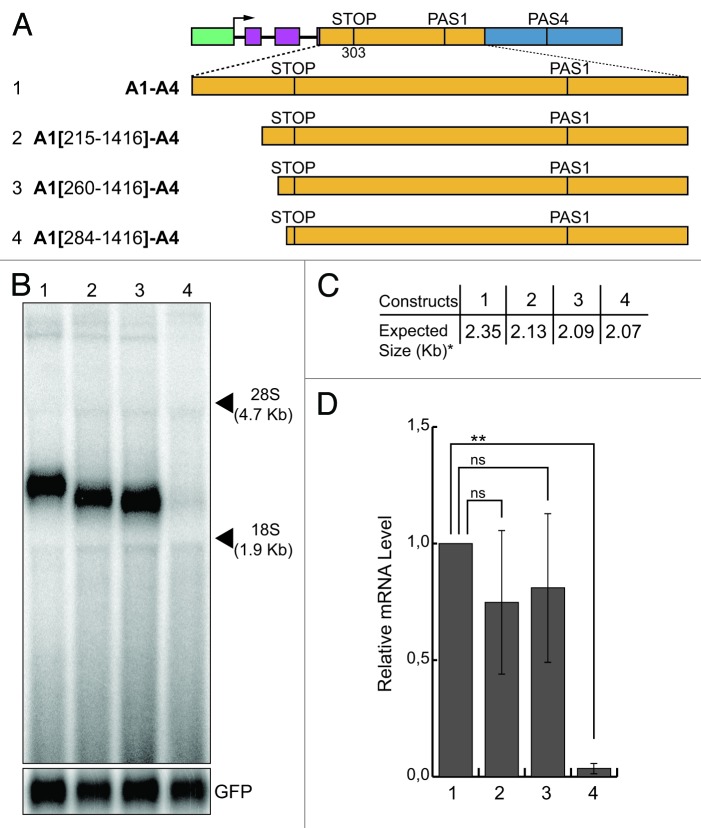
To map the active sequences of the enhancer element located upstream of the stop codon, we adopted an approach based on serial deletions starting from the 5′ end of the A1 region, thus partially eliminating the coding portion of the A1 (Fig. 5A). All deletions were done without altering the reading frame of the coding sequence. We replaced the A1 region from the A1-A4 construct by these deleted segments (Fig. 5A), expecting that deletion of the enhancer-active sequences should result in a reduction in the levels of minigene-derived transcripts in the northern blot analysis of the transfected cells. All tested constructs generated the expected bands, except for construct A1(284-1,416)-A4 (Fig. 5B, lane 4) where no band was observed. The only difference between construct A1(260-1,416)-A4, which still maintained pre-mRNA processing activity, and construct A1[(284-1,416)-A4, which was inactive since no band was generated, consisted in the presence of 24 bp in construct A1(260-1,416)-A4 (Fig. 5B, lane 3). Therefore, the presence of these 24 nt was essential to confer pre-mRNA processing activity and were considered as the upstream border of the enhancer (Fig. S4).
Figure 5. Mapping of the enhancer element present in the A1 region: sequences upstream of the stop codon. (A) Schematic representation of the chimeric minigene constructs: A1-A4 (1), A1(215-1,416)-A4 (2), A1(260-1,416)-A4 (3) and A1(284-1,416)-A4 (4). (B) Northern blot analysis of total RNA prepared from HeLa cells previously transfected with the constructs shown in (A); the α globin fragment used as a probe for the northern blots is indicated in Figure 1; the p-EGFP-C2 plasmid was used for the normalization of the transfection efficiency; the positions of the 28S and 18S rRNAs are indicated on the right; (C) the expected sizes of the mRNAs of the constructs shown in (A), up to the predicted PAS4 (i.e., the asterisk indicates without the pA tail). (D) the relative mRNA levels were determined as described for Figure 1; the mean +/− SD of three independent experiments is shown; data were analyzed using one-way ANOVA and Tukey's multiple comparison test; *** and ns indicate p < 0.0001 and not significant, respectively.
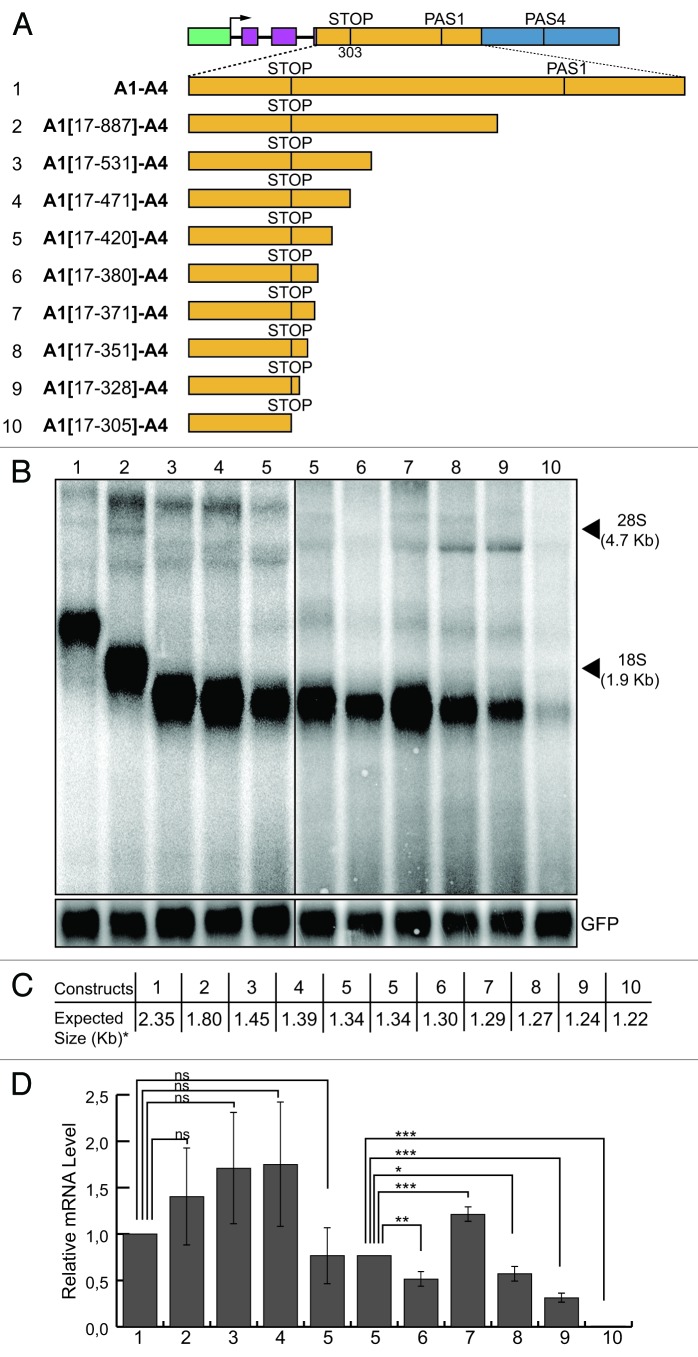
Following the same strategy to more finely map the active elements located downstream of the stop codon, we created serial deletions starting from the 3′end of the A1 region. The fragments containing these deletions were inserted at the place of the whole A1 region in the A1-A4 construct (Fig. 6A) and the specific signals monitored by northern blot analysis. As expected, the size of the detected mRNA inversely correlated with the length of the deletion performed in each construct (Fig. 6B). Similarly to the approach adopted for the upstream enhancer, the absence or important reduction of mRNA in the northern blot analysis was considered as the consequence of the loss of downstream enhancer elements. All constructs generated a strong signal in the northern blot, except for the faint signal observed for the A1(17-305)-A4 construct (Fig. 6B and D, lane 10). The difference between the constructs A1(17-305)-A4 and A1(17-328)-A4 was the deletion of just 23 bp in the former construct. Therefore, the presence of these 23 nt in the A1(17-328)-A4 construct were necessary to provide pre-mRNA 3′ end processing activity at the PAS4 and were considered as the downstream border of the enhancer (Fig. S4).
Figure 6. Mapping of the enhancer element present in the A1 region: sequences downstream of the stop codon. (A) Schematic representation of the chimeric minigene constructs: A1-A4 (1), A1(17–887)-A4 (2), A1(17–531)-A4 (3), A1(17-471)-A4 (4), A1(17-420)-A4 (5), A1(17-380)-A4 (6), A1(17-371)-A4 (7), A1(17-351)-A4 (8), A1(17-328)-A4 (9) and A1(17-305)-A4 (10). (B) northern blot analysis of total RNA prepared from HeLa cells previously transfected with the constructs shown in (A); the α globin fragment used as a probe for the northern blots is indicated in Figure 1; the p-EGFP-C2 plasmid was used for the normalization of the transfection efficiency; the positions of the 28S and 18S rRNAs are indicated on the right. (C) The expected sizes of the mRNAs of the constructs shown in (A), up to the predicted PAS4 (i.e., the asterisk indicates without the pA tail). (D) The relative mRNA levels were determined as described for Figure 1; the mean +/− SD of three independent experiments is shown; data were analyzed using one-way ANOVA and Tukey's multiple comparison test; *** p < 0.001, ** p < 0.01, * p < 0.05, ns, not significant.
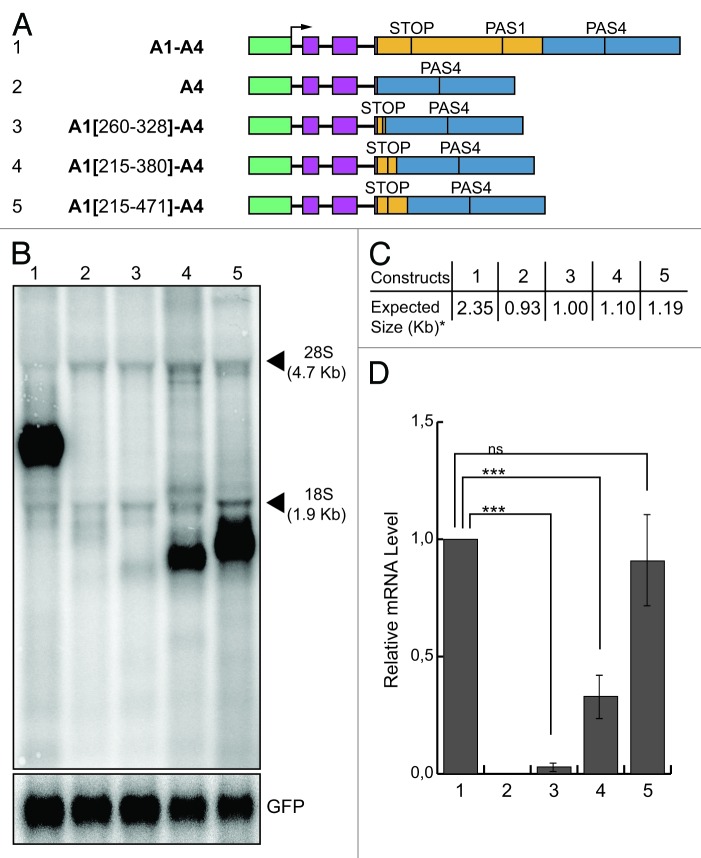
Taking both analyses together, it was possible to define a minimum composite 69 nt bases-long enhancer element, centered at the stop codon of the β-adducin gene, comprising the upstream and downstream elements. To test the functionality of the putative enhancer element, we added increasing-length fragments containing the enhancer elements to the A4 construct (Fig. 7A). When we placed the 69 bases-long enhancer element upstream of the A4 region [A1(260-328)-A4 construct, Fig. 7A] a very faint signal was observed in the northern blot analysis, after transfecting the construct into HeLa cells (Fig. 7B, lane 3). This results indicates that the 69 bases-long enhancer was able to activate pre-mRNA processing of the A1(260-328)-A4 transcript, but was not enough to confer full activity, suggesting that additional elements were needed. Therefore, we placed a 166 bp segment upstream of the A4 region, which contained about 50 bp of additional sequences at each side of the 69 nt-long element [Fig. 7A, construct A1(215-380)-A4]. The 166 bases-long segment was able to enhance the processing at the PAS4 and to rescue the signal in the northern blot (Fig. 7B, lane 4). However, the detected signal did not reach the levels of the signal obtained from the A1-A4 construct (Fig. 7D, lane 4 compared with 1). This difference was not due to the differences in the stability of the mRNAs derived from A1-A4 and A1[215–380]-A4 constructs (Fig. S2), suggesting that other cis-acting sequences may still be necessary. In fact, when we tested a construct containing a 257 nt-long fragment, we observed a signal similar to that obtained with the A1-A4 construct (Fig. 7B and D, lane 5 compared with 1), confirming the previous experiments which suggested that other elements cooperate with the minimal enhancer in activating polyadenylation of the Add2 PAS4.
Figure 7. Two hundred and fifty-seven nt-long enhancer element is enough to restore activity at the PAS4. (A) Schematic representation of the chimeric minigene constructs: A1-A4 (1), A4 (2), A1(260-328)-A4 (3), A1(215-380)-A4 (4), A1(215-471)-A4 (5); (B) northern blot analysis of total RNA prepared from HeLa cells previously transfected with the constructs shown in (A); the α globin fragment used as a probe for the northern blots is indicated in Figure 1; the p-EGFP-C2 plasmid was used for the normalization of the transfection efficiency; the positions of the 28S and 18S rRNAs are indicated on the right; (C) the expected sizes of the mRNAs of the constructs shown in panel A up to the predicted PAS4, without pA tail (indicated by an asterisk). (D) The relative mRNA levels calculated as the ratio between analyzed mRNAs and GFP mRNA control represented in (B); quantification of the northern blot signals has been performed as described above; the mean +/− SD of four independent experiments is shown; data were analyzed using one-way ANOVA and Tukey's multiple comparison test; *** p < 0.001, ns, not significant.
Discussion
The presented results underscore the complexity of the mechanisms regulating cleavage and polyadenylation of the mouse Add2 pre-mRNA. We first predicted the canonical cis-acting elements (hexanucleotide motif, DSE and USE) of the three alternative polyadenylation regions of the Add2 gene.17 Then, using a minigene-based approach, we identified and functionally tested the core elements, which were necessary for the correct definition and precise pre-mRNAs processing at the distal polyadenylation site (PAS4) of Add2.16 In the present study, we identified novel non-canonical long-distance upstream cis-acting elements regulating 3′end processing at the Add2 PAS4.
According to some bioinformatic studies, the most-distal PAS tends to be the strongest and most efficiently used PAS in genes containing an array of tandem PASs.23,24 When we transfected our minigene model containing all three Add2 polyadenylation sites into HeLa cells, we observed the preferential use of the distal PAS4. The use of distal PAS in HeLa cells has already been observed,25 although distal PAS are more frequently used in non-dividing tissues such as brain than in replicating cells, which preferentially use proximal PAS.26
Although the core cis-acting elements are critical for pre-mRNA processing at the PAS4,16 we showed here that they were not sufficient to efficiently trigger cleavage and polyadenylation at the PAS4. In fact, the presence of the upstream 1.4 kb-long A1 region appeared to be a requisite for 3′end processing at the PAS4, as no mature mRNAs were detected in its absence (Fig. 1, construct A4, lane 2). Mapping experiments showed that a long-distance upstream enhancer element was responsible for activation of the PAS4. This enhancer was contained within a 257 nt sequence and was able to activate the usage of the PAS4 with the same efficiency as the full-length 1.4 Kb A1 region [Fig. 7, construct A1(215-471)-A4]. These 257 nt comprise the last 88 nucleotides of the coding region, the stop codon and the first 166 nt of the 3′UTR, spanning from base 215 to base 471 of the mouse Add2 terminal exon. Shorter fragments of 69 nt and 166 nt also showed enhancer activity, but they were less efficient to activate the PAS4, without reaching the same levels obtained with the full A1 region. The coding region of the enhancer shows a remarkably high conservation across species (Figs. S4 and S5), as this region codes for the very conserved MARKS domain of the protein.27 Conservation in the region located downstream of the stop codon is lower, although islands of high conservation are also present, supporting the hypothesis that it may also be functional in other species (Fig. S5). Several characteristics distinguish this polyadenylation upstream enhancer element from the more “classical” USEs described so far and are discussed below.
The Add2 enhancer element is located more than 2 Kb upstream of the PAS4 in the minigene context (construct A1-A23-A4). In the mouse Add2 gene, the distance is more than 5 Kb. Even if we have not yet checked the functionality of the enhancer element in this context, we expect it will be functional because its activity was not affected by the distance between the enhancer and the PAS4, as verified with all tested constructs. The presence of a long-distance upstream polyadenylation enhancer element is quite peculiar for eukaryotic polyadenylation signals. In general, USEs are positioned immediately upstream of the hexanucleotide motif. Interestingly, a long-distance upstream polyadenylation enhancer element has been observed for the Rous sarcoma virus (RSV) polyadenylation signal.8,9 This element is called negative regulator of splicing (NRS) and is located in the gag gene, about 8 Kb upstream of the RSV polyadenylation signal. Binding of the SR proteins SRSF1, SRSF3 and SRSF7 (former names SF2/ASF, SRp20 and 9G8, respectively) to the NRS stimulates polyadenylation at the RSV PAS.28 The SR proteins are in competition for the binding to the NRS with hnRNP H, which inhibits polyadenylation at the RSV PAS.9 Some well-known upstream enhancers are described for the SV40 late, human C2 complement, lamin B2, COX-2 and collagen polyadenylation signals.14,29-32 In all of these cases, mutation and/or deletion of the U-rich USEs led to the inefficient 3′end processing. These elements map close to the AAUAAA sequence,33 and the involvement of far-upstream enhancers of polyadenylation has not been reported so far in non-viral genes. However, a long-distance downstream enhancer has been found to stimulate cleavage efficiency of the human melanocortin receptor 1 (MC1R) pre-mRNA.7 This enhancer is G-rich and is located 440 nt downstream of the cleavage and polyadenylation site. In the same study, another G-rich enhancer, placed immediately downstream of the PAS, has been identified to be essential for the 3′ end processing at MC1R PAS. Absence of either of these two downstream enhancers led to the reduced cleavage efficiency in the RNase protection analysis.7
In addition, the novel upstream enhancer element is present in a context of tandem alternative polyadenylation sites specifically influencing the usage of PAS4. The activity of PAS1 and PAS2-3, also located downstream of the enhancer, seem not to be affected even in the absence of the A4 region (data not shown). Thus, it seems unlikely that this element acts as a general enhancer of polyadenylation.
The non-canonical upstream enhancer partially spans throughout the coding region, a characteristic not present in the reported USEs. Being positioned in the coding region, the sequence composition has also to respond to evolutionary constrains dictated by the encoded protein and, thus, is not similar to that of the ”standard” USEs described so far. Usually, USEs are represented by U-rich sequences, while the coding region of the non-canonical enhancer element is purine-rich. It is known that purine-rich regions in pre-mRNAs are potential landing pads for RNA-binding proteins of the SR family, such as SRSF1 and SRSF10. This family of proteins participates in constitutive and alternative splicing of pre-mRNA, generally as enhancers of the splicing reaction,34 but in some cases they can also enhance 3′end processing, as mentioned above for the RSV polyadenylation.28,35-37
The Add2 non-canonical enhancer element is positioned in the vicinity of the 3′splice site of the terminal exon. It is well known that terminal-exon definition relies on the crosstalk between splicing and 3′end processing events.38 Thus, the non-canonical enhancer element could function in the activation of the PAS4 site through direct stimulation of the terminal exon definition and splicing of the last intron. Preliminary data showed that removal of all introns from the A1-A23-A4 construct abolished its expression, as no signal was detected by northern blot analysis (A. Iaconcig and L. Costessi, unpublished data). Therefore, it is probable that the mechanism of the non-canonical enhancer element to activate polyadenylation of the PAS4 could be associated to splicing of the terminal exon, supporting the hypothesis that key factors recognizing the enhancer could belong to the SR-protein family.
In addition to the non-canonical upstream polyadenylation enhancer element, we also showed the presence of another upstream polyadenylation regulatory element which inhibited processing at the mouse Add2 PAS4. Our experiments suggested that this non-canonical silencer element was included within a 200 nt-long segment, which spanned from base 1,581 to base 1,781 of the Add2 3′UTR, respective to the first base of the terminal exon. Removal of this 200 nt segment led to about 3-fold increase in mRNA levels. The possibility that the silencer element is affecting the half-life of the mRNA was ruled out by mRNA stability assays. In fact, the stability of the mRNA derived from the A1-A23-A4 construct, thus containing the silencer element, was similar to that of the A1-A4 construct, which lacks this element. Furthermore, the RNase protection analysis strongly suggests that the negative cis-acting element within the A23 region affects cleavage efficiency. Additional experiments are needed to determine the molecular mechanisms involved.
As observed for the enhancer element, also the non-canonical silencer element represents a long-distance polyadenylation regulatory element. In the minigene context, the distance from the silencer element to the PAS4 is about 800 bases, and in the natural context of the mouse Add2 gene it is about 4 Kb. Another interesting observation is that USEs are usually enhancers of polyadenylation.29,31,32 The presence of an upstream polyadenylation silencer element has also been described for the U1A pre-mRNA and is involved in U1A protein autoregulation.39 This silencer element is ~50 nt long and is located 19 nt upstream of the hexanucleotide motif of the U1A pre-mRNA.
All constructs were transcriptionally active suggesting that the absence of mature transcript from transcripts lacking the enhancer element was not caused by a block in transcription. We ruled out some of the possible mechanisms of degradation such as differences in mRNA stability and mRNA degradation by the surveillance machinery, with experimental data pointing to defects in the cleavage reaction. Transcripts originating from constructs lacking the enhancer element were undetectable even after treatment of the transfected cells with cycloheximide, a drug known to block translation and mRNA degradation associated to NMD (and other surveillance pathways).22 Yet, we cannot completely rule out the possibility that the enhancer element may participate in mRNA stability in a mechanism in which the main regulator of NMD is the distance between the stop codon and the PAS of the mRNA.40
Taken together, in the present study we identified novel long-distance upstream non-canonical polyadenylation-regulatory elements that enhance or silence 3′end processing of the mouse Add2 PAS4. The non-canonical upstream enhancer element is indispensable for the efficient use of the PAS4. On the contrary, the non-canonical upstream silencer element inhibits the use of the PAS4. These results underscore the complex mechanisms regulating polyadenylation of the Add2 gene.
Materials and Methods
Generation of minigene constructs
The mouse β adducin genomic regions containing PASs (A1, A23 and A4 regions) were cloned instead of α globin PAS to create A1-A23-A4 construct as previously described.16 The A4, A23-A4 and A1-A4 constructs were obtained by removing the A1 and A23 region, only A1 region and only A23 region from the original A1-A23-A4 construct, respectively. The INA1-A4 construct was obtained by cloning the A1 region in the antisense orientation in the A4 construct. The different subregions of the A1 and A23 regions were PCR amplified, subcloned into pUC19, sequenced and cloned into A4 or A1-A4 constructs to obtain different A1(x-y)-A4constructs, respectively [numbers in the brackets determine from which nt (x) to which nt (y) the subregion spans, in reference to the first nt of the last mouse β adducin exon].
Cell culture, transfections and RNA preparation
HeLa cells were grown in Dulbecco’s modified Eagle medium (DMEM) with GlutaMAX, D-glucose and Pyruvate, supplemented with 10% of Fetal Calf Serum (FCS) and 100 mg/ml of Normicin (antibiotic/anti-mycotic). Transient transfections of HeLa cells were performed using Lipofectamine 2000 reagent (Invitrogen), according to the manufacturer’s instructions. The transfections were performed in 35 mm Petri dish or 6-well plates. Along with the test minigene construct, each plasmid DNA mix contained the plasmid expressing SV40 T-antigen to stimulate SV40 promoter activity and the pEGFP-C2 or an intron-containing variant of the phRG-TK vector for checking transfection efficiency. Twenty-four hours after the transfections (except when treated with the cycloheximide or the actinomycin D), the cells were collected and total RNA was extracted using EuroGold TriFast reagent (EuroClone), according to the manufacturer’s instructions.
RT-PCR and RT-qPCR
cDNAs were prepared by reverse transcription (RT) from DNase treated total RNA isolated from the transiently transfected HeLa cells, using M-MLV Reverse Transcriptase (Invitrogen).
RT-PCRs were performed using following GAPDH and TDP-43 primer pairs: qGAPDH81Dir -5′ AAGGTGAAGGTCGGAGTCAA 3′; qPCR GAPDH 188 R - 5′ AATGAAGGGGTCATTGATGG 3′; nwXbaBglx5FW - 5′ CTAGTCTAGAAGATCTCGCAAAGCCAAGATGAGCCTTTGAG 3′; hTDPx6RV2 - 5′ ATTAACTGCTATGAATTCTTTGCATTCAG 3′.
RT-qPCRs were performed using IQ Custom Syber Green Supermix 2x (BioRad). The following primer pairs were used for the qPCRs: agloEX1 real DIR - 5′ GCCGACAAGACCAACGTCAA 3′; aGloex2 Real REV - 5′ AGGTCGAAGTGCGGGAAGTA 3′; qREN1436D - 5′ CTGGAGCCATTCAAGGAGAA 3′; qREN1554R - 5′ GCGTTGTAGTTGCGGACAAT 3′; α Glo Ex2 real DIR - 5′ TGGACGACATGCCCAAC 3′; α Glo Int2 real REV - 5′ AACCCGCGTGATCCTCT 3′; GFP 1004 FW - 5′ TCAAGGAGGACGGCAACATC 3′; GFP 1124 RV - 5′ TTGTGGCGGATCTTGAAGTTC 3′. The reactions were performed in duplicates for each sample. A Bio-Rad C1000 thermo-cycler coupled with a CFX96 real-time system was used for the amplification. The amplification reaction steps were as follows: 98°C for 30 s, then 40 cycles at 95°C for 5 s and 58°C for 25 s. The ΔΔCt between the amplicon of interest and reference amplicon was calculated using the Bio-Rad software.
Northern blot analysis
The northern blot experiments were performed as described in Costessi et al.17 Five μg of total RNA prepared from transfected HeLa cells were run on a 1.4% denaturing agarose gel and then transferred to nitrocellulose membrane (Hybond-N+, Amersham Pharmacia biotech). The membrane was pre-hybridized at 42°C for 1 h in ultrasensitive hybridization solution (Ambion), followed by the ON hybridization to the α globin or the GFP probe. The DNA probes were labeled with (α-32P) dCTP using RediprimeII DNA labeling System (GE Healthcare) and purified using Illustra nick columns (GE Healthcare). The pUC19 plasmid containing sequences of the α globin exon 1 and 2 (from Dr. Laura de Conti, Molecular Pathology Group, ICGEB) was cut with BamHI and EcoRI to create the α globin probe. The GFP probe was generated by a PCR reaction using the pEGFP-C2 plasmid as template and primer pair: GFP813FW - 5′ CGGCGTGCAGTGCTTCAGCCGCTAC 3′ GFPendRV - 5′ CTTGTACAGCTCGTCCATGCCGAGAG 3′.
RNase protection assay
The RNase protection assays were performed as described in Dalziel et al.7 RNA samples were prepared from total RNA of transfected HeLa cells. Total RNA was treated with DNase, acid phenol-chloroform purified, ethanol precipitated and resuspended in R-loop buffer. The quantity of each RNA sample used for the RNase protection assay was normalized for the transfection efficiency. Four hundred ng of one RNA sample were used for the assay and this sample was chosen as reference in the normalization calculations. Ten μg of mouse brain total RNA were used as a control for the correct cleavage protected fragment. Total RNA from mouse brain was prepared using EuroGold TriFast reagent (EuroClone) and resuspended in R-loop buffer.
For the RNA probe preparation, the fragment upstream and downstream of the PAS4 was amplified using following primer pair: mXhoI dir 2 - 5′ CCGCTCGAGATTTATGGGTTGGTTTA 3′ mADD2 BR short rev - 5′ CGGGATCCCCGCGGGAATTCCCTACATTTC 3′. The amplicon was cloned in the antisense orientation downstream of the T7 promoter in the PBS II KS plasmid (Stratagene), in vitro transcribed (T7 RNA polymerase, Stratagene) using (α-32P) UTP and gel purified through 6% polyacrylamide 8M urea gel. Approximately 500 cps of the probe were used for the hybridization reactions.
One Kb DNA Ladder (Invitrogen) has been labeled with (α-32P) dCTP according to the manufacturer’s T4 DNA polymerase labeling protocol.
Supplementary Material
Acknowledgments
The authors are thankful to Dr Marcos Morgan (EMBL) for helpful discussions and suggestions.
Glossary
Abbreviations:
- PAS
cleavage and polyadenylation site
- DSE
downstream sequence element
- CPSF
cleavage and polyadenylation specificity factor
- CstF
cleavage stimulatory factor
- auxDSE
auxiliary DSE
- USE
upstream sequence element
- GRS
G-rich sequence
- RSV
Rous sarcoma virus
- PAS4
distal brain-specific Add2 PAS
- PAS1
first proximal erythroid-specific Add2 PAS
- PAS2-3
second proximal erythroid-specific Add2 PAS
- 3′UTR
3′ untranslated region
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/23855
References
- 1.Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–82. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–45. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Custódio N, Carmo-Fonseca M, Geraghty F, Pereira HS, Grosveld F, Antoniou M. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 1999;18:2855–66. doi: 10.1093/emboj/18.10.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–42. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- 5.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–12. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zarudnaya MI, Kolomiets IM, Potyahaylo AL, Hovorun DM. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 2003;31:1375–86. doi: 10.1093/nar/gkg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalziel M, Nunes NM, Furger A. Two G-rich regulatory elements located adjacent to and 440 nucleotides downstream of the core poly(A) site of the intronless melanocortin receptor 1 gene are critical for efficient 3′ end processing. Mol Cell Biol. 2007;27:1568–80. doi: 10.1128/MCB.01821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fogel BL, McNally LM, McNally MT. Efficient polyadenylation of Rous sarcoma virus RNA requires the negative regulator of splicing element. Nucleic Acids Res. 2002;30:810–7. doi: 10.1093/nar/30.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilusz JE, Beemon KL. The negative regulator of splicing element of Rous sarcoma virus promotes polyadenylation. J Virol. 2006;80:9634–40. doi: 10.1128/JVI.00845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown PH, Tiley LS, Cullen BR. Efficient polyadenylation within the human immunodeficiency virus type 1 long terminal repeat requires flanking U3-specific sequences. J Virol. 1991;65:3340–3. doi: 10.1128/jvi.65.6.3340-3343.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carswell S, Alwine JC. Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol Cell Biol. 1989;9:4248–58. doi: 10.1128/mcb.9.10.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall-Pogar T, Liang S, Hague LK, Lutz CS. Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3′-UTR. RNA. 2007;13:1103–15. doi: 10.1261/rna.577707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley JL, Proudfoot NJ. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 1998;12:2522–34. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Natalizio BJ, Muniz LC, Arhin GK, Wilusz J, Lutz CS. Upstream elements present in the 3′-untranslated region of collagen genes influence the processing efficiency of overlapping polyadenylation signals. J Biol Chem. 2002;277:42733–40. doi: 10.1074/jbc.M208070200. [DOI] [PubMed] [Google Scholar]
- 15.Valsamakis A, Zeichner S, Carswell S, Alwine JC. The human immunodeficiency virus type 1 polyadenylylation signal: a 3′ long terminal repeat element upstream of the AAUAAA necessary for efficient polyadenylylation. Proc Natl Acad Sci USA. 1991;88:2108–12. doi: 10.1073/pnas.88.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costessi L, Porro F, Iaconcig A, Nedeljkovic M, Muro AF. Characterization of the distal polyadenylation site of the β-adducin (ADD2) pre-mRNA. PLoS One. 2013 doi: 10.1371/journal.pone.0058879. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costessi L, Devescovi G, Baralle FE, Muro AF. Brain-specific promoter and polyadenylation sites of the beta-adducin pre-mRNA generate an unusually long 3′-UTR. Nucleic Acids Res. 2006;34:243–53. doi: 10.1093/nar/gkj425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilligan DM, Lozovatsky L, Gwynn B, Brugnara C, Mohandas N, Peters LL. Targeted disruption of the beta adducin gene (Add2) causes red blood cell spherocytosis in mice. Proc Natl Acad Sci USA. 1999;96:10717–22. doi: 10.1073/pnas.96.19.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muro AF, Marro ML, Gajović S, Porro F, Luzzatto L, Baralle FE. Mild spherocytic hereditary elliptocytosis and altered levels of alpha- and gamma-adducins in beta-adducin-deficient mice. Blood. 2000;95:3978–85. [PubMed] [Google Scholar]
- 20.Porro F, Rosato-Siri M, Leone E, Costessi L, Iaconcig A, Tongiorgi E, et al. beta-adducin (Add2) KO mice show synaptic plasticity, motor coordination and behavioral deficits accompanied by changes in the expression and phosphorylation levels of the alpha- and gamma-adducin subunits. Genes Brain Behav. 2010;9:84–96. doi: 10.1111/j.1601-183X.2009.00537.x. [DOI] [PubMed] [Google Scholar]
- 21.Bednarek E, Caroni P. β-Adducin is required for stable assembly of new synapses and improved memory upon environmental enrichment. Neuron. 2011;69:1132–46. doi: 10.1016/j.neuron.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 22.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–26. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 23.Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–10. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Legendre M, Gautheret D. Sequence determinants in human polyadenylation site selection. BMC Genomics. 2003;4:7. doi: 10.1186/1471-2164-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubo T, Wada T, Yamaguchi Y, Shimizu A, Handa H. Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3′-UTRs. Nucleic Acids Res. 2006;34:6264–71. doi: 10.1093/nar/gkl794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayr C, Bartel DP. Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–84. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuoka Y, Li X, Bennett V. Adducin: structure, function and regulation. Cell Mol Life Sci. 2000;57:884–95. doi: 10.1007/PL00000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maciolek NL, McNally MT. Serine/arginine-rich proteins contribute to negative regulator of splicing element-stimulated polyadenylation in rous sarcoma virus. J Virol. 2007;81:11208–17. doi: 10.1128/JVI.00919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brackenridge S, Proudfoot NJ. Recruitment of a basal polyadenylation factor by the upstream sequence element of the human lamin B2 polyadenylation signal. Mol Cell Biol. 2000;20:2660–9. doi: 10.1128/MCB.20.8.2660-2669.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall-Pogar T, Zhang H, Tian B, Lutz CS. Alternative polyadenylation of cyclooxygenase-2. Nucleic Acids Res. 2005;33:2565–79. doi: 10.1093/nar/gki544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreira A, Wollerton M, Monks J, Proudfoot NJ. Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J. 1995;14:3809–19. doi: 10.1002/j.1460-2075.1995.tb00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schek N, Cooke C, Alwine JC. Definition of the upstream efficiency element of the simian virus 40 late polyadenylation signal by using in vitro analyses. Mol Cell Biol. 1992;12:5386–93. doi: 10.1128/mcb.12.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11:1485–93. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blencowe BJ. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci. 2000;25:106–10. doi: 10.1016/S0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- 35.Lou H, Neugebauer KM, Gagel RF, Berget SM. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol Cell Biol. 1998;18:4977–85. doi: 10.1128/mcb.18.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lou H, Yang Y, Cote GJ, Berget SM, Gagel RF. An intron enhancer containing a 5′ splice site sequence in the human calcitonin/calcitonin gene-related peptide gene. Mol Cell Biol. 1995;15:7135–42. doi: 10.1128/mcb.15.12.7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCracken S, Longman D, Johnstone IL, Cáceres JF, Blencowe BJ. An evolutionarily conserved role for SRm160 in 3′-end processing that functions independently of exon junction complex formation. J Biol Chem. 2003;278:44153–60. doi: 10.1074/jbc.M306856200. [DOI] [PubMed] [Google Scholar]
- 38.Martinson HG. An active role for splicing in 3′-end formation. Wiley Interdiscip Rev RNA. 2011;2:459–70. doi: 10.1002/wrna.68. [DOI] [PubMed] [Google Scholar]
- 39.Boelens WC, Jansen EJ, van Venrooij WJ, Stripecke R, Mattaj IW, Gunderson SI. The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell. 1993;72:881–92. doi: 10.1016/0092-8674(93)90577-D. [DOI] [PubMed] [Google Scholar]
- 40.Brogna S, Wen J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat Struct Mol Biol. 2009;16:107–13. doi: 10.1038/nsmb.1550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.