Abstract
Feline immunodeficiency virus (FIV) is a lentivirus that causes AIDS-like immunodeficiency disease in domestic cats. Free-ranging lions, Panthera leo, carry a chronic species-specific strain of FIV, FIV-Ple, which so far has not been convincingly connected with immune pathology or mortality. FIV-Ple, harboring the three distinct strains A, B, and C defined by pol gene sequence divergences, is endemic in the large outbred population of lions in the Serengeti ecosystem in Tanzania. Here we describe the pattern of variation in the three FIV genes gag, pol-RT, and pol-RNase among lions within 13 prides to assess the occurrence of FIV infection and coinfection. Genome diversity within and among FIV-Ple strains is shown to be large, with strain divergence for each gene approaching genetic distances observed for FIV between different species of cats. Multiple in fections with two or three strains were found in 43% of the FIV-positive individuals based on pol-RT sequence analysis, which may suggest that antiviral immunity or interference evoked by one strain is not consistently protective against infection by a second. This comprehensive study of FIV-Ple in a free-ranging population of lions reveals a dynamic transmission of virus in a social species that has historically adapted to render the virus benign.
Feline immunodeficiency virus (FIV) is a lentivirus that infects both wild and domestic feline species and is closely related to human and simian immunodeficiency viruses (HIV and SIV, respectively) (26). Species-specific strains of FIV have been isolated from domestic cat (Felis catus), puma (Puma concolor), lion (Panthera leo), leopard (Panthera pardus), and Pallas' cat (Otocolobus manul) (termed FIV-Fca, FIV-Pco, FIV-Ple, FIV-Ppa, and FIV-Oma, respectively) (4, 6, 8, 9, 25, 51). In domestic cats, viral infection results in an AIDS-like pathology typified by a period of latency followed by CD4 depletion, immune suppression, a host of subsequent secondary infections, and high mortality (54). Because of its striking similarity to the pattern of disease progression observed with HIV infection in humans, FIV-Fca has been an ideal model system for understanding many clinical aspects of retroviral infection (54).
Although antibodies that cross-react to FIV are found in many species of wild cats in the Americas and Africa and a few species in Europe and Asia (7, 18, 27, 30, 33, 55), there is no documented disease association. Therefore, the clinical implications of FIV infection in populations of wild feline species are still unclear. Seroprevalence of FIV in free-ranging populations of African lions within east and south Africa exceeds those seen in any feline species to date (6, 7, 18, 33). Phylogenetic analyses of FIV genome sequences circulating in free-ranging populations of lions display high levels of sequence divergence (6), suggesting that FIV infection in these animals may be an ancient event (6, 7). Three highly divergent strains (or subtypes) of FIV-Ple have been described based on a fragment of the reverse transcriptase (RT) region of the pol gene (6). The genetic divergence between these three strains approached the level of difference seen between FIV strains isolated from different Felidae species, suggesting that the three strains evolved in geographically distant populations or possibly in different African felid species. However, the occurrence of all three strains in the lions of Tanzania's Serengeti National Park indicates that the strains have recently converged within the same population, and they have therefore been designated FIV-Ple A, FIV-Ple B, and FIV-Ple C.
This population of FIV-Ple-infected Serengeti lions provides an interesting parallel to studies of primate lentiviruses. To date, no known wild population of simians contains multiple SIV subtypes (44), yet subtype dynamics are an important aspect of HIV epidemiology (28). The high incidence of FIV infection in the free-ranging lions of the Serengeti National Park (90%), combined with the presence of three divergent FIV-Ple subtypes, provides a novel opportunity to examine the population dynamics, evolution, and interactions between subtypes in a natural setting.
The Serengeti National Park contains a large, genetically diverse outbred population (n = 3,000) of east African lions (16). Extensive genetic, demographic, and behavioral information exists for these lions (16, 35, 36), as well as serological and genetic data for FIV and outbreaks of other viruses (6, 18, 42). In this study, we analyze the patterns of proviral strain diversity and occurrence among members of 13 prides, examining three distinct FIV genes-gag, pol-RT, and pol-RNase. Analysis of these three gene regions shows that all three FIV-Ple strains circulate freely in the population. Distinct strains are observed within the same pride and frequently within the same individual. While sequences of cloned PCR products from several individual lions demonstrate the monophyly of viral quasispecies within individual lions consistent with a clonal expansion after an initial infection, we also describe a high number of individuals that are infected with more than one highly divergent FIV-Ple subtype.
MATERIALS AND METHODS
Samples for seroprevalence and DNA analysis.
Blood samples were collected from 349 free-ranging lions representing 13 known prides within the Serengeti National Park, Tanzania (Fig. 1B). Serum or plasma from Serengeti lion samples was tested for the presence of FIV-reactive antibodies by Western blot analysis. Samples were tested against viral antigens from domestic cat FIV-Petaluma strain and lion FIV-Ple strain. Immunoblotting was performed as described previously (5).
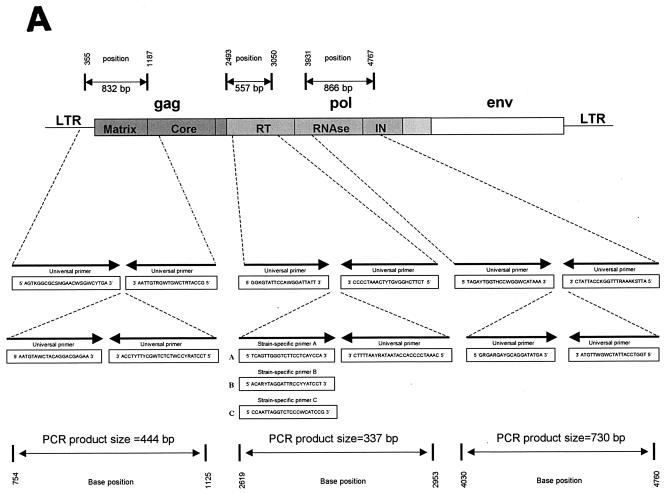
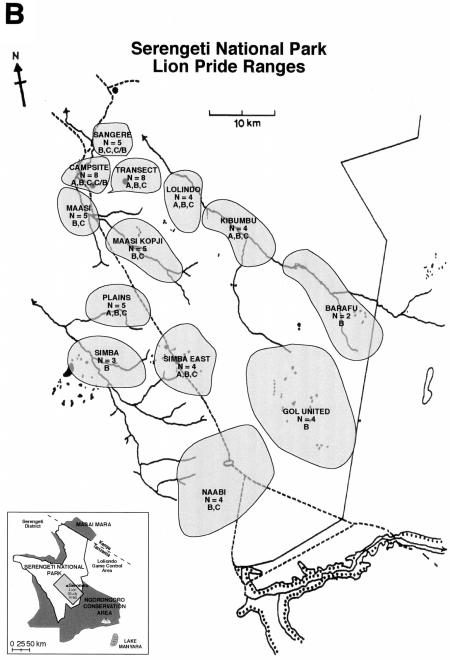
FIG. 1.
(A) Map of external and internal primers on the FIV genome. Positions are homologous to the FIV-Fca sequence (GenBank accession no. U11820). (B) Approximate pride ranges of the Serengeti lions during the dry season in 1994 (unpublished data). Numbers indicate how many individuals from each pride are included in this study, and letters refer to FIV-Ple subtypes. Pride ranges fluctuate with the seasons and over the years; prides remain discrete for many years.
PCR amplification of proviral DNA.
Genomic DNA was isolated from the white blood cells of 68 FIV-seropositive lions and 1 seronegative lion by proteinase K digestion followed by a standard phenol-chloroform extraction. Nested PCR using primers designed from the conserved RT region of pol, the RNase region of pol, and the p17/p26 region of gag were used to amplify the FIV sequence (Fig. 1A). First-round primers were designed from published sequences of FIV-Fca (GenBank accession no. M25381 and U11820), FIV-Pco (accession no. U03982), and FIV-Oma (accession no. U56928). For the pol-RT region, second-round primers were specifically designed for three previously described (6) RT subtypes (A, B, and C). For pol-RNase and gag, second-round primers were designed from the alignment of the FIV-Fca, FIV-Pco, and FIV-Oma sequences. All first-round PCRs were performed by using 100 ng of genomic DNA in a 50-μl reaction with 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 2.5 mM MgCl2, 0.25 mM concentrations of dATP, dCTP, dGTP, and dTTP, 2 mM concentrations of each primer, and 2 U of AmpliTaq DNA polymerase from Applied Biosystems. PCR cycling conditions were as follows: 3 min at 94°C followed by 45 cycles of 15 s at 94°C, 30 s at 52°C or 50°C, and 45 s at 72°C, with a final extension of 10 min at 72°C. Second-round PCRs were done under the same conditions with 1 μl of product from the first-round reaction used as a template. These amplifications were performed in a Perkin-Elmer 9700 system and visualized on a 1% agarose gel. Samples were also run in a Biometra TGradient machine with an annealing temperature gradient from 48 to 60°C. All other cycling conditions were the same as those described above. Second-round PCR products were sequenced by using standard ABI BigDye terminator reactions.
Representative PCR products were subsequently cloned and sequenced. For the pol-RNase sequence, products were cloned from 21 lions, representing both singly infected (Ple-624) and coinfected individuals (Table 1). For the gag sequence, products were also cloned from 21 lions: 3 singly infected, 2 multiply infected with all three RT subtypes, 17 coinfected with RT subtype B and C, and 3 with both A and B RT subtypes. Cloning was performed with a TOPO-TA cloning kit (Invitrogen) according to the manufacturer's instructions. DNA was isolated from 8 to 12 clones from each reaction product by using a QIAGEN miniprep kit. Sequences were obtained from clones with the correct insert by using standard ABI BigDye terminator reactions.
TABLE 1.
FIV subtype distribution for 68 Serengeti lions based on gene sequence analysis
| Liona | Pride affiliation | Year of sample | FIV type(s) detected at indicated gene regionb
|
||||
|---|---|---|---|---|---|---|---|
|
pol-RT (337 bp)
|
pol-RNase (730 bp), universal primer | gag (444 bp), universal primer | |||||
| Primer type A | Primer type B | Primer type C | |||||
| Ple-531 | BARAFU | 1989 | - | B | - | B | - |
| Ple-583 | BARAFU | 1994 | - | B | - | B | B |
| PLE-241 | CAMPSITE | 1985 | - | B | C | B | cB, cC |
| PLE-254 | CAMPSITE | 1985 | - | B | C | cB | cB, cC |
| PLE-262 | CAMPSITE | 1994 | A | B | C | cB, cC | cB, cC |
| PLE-272 | CAMPSITE | 1986 | - | B | C | cB | cB |
| PLE-276d | CAMPSITE | 1991 | - | - | C | B | B |
| PLE-355 | CAMPSITE | 1987 | - | - | C | - | C |
| PLE-923 | CAMPSITE | 1998 | - | - | C | - | C |
| PLE-268 | CS-NOMAD | 1994 | - | B | C | cB | cB, cC |
| PLE-373 | GOL UNITED | 1994 | - | B | - | B | B |
| PLE-593 | GOL UNITED | 1994 | - | B | - | B | B |
| PLE-608 | GOL UNITED | 1996 | - | B | - | B | B |
| PLE-609 | GOL UNITED | 1994 | - | B | - | B | - |
| PLE-385 | KIBUMBU | 1987 | A | B | - | cB | cA, cB |
| PLE-388 | KIBUMBU | 1994 | - | - | - | - | - |
| PLE-389 | KIBUMBU | 1986 | - | B | C | cB, cC | B, C |
| PLE-916 | KIBUMBU | 1998 | - | B | - | - | - |
| PLE-342d | LOLINDO | 1987 | - | - | C | - | B |
| PLE-458 | LOLINDO | 1997 | - | B | - | - | - |
| PLE-597 | LOLINDO | 1994 | A | B | - | cB | cB |
| PLE-610 | LOLINDO | 1994 | - | B | C | - | - |
| PLE-336 | MAASI | 1987 | - | - | - | B | B |
| PLE-611 | MAASI | 1994 | - | B | - | B | B |
| PLE-612 | MAASI | 1994 | - | B | C | cB, cC | cB, cC |
| PLE-613 | MAASI | 1994 | - | B | - | - | - |
| PLE-614 | MAASI | 1994 | - | B | - | B | B |
| PLE-475d | MAASI KOPJE | 1986 | - | - | C | B | - |
| PLE-486 | MAASI KOPJE | 1984 | - | B | - | B | - |
| PLE-603 | MAASI KOPJE | 1994 | - | B | - | B | B |
| PLE-605 | MAASI KOPJE | 1994 | - | B | - | B | B |
| PLE-606 | MAASI KOPJE | 1994 | - | B | - | B | B |
| PLE-591 | NAABI | 1994 | - | B | C | B | B |
| PLE-598 | NAABI | 1994 | - | - | - | - | B |
| PLE-599 | NAABI | 1994 | - | B | - | B | B |
| PLE-601 | NAABI | 1994 | - | B | C | cB | cB, cC |
| PLE-513 | PLAINS | 1994 | A | B | - | cB | cA, cB |
| PLE-622 | PLAINS | 1994 | - | - | - | - | - |
| PLE-623 | PLAINS | 1994 | - | B | - | B | cB |
| PLE-624 | PLAINS | 1994 | - | - | C | C | cC |
| PLE-924 | PLAINS | 1998 | A | - | - | - | - |
| PLE-225 | SANGERE | 1987 | - | B | - | B | - |
| PLE-506 | SANGERE | 1993 | - | B | C | - | - |
| PLE-618 | SANGERE | 1994 | - | - | C | cB, cC | cB, cC |
| PLE-619d | SANGERE | 1994 | - | - | C | B | B |
| PLE-621 | SANGERE | 1994 | - | - | C | C | C |
| PLE-584 | SIMBA | 1994 | - | B | - | B | cB |
| PLE-630 | SIMBA | 1994 | - | B | - | B | B |
| PLE-650 | SIMBA | 1994 | - | B | - | - | B |
| PLE-626 | SIMBA EAST | 1994 | A | - | C | - | - |
| PLE-627 | SIMBA EAST | 1994 | - | B | - | - | - |
| PLE-631 | SIMBA EAST | 1994 | - | - | - | - | B |
| PLE-648 | SIMBA EAST | 1995 | - | B | C | cB, cC | cB, cC |
| PLE-592 | SIMBA WEST | 1994 | - | - | - | - | - |
| PLE-346 | TRANSECT | 1994 | - | B | C | cB | cB |
| PLE-349 | TRANSECT | 1987 | - | B | - | B | B |
| PLE-350 | TRANSECT | 1987 | A | B | C | cB | cA, cB, cC |
| PLE-569 | TRANSECT | 1994 | - | B | C | cB | cB, cC |
| PLE-633 | TRANSECT | 1994 | - | B | C | cB, cC | cC |
| PLE-634 | TRANSECT | 1994 | - | B | C | cB, cC | cC |
| PLE-636 | TRANSECT | 1994 | - | B | C | cC | cB cC |
| PLE-637 | TRANSECT | 1994 | - | B | C | cB | cB |
| PLE-913 | unknown | 1997 | - | - | - | - | A |
| PLE-914 | unknown | 1997 | - | - | - | C | C |
| PLE-915 | unknown | 1997 | - | B | - | B | B |
| PLE-941 | unknown | 1997 | - | B | C | - | - |
| PLE-942 | unknown | 1999 | - | B | C | B | B, C |
| PLE-943 | unknown | 1999 | - | B | C | - | - |
Sample collection and pride designation are by C. Packer and M. E. Roelke (see Fig. 1B).
Letters refer to PCR product with sequence that is defined as FIV-Ple A, FIV-Ple B, or FIV-Ple C based on clade position in Fig. 3. Bold type indicates a reference sequence (demonstrates consistent single strain infection). Dash indicates absence of a PCR product with the indicated primer pair.
Sequences obtained from cloned PCR products. Others are direct sequences.
Inconsistent results at different loci.
Anticontamination measures were taken at all steps of PCR amplification and post-PCR processing. Pre-PCR setup was performed in a laminar flow hood, DNA was added in a free-standing containment hood in a separate room, and all post-PCR manipulations were performed under a fume hood in a third room. All surfaces were washed with a 10% bleach solution, and each hood was exposed to UV light for 30 min before and after use. PCR tubes, rather than 96-well plates, were used and kept closed except when reagents and DNA were being added or aliquots were extracted for use. Tubes were only opened in their designated hoods, and, to avoid cross-contamination, tubes were never open simultaneously. Three negative controls were run with every reaction: a naïve cell line, an FIV-negative lion sample, and a water blank. Two positive controls of known sequence were also used in each PCR and sequencing reaction: one from an FIV-positive cell line and one from Ple-624. Finally, sequence analysis confirmed unique nucleotide sequences for most PCR products.
Phylogenetic analysis.
The data sets from pol-RT, pol-RNase, and gag were examined separately. Nucleotide sequences were compiled and aligned for subsequent phylogenetic analysis by ClustalX (48) and verified visually. Phylogenetic reconstruction was performed with the PAUP version 4.0 software (45) by the following methods: minimum evolution estimated by neighbor joining, maximum parsimony, and maximum likelihood. ModelTest (39) was used to estimate the optimal model of sequence evolution, and these settings were incorporated into subsequent analyses. Minimum evolution trees were constructed from models of substitution specified by ModelTest, with starting trees obtained by neighbor joining followed by application of a tree-bisection-reconnection (TBR) branch-swapping algorithm during a heuristic search for the optimal tree. Maximum parsimony analysis employed a heuristic search of starting trees obtained by stepwise addition followed by TBR. Maximum likelihood parameters specified by ModelTest selected for the general time-reversible model of substitution included empirical base frequencies and estimated rate matrix and corrected for among-site rate variation (gamma distribution). A bootstrap analysis using 1,000 iterations was performed with each method (45).
Amino acid distances were determined by using the PHYLIP program with the Dayhoff PAM matrix (11) for weighing probability of substitutions. These distances were subsequently imported into the PAUP program for phylogenetic analysis using a heuristic searching TBR branch-swapping algorithm. The ratio of nonsynonymous to synonymous substitutions (Ka/Ks) was calculated by Sequencer 6.1.0 (B. Kessing, personal communication) using the algorithm of Li (26) and Pamilo and Bianchi (37). Phylogenetic trees were rooted by using the published sequences from two of the most divergent strains of FIV-Fca (GenBank accession numbers M25381 and U11820) as outgroups.
Nucleotide sequence accession numbers.
The nucleotide sequences of pol-RT, pol-RNase, and gag have been deposited in the GenBank database under accession numbers AY549217 to AY549304, AY552614 to AY552683, and AY552684 to AY552748.
RESULTS
The large outbred Serengeti lion population consists of approximately 3,000 lions that display a very high frequency of FIV antibody-positive individuals. A recent analysis of 349 serum samples by a Western blot assay with FIV isolated from domestic cat (FIV-Fca), puma (FIV-Pco), and African lions (FIV-Ple) as target antigens revealed that 90% were FIV antibody positive (6, 18; J. L. Troyer et al., unpublished data). A previous phylogenetic study of 375 bp of the pol sequence suggested the existence of three distinct strains or subtypes of FIV-Ple among the Serengeti lions (6).
We have designed PCR primers specific for FIV-Ple A, FIV-Ple B, and FIV-Ple C strains within the same pol region, plus additional primer sequences to amplify the gag and pol-RNase regions of FIV (Fig. 1A). DNA from peripheral blood mononuclear cells collected from 68 lions was tested with PCR primers for all three proviral FIV genes by using strain-specific primers for FIV-Ple A, FIV-Ple B, and FIV-Ple C in the pol-RT region and universal primers for the pol-RNase and gag regions (Fig. 1A). The specimens, collected at different times from 1985 to 1999, were from lions selected to sample each of 13 lion prides as well as from nomadic males (wandering males not associated with prides) (Fig. 1B and Table 1).
Nucleotide sequences for each proviral gene amplified from the lions were aligned and analyzed for phylogenetic relationship by using minimum evolution, maximum parsimony, and maximum likelihood algorithms as described in Materials and Methods. We also present an amino acid alignment derived from computationally translated nucleotide sequences obtained from each lion by PCR primers to detect pol-RT (337 bp) (Fig. 2a) pol-RNase (730 bp) (Fig. 2b), and gag (444 bp) (Fig. 2c). Some sequences were direct PCR sequences; however, pol-RNase PCR products from 21 lions and gag PCR products from 22 lions were cloned to sample diversity within individual lions (Table 1 and Fig. 2 and 3). In Fig. 3, maximum likelihood phylogenetic trees are presented for nucleotide sequences obtained for each of the three viral genes.
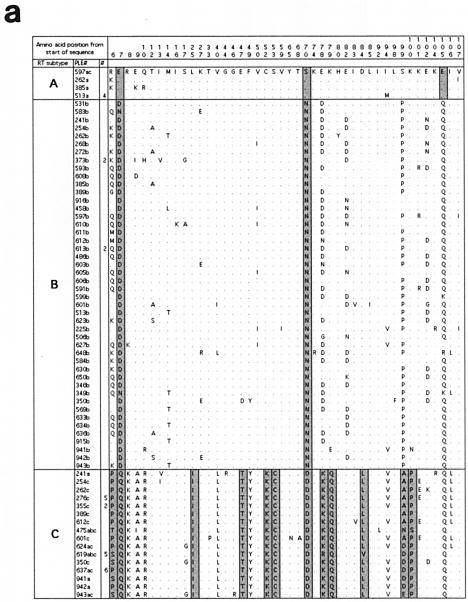
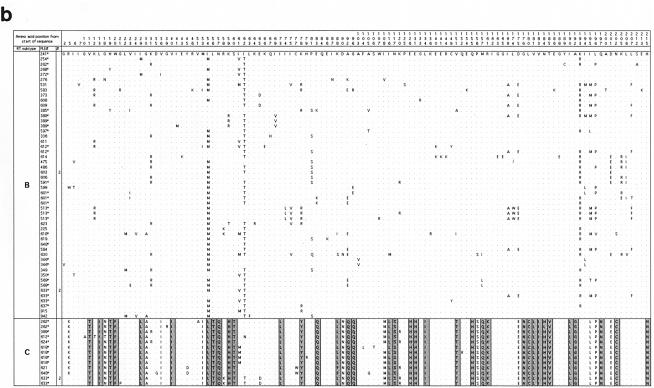
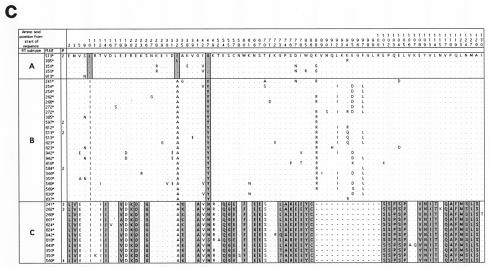
FIG. 2.
Alignments of the predicted amino acid translation products of FIV-Ple sequences from individual lions. Single-letter amino acid code was used. Only variable amino acids are included, and residues differing from the top sequence are shown. A dot designates identity with the top sequence. Dashed lines indicate gaps introduced to optimize the alignment. Sites that distinguish FIV-Ple RT-subtypes from each other are boxed, and fixed differences are shaded. In cases where more than one lion has identical FIV-Ple sequences, the number of lions (#) with that sequence is indicated. Cloned sequences are followed by an asterisk. (a) pol-RT translation products. The following lions had identical amino acid sequences: Ple-513, Ple-924, Ple-350, and Ple-626 (clade A); Ple-373 and Ple-609 (clade B); Ple-613 and Ple-614 (clade B); Ple-633 and Ple-637 (clade B); Ple-262 and Ple-610 (clade C); Ple-626, Ple-276, Ple-272, Ple-268, and Ple-923 (clade C); Ple-619, Ple-648, Ple-621, Ple-506, Ple-591, and Ple-618 (clade C); Ple-637, Ple-346, Ple-633, Ple-636, Ple-634, and Ple-569 (clade C); and Ple-355 and Ple-342 (clade C). (b) pol-RNase translation products. The following lions had identical amino acid sequences: Ple-603 and Ple-605 (clade B); Ple-633 and Ple-634 (clade B); and Ple-633, Ple-634, and Ple-636 (clade C). (c) gag translation products. The following lions had identical amino acid sequences: Ple-513 and Ple-913 (clade A); Ple-597 and Ple-601 (clade B); Ple-513 and Ple-623 (clade B); Ple-584 and Ple-648 (clade B); Ple-268, Ple-262, Ple-254, Ple-612, and Ple-618 (Clade C); and Ple-569, Ple-636, Ple-634, and Ple-633 (clade C).
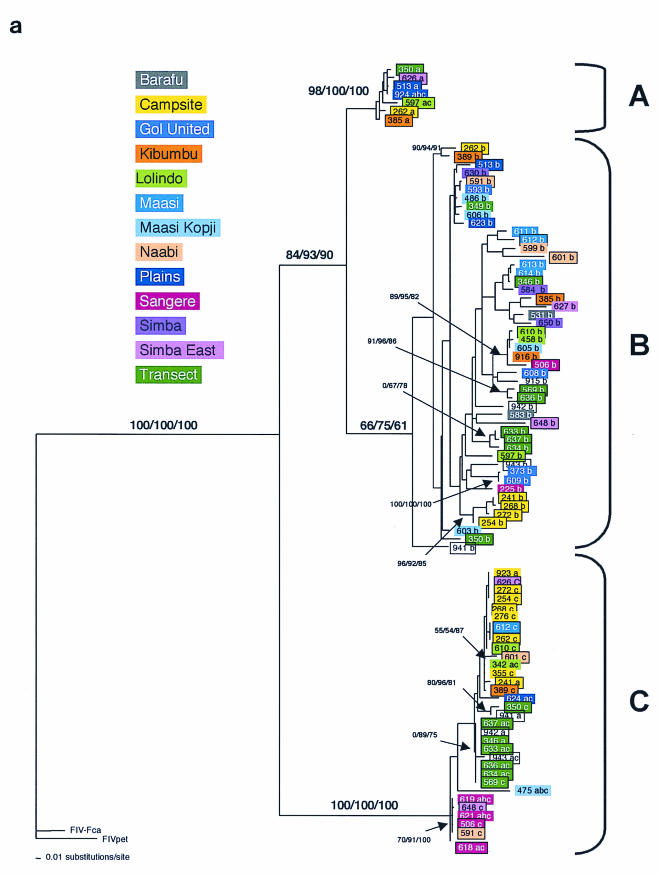
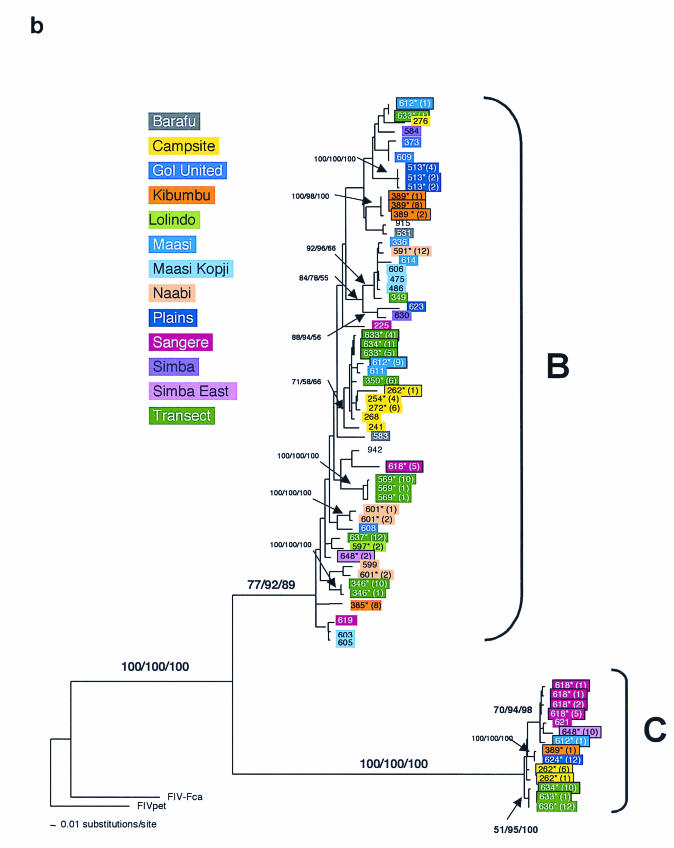
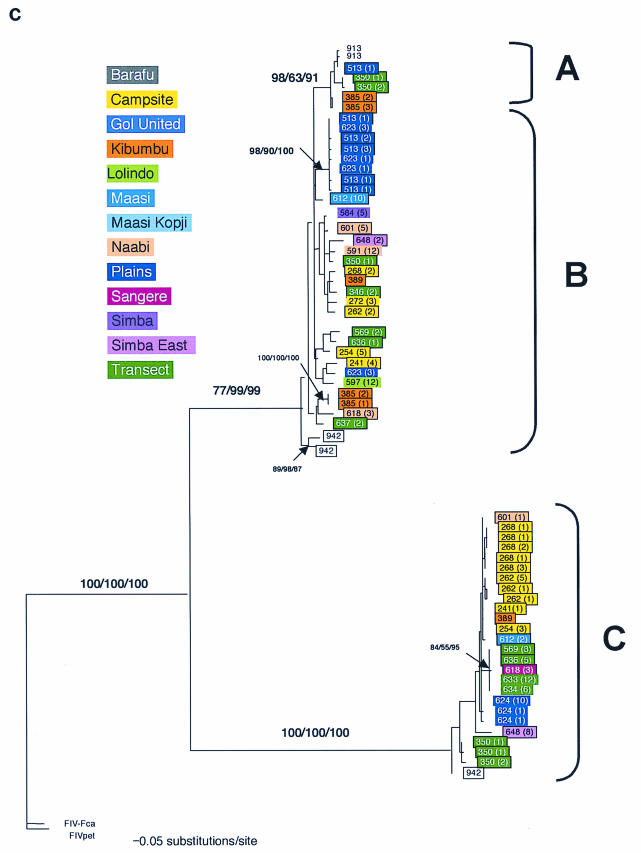
FIG.3.
Phylogenetic tree for FIV-Ple nucleotide sequences. Shown here in each case is the single maximum likelihood (ML) tree. Branch lengths represent percent sequence divergence based on the model of sequence evolution specified by ModelTest (39). Minimum evolution estimated by neighbor-joining (NJ) and maximum parsimony (MP) trees gave similar topologies, and bootstrap values are included at all nodes with bootstrap support of >70 (ML/NJ/MP). Taxa are designated with the source lion identification number. Boxed taxa indicate individuals with FIV sequences that fall into more than one clade. Cloned sequences are followed by an asterisk, and the number of clones with that sequence is given in parentheses. Pride affiliations are designated by color. (a) pol-RT, 337 bp. Maximum likelihood tree under the HKY+I+G model of sequence evolution with an estimated shape parameter of 0.6272, an estimated transition/transversion ratio of 4.0911, and an estimated proportion of invariant sites of 0.3750 (−ln likelihood = 3,978.9614). Taxa are followed by a lowercase letter indicating the internal subtype-specific primer used (a, b, or c) (Fig. 1A). In most instances, an RT-subtype “a” primer set produced an RT-subtype A sequence, RT-subtype “b” primers produced an RT-subtype B sequence, and “c” primers produced a C sequence. Reference sequences for the three previously identified FIV-Ple subtypes are as follows: A (Ple-241 and Ple-385), B (Ple-254 and Ple-272), and C (Ple-355 and Ple-389). (b) pol-RNase, 730 bp. The maximum likelihood tree under the GTR+I+G model of sequence evolution has an estimated shape parameter of 0.4274 and an estimated substitution matrix as follows: A/C = 2.0073, A/G = 7.902, A/T = 1.4491, C/G = 2.6080, C/T = 14.7031, and an estimated proportion of invariant sites of 0.1153 (−ln likelihood = 6,951.6733). (c) gag cloned sequences, 444 bp. Boxed taxa with no numbers in parentheses are the result of different sequences obtained at different temperatures in a gradient. The maximum likelihood tree under the GTR+G model of sequence evolution has an estimated shape parameter of 0.3584 and an estimated substitution matrix as follows: A/C = 2.3398, A/G = 9.8759, A/T = 2.6700, C/G = 2.8285, C/T = 16.5121 (−ln likelihood = 3,582.6128). Deletion of 13 amino acid residues in FIV-Ple C is excluded in this analysis (see text).
A proviral FIV sequence from at least one gene region was obtained from 95.6% of the individuals examined. However, all primer sets had a significant failure rate (12% for strain-specific pol-RT primers, 29% for universal pol-RNase primers, and 25% for universal pol-RT primers). This failure rate is most likely due to an interaction between viral load and the extreme sequence variation of FIV, even in relatively conserved primer sites. Nested PCR may contribute to the failure rate, since four primers are required to retain sufficient sequence similarity for amplification to occur. The three individuals that did not amplify from any gene region may be false positives or may have extremely low viral load. Despite PCR dropout, sequences were obtained from all three gene regions for 41 individuals.
The results of the pol-RT gene phylogenetic analysis reveal appreciable diversity within and between the A, B, and C clade strains (or subtypes) amplified from different individual lions (Fig. 3a). The three FIV-Ple strains are widely dispersed among the different prides. Sixty lions produced FIV-Ple sequences based on the pol-RT region, and of these, 26 (43%) carried two or three FIV-Ple subtypes (summarized in Table 1 under pol-RT primers). Of 34 lions infected with only one strain, 1 had FIV-Ple A, 24 had FIV-Ple B, and 9 had FIV-Ple C (Table 1).
The phylogenetic analysis (Fig. 3a) also shows that although the FIV-Ple strain-specific PCR primers generally amplify the predicted viral clade, there are several exceptions, indicating the imperfect clade specificity of these primers under the amplification conditions used. For example, the lion Ple-924 pol-RT strain A sequence was amplified by primers specific for A, B, or C (Fig. 3a, lowercase letters a, b, and c). Similarly, the pol-RT clade FIV-Ple C sequences in Fig. 3a were often obtained by primers designed to be specific for FIV-Ple A, FIV-Ple B, or FIV-Ple C. By contrast, all pol-RT clade B sequences came only from clade B-specific primers (Fig. 3a). Because of the primer overlap, FIV-Ple strain designations (Fig. 3 and Table 1) were based on sequence phylogenetic placement rather than starting PCR primer. Finally, the genetic distance between the three clades was not the same, with clade C showing significantly greater genetic distance from clade A or B than the distance between clade A and B (Fig. 3a).
The analysis of the pol-RNase region also showed considerable diversity among FIV-Ple genome sequences (Fig. 3b). Two clades were detected that are imputed to represent FIV-Ple B and FIV-Ple C. This conclusion is reached by identifying individual lions that retain a single clade based on the pol-RT sequence and matching the sequence derived for pol-RNase. Thus, two lions, Ple-621 and Ple-624, have only FIV-Ple C based on the pol-RT sequence (Fig. 3a and Table 1), and both of these individuals show a single sequence that falls in the C clade of pol-RNase (Fig. 3b). Similarly, 19 lions that are singly infected with clade B based on pol-RT results (Fig. 3a and Table 1) are all in the imputed FIV-Ple B clade of the pol-RNase region (Fig. 3b and Table 1). Further, lions infected by both FIV-Ple B and FIV-Ple C according to the pol-RNase sequence (Ple-262, Ple-389, Ple-612, Ple-633, Ple-634, and Ple-648) are also doubly infected with FIV-Ple B and FIV-Ple C by pol-RT assessment (Table 1). There is no recovery of FIV-Ple A with pol-RNase-specific PCR primers since lions infected with strains FIV-Ple A and FIV-Ple B or FIV-Ple C (Table 1, pol-RT region) reveal only B or C pol-RNase clades. This failure to resolve FIV-Ple A likely reflects a mutational divergence of pol-RNase primer binding sites in strain A virion genomes but may also indicate that FIV-Ple A is a truncated or recombinant type in the pol-RNase region.
The analysis of gag sequences also recapitulates the three FIV-Ple clades which can be imputed as A, B, and C by similar lion-FIV strain association (Fig. 3c). Thus, lions that retain only FIV-Ple B in the pol-RT and pol-RNase regions (Ple-349, Ple-373, Ple-583, Ple-584, Ple-593, Ple-599, Ple-603, Ple-605, Ple-606, Ple-608, Ple-611, Ple-614, Ple-623, Ple-630, Ple-915) all show a single FIV-Ple B in gag sequences as well. The same concordance for pol-RT and gag sequences is true among lions singly infected with FIV-Ple C (Ple-355, Ple-621, Ple-624 and Ple-923), those coinfected with FIV-Ple A and FIV-Ple B (Ple-385 and Ple-513), those coinfected with FIV-Ple B and FIV-Ple C (Ple-241, Ple-254, Ple-268, Ple-389, Ple-569, Ple-601, Ple-612, Ple-636, Ple-648, and Ple-942), and one lion infected with all three strains (Ple 350) (Table 1 and Fig. 3c).
With four exceptions (Ple-276, Ple-342, Ple-475, and Ple-619), there is a striking concordance of clade disposition in the 40 lions sampled, suggesting that strain or subtype recognition generally extends across the three FIV-Ple gene regions. These four exceptions retain the strain C pol-RT sequence but strain B sequences in the pol-RNase and/or gag regions (Table 1 and Fig. 3a). These lions may contain rare recombinants or, alternatively, the strain discrepancies may be a result of disparate primer affinities for the different strains, leading to inconsistent PCR results.
The infection of lions with multiple FIV-Ple strains is apparent in all three gene regions (Table 1) and is most clearly illustrated by the pol-RT sequencing results. Forty-three percent of the pol-RT PCR-positive lions were infected with more than one FIV-Ple strain, of which two individuals (Ple-262 and Ple-350) were infected with all three subtypes (Table 1). In this sample set, FIV-Ple B was most common (infecting 82% of PCR-positive lions), while FIV-Ple A was rare (infecting only 12% of PCR-positive lions) and FIV-Ple C had intermediate prevalence (infecting 53% of PCR-positive lions). Based on the frequency of each FIV-Ple strain in the population, observed coinfections (2 individuals with all three strains, 3 with strains A and B, 20 with strains B and C, and 1 with strains A and C) were not significantly different by Chi-square analysis than expected values (0.995 > P > 0.975).
Demonstrating coinfection was more difficult in the pol-RNase and gag regions because of the use of universal primers. In most cases, one subtype was amplified preferentially over the others. This result could be due either to differences in viral load between the subtypes present in the individual or to differences in primer affinity to the different strains. Direct sequencing reflected the best-amplified FIV-Ple subtype, and cloning was needed to isolate secondary FIV-Ple subtypes. This preferential amplification also resulted in a skewed distribution of cloned sequences, where the majority of clones from an individual were one subtype, and only one or two clones contained sequences from the second subtype (Fig. 3b and c). This effect was particularly pronounced in the longer pol-RNase amplification product, which was more difficult both to amplify and to clone. In this region, seven individuals were confirmed to have multiple infections. Biased amplification also explains why the phenomenon of coinfection was not observed in earlier studies, where only four clones from each of 15 animals in this population were sequenced (6). Despite these difficulties, coinfection was confirmed at more than one gene region for 17 lions (Table 1).
Genetic divergence between FIV-Ple strains.
The level of phylogenetic distinctiveness among clades A, B, and C is apparent in several comparative aspects of the three FIV-Ple gene segments. First, there is very high bootstrap support in the separation of the three clades in the pol-RT and gag regions (Fig. 3a and c) and for FIV-Ple B versus FIV-Ple C in the pol-RNase region (Fig. 3b). The C strain is the basal lineage in all three phylogenies, with FIV-Ple A being 1.5 to 4 times closer to FIV-Ple B sequences than either is to FIV-Ple C, based on the phylogenetic analyses of FIV genes (Fig. 3a and c; Tables 2 to 4).
TABLE 2.
Mean percent nucleotide and amino acid sequence differences between FIV-Ple strains in the pol-RT gene region (337 bp)
| Strain (no. of sequences) | Mean percent (range) sequence differences of viral strains in the pol-RT regiona
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FIV-Ple A
|
FIV-Ple B
|
FIV-Ple C
|
FIV-Fca
|
FIV-Pco
|
||||||
| aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | |
| FIV-Ple A (7) | 2 (0-4) | 3 (0-4) | 19 (15-22) | 28 (24-31) | 32 (30-34) | 30 (28-32) | ||||
| FIV-Ple B (31) | 7 (1-13) | 4 (0-12) | 10 (1-17) | 27 (18-34) | 33 (29-39) | 28 (28-24) | ||||
| FIV-Ple C (26) | 24 (18-27) | 24 (17-31) | 4 (0-8) | 5 (0-9) | 32 (30-35) | 35 (31-35) | ||||
| FIV-Fca (2) | 31 (29-33) | 32 (28-36) | 38 (31-45) | 7 | 13.7 | 29 (28-30) | ||||
| FIV-Pco (1) | 27 (25-29) | 28 (23-32) | 34 (33-37) | 26 (23-28) | ||||||
Distances for FIV-Fca are calculated across subtypes A and D of FIV-Fca. FIV-Pco distances are based on PLV-14. aa, amino acids; nt, nucleotides.
TABLE 4.
Mean percent nucleotide and amino acid sequence differences between FIV-Ple strains in the gag gene region (444 bp)
| Strain (no. of sequences) | Mean percent (range) sequence differences of viral strains in the gag regiona
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FIV-Ple A
|
FIV-Ple B
|
FIV-Ple C
|
FIV-Fca
|
FIV-Pco
|
||||||
| aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | |
| FIV-Ple A (7) | 3 (1-5) | 4 (1-5) | 11 (8-15) | 46 (44-51) | 44 (42-46) | 69 (67-71) | ||||
| FIV-Ple B (31) | 7 (3-11) | 5 (0-12) | 11 (0-28) | 35 (29-45) | 33 (27-40) | 68 (63-72) | ||||
| FIV-Ple C (26) | 37 (34-41) | 53 (50-59) | 2 (0-5) | 4 (0-7) | 41 (37-44) | 65 (62-71) | ||||
| FIV-Fca (2) | 60 (57-62) | 61 (58-68) | 62 (60-67) | 11 | 23 | 69 (66-71) | ||||
| FIV-Pco (1) | 93 (91-94) | 91 (85-97) | 94 (91-97) | 97 (94-99) | ||||||
Distances for FIV-Fca are calculated across subtypes A and D of FIV-Fca. FIV-Pco distances are based on PLV-14. aa, amino acids; nt, nucleotides.
The mean percent difference between FIV-Ple clade sequences in terms of both nucleotide and amino acid pairwise alignments is presented in Tables 2 to 4. For each gene the C-A or C-B strain divergence is nearly as great as the genetic distances that occur between FIV sequences from any lion versus domestic cat (or puma) FIV gene sequences. This is true for both nucleotide and amino acid comparisons for each gene. For example, in the gag region, FIV-Ple A and FIV-Ple B differ on average by 11% (nucleotide) and 7% (amino acid). Nucleotide differences from FIV-Ple A to FIV-Ple C and FIV-Ple B to FIV-Ple C are 46 and 35%, respectively, which is similar to the difference seen between FIV-Ple and FIV-Fca (33 to 44%). Amino acid differences from FIV-Ple A to FIV-Ple C (37%) and FIV-Ple B to FIV-Ple C (53%) are also much higher, approaching those from FIV-Ple to FIV-Fca (60 to 62%). Similar trends are seen in the pol-RT and pol-RNase genes. Results from pol-RT sequencing confirm those seen earlier with a smaller data set (6). However, both the pol-RNase and gag sequences reveal a greater genetic distance between FIV-Ple B and FIV-Ple C than that seen with the pol-RT sequence. Conversely, the gag sequence shows a much smaller distance between FIV-Ple A and FIV-Ple B than that seen with the pol-RT sequence.
The striking divergence among FIV-Ple clades is also apparent when we examine the number of fixed amino acid substitutions between clades (Fig. 2). Strain A differs from strain B by three fixed differences in the pol-RT region, confirming earlier results (6), and three fixed differences in the gag region (strain A is unresolved in the pol-RNase region) (Fig. 2b). By contrast, there are 18 and 45 fixed amino acid differences between FIV-Ple A and FIV-Ple C in the pol-RT and gag sequences, respectively, and 12, 37, and 39 fixed amino acid differences in the pol-RT, pol-RNase, and gag sequences between strains B and C. Five of the differences in the pol-RT sequence (at positions 47, 78, 84, 99, and 100) (Fig. 2a) represent nonconservative changes in amino acid charge or polarity. Clearly, the C strain is basal, older, and reflective of the greatest evolutionary divergence of the three FIV-Ple strains.
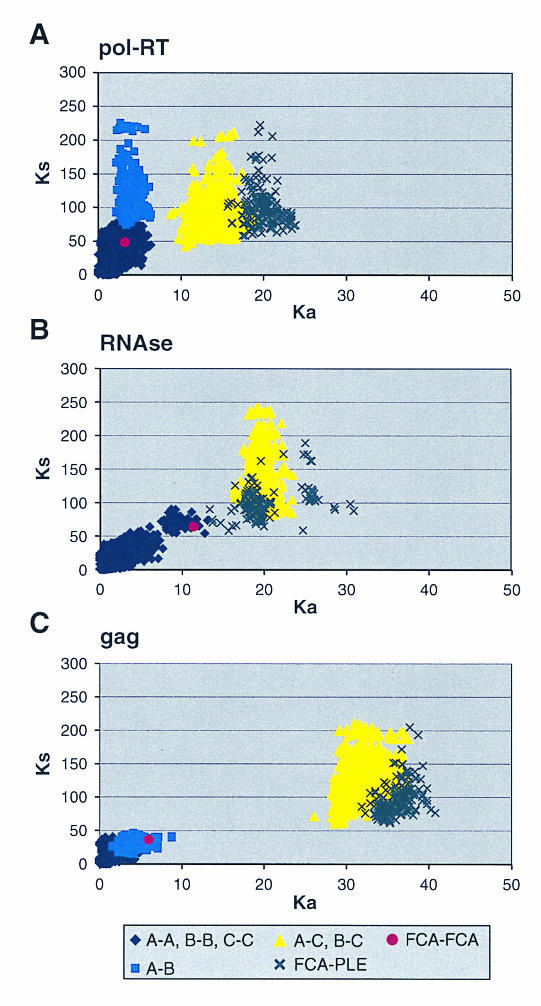
The breadth of evolutionary divergence is also illustrated by comparing the extent of synonymous and nonsynonymous nucleotide substitutions apparent within the FIV-Ple strains, between strains, and between the three FIV genes. In Fig. 4, the plot of Ka/Ks distances illustrates the quantitative distinctions that define sequence divergence. The pol-RT analysis illustrates the patterns seen with all three genes (Fig. 4A). Within clades a swarm of sequence comparisons within each FIV-Ple strain shows small distances, while sequence comparisons of FIV-Ple A to FIV-Ple B show higher synonymous substitution but less nonsynonymous divergence. The comparisons of FIV-Ple C to FIV-Ple A or FIV-Ple C to FIV-Ple B show a substantial increase in nonsynonymous amino acid-altering substitution, again approaching the divergence of FIV comparisons between different cat species.
FIG. 4.
Synonymous and nonsynonymous distances between pairs of FIV sequences. Each point represents one pairwise comparison. Synonymous and nonsynonymous distances were calculated by the algorithm of Li (26) and Pamilo and Bianchi (37). (A) pol-RT. (B) pol-RNase. (C) gag.
In contrast to the patterns of divergence observed in the pol-RT gene sequence, comparison of nonsynonymous and synonymous changes in the gag region indicates that a greater proportion of nucleotide substitutions encode amino acid changes for subtype A and subtype B than for subtype C. FIV-Ple A and FIV-Ple B are distinguished from FIV-Ple C by a shared 39-bp (13-amino-acid) insertion (position 88 to 100) (Fig. 2c), as well as by multiple fixed sequence differences (Fig. 2c). Seven of the polymorphic amino acids seen within the FIV-Ple B sequences and three polymorphic sites in FIV-Ple A occur within the shared insertion (Fig. 3c), partially explaining the lower diversity in FIV-Ple C, which lacks these amino acids. The 30 residues following the insertion, representing the transition to the core protein (p26) region of gag, are highly conserved within each strain but quite divergent between FIV-Ple C and the other two strains (15 fixed differences, 7 of which represent changes in either charge or polarity). As in pol-RT, the majority of changes occur between FIV-Ple C and the other two strains (a total of 32 fixed differences) with only 2 fixed differences between FIV-Ple A and FIV-Ple B.
Population dynamics of FIV-Ple subtypes.
Diverse patterns of FIV transmission among lions are evident within the viral phylogenies. First, geographic concordance is demonstrated by monophyletic FIV lineages within the Campsite pride individuals. Specifically, eight lions from the Campsite pride are infected with a pol-RT strain C that forms a monophyletic lineage within the pol-RT phylogeny (Fig. 3a). In addition, four of eight Campsite lions are coinfected with pol-RT B sequences that form a common lineage, suggesting a shared transmission event for both FIV-Ple B and FIV-Ple C within this pride. As observed in the pol-RT phylogeny, the clustering of these five Campsite lions (Ple-241, Ple-254, Ple-268, Ple-272, and Ple-262) within clade B is recapitulated by the pol-RNase sequences (Fig. 3b).
Second, the phylogenies show that lions with the most closely related FIV sequences may also be related to each other. For example, three sibling pairs, Ple-619 and Ple-621 (Sangere pride), Ple-633 and Ple-634 (Transect pride), and Ple-613 and Ple-614 (Maasi pride), each share nearly identical pol-RT sequences (Fig. 3a), indicating a common origin of the infecting strains. A similar level of identity is found in pol-RT, pol-RNase, and gag sequences of the mother-daughter pair Ple-373 and Ple-609 (Gol United pride) (Fig. 3a and b; data not shown for gag). In contrast, we also observed closely related FIV strains in individuals from different prides. For example, the FIV-Ple C sequences of Ple-621 and Ple-619 from the Sangere pride also group closely with those of Ple-506 and Ple-618, also from the Sangere pride, Ple-648, a lion from Simba East pride, and Ple-591, a lion from the Naabi pride. The close relationship of the Ple-621 and Ple-648 clade C sequences is maintained in all three gene regions (Fig. 3a and b; data not shown for gag). All three strains are observed across multiple prides, and FIV-Ple from all three divergent FIV-Ple strains can be found in 6 of the 13 prides (Campsite, Lolindo, Kibumbu, Plains, Simba East, and Transect).
DISCUSSION
Lions within the Serengeti National Park, Tanzania, 90% of which are infected with FIV, offer a unique opportunity to examine patterns of lentiviral evolution, transmission, and dissemination in the wild. Analyses of three gene regions of the FIV-Ple genome indicate that distinct FIV-Ple strains circulate freely within the population. Moreover, an unusually high number of lions (43%) are coinfected with these distinct subtypes.
Phylogenetic analyses of FIV-Ple sequences within this population provide evidence for the transmission of infection both between and within prides. Even with small within-pride sample sizes, all three FIV-Ple strains are observed across multiple prides, and FIV-Ple A, FIV-Ple B, and FIV-Ple C are all circulating in at least 6 of the 13 prides (Campsite, Lolindo, Kibumbu, Plains, Simba East, and Transect). Only the Campsite pride showed monophyletic clustering of FIV-Ple sequences, and this was seen in clades B and C. However, FIV-Ple sequences from other prides were also included with those of the Campsite lions. In three cases, sibling pairs had the most closely related FIV-Ple sequences, either reflecting transmission between cubs or infection from a common third source. In addition, one mother-offspring pair was observed to have closely related FIV-Ple sequences, suggesting the possibility of vertical transmission, although infection from a common third source cannot be ruled out.
Among pol-RT PCR-positive lions in this sample set, FIV-Ple A was rare (12%), FIV-Ple B was most common (82%), and FIV-Ple C had intermediate prevalence (53%). FIV-Ple B strains had the most nucleotide diversity in all three gene regions: 10% in pol-RT (Table 2), 8% in pol-RNase (Table 3), and 11% in gag (Table 4). FIV-Ple subtype A can be identified in two gene regions, pol-RT and gag, and has less nucleotide and amino acid diversity than the other two strains (Tables 2 and 4). Given that subtype A is most distinct from subtype B in the pol-RT region, fairly similar to subtype B in the gag region, not found with these primers in the pol-RNase region, and only appears as a coinfection with FIV-Ple B and/or FIV-Ple C, three possibilities for its origin exist. It is possible that FIV-Ple A is a recombinant form, a truncated form, or merely a monophyletic clade within the highly diverse subtype B. Full-length sequences as well as sequences from the more variable env region (not yet available for FIV-Ple) will help to clarify subtype designations and define the levels of strain divergence.
TABLE 3.
Mean percent nucleotide and amino acid sequence differences between FIV-Ple strains in the pol-RNase gene region (730 bp)
| Strain (no. of sequences) | Mean percent (range) sequence differences of viral strains in the pol-RNase regiona
|
|||||||
|---|---|---|---|---|---|---|---|---|
| FIV-Ple B
|
FIV-Ple C
|
FIV-Fca
|
FIV-Pco
|
|||||
| aa | nt | aa | nt | aa | nt | aa | nt | |
| FIV-Ple B (55) | 4 (0-12) | 8 (0-14) | 43 (36-49) | 43 (39-48) | 54 (43-67) | |||
| FIV-Ple C (14) | 35 (28-45) | 4 (0-10) | 4 (0-14) | 47 (44-51) | 61 (58-65) | |||
| FIV-Fca (2) | 30 (18-27) | 47 (43-59) | 20 | 15 | 56 (55-57) | |||
| FIV-Pco (1) | 45 (38-53) | 51 (47-54) | 51 | |||||
Distances for FIV-Fca are calculated across subtypes A and D of FIV-Fca. FIV-Pco distances are based on PLV-14. aa, amino acids; nt, nucleotides.
Observed patterns of nucleotide and amino acid divergence suggest different natural histories among FIV-Ple strains despite concordant distributions within Serengeti lions. By all measures, FIV-Ple C is most divergent, as differences between FIV-Ple C and FIV-Ple A or FIV-Ple B approach those seen between FIV sequences isolated from different species. These comparisons, affirmed by comparisons of synonymous and nonsynonymous nucleotide substitutions within and between strains, illustrate the stepwise evolution of viral strains, with synonymous substitution becoming saturated or “blinking” earlier within strains, while the slower time-dependent accumulation of amino acid-altering variants continues to increase to a maximum level seen in FIV isolated from separate species (7). The simplest interpretation of this pattern is that the three FIV-Ple strains likely derive from separate lion populations, although it cannot be excluded that they originally evolved within a distinct African cat species (e.g., leopard or cheetah) also known to harbor their own species-specific FIV strain (5; J. L. Troyer et al., unpublished data) to emerge recently in the Serengeti lions.
In the Serengeti lions, infection with one FIV-Ple subtype may not be protective against secondary infection with a different FIV-Ple subtype (i.e., the probability of coinfection with multiple strains is directly correlated with the product of the frequencies of those strains in the population). This data set does not include time point data that would confirm that superinfection, rather than simultaneous coinfection, is occurring. However, many pairs of lions share a common sequence for one subtype but have quite divergent sequences in another subtype, suggesting that distinct transmission events have taken place. These include lions with similar FIV-Ple C sequences but having extremely distinct FIV-Ple B sequences for pol-RT: Ple-942 and Ple-943, Ple-350 and Ple-941, and Ple-241 and Ple-389 (Fig. 3a). Further, Ple-262 and Ple 626 share similar FIV-Ple A and FIV-Ple C sequences, but Ple-262 has a unique FIV-Ple B sequence not found in Ple-626, raising the prospect of superinfection of Ple-262.
Multiple infection with divergent FIV strains is not unprecedented: coinfection with highly divergent strains of FIV-Pco has been demonstrated in two free-ranging pumas (8), and FIV-Fca subtypes A and B have been shown to coinfect domestic cats in both wild (24) and laboratory (20, 32) settings. However, the high incidence of coinfection seen in this population of lions is unprecedented, with potential implications for patterns of HIV type 1 (HIV-1) strain dispersal. Infection with multiple HIV-1 subtypes, once considered to be a rare occurrence (41, 52), is now clearly established as a common, albeit low-frequency, event (21, 41). In addition, coinfections with HIV-1 and HIV-2 are reported in surprisingly high frequency in Guinea-Bissau and Cote d'Ivoire and in Brazil (1, 15, 38).
The high genetic diversity between FIV-Ple subtypes (19 to 28% in the conserved pol-RT region, which is similar to that seen between HIV-1 and HIV-2 (1, 6), combined with 90% FIV-Ple seroprevalence in the population, may relate to the high incidence of multiple infection in this population (43%). For superinfection to occur, genetic divergence of the second viral strain must allow it to evade the host immune responses mounted against an initial infection. Although it is not clear what genetic changes allow this evasion, studies of coinfection and superinfection in wild populations will help define the genetic divergence limitations for effective HIV inoculations in human populations.
Genetic diversity is an acknowledged barrier to vaccine development for HIV (21, 31, 34), and it has been suggested that vaccination with one HIV subtype will not protect against infection with another subtype (13, 17, 31). This parallels laboratory vaccine trials for FIV-Fca and SIV and HIV vaccine trials in simian models, where vaccines may be successful against homologous challenge but ineffective against heterologous challenges. For example, in domestic cat a single subtype vaccine is protective against homologous challenge but ineffective against heterologous challenge with a different subtype (19). Additionally, a dual-subtype vaccine was completely protective against homologous challenge but only partially protective against heterologous challenge (40). In simian models, vaccine results have varied from partially protective to ineffective against heterologous HIV and SIV challenges (13, 17, 22).
A possible outcome of coinfection is recombination between subtypes. HIV studies show that recombination between divergent viral subtypes occurs frequently and contributes significantly to viral diversity and to increased virulence (3, 12, 21, 41, 43, 53, 57). Eleven HIV-1 circulating recombinant forms have been defined, and in some countries these contribute significantly to the AIDS epidemic (1, 2, 3, 10, 14, 23, 29, 41, 46, 47, 49, 50, 56). While we present no data here that directly addresses recombination in FIV-Ple, there are indications that it occurs. In four lions (Ple-276, Ple-342, Ple-475, and Ple-619), results from gag and pol-RNase sequencing do not correspond to those seen in pol-RT sequences, suggesting that these genes may harbor a circulating recombinant form of FIV-Ple. It is also possible that subtype A is itself a recombinant form.
While the effect of FIV-Ple infection on the health of Serengeti lions has not yet been determined, the high sequence divergence between FIV-Ple strains indicates an ancient association between the virus and its host species that likely predates the human-HIV association (7, 8). Thus, it is hypothesized that coevolution between lion immune systems and FIV-Ple has led to partial adaptation between the virus and its host (6, 7). Whether this adaptation is complete for each of the circulating subtypes is, as yet, unknown. The different patterns of nucleotide and amino acid divergence between FIV-Ple strains, the multiple infection status of several lions within the population, and the ability of these viruses to transmit freely between prides, suggest a dynamic evolutionary milieu for FIV-Ple and its host species.
Acknowledgments
We are grateful to Warren Johnson for critical reading and discussion of the manuscript, Stanley Cevario for technical assistance, and Jan Martenson for help and advice on many aspects of this project. Samples were collected in full compliance with specific federal permits (CITES; Endangered and Threatened Species) issued to the National Cancer Institute, principal investigator S. J. O'Brien, by the U.S. Fish and Wildlife Service of the Department of the Interior.
This study has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract number NO1-CO-12400.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the U.S. Government.
REFERENCES
- 1.Andersson, S., H. Norrgren, F. Dias, G. Biberfeld, and J. Albert. 1999. Molecular characterization of human immunodeficiency virus (HIV)-1 and -2 in individuals from Guinea-Bissau with single or dual infections: predominance of a distinct HIV-1 subtype A/G recombinant in West Africa. Virology 262:312-320. [DOI] [PubMed] [Google Scholar]
- 2.Balrich-Rubio, E., S. Anagonou, K. Stirrups, E. Lafia, D. Candotti, H. Lee, and J. P. Allain. 2001. A complex human immunodeficiency virus type 1 A/G/J recombinant virus isolated from a seronegative patient with AIDS from Benin, West Africa. J. Gen. Virol. 82:1095-1106. [DOI] [PubMed] [Google Scholar]
- 3.Barlow, K. L., I. D. Tatt, P. A. Cane, D. Pillay, and J. P. Clewley. 2001. Recombinant strains of HIV type 1 in the United Kingdom. AIDS Res. Hum. Retrovir. 17:467-474. [DOI] [PubMed] [Google Scholar]
- 4.Barr, M. C., L. Zou, F. Long, W. A. Hoose, and R. J. Avery. 1997. Proviral organization and sequence analysis of feline immunodeficiency virus isolated from a Pallas' cat. Virology 228:84-91. [DOI] [PubMed] [Google Scholar]
- 5.Brown, E. W., S. Miththapala, and S. J. O'Brien. 1993. Prevalence of exposure to feline immunodeficiency virus is exotic felid species. J. Zool. Wildl. Med. 20:265-272. [Google Scholar]
- 6.Brown, E. W., N. Yuhki, C. Packer, and S. J. O'Brien. 1994. A lion lentivirus related to feline immunodeficiency virus: epidemiologic and phylogenetic aspects. J. Virol. 68:5953-5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpenter, M. A., and S. J. O'Brien. 1995. Coadaptation and immunodeficiency virus: lessons from the Felidae. Curr. Opin. Genet. Dev. 5:739-745. [DOI] [PubMed] [Google Scholar]
- 8.Carpenter, M. A., E. W. Brown, M. Culver, W. E. Johnson, J. Pecon Slattery, D. Brousset, and S. J. O'Brien. 1996. Genetic and phylogenetic divergence of feline immunodeficiency virus in the puma (Puma concolor). J. Virol. 70:6682-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpenter, M. A., E. W. Brown, D. W. MacDonald, and S. J. O'Brien. 1998. Phylogeographic patterns of feline immunodeficiency virus genetic diversity in the domestic cat. Virology 251:234-243. [DOI] [PubMed] [Google Scholar]
- 10.Carr, J. K., J. N. Torimiro, N. D. Wolfe, M. N. Eitel, B. Kim, E. Sanders-Buell, L. L. Jagodzinski, D. Gotte, D. S. Burke, D. L. Birx, and F. E. McCutchan. 2001. The AG recombinant IbNG and novel strains of group M HIV-1 are common in Cameroon. Virology 286:168-181. [DOI] [PubMed] [Google Scholar]
- 11.Dayhoff, M. O. 1978. Atlas of protein sequence and structure, vol. 5, suppl. 3. National Biomedical Research Foundation, Silver Spring, Md.
- 12.Diaz, R. S., E. C. Sabino, A. Mayer, J. W. Mosley, M. P. Busch, and the Transfusion Safety Study Group. 1995. Dual human immunodeficiency virus type 1 infection and recombination in a dually exposed transfusion recipient. J. Virol. 69:3273-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuller, D. H., P. A. Rajakumar, L. A. Wilson, A. M. Trichel, J. T. Fuller, T. Shipley, M. S. Wu, K. Weis, C. R. Rinaldo, J. R. Haynes, and M. Murphy-Corb. 2002. Induction of mucosal protection against primary, heterologous simian immunodeficiency virus by a DNA vaccine. J. Virol. 76:3309-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, F., D. L. Robertson, S. G. Morrison, H. Hui, S. Craig, J. Decker, P. N. Fultz, M. Girard, G. M. Shaw, B. H. Hahn, and P. M. Sharp. 1996. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J. Virol. 70:7013-7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.George, J. R., C.-Y. Ou, B. Parekh, K. Brattegaard, V. Brown, E. Boateng, and K. M. Decock. 1992. Prevalence of HIV-1 and HIV-2 mixed infections in Côte d'Ivoire. Lancet 340:337-339. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert, D. A., C. Packer, A. E. Pusey, J. C. Stephens, and S. J. O'Brien. 1991. Analytical DNA fingerprinting in lions: parentage, genetic diversity, and kinship. J. Hered. 82:378-386. [DOI] [PubMed] [Google Scholar]
- 17.Girard, M., L. Yue, F. Barre-Sinoussi, E. van der Ryst, B. Meignier, E. Muchmore, and P. N. Fultz. 1996. Failure of a human immunodeficiency virus type 1 (HIV-1) subtype B-derived vaccine to prevent infection of chimpanzees by an HIV-1 subtype E strain. J. Virol. 70:8229-8233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann-Lehmann, R., D. Fehr, M. Grob, M. Elgizoli, C. Packer, J. S. Martenson, S. J. O'Brien, and H. Lutz. 1996. Prevalence of antibodies to feline parvovirus, calcivirus, herpesvirus, coronavirus, and immunodeficiency virus and of feline leukemia virus antigen and the interrelationship of these viral infections in free-ranging lions in East Africa. Clin. Diagn. Lab. Immunol. 3:554-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hohdatsu, T., S. Okada, K. Motokawa, C. Aizawa, J. K. Yamamoto, and H. Koyama. 1997. Effect of dual-subtype vaccine against feline immunodeficiency virus infection. Vet. Microbiol. 58:155-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hohdatsu, T., T. Sasagawa, A. Yamazaki, K. Motokawa, H. Kuusuhara, T., Kaneshima, and H. Koyama. 2002. CD8+ T cells from feline immunodeficiency virus (FIV) infected cats suppress exogenous FIV replication of their peripheral blood mononuclear cells in vitro. Arch. Virol. 147:1517-1529. [DOI] [PubMed] [Google Scholar]
- 21.Holmes, E. C. 2001. On the origin and evolution of the human immunodeficiency virus. Biol. Rev. 76:239-254. [DOI] [PubMed] [Google Scholar]
- 22.Horton, H., T. U. Vogel, D. K. Carter, K. Vielhuber, D. H. Fuller, T. Shipley, J. T. Fuller, K. J. Kunstman, G. Sutter, D. C. Montefiori, S. M. Wolinsky, C. Wang, D. B. Allison, and D. I. Watkins. 2002. Immunization of rhesus macaques with a DNA prime/modified vaccinia virus Ankara boost regimen induces broad simian immunodeficiency virus (SIV)-specific T-cell responses and reduces initial viral replication but does not prevent disease progression following challenge with pathogenic SIVmac239. J. Virol. 76:7187-7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koulinska, I. N., T. Ndung'u, D. Mwakagile, G. Msamanga, C. Kagoma, W. Fawzi, M. Essex, and B. Renjifo. 2001. A new human immunodeficiency virus type 1 circulating recombinant form from Tanzania. AIDS Res. Hum. Retrovir. 17:423-431. [DOI] [PubMed] [Google Scholar]
- 24.Kyaw-Tanner, M. T., and W. F. Robinson. 1996. Quasispecies and naturally occurring superinfection in feline immunodeficiency virus infection. Arch. Virol. 141:1703-1713. [DOI] [PubMed] [Google Scholar]
- 25.Langley, R. J., V. M. Hirsch, S. J. O'Brien, D. Adger-Johnson, R. M. Goeken, and R. Olmsted. 1994. Nucleotide sequence analysis of puma lentivirus (PLV-14): genomic organization and relationship to other lentiviruses. Virology 202:853-864. [DOI] [PubMed] [Google Scholar]
- 26.Li, W. H. 1993. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J. Mol. Evol. 36:96-99. [DOI] [PubMed] [Google Scholar]
- 27.Lutz, H., R. Hofmann-Lehmann, D. Fehr, C. Leutenegger, M. Hartmann, P. Ossent, M. Grob, M. Elgizoli, and P. Weilenmann. 1996. Liberation into the wild of wild felines—danger of the release of virus infections. Schweiz. Arch. Tierheilkd. 138:579-585. [PubMed] [Google Scholar]
- 28.Moore, J. P., P. W. Parren, and D. R. Burton. 2001. Genetic subtypes, humoral immunity, and human immunodeficiency virus type 1 vaccine development. J. Virol. 75:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Motomura, K., S. Kusagawa, K. Kato, K. Nohtomi, H. H. Lwin, K. M. Tun, M. Thwe, K. Y. Oo, S. Lwin, O. Kyaw, M. Zaw, Y. Nagai, and Y. Takebe. 2000. Emergence of new forms of human immunodeficiency virus type 1 intersubtype recombinants in central Myanmar. AIDS Res. Hum. Retrovir. 16:1831-1843. [DOI] [PubMed] [Google Scholar]
- 30.Nishimura, Y., Y. Goto, K. Yoneda, Y. Endo, T. Mzuno, M. Hamachi, H. Maruyama, H. Kinoshita, S. Koga, M. Komori, S. Fushuku, K. Ushinohama, M. Akuzawa, T. Watari, A. Hasegawa, and H. Tsujimoto. 1999. Interspecies transmission of feline immunodeficiency virus from domestic cat to the Tsushima cat (Felis begalensis euptilura) in the wild. J. Virol. 73:7916-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novitsky, V., U. R. Smith, P. Gilbert, M. F. McLane, P. Chigwerdere, C. Williamson, T. Ndung'u, I. Klein, S. Y. Chang, T. Peter, I. Thior, B. T. Foley, S. Gaolekwe, N. Rybak, S. Gaseitsiwe, F. Vannberg, R. Marlink, T. H. Lee, and M. Essex. 2002. Human immunodeficiency virus type 1 subtype C molecular phylogeny: consensus sequence for an AIDS vaccine design. J. Virol. 76:5435-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okada, S., R. Pu, E. Young, W. V. Stoffs, and J. K. Yamamoto. 1994. Superinfection of cats with feline immunodeficiency virus subtypes A and B. AIDS Res. Hum. Retrovir. 10:1739-1746. [DOI] [PubMed] [Google Scholar]
- 33.Olmstead, R. A., R. Langley, M. E. Roelke, R. M. Goeken, D. Adger-Johnson, J. P. Goff, J. P. Albert, C. Packer, M. K. Laurenson, T. M. Caro, L. Scheepers, D. E. Wildt, M. Bush, J. S. Martenson, and S. J. O'Brien. 1992. Worldwide prevalence of lentivirus infection in wild feline species: epidemiologic and phylogenetic aspects. J. Virol. 66:6008-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otten, R. A., D. L. Ellenberger, D. R. Adams, C. A. Fridlund, E. Jackson, D. Pieniazek, and M. A. Rayfield. 1999. Identification of a window period for susceptibility to dual infection with two distinct human immunodeficiency virus type 2 isolates in a Macaca nemestrina (pig-tailed macaque) model. J. Infect. Dis. 180:673-684. [DOI] [PubMed] [Google Scholar]
- 35.Packer, C., D. A. Gilbert, A. E. Pusey, and S. J. O'Brien. 1991. A molecular genetic-analysis of kinship and cooperation in African lions. Nature 351:562-565. [Google Scholar]
- 36.Packer, C., A. E. Pusey, H. Rowley, D. A. Gilbert, J. Martenson, S. J. O'Brien. 1991. Case-study of a population bottleneck-lions of the Ngorongoro Crater. Conserv. Biol. 5:219-230. [Google Scholar]
- 37.Pamilo, P., and N. O. Bianchi. 1993. Evolution of the ZFX and ZFY genes: rates and interdependence between the genes. Mol. Biol. Evol. 110:271-281. [DOI] [PubMed] [Google Scholar]
- 38.Pieniazek, D., J. M. Peralta, J. A. Ferreira, J. W. Krebs, S. M. Owen, F. S. Sion, F. R. Celso, A. B. Sereno, C. A. M. Desa, B. G. Weniger, W. L. Heyward, C. Y. Ou, N. J. Pieniazek, G. Schochetman, and M. A. Rayfield. 1991. Identification of mixed HIV-1/HIV-2 infections in Brazil by polymerase chain-reaction. AIDS 5:1293-1299. [DOI] [PubMed] [Google Scholar]
- 39.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 40.Pu, R., J. Coleman, M. Omori, M. Arai, T. Hohdatsu, C. Huang, T. Tanabi, and J. K. Yamamoto. 2001. Dual-subtype FIV vaccine protects cats against in vivo swarms of both homologous and heterologous subtype FIV isolates. AIDS 15:1225-1237. [DOI] [PubMed] [Google Scholar]
- 41.Robertson, D. L., P. M. Sharp, F. E. McCutchan, and B. H. Hahn. 1995. Recombination in HIV-1. Nature 374:124-126. [DOI] [PubMed] [Google Scholar]
- 42.Roelke-Parker, M. E., L. Munson, C. Packer, R. Kock, S. Cleveland, M. Carpenter, S. J. O'Brien, A. Popsichil, R. Hofmann-Lehmann, H. Lutz, G. L. M. Mwamengele, M. N. Mgasa, G. A. Machange, B. A. Summers, and M. J. G. Appel. 1996. A canine distemper virus epidemic in Serengeti lions (Panthera leo). Nature 379:441-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salminen, M. O., J. K. Carr, D. L. Robertson, P. Hegerich, D. Gotte, C. Koch, E. Sanders-Buell, F. Gao, P. M. Sharp, B. H. Hahn, D. S. Burke, and F. E. McCutchan. 1997. Evolution and probable transmission of intersubtype recombinant human immunodeficiency virus type 1 in a Zambian couple. J. Virol. 71:2647-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Souquiere, S., F. Bibollet-Ruche, D. L. Robertson, M. Makuwa, C. Apetrei, R. Onanga, C. Kornfeld, J. C. Plantier, F. Gao, K. Abernethy, L. J. White, W. Karesh, P. Telfer, E. J. Wickings, P. Mauclere, P. A. Marx, F. Barre-Sinoussi, B. H. Hahn, M. C. Muller-Trutwin, and F. Simon. 2001. Wild Mandrillus sphinx are carriers of two types of lentivirus. J. Virol. 75:7086-7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swofford, D. L. 1998. PAUP*: phylogenetic analysis using parsimony (*and other methods). Sinauer, Sunderland, Mass.
- 46.Takahoko, M., M. Tobiume, K. Ishikawa, W. Ampofo, N. Yamamoto, M. Matsuda, and M. Tatsumi. 2001. Infectious DNA clone of HIV type 1 A/G recombinant (CRF02_AG) replicable in peripheral blood mononuclear cells. AIDS Res. Hum. Retrovir. 17:1083-1087. [DOI] [PubMed] [Google Scholar]
- 47.Takehisa, J., L. Zekeng, E. Ido, Y. Yamaguchi-Kabata, I. Mboudjeka, Y. Harada, T. Miura, L. Kaptu, and M. Hayami. 1999. Human immunodeficiency virus type 1 intergroup (M/O) recombination in Cameroon. J. Virol. 73:6810-6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmough, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomson, M. M., E. Delgado, N. Manjon, A. Ocampo, M. L. Villahermosa, A. Marino, I. Herrero, M. T. Cuevas, E. Vazquez-de Parga, L. Perez-Alvarez, L. Medrano, J. A. Taboada, and R. Najera. 2001. HIV-1 genetic diversity in Galicia, Spain: BG intersubtype recombinant viruses circulating among injecting drug users. AIDS 15:509-516. [DOI] [PubMed] [Google Scholar]
- 50.Tovanabutra, S., V. Polonis, M. De Souza, R. Trichavaroj, P. Chanbancherd, B. Kim, E. Sanders-Buell, S. Nitayaphan, A. Brown, M. R. Robb, D. L. Birx, F. E. McCutchan, and J. K. Carr. 2001. First CRF01_AE/B recombinant of HIV-1 is found in Thailand. AIDS 15:1063-1065. [DOI] [PubMed] [Google Scholar]
- 51.VandeWoude, S., C. A. Hageman, S. J. O'Brien, and E. A. Hoover. 2002. Nonpathogenic lion and puma lentiviruses impart resistance to superinfection by virulent feline immunodeficiency virus. J Acq. Immun. Def. Synd. 29:1-10. [DOI] [PubMed] [Google Scholar]
- 52.Vernazza, P. L., E. Bernasconi, and B. Hirschel. 2000. HIV superinfection: myth or reality? Schweiz. Med. Wochenschr. 130:1101-1104. [PubMed] [Google Scholar]
- 53.Wei, Q., and P. N. Fultz. 1998. Extensive diversification of human immunodeficiency virus type 1 subtype B strains during dual infection of a chimpanzee that progressed to AIDS. J. Virol. 72:3005-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willett, B. J., J. N. Flynn, and M. J. Hosie. 1999. FIV infection in domestic cats: an animal model for AIDS. Immunol. Today 18:182-189. [DOI] [PubMed] [Google Scholar]
- 55.Yamaguchi, N., D. W. MacDonald, W. C. Passanisi, D. A. Harbour, and C. D. Hopper. 1996. Parasite prevalence in free-ranging farm cats, Felis silvestris catus. Epidemiol. Infect. 116:217-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu, X. F., W. Liu, J. Chen, W. Kong, B. Liu, J. Yang, F. Liang, F. McCutchan, S. Piyasirisilp, and S. Lai. 2001. Rapid dissemination of a novel B/C recombinant HIV-1 among injection drug users in southern China. AIDS 15:523-525. [DOI] [PubMed] [Google Scholar]
- 57.Zhu, T., N. Wang, A. Carr, S. Wolinsky, and D. D. Ho. 1995. Evidence for coinfection by multiple strains of human immunodeficiency virus type 1 subtype B in an acute seroconverter. J. Virol. 69:1324-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]