Abstract
An RNA secondary structure model for the complete HIV-1 genome has recently been published based on SHAPE technology. Several well-known RNA motifs such as TAR and RRE were confirmed and numerous new structured motifs were described that may play important roles in virus replication. The 9 kb viral RNA genome is densely packed with many RNA hairpin motifs and the collective fold may play an important role in HIV-1 biology. We initially focused on 16 RNA hairpin motifs scattered along the viral genome. We considered conservation of these structures, despite sequence variation among virus isolates, as a first indication for a significant function. Four relatively small hairpins exhibited considerable structural conservation and were selected for experimental validation in virus replication assays. Mutations were introduced into the HIV-1 RNA genome to destabilize individual RNA structures without affecting the protein-coding properties (silent codon changes). No major virus replication defects were scored, suggesting that these four hairpin structures do not play essential roles in HIV-1 replication.
Keywords: RNA, SHAPE, HIV-1, RNA structure, hairpin
Introduction
High-throughput RNA structure probing by means of selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE) technology was first described in 2005.1 SHAPE has revolutionized the analysis of structured RNA molecules. This technology allows one to determine the reactivity of nearly all nucleotides (nts) in a lengthy RNA molecule such as the 9 kb HIV-1 genome.2 The SHAPE reactivities provide information on RNA structure: regions with low reactivity signify domains with substantial base paired RNA secondary structure and high SHAPE reactivity marks regions with largely unstructured nts.1-5 The availability of detailed RNA structure information is important for addressing the biological function of small RNA domains or complete RNA molecules. In particular, the genomes of certain RNA viruses have been proposed to adopt a genome-scale ordered RNA structure (GORS) that is critical for virus replication, e.g., to resist the action of intracellular RNases or to allow the melting of double-stranded RNA replication intermediates.6,7 This aspect has not yet been studied for the RNA genome of retroviruses like the human immunodeficiency virus type 1 (HIV-1).
RNA structure information is also important for certain applications. For instance, antiviral gene therapy based on the mechanism of RNA interference (RNAi) uses short hairpin RNA (shRNA) inhibitors that target the viral RNA genome.8,9 We previously demonstrated that RNAi attack is influenced by the local RNA structure of the HIV-1 RNA target sequence10,11 and more recently reported that the design of shRNA inhibitors can be significantly improved by the exclusive targeting of accessible HIV-1 RNA domains as determined by SHAPE technology.12
The new structure model of the HIV-1 RNA genome revealed many local folds like stem-loop or hairpin motifs that may play a distinct role in the virus replication cycle. Several well-known and important RNA structures were confirmed, such as the TAR hairpin and the RRE domain that serve as binding sites for the essential HIV-1 regulatory proteins Tat and Rev, respectively. The TAR hairpin is present at the extreme 5′ end of all HIV-1 transcripts and facilitates binding of Tat and host cell co-factors to stimulate transcription from the LTR promoter.13-16 The RRE structure binds Rev to support the nuclear export of unspliced transcripts.17 Other RNA structures were also confirmed, including the hairpin of the translational frameshift signal that facilitates Pol translation18,19 and hairpins that regulate the accessibility of polyadenylation20 and splicing signals.21 Other RNA motifs that cluster in the 5′ untranslated region (5′ UTR) have been implicated in dimerization and packaging of the RNA genome in virion particles and the subsequent process of reverse transcription.21-31 In addition, several long-distance base pairing interactions have been proposed.32-34
The SHAPE-derived HIV-1 RNA structure model revealed a multitude of novel structures that may play a role in virus replication. For instance, HIV-1 genomic RNA structures were recently proposed to orchestrate virus recombination events.35 In this study, we set out to test the functional significance of some individual small hairpin RNA structures.
Results
Phylogenetic analysis of structured RNA motifs
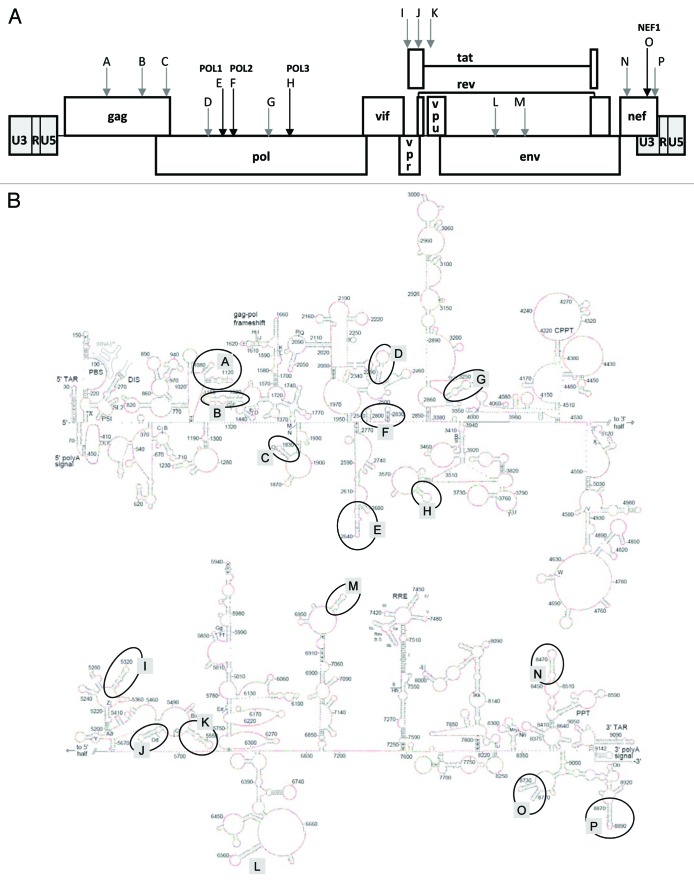
We selected 16 candidate hairpin motifs across the SHAPE-derived RNA secondary structure model of the genome of the HIV-1 NL4-3 isolate for initial inspection (motifs A through P in Fig. 1).2 RNA structures that play important biological functions usually demonstrate robust structural conservation despite the presence of sequence variation in diverse virus isolates.2,15,36-38 The best example of such structural conservation is the occurrence of base pair co-variations, e.g., A-U converted into G-C in another virus isolate. We therefore set out to focus on phylogenetically conserved RNA motifs as the most likely candidates to fulfill an important function in HIV-1 biology.
Figure 1. Position of the 16 structured RNA motifs in the HIV-1 genome. (A) Motifs A through P on the HIV-1 DNA map. (B) SHAPE-derived model of the HIV-1 RNA secondary structure.2 The 16 RNA motifs are marked.
We analyzed the folding of the proposed hairpins with 5′ and 3′ flanks of maximal 5 nts in diverse HIV-1 isolates for which the full-length genome sequence is known. Isolates belonging to the major HIV-1 subtypes A, B, C and D, for which multiple full-length genome sequences are available, were aligned with the QuickAlign Analysis Tool (www.hiv.lanl.gov/) (July 2010). For each of the 16 hairpin fragments, we selected the most prevalent sequence per subtype as the prototype sequence. For example, we picked a subtype B sequence for hairpin H that occurs in 59 out of 235 sequences (Table 1, fourth column). The 64 prototype sequences were subsequently probed by the mfold program to allow the large scale analyses of predicted RNA secondary structures (Fig. 2). First, we analyzed the mfold prediction for the HIV-1 subtype B NL4-3 isolate that was used to generate the SHAPE-directed RNA structure model. Only 10 of the 16 hairpins are predicted to fold according to the SHAPE model, consistent with the observation that inclusion of experimental data tends to yield significantly different structures than those predicted by mfold-type algorithms alone (listed in Table 1, last column as “+” for SHAPE-like or “-“ for non-SHAPE with the thermodynamic stability in kcal/mol). When a non-SHAPE structure was predicted, the SHAPE-structure was sometimes seen as sub-optimal RNA fold, but frequently not predicted within the 5% energy window set by the mfold program.
Table 1. Phylogenetic analysis of 16 RNA structures in the HIV-1 genome.
| name | positiona | subtype | Prototype sequenceb | nt differencesc | RNA structure | |
|---|---|---|---|---|---|---|
|
ΔG (kcal/mol)d |
Conservatione |
|||||
| A |
1076–1142 |
NL4–3 |
|
|
-22.9 |
+ |
| gag, p24 |
A |
2/6 (33%) |
4 |
-22.9 |
+ |
|
| |
B |
23/235 (10%) |
1 |
-22.9 |
+ |
|
| |
C |
12/397 (3%) |
9 |
-16.1 |
- |
|
| |
D |
6/49 (12%) |
4 |
-21.3 |
+ |
|
| B |
1459–1522 |
NL4–3 |
|
|
-12.5 |
- |
| gag, p2 |
A |
1/6 (17%) |
13 (3 del) |
-8.0 |
- |
|
| |
B |
7/371 (2%) |
2 |
-15.0 |
- |
|
| |
C |
3/399 (1%) |
23 (4 del) |
-11.3 |
- |
|
| |
D |
2/51 (4%) |
15 |
-11.3 |
- |
|
| C |
1823–1859 |
NL4–3 |
|
|
-19.1 |
+ |
| gag, p6 |
A |
2/6 (33%) |
11 |
-7.9 |
1/2Bf |
|
| pol, prot |
B |
41/371 (11%) |
2 |
-18.1 |
+ |
|
| |
C |
11/399 (3%) |
10 |
-17.1 |
- |
|
| |
D |
3/51 (6%) |
5 |
-17.5 |
+ |
|
| D |
2388–2484 |
NL4–3 |
|
|
-23.1 |
- |
| pol, RT |
A |
1/6 (17%) |
10 |
-34.1 |
- |
|
| |
B |
7/235 (3%) |
2 |
-21.7 |
- |
|
| |
C |
10/397 (3%) |
8 |
-27.5 |
- |
|
| |
D |
1/49 (2%) |
6 |
-21.4 |
- |
|
| E POL1 |
2619–2686 |
NL4–3 |
|
|
-20.7 |
+ |
| pol, RT |
A |
1/6 (17%) |
6 |
-9.2 |
+ |
|
| |
B |
39/235 (17%) |
1 |
-18.5 |
+ |
|
| |
C |
26/397 (7%) |
4 |
-17.4 |
1/2Bf |
|
| |
D |
6/49 (12%) |
3 |
-15.0 |
1/2Bf |
|
| |
CPZ |
1/9 (9%) |
26 |
-12.4 |
+ |
|
| F POL2 |
2781–2835 |
NL4–3 |
|
|
-23.2 |
+ |
| pol, RT |
A |
2/6 (33%) |
6 |
-14.5 |
1/2Af |
|
| |
B |
53/235 (21%) |
0 |
-23.2 |
+ |
|
| |
C |
122/397 (31%) |
4 |
-20.2 |
+ |
|
| |
D |
8/49 (16%) |
3 |
-17.6 |
+ |
|
| |
CPZ |
8/49 (16%) |
3 |
-17.6 |
+ |
|
| G |
3285–3358 |
NL4–3 |
|
|
-23.4 |
- |
| pol, RT |
A |
1/6 (17%) |
8 |
-25.2 |
- |
|
| |
B |
25/235 (11%) |
1 |
-23.9 |
- |
|
| |
C |
55/396 (14%) |
5 |
-22.5 |
1/2Bf |
|
| |
D |
4/49 (8%) |
3 |
-23.9 |
- |
|
| H POL3 |
3606–3639 |
NL4–3 |
|
|
-5.8 |
+ |
| pol, RNase H |
A |
1/6 (17%) |
6 |
-5.1 |
+ |
|
| |
B |
59/235 (25%) |
2 |
-2.0 |
+ |
|
| |
C |
140/395 (35%) |
1 |
-5.8 |
+ |
|
| |
D |
17/49 (35%) |
1 |
-5.8 |
+ |
|
| |
CPZ |
17/49 (35%) |
1 |
-5.8 |
+ |
|
| I |
5303–5343 |
NL4–3 |
|
|
-12.3 |
- |
| vpr |
A |
3/6 (50%) |
5 |
-3.9 |
- |
|
| |
B |
35/234 (15%) |
1 |
-9.8 |
- |
|
| |
C |
78/397 (20%) |
6 |
-4.5 |
- |
|
| |
D |
6/49 (12%) |
3 |
-4.5 |
- |
|
| J |
5530–5581 |
NL4–3 |
|
|
-12.4 |
+ |
| tat(1), rev(1) |
A |
1/6 (17%) |
14 |
-14.7 |
- |
|
| |
B |
6/371 (2%) |
4 |
-10.1 |
- |
|
| |
C |
17/399 (4%) |
12 |
-14.9 |
- |
|
| |
D |
2/51 (4%) |
11 |
-6.9 |
- |
|
| K |
5600–5645 |
NL4–3 |
|
|
-7.6 |
- |
| |
A |
1/6 (17%) |
21 |
-6.5 |
- |
|
| |
B |
2/235 (1%) |
7 |
-8.8 |
- |
|
| |
C |
3/397 (1%) |
30 |
-0.4 |
- |
|
| |
D |
2/49 (4%) |
8 |
-8.8 |
- |
|
| L |
6536–6598 |
NL4–3 |
|
|
-17.8 |
- |
| env, gp120 |
A |
1/6 (17%) |
11 |
-10.3 |
- |
|
| |
B |
6/371 (2%) |
4 |
-8.8 |
- |
|
| |
C |
5/398 (1%) |
13 |
-8.4 |
- |
|
| |
D |
2/51 (4%) |
9 |
-11.6 |
- |
|
| M |
6982–7016 |
NL4–3 |
|
|
-7.9 |
+ |
| env, gp120 |
A |
1/6 (17%) |
17 |
-1.3 |
- |
|
| |
B |
1/235 (< 1%) |
14 |
0.7 |
- |
|
| |
C |
1/397 (< 1%) |
17 (7 del) |
-0.6 |
- |
|
| |
D |
1/49 (2%) |
13 |
-1.7 |
- |
|
| N |
8455–8505 |
NL4–3 |
|
|
-18.3 |
+ |
| nef, LTR |
A |
1/6 (17%) |
11 |
-11.8 |
- |
|
| |
B |
24/370 (7%) |
partial del |
-1.6 |
- |
|
| |
C |
8/399 (2%) |
13 |
-15.0 |
+ |
|
| |
D |
4/51 (8%) |
7 |
-13.4 |
- |
|
| O NEF1 |
8723–8773 |
NL4–3 |
|
|
-26.6 |
+ |
| nef, LTR |
A |
1/6 (17%) |
8 |
-13.3 |
1/2Bf |
|
| |
B |
16/219 (7%) |
2 |
-19.0 |
+ |
|
| |
C |
49/396 (12%) |
4 |
-25.1 |
+ |
|
| |
D |
9/48 (19%) |
4 |
-19.6 |
+ |
|
| |
CPZ |
1/11 (9%) |
10 |
-17.5 |
+ |
|
| P | 8867–8906 |
NL4–3 |
|
|
-19.5 |
+ |
| nef, LTR |
A |
3/6 (50%) |
13 |
-11.0 |
+ |
|
| |
B |
15/59 (25%) |
6 |
-7.1 |
+ |
|
| |
C |
2/25 (8%) |
14 |
-6.0 |
- |
|
| D | 1/4 (25%) | 11 | -3.9 | - | ||
a) NL4-3 coordinates
b) Number of virus isolates that match the exact prototype sequence, which was identified by PrimAlign (Los Alamos HIV database)
c) Compared with NL4-3
d) Calculated by mfold for the prototype sequence
e) Structural similarity between SHAPE model2 and predicted mfold structure; structure maintained = +; structure changed = -
f) 1/2A = only first domain maintained, 1/2B = only second domain maintained.
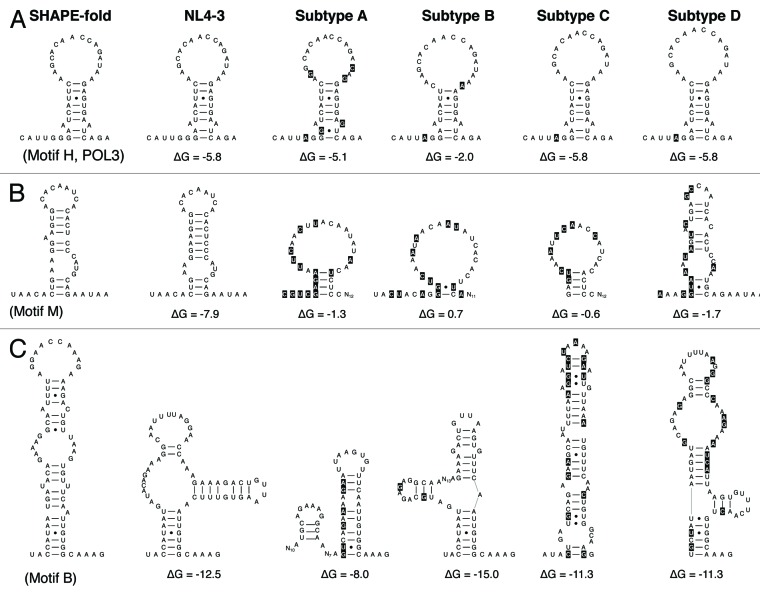
Figure 2. Probing RNA structure conservation among the subtypes. The original SHAPE-directed model (left) is compared with the mfold prediction for the subtype B isolate NL4-3 and the prototype sequence of subtypes A, B, C and D. Sequence differences compared with the NL4-3 sequence are highlighted in black. Structural conservation is illustrated for POL3, partial conservation for motif M and a lack of conservation for motif B. See Table 1 for more details. Single-stranded RNA segments that lack mutations compared with the NL4-3 sequence are indicated as Nx, with x indicating the number of nucleotides.
Mfold analyses of the 16 hairpin segments among the 4 HIV-1 subtypes provided a wide range of results. Some SHAPE-like hairpins were recapitulated by mfold, not only for subtype B but for all four subtypes (Table 1, motifs E, F, H and O). For example, we show the predicted structure for motif H (Fig. 2A). Figure 2B and C show some of the 14 structured motifs (M and B, respectively) that are not supported by this phylogenetic survey (marked in gray in Table 1) and that will be discussed below. The structural conservation of the motifs E, F, H and O is remarkable because there is considerable sequence variation among the isolates and especially the subtypes. For instance, we scored 4‒8 nt changes in motif O in the non-B subtypes compared with the NL4-3 strain (Table 1, motif O). This provides some phylogenetic support for structural conservation, which hints at a biological function of the candidate RNA structures. On the other hand, it is clear that the new candidate hairpins are not as stable stem-loop structures as the well-known TAR and RRE motifs. Based on these results, we selected the most promising small hairpin motifs—E, F, H and O—for further functional analysis. Although we assume that these relatively small RNA hairpins were conserved during HIV-1 evolution and diversification, thus suggesting a role in HIV-1 biology, it cannot formally be excluded that they arose by convergent evolution. These four structures were renamed POL1, POL2, POL3 and NEF1, respectively, according to their position in the HIV-1 genome (Fig. 1A). POL3 is a small hairpin and the other motifs represent tandem hairpin structures with two subdomains, termed A and B. We also probed these four structures in the more divergent simian immunodeficiency virus chimpanzee isolate (SIV-cpz), which confirmed the SHAPE-like folding (Table 1).
All other 12 hairpin candidates were dismissed because no SHAPE-like structure was consistently predicted by mfold (marked in gray in Table 1). For five hairpin motifs, none of the prototype sequences of different HIV-1 subtypes adopt the SHAPE-proposed folding as the most favorite RNA conformation (Table 1, motifs B, D, I, K, L). For example, a wide range of structural presentations are apparent for motif B (Fig. 2C). For seven hairpin motifs, an intermediate result was obtained, i.e., prediction of a non-SHAPE structure for at least one subtype (Table 1, motifs A, C, G, J, M, N and P). For instance, motif M adopts SHAPE-like structures only for the NL4-3 isolate (Fig. 2B). We realize that this in silico mfold analysis is certainly not appropriate to formally rule out some of the predicted hairpin motifs. This information was however used to select the SHAPE-determined RNA structures that have the strongest phylogenetic support.
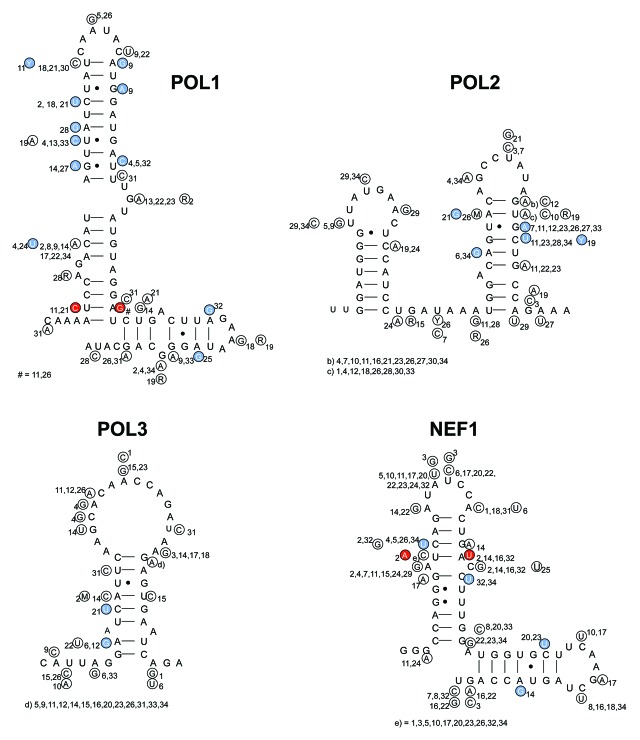
As a next test for the four conserved hairpins that were selected, we probed the sequence variation in all virus isolates for the presence of base pair co-variations that could provide phylogenetic support for the proposed base pairing schemes. We used the sequence information present in the HIV-1 2010 compendium (subtype A with 16 isolates, B = 34, C = 21, D = 11 and SIV-cpz = 5).39 We checked whether the available sequence variation does maintain or destabilize the proposed hairpin structure. We first performed this phylogenetic analysis within each subtype. Figure 3 illustrates this analysis for subtype B for the four structures POL1, POL2, POL3 and NEF1. Analyses of all inspected subtypes A, B, C, D and SIV-cpz are presented in the Figure S1. Of special interest are structure-conservative mutations on one strand (e.g., G-U to G-C, marked blue in Fig. 3) or on both strands of the base paired stem (e.g., A-U to G-C co-variation, marked red in Fig. 3). We identified several structure-conservative mutations in these RNA structures and co-variations were detected in the POL1 and NEF1 hairpins (Fig. 3). In total, the four SHAPE-identified hairpin motifs POL1, POL2, POL3 and NEF1 harbor a fair degree of structural conservation, although much more extensive phylogenetic evidence was previously reported for TAR hairpin.15
Figure 3. The impact of natural sequence variation on the POL1, POL2, POL3 and NEF1 structures for subtype B sequences derived from the HIV-1 compedium 2010. The majority sequence is depicted for each subtype with variations in small circles with a number that refers to the particular isolate. Blue circles mark changes that conserve the structure, red circles represent base pair co-variation. Isolate code: Number = isolate name: 1 = B.AR.04.04AR143170; 2 = B.AU.04.PS1038_Day174; 3 = B.BO.99.BOL0122; 4 = B.BR.05.BREPM1081; 5 = B.CA.97.CANB3FULL; 6 = B.CN.05.05CNHB_hp3; 7 = B.CO.01.PCM001; 8 = B.CU.99.Cu19; 9 = B.CY.06.CY165; 10 = B.DE.86.D31; 11 = B.DK.04.PMVL_012; 12 = B.DO.05.05DO_160884; 13 = B.EC.89.EC003; 14 = B.ES.07.X2231; 15 = B.FR.92.92FR_BX08; 16 = B.GA.88.OYI; 17 = B.GB.86.GB8_46R; 18 = B.GE.03.03GEMZ004; 19 = B.HT.05.05HT_129389; 20 = B.IN.x.11807; 21 = B.JM.05.05JM_KJ108; 22 = B.JP.05.DR6538; 23 = B.KR.04.04KMH5; 24 = B.MM.99.mSTD101; 25 = B.NL.00.671_00T36; 26 = B.RU.04.04RU128005; 27 = B.TH.00.00TH_C3198; 28 = B.TT.01.01TT_CRC50069; 29 = B.TW.94.TWCYS; 30 = B.UA.01.01UAKV167; 31 = B.US.07.CR0027M; 32 = B.UY.01.01UYTRA1179; 33 = B.YE.02.02YE507; 34 = B.ZA.03.03ZAPS045MB2.
Mutational disruption of selected hairpin motifs
To investigate the biological relevance of the four selected hairpin motifs, we destabilized the individual hairpins by introducing mutations in the genome of the primary CXCR4 using HIV-1 LAI isolate. We designed silent codon changes in the underlying pol and nef genes to exclude adverse effects due to amino acid substitutions in the encoded proteins. Additionally, we ensured that the introduced mutations did not modify known RNA signals such as splice donor/acceptor sites. For motifs POL1, POL2 and NEF1 that consist of two hairpin domains, we chose to destabilize the stem region that contributes most to the stability of the motif, as determined by mfold analyses (results not shown). All possible silent codon changes were analyzed using mfold to determine which ones were most disruptive to the RNA structure. We designed two to six mutations per motif to trigger the formation of an alternative, less stable RNA fold (Fig. 4). In addition, we made several combination mutants: Double (POL1+POL3), Triple (POL1-3) and Quadruple (POL1-3, NEF1).
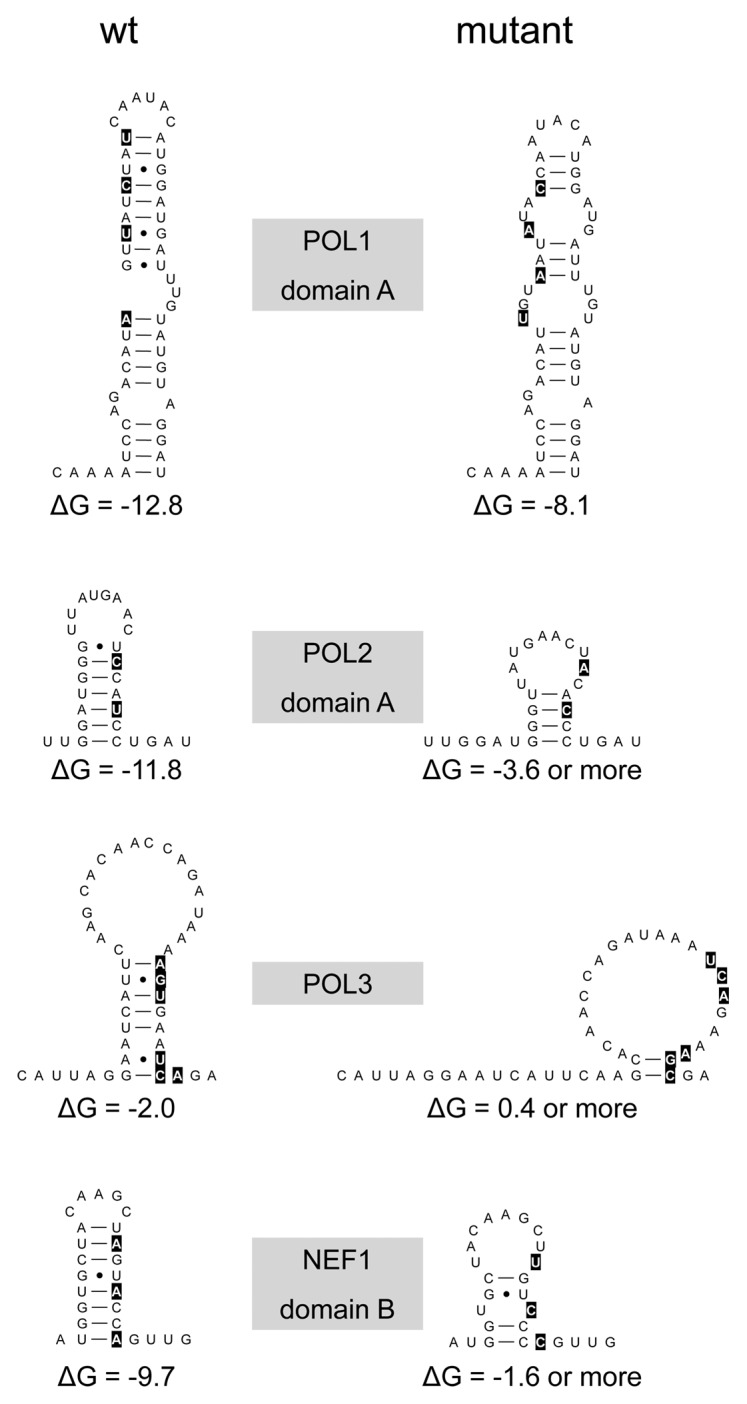
Figure 4. Mutational disruption of the POL1, POL2, POL3 and NEF1 structures. We marked the nts that were altered in wt (left) to create the mutant (right) structures. For POL1, POL2 and NEF1 we changed a single stem domain.
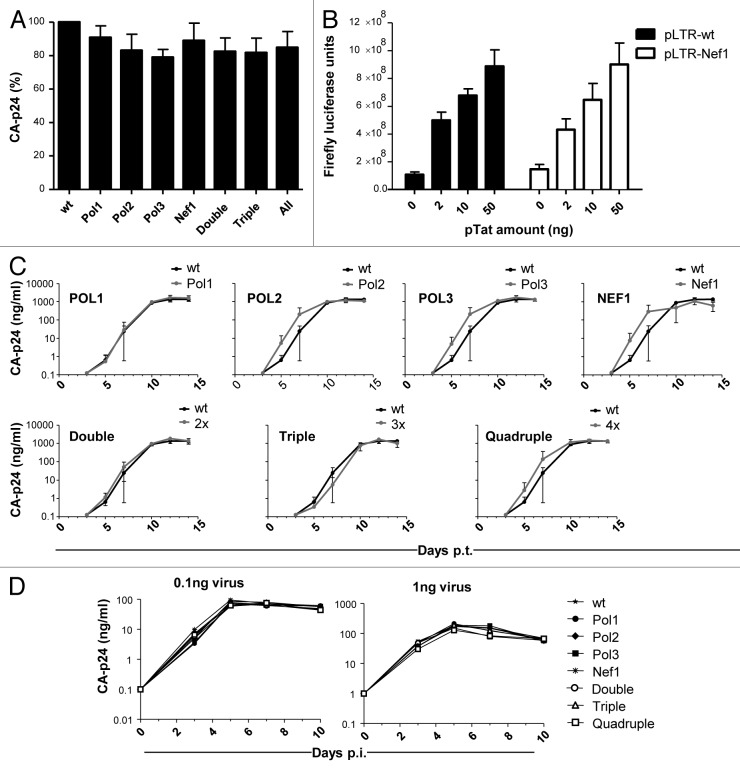
We first tested the wild-type (wt) and mutant HIV-1 LAI-based DNA constructs for their ability to produce viral proteins. 293T cells were transfected and CA-p24 production was measured in the culture medium after 2 d. All mutants, including the combination mutants, exhibited similar CA-p24 production as the wt construct (Fig. 5A). These results imply that the RNA mutations do not have a significant impact on HIV-1 gene expression, including the processes of transcription, splicing and translation, but also mRNA stability.
Figure 5. Phenotype tests of the wt and mutant pLAI constructs. (A) CA-p24 production in 293T cells transfected with pLAI variants. (B) LTR-luc constructs for wt and Nef1 were tested for promoter activity in transfected 293T cells. (C) Virus replication in SupT1 T cells monitored by CA-p24 production. Cells were transfected with the indicated constructs at day 0 (x-axis: days post-transfection). (D) Virus replication in PBMCs monitored by CA-p24 production. Cells were infected with the indicated HIV-1 variants at day 0 (x-axis: days post-infection) (left: 0.1 ng CA-p24, right: 1 ng CA-p24).
It is important to discuss the NEF1 mutant in more detail. The NEF1 hairpin is located within the nef gene, more particular in the U3 region encoded by the 3′LTR. We introduced the NEF1 mutation in the 3′LTR of the LAI molecular clone, but these U3 sequences are copied into the 5′LTR during reverse transcription. Thus, the viral progeny will carry this mutation in the 3′ end of the HIV-1 RNA genome and in the 5′LTR that serves as the transcriptional promoter. The latter modification may also have an impact on virus replication. To test for such a transcriptional effect, we made LTR-luciferase constructs with the wt and Nef1 mutant sequences. These constructs were transfected into 293T cells without or with an increasing amount of pTat expression plasmid. We measured no differences in luciferase expression level between the wt and Nef1 constructs, both in basal activity and Tat-induced expression (Fig. 5B). Thus, the U3 changes introduced in the Nef1 mutant do not adversely affect LTR promoter activity.
We next performed replication studies with the wt and mutant HIV-1 viruses on the CXCR4-expressing SupT1 T cell line that supports efficient replication of the LAI strain. We first infected SupT1 T cells with equal amounts of virus (based on CA-p24) that was produced in 293T cells. The cell cultures were monitored on a daily basis to score HIV-1 induced cytopathic effects and cell culture supernatants were regularly collected to quantify the accumulation of CA-p24 as a sign of virus replication. We did not observe significant differences in replication capacity between the wt and mutant viruses (Fig. 5C). Similar results were obtained in replication assays that were started by electroporation of the viral DNA constructs into SupT1 cells (results not shown). We next tested virus replication on peripheral blood mononuclear cells (PBMCs) because some defects, e.g., the Nef protein minus phenotype, become apparent only in primary cells and not in transformed T cell lines.40 We pooled PBMCs of four healthy donors and infected them with the wt and mutant HIV-1 variants. Again, all HIV-1 mutants including the combination mutants replicated as efficiently as wt virus (Fig. 5D).
We finally applied the ultra-sensitive virus competition assay to document whether the mutants are truly equal in replication capacity to the wt virus. This method of direct virus competition was specifically designed to reveal very subtle differences in virus replication rates.41 We made an equimolar mixture of two HIV-1 plasmids (wt and mutant) and transfected SupT1 cells via electroporation to start the competitive virus infections. We took cell samples at several time intervals (2, 3, 5 and 8 wk post-transfection) and analyzed the proviral DNA sequences as a measure of the wt:mutant ratio in order to observe outgrowth of the fittest virus. To do so, we isolated the proviral DNA and sequenced the relevant DNA segments. The input virus population was visible as a mixed population sequence, but this will change over time once the fittest virus becomes the dominant species. We detected only insignificant replication defects for some of the mutants upon extended culturing (results not shown). Thus, these mutations have no or only a negligible impact on HIV-1 replication in vitro.
Discussion
The SHAPE-determined structure of the complete 9 kb HIV-1 RNA genome does reveal an intense collection of structured RNA elements, mostly local hairpin structures.2 Other studies revealed additional long-range RNA interactions.32,33 This may suggest an important role for the genome-wide HIV-1 RNA structure in virus replication, which could include protection against RNases and modulation of the innate immune responses. We set out to experimentally probe the importance of several small RNA motifs across the viral genome. A total of 16 hairpin motifs were examined in silico for conservation among different HIV-1 isolates and subtypes, and we selected the four most promising hairpin motifs for experimental validation. We were not able to score a significant replication deficit of these mutants, even in primary T cells and in ultra-sensitive virus competition assays. We nevertheless believe that the presence of many structured RNA motifs in the HIV-1 RNA genome does likely reflect the presence of a minor evolutionary pressure that shaped the viral genome over extended times, but many of these structured motifs may not have a direct and critical replicative function. This result does obviously not exclude an important function for other SHAPE-determined RNA-structures in HIV-1 biology. These results would argue against the notion that the HIV-1 retrovirus belongs to the group of RNA viruses with a global genome-wide RNA conformation (GORS).
The RNA genome of several but not all RNA viruses is highly structured. One of the most intensively studied examples is the RNA genome of Escherichia coli RNA phages MS2 and Qβ. This RNA is highly structured with 75% of the nts estimated to take part in base pairing. Detailed RNA structure models have been built based on enzymatic and chemical sensitivity of nts, phylogenetic sequence comparison and the phenotypes of constructed mutants.42,43 The RNA folds into an array of mostly irregular helices and is further condensed by several long-distance interactions. Substantial conservation of these helices between the related coliphages attests to the relevance of discrete RNA folding. Certain structured RNA motifs fulfill important roles in virus replication, e.g., as protein binding site or to regulate translation and replication.44,45 This genome-wide secondary structure is thought to prevent permanent annealing of the plus and minus strand RNA strands during virus replication and/or to confer protection against RNases. Unlike positive-stranded RNA viruses, HIV-1 is not likely to require RNA structure to prevent permanent annealing of plus and minus strands during virus replication because retroviruses replicate through an RNA-DNA intermediate, of which the RNA strand is actively removed by RNase H activity of the elongating reverse transcriptase enzyme. The minus strand DNA is subsequently copied into double-stranded DNA that integrates into the host cell genome.
RNA structures also play a crucial role in the innate defense mechanism that protects cells against invading viral pathogens. Viral infection results in the generation of non-self RNA species in the cell, which are recognized by retinoic acid inducible gene-I-like receptors (RLRs) to initiate innate antiviral responses, including the production of proinflammatory cytokines and type I interferon.46 Much research is ongoing to specify the non-self RNA patterns that are recognized by the defense machinery of the host cell. The double-stranded nature of the genome of eukaryotic viruses does relate to the presence of innate defense mechanisms of the host cell. Viral RNA genomes may use the extensive double-stranded character for interaction with a family of structurally related dsRNA-binding proteins (DRBPs) that are coupled to a wide range of antiviral effector pathways.47-49 The dsRNA-dependent protein kinase (PKR) induces apoptosis and modulates the interferon response pathways, and activation of oligoadenylate synthetase results in RNase L production and cytoplasmic RNA cleavage.50 Dicer-mediated cleavage of dsRNA produces small interfering RNAs (siRNA) that target complementary RNA sequences for destruction by the RNA-induced silencing complex (RISC) of the RNAi mechanism, although this option does not hold for retroviruses like HIV-1 that lack a dsRNA replication intermediate. Furthermore, viral RNA genomes may encode hairpin motifs that resemble microRNA (miRNA) precursors, which are cleaved by the Drosha endonuclease. Such antiviral siRNAs and miRNAs have recently also been detected in HIV-infected cells.51-54
The ability of HIV-1 to persist in the host may relate to its ability to circumvent these innate defense systems by having a highly structured RNA genome. Using bioinformatics, GORS were identified in several positive-strand animal and plant RNA viruses.6 Using atomic force microscopy, hepatitis C virus RNA was visualized as tightly compacted spheroids, while under the same experimental conditions the predicted unstructured poliovirus and rubella virus RNA were pleomorphic with an extensive single-stranded signature.7 There was remarkable variability between the virus genera that possess this characteristic; e.g., some viruses show evidence for extensive base pairing throughout the protein-coding sequences that was absent in other viruses. The presence of a GORS-like RNA genome seems to correlate with the capacity of the corresponding viruses to cause a persistent infection in their natural hosts.6 This raises the intriguing possibility of a role for GORS in the modulation of innate intracellular defense mechanisms triggered by double-stranded RNA.55,56 Our in vitro virus replication studies may not be appropriate to test for such effects. The study by Simmonds et al. also revealed that persistent and non-persistent viruses differ in the extent of their dinucleotide frequency biases.6 In this respect, the HIV-1 RNA genome has a highly biased composition with up to 40% A nts.57,58 It is likely that both sequence and structure elements of the viral RNA genome control the functional interactions with the host cell.
Materials and Methods
In silico analysis
Sixteen RNA structures in the SHAPE-derived HIV-1 RNA model were selected for this study.2 To determine phylogenetic conservation of RNA structure motifs, the four prototype sequences representing the four dominant HIV-1 subtypes (A, B, C and D, and in some cases also SIV-chimpanzee sequences) were determined with the QuickAlign Analysis Tool (Los Alamos HIV database, July 2010). The prototype sequences were used to perform mfold RNA structure prediction using standard settings.59 Four RNA motifs revealed high structural conservation despite sequence variation. The sequence variation in natural HIV-1 isolates is shown in Figure S1 (number of sequences per subtype: A = 16, B = 34, C = 21, D = 11 and SIV-cpz = 5).
HIV-1 DNA constructs
To investigate the potential biological function of the 4 selected RNA motifs, silent mutations were introduced into the subtype B molecular clone pLAI with oligonucleotides (Table 2). We used mfold to determine which silent codon mutations have the greatest structural impact. For mutants POL1, POL2 and POL3, the QuikChange Site-Directed Mutagenesis kit (Stratagene) was used following the manufacturer’s instructions including the Pfu Turbo DNA polymerase. For mutant NEF1, a 2-step PCR method was applied using the Phusion High Fidelity DNA polymerase (Finnzymes) as this sequence appears twice in pLAI in the 5′- and 3′LTR sequences. The NEF1-PCR fragment was subsequently cloned into the 3′LTR pLAI via the restriction sites Aat2 and BamH1. During HIV-1 replication, the Nef1 mutation in the 3′LTR will be copied in the 5′LTR promoter during the process of reverse transcription. To test for an effect on promoter activity, the LTR was fused to the firefly luciferase gene in pLTR-luc plasmid.60 The Nef1 mutations were introduced to generate the pLTR-Nef1 luciferase construct using the QuikChange Site-Directed Mutagenesis kit. All mutations were confirmed via sequencing.
Table 2. Oligonucleotides used for mutagenesis, PCR amplification and sequencing.
| Name | Positiona | Sequenceb |
|---|---|---|
| POL1 fw |
2651–2696 |
CAAAATCCAGACATTGTAATATACCAATACATGGATGATTTGTATG |
| POL1 rev |
|
CATACAAATCATCCATGTATTGGTATATTACAATGTCTGGATTTTG |
| POL2 fw |
2816–2858 |
GGATGGGTTATGAACTACACCCTGATAAATGGACAGTACAGCC |
| POL2 rev |
|
GGCTGTACTGTCCATTTATCAGGGTGTAGTTCATAACCCATCC |
| POL3 fw |
3650–3696 |
CAAGCACAACCAGATAAATCAGAAAGCGAGTTAGTCAATCAAATAAT |
| POL3 rev |
|
ATTATTTGATTGACTAACTCGCTTTCTGATTTATCTGGTTGTGCTTG |
| NEF1 fw |
8803–8846 |
CTTTGGATGGTGCTACAAGCTTGTCCCCGTTGAGCCAGATAAGG |
| NEF1 rev |
|
CCTTATCTGGCTCAACGGGGACAAGCTTGTAGCACCATCCAAAG |
| FGSA-32-Fb |
2289–2308 |
AAATCC ATACAATACTCCAG |
| TA051 |
3471–3488 |
CAGGGAGACTAAATTAGG |
| RT-Pfo-as |
3973–3997 |
GTTGCCATATTCCTGGACTACAGTC |
| 5′NEF-1 |
8277–8300 |
GCAGTAGCTGAGGGGACAGATAGG |
| 5′NEF-LTR01 |
8612–8633 |
CTTTAAGACCAATGACTTACAA |
| tTA-rev1-AD | 8925–8946 | GTCAAACCTCCACTCTAACACT |
a) HIV LAI
b) Silent mutations marked.
Mutations were combined as follows: pLAI-POL1 was used for a second round of site-directed mutagenesis using oligonucleotides POL3 fw and rev to create the double mutant, which was used to create the triple mutant with the POL2 oligonucleotides. Finally, the region encompassing the NEF1 motif was cloned into the triple pLAI mutant via the restriction sites Aat2 and BamH1 to make the quadruple mutant.
Cell culture
293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) with 100 U/ml penicillin, 100 µg/ml streptomycin, 10% fetal calf serum (FCS, Hydrobond) and minimal essential medium nonessential amino acids (DMEM/10% FCS). The SupT1 cell line was maintained in Advanced RPMI (Gibco) supplemented with L-glutamine, 1% FCS, 30 U/ml penicillin and 30 µg/ml streptomycin. PBMCs were obtained from fresh buffy coats (Central Laboratory Blood Bank) of different HIV-1 negative donors using Ficoll-Hypaque gradient. Cells of four donors were pooled and stored at -150°C. One week prior to infection, cells were thawed, and stimulated for 2 d with phytohemagglutinin (PHA, 4 µg/ml) and Interleukin-2 (IL-2, 100 U/ml) as described previously.61 PBMCs were cultured in RPMI medium supplemented with 10% FCS, IL-2 and without antibiotics. All cells were kept at 37°C and 5% CO2.
Luciferase assay
293T cells were seeded one day prior to transfection in the 24-well plate format. Eighty ng of pLTR-luc or pLTR-NEF1-luc were mixed with 0–50 ng of pTat to activate the LTR promoter. pBluescript was added to 250 ng of total DNA amount. Transfection was performed with Lipofectamine 2000 following the manufacturer’s instructions. Cells were lysed 48 h post-transfections with 150 µl of Passive Lysis Buffer (Promega) and Firefly expression was measured as described previously.62
HIV-1 production, virus replication and virus competition studies
Virus production was measured in 293T cells that were seeded in the 24-well plate format and transfected with 200 ng of the pLAI variants with the Lipofectamine 2000 protocol. Culture supernatants were collected after 48 h and virus production was scored by CA-p24 ELISA. These experiments were conducted in duplicate or triplicate and performed on three occasions. Variation between the three independent experiments was corrected using the factor correction program.63
HIV-1 virus stocks were produced on 293T cells, which were seeded in a 6-well plate format one day prior to transfection. Four µg of wt or mutant pLAI was transfected following the Lipofectamine 2000 protocol and the supernatant was harvested at 48 h post transfection. Cells were removed by centrifugation (4,000 × g) and the supernatant was aliquoted and stored at -80°C. Virus production was quantified by CA-p24 ELISA. HIV-1 (0.001 ng of CA-p24) was used to infect 200,000 SupT1 cells in the 24-well plate format, with three or six parallel cultures. PBMC infections were performed in quadruplicate in the 96-well plate format with 200,000 PBMCs and HIV-1 (0.1–1 ng CA-p24)-. Supernatants of the SupT1 and PBMC cultures were collected three times a week to measure CA-p24, and cultures were monitored for cytopathic effects using light microscopy.
Virus competition studies were performed to detect subtle replication differences. One hundred and fifty ng of the mutant plasmids (Pol1, Pol2, Pol3, Nef1, double, triple and quadruple) were individually mixed with an equal amount (150 ng) of wt pLAI. Input plasmid mixtures were sequenced to verify the equimolar input, which is displayed by a mixed sequence with equally high peaks. SupT1 cells were transfected with this equimolar plasmid mixture via electroporation (250V, 975 µF) using the Bio-Rad Gene Pulser II.21 Cells were split once or twice a week and cell-free virus was passaged onto fresh SupT1 cells when massive virus-induced cytopathic effects were detected for up to 8 wk.21,64 This was performed in three parallel cultures. The dominant variant in each culture was determined by isolation of the total cellular DNA, PCR amplification of the mutated region in integrated HIV-1 proviruses and determination of the population sequence. Total cellular DNA was isolated using proteinase K treatment as previously described,65 and the regions encompassing the altered motifs were PCR amplified and sequenced. For POL1, POL2 and POL3, oligonucleotides FGSA-32-Fb and RT-Pfo-as were used to amplify a 1.7 kb Pol fragment. For NEF1, primers 5′NEF-1 and tTA-rev1-AD amplified the respective part of the nef/U3 region. The quality of PCR products was checked on agarose gel. Motifs POL1 and POL2 were subsequently sequenced with FGSA-32-Fb, POL3 with TA051 and NEF1 with 5′NEF-LTR01. All oligonucleotides are listed in Table 2.
Supplementary Material
Acknowledgments
We are grateful to Atze Das and Alex Harwig for the generous gift of LTR-luciferase constructs, Ying Poi Liu for manuscript editing and Monique Vink for cell culture advice. We thank Emily Mouser and Thijs van Montfort for providing PBMC samples and Stephan Heynen for performing CA-p24 ELISA. We thank Kevin M. Weeks for critical reading of the manuscript. The study was promoted by a NWO TOP grant to B.B., S.A.K. was supported by the DAAD (German Academic Exchange Service).
Glossary
Abbreviations:
- SHAPE
selective 2’-hydroxyl acylation analyzed by primer extension
- nt(s)
nucleotide(s)
- GORS
genome-scale ordered RNA structure
- HIV-1
human immunodeficiency virus type 1
- RNAi
RNA interference
- shRNA
short hairpin RNA
- TAR
trans-activation response
- RRE
rev-responsive element
- 5′ UTR
5′ untranslated region
- wt
wild-type
- PBMCs
peripheral blood mononuclear cells
- RLRs
retinoic acid inducible gene-I-like receptors
- DRBPs
dsRNA-binding proteins
- siRNA
small-interfering RNA
- RISC
RNA-induced silencing complex
- miRNA
micro RNA
- PHA
phytohemagglutinin
- IL-2
interleukin-2
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/24133
References
- 1.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–31. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 2.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr., Swanstrom R, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–6. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc. 2007;129:4144–5. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 4.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–90. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–6. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 6.Simmonds P, Tuplin A, Evans DJ. Detection of genome-scale ordered RNA structure (GORS) in genomes of positive-stranded RNA viruses: Implications for virus evolution and host persistence. RNA. 2004;10:1337–51. doi: 10.1261/rna.7640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis M, Sagan SM, Pezacki JP, Evans DJ, Simmonds P. Bioinformatic and physical characterizations of genome-scale ordered RNA structure in mammalian RNA viruses. J Virol. 2008;82:11824–36. doi: 10.1128/JVI.01078-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Eije KJ, Berkhout B. RNA-interference-based gene therapy approaches to HIV type-1 treatment: tackling the hurdles from bench to bedside. Antivir Chem Chemother. 2009;19:221–33. doi: 10.1177/095632020901900602. [DOI] [PubMed] [Google Scholar]
- 9.Knoepfel SA, Centlivre M, Liu YP, Boutimah F, Berkhout B. Selection of RNAi-based inhibitors for anti-HIV gene therapy. World Journal of Virology. 2012;1:79–90. doi: 10.5501/wjv.v1.i3.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33:796–804. doi: 10.1093/nar/gki220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westerhout EM, Berkhout B. A systematic analysis of the effect of target RNA structure on RNA interference. Nucleic Acids Res. 2007;35:4322–30. doi: 10.1093/nar/gkm437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Low JT, Knoepfel SA, Watts JM, ter Brake O, Berkhout B, Weeks KM. SHAPE-directed discovery of potent shRNA inhibitors of HIV-1. Mol Ther. 2012;20:820–8. doi: 10.1038/mt.2011.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aboul-ela F, Karn J, Varani G. The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein. J Mol Biol. 1995;253:313–32. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 14.Aboul-ela F, Karn J, Varani G. Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucleic Acids Res. 1996;24:3974–81. doi: 10.1093/nar/24.20.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berkhout B. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic Acids Res. 1992;20:27–31. doi: 10.1093/nar/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dingwall C, Ernberg I, Gait MJ, Green SM, Heaphy S, Karn J, et al. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci USA. 1989;86:6925–9. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–7. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 18.Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–3. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 19.Wilson W, Braddock M, Adams SE, Rathjen PD, Kingsman SM, Kingsman AJ. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988;55:1159–69. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- 20.Klasens BIF, Thiesen M, Virtanen A, Berkhout B. The ability of the HIV-1 AAUAAA signal to bind polyadenylation factors is controlled by local RNA structure. Nucleic Acids Res. 1999;27:446–54. doi: 10.1093/nar/27.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abbink TE, Berkhout B. RNA structure modulates splicing efficiency at the HIV-1 major splice donor. J Virol. 2007;82:3090–8. doi: 10.1128/JVI.01479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aldovini A, Young RA. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–6. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baudin F, Marquet R, Isel C, Darlix JL, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J Mol Biol. 1993;229:382–97. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 24.Clavel F, Orenstein JM. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J Virol. 1990;64:5230–4. doi: 10.1128/jvi.64.10.5230-5234.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harrison GP, Lever AML. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992;66:4144–53. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laughrea M, Jetté L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–74. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 27.Lever A, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–7. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Reilly MM, McNally MT, Beemon KL. Two strong 5′ splice sites and competing, suboptimal 3′ splice sites involved in alternative splicing of human immunodeficiency virus type 1 RNA. Virology. 1995;213:373–85. doi: 10.1006/viro.1995.0010. [DOI] [PubMed] [Google Scholar]
- 29.Purcell DFJ, Martin MA. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol. 1993;67:6365–78. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heng X, Kharytonchyk S, Garcia EL, Lu K, Divakaruni SS, LaCotti C, et al. Identification of a minimal region of the HIV-1 5′-leader required for RNA dimerization, NC binding, and packaging. J Mol Biol. 2012;417:224–39. doi: 10.1016/j.jmb.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakuragi J, Ode H, Sakuragi S, Shioda T, Sato H. A proposal for a new HIV-1 DLS structural model. Nucleic Acids Res. 2012;40:5012–22. doi: 10.1093/nar/gks156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abbink TEM, Berkhout B. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J Biol Chem. 2003;278:11601–11. doi: 10.1074/jbc.M210291200. [DOI] [PubMed] [Google Scholar]
- 33.Ooms M, Abbink TE, Pham C, Berkhout B. Circularization of the HIV-1 RNA genome. Nucleic Acids Res. 2007;35:5253–61. doi: 10.1093/nar/gkm564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, et al. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science. 2011;334:242–5. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon-Loriere E, Martin DP, Weeks KM, Negroni M. RNA structures facilitate recombination-mediated gene swapping in HIV-1. J Virol. 2010;84:12675–82. doi: 10.1128/JVI.01302-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malim MH, Tiley LS, McCarn DF, Rusche JR, Hauber J, Cullen BR. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990;60:675–83. doi: 10.1016/0092-8674(90)90670-A. [DOI] [PubMed] [Google Scholar]
- 37.Peleg O, Trifonov EN, Bolshoy A. Hidden messages in the nef gene of human immunodeficiency virus type 1 suggest a novel RNA secondary structure. Nucleic Acids Res. 2003;31:4192–200. doi: 10.1093/nar/gkg454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acid Res Mol Biol. 1996;54:1–34. doi: 10.1016/S0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 39.Kuiken C, Foley B, Leitner T, Apetrei C, Hahn B, Mizrachi I, et al. HIV Sequence Compendium 2010. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory. 2010. [Google Scholar]
- 40.Olivetta E, Pugliese K, Bona R, D’Aloja P, Ferrantelli F, Santarcangelo AC, et al. cis expression of the F12 human immunodeficiency virus (HIV) Nef allele transforms the highly productive NL4-3 HIV type 1 to a replication-defective strain: involvement of both Env gp41 and CD4 intracytoplasmic tails. J Virol. 2000;74:483–92. doi: 10.1128/JVI.74.1.483-492.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koken SEC, van Wamel JL, Goudsmit J, Berkhout B, Geelen JL. Natural variants of the HIV-1 long terminal repeat: analysis of promoters with duplicated DNA regulatory motifs. Virology. 1992;191:968–72. doi: 10.1016/0042-6822(92)90274-S. [DOI] [PubMed] [Google Scholar]
- 42.Beekwilder MJ, Nieuwenhuizen R, van Duin J. Secondary structure model for the last two domains of single-stranded RNA phage Q β. J Mol Biol. 1995;247:903–17. doi: 10.1006/jmbi.1995.0189. [DOI] [PubMed] [Google Scholar]
- 43.Skripkin EA, Adhin MR, de Smit MH, van Duin J. Secondary structure of the central region of bacteriophage MS2 RNA. Conservation and biological significance. J Mol Biol. 1990;211:447–63. doi: 10.1016/0022-2836(90)90364-R. [DOI] [PubMed] [Google Scholar]
- 44.Berkhout B, van Duin J. Mechanism of translational coupling between coat protein and replicase genes of RNA bacteriophage MS2. Nucleic Acids Res. 1985;13:6955–67. doi: 10.1093/nar/13.19.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt BF, Berkhout B, Overbeek GP, van Strien A, van Duin J. Determination of the RNA secondary structure that regulates lysis gene expression in bacteriophage MS2. J Mol Biol. 1987;195:505–16. doi: 10.1016/0022-2836(87)90179-3. [DOI] [PubMed] [Google Scholar]
- 46.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–5. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 47.Levy DE, García-Sastre A. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 2001;12:143–56. doi: 10.1016/S1359-6101(00)00027-7. [DOI] [PubMed] [Google Scholar]
- 48.Girardin SE, Sansonetti PJ, Philpott DJ. Intracellular vs extracellular recognition of pathogens--common concepts in mammals and flies. Trends Microbiol. 2002;10:193–9. doi: 10.1016/S0966-842X(02)02334-X. [DOI] [PubMed] [Google Scholar]
- 49.Saunders LR, Barber GN. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 2003;17:961–83. doi: 10.1096/fj.02-0958rev. [DOI] [PubMed] [Google Scholar]
- 50.Player MR, Torrence PF. The 2-5A system: modulation of viral and cellular processes through acceleration of RNA degradation. Pharmacol Ther. 1998;78:55–113. doi: 10.1016/S0163-7258(97)00167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, et al. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol Biol. 2007;8:63. doi: 10.1186/1471-2199-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ouellet DL, Plante I, Landry P, Barat C, Janelle ME, Flamand L, et al. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 2008;36:2353–65. doi: 10.1093/nar/gkn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lefebvre G, Desfarges S, Uyttebroeck F, Muñoz M, Beerenwinkel N, Rougemont J, et al. Analysis of HIV-1 expression level and sense of transcription by high-throughput sequencing of the infected cell. J Virol. 2011;85:6205–11. doi: 10.1128/JVI.00252-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schopman NC, Willemsen M, Liu YP, Bradley T, van Kampen A, Baas F, et al. Deep sequencing of virus-infected cells reveals HIV-encoded small RNAs. Nucleic Acids Res. 2012;40:414–27. doi: 10.1093/nar/gkr719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–44. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 56.Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berkhout B, van Hemert FJ. The unusual nucleotide content of the HIV RNA genome results in a biased amino acid composition of HIV proteins. Nucleic Acids Res. 1994;22:1705–11. doi: 10.1093/nar/22.9.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Hemert FJ, Berkhout B. The tendency of lentiviral open reading frames to become A-rich: constraints imposed by viral genome organization and cellular tRNA availability. J Mol Evol. 1995;41:132–40. doi: 10.1007/BF00170664. [DOI] [PubMed] [Google Scholar]
- 59.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Das AT, Harwig A, Berkhout B. The HIV-1 Tat protein has a versatile role in activating viral transcription. J Virol. 2011;85:9506–16. doi: 10.1128/JVI.00650-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Montfort T, Thomas AA, Pollakis G, Paxton WA. Dendritic cells preferentially transfer CXCR4-using human immunodeficiency virus type 1 variants to CD4+ T lymphocytes in trans. J Virol. 2008;82:7886–96. doi: 10.1128/JVI.00245-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Knoepfel SA, Abad A, Abad X, Fortes P, Berkhout B. Design of modified U1i molecules against HIV-1 RNA. Antiviral Res. 2012;94:208–16. doi: 10.1016/j.antiviral.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 63.Ruijter JM, et al. Factor correction as a tool to eliminate between-session variation in replicate experiments: application to molecular biology and retrovirology. Retrovirology. 2006;3:1–8. doi: 10.1186/1742-4690-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klaver B, Berkhout B. Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. EMBO J. 1994;13:2650–9. doi: 10.1002/j.1460-2075.1994.tb06555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beerens N, Berkhout B. The tRNA primer activation signal in the HIV-1 genome is important for initiation and processive elongation of reverse transcription. J Virol. 2002;76:2329–39. doi: 10.1128/jvi.76.5.2329-2339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.