Abstract
The RNA-binding protein TIAR is an mRNA-binding protein that acts as a translational repressor, particularly important under conditions of cellular stress. It binds to target mRNA and DNA via its RNA recognition motif (RRM) domains and is involved in both splicing regulation and translational repression via the formation of “stress granules.” TIAR has also been shown to bind ssDNA and play a role in the regulation of transcription. Here we show, using surface plasmon resonance and nuclear magnetic resonance spectroscopy, specific roles of individual TIAR domains for high-affinity binding to RNA and DNA targets. We confirm that RRM2 of TIAR is the major RNA- and DNA-binding domain. However, the strong nanomolar affinity binding to U-rich RNA and T-rich DNA depends on the presence of the six amino acid residues found in the linker region C-terminal to RRM2. On its own, RRM1 shows preferred binding to DNA over RNA. We further characterize the interaction between RRM2 with the C-terminal extension and an AU-rich target RNA sequence using NMR spectroscopy to identify the amino acid residues involved in binding. We demonstrate that TIAR RRM2, together with its C-terminal extension, is the major contributor for the high-affinity (nM) interactions of TIAR with target RNA sequences.
Keywords: RNA-binding protein, TIAR, TIA-1, RRM, C-terminal extension, translational regulation, surface plasmon resonance (SPR), NMR
Introduction
TIA-1 (T cell restricted intracellular antigen-1) and TIAR (TIA-1 related protein) are nucleo-cytoplasmic shuttling, multi-functional RNA-binding proteins (RBPs) that play important roles in the regulation of gene expression.1,2 In the nucleus, they have been shown to be involved in both the regulation of transcription by binding to single-stranded T-rich DNA (ssDNA) targets and alternative pre-mRNA splicing by binding to U-rich RNA targets and promoting the recognition of atypical 5′ splice sites.1,3-10 In the cytoplasm, they are involved in translational repression when cells are under stress by binding to specific RNA elements (sometimes rich in specific residues, such as U, CU, AU, etc.) of target mRNAs and sequestering the bound mRNA into stress granules, where mRNAs are generally stable and translation is suppressed.11-15 RNA regulatory elements are commonly found in the 3′ and 5′ untranslated regions (UTRs) of mRNAs subject to rapid changes in stability and translation, including those that encode cytokines, pro-inflammatory mediators, stress-response proteins, extracellular matrix remodeling enzymes and oncoproteins.16-19 Most recently, TIA-1 and TIAR have also been found to arrest translation at the initiation step, through the binding of 5′-oligopyrimidine (TOP) elements under periods of stress. TOP elements occur predominantly in mRNA encoding ribosomal proteins and translation factors that are not required during stress.20
There has thus been much interest in characterizing the molecular basis for substrate recognition by TIA-1 and TIAR. Investigations of its ability to bind to DNA suggest that TIA-1 and TIAR are recruited to transcriptionally active T-rich DNA from where they would be positioned to form complexes with target mRNA.10 This could assist their binding to mRNA target sequences preferentially over other RBPs with similar affinities, such as AU-binding factor 1 (AUF1), Tristetraprolin (TTP) and Human antigen R (HuR) to dictate the final outcome for the mRNA. While TIA protein binding to target mRNAs can lead to translational repression, AUF1 or TTP binding leads to rapid decay of the target mRNA.12,15,21-25 Alternatively, HuR-mRNA interactions can lead to stabilization of the mRNA targets26,27 or, in some cases, to reduced translation.28 This interplay is particularly important in fine-tuning the innate immune response where slightly prolonged expression of inflammatory genes leads to chronic inflammation.29 It is proposed that TIAR plays a role in preventing the pathological overexpression of inflammatory mediators such as TNF-α, IL-1β and COX2 as occurs in rheumatoid arthritis.29 Previously, we have reported on the ability of TIAR to bind to C-rich sequences30 and on the similarities and fundamental differences between TIAR-mRNA and HuR-mRNA interactions in terms of their binding affinities, kinetics and mode of interactions.31 Here we further characterize the roles of different regions of TIAR in binding to substrate RNAs to better understand how exactly it exerts its strong binding to mRNAs that bear these sequences.
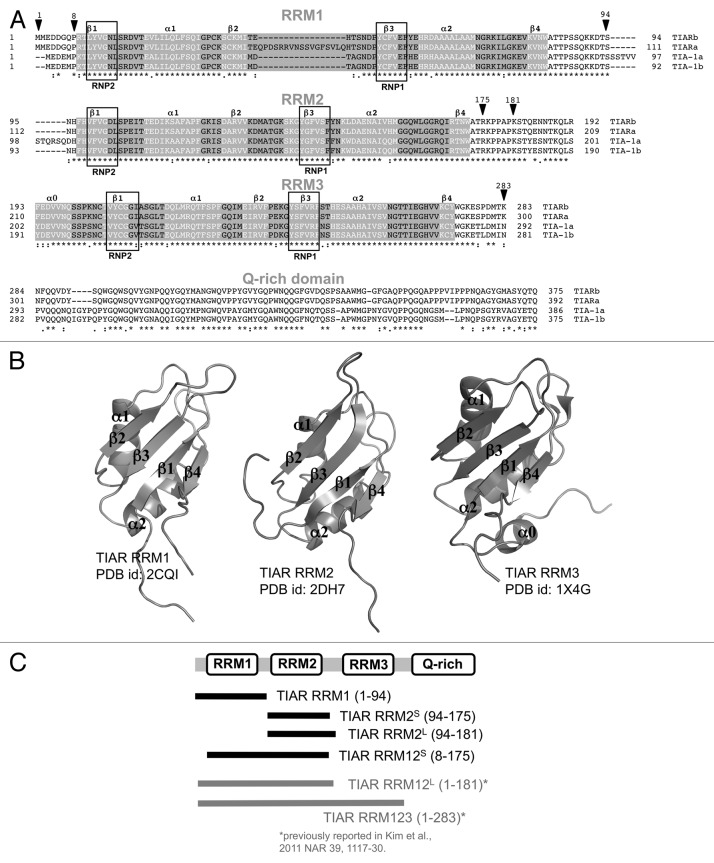
TIAR is a 375-amino acid protein belonging to the RNA-recognition motif (RRM) containing a family of RNA-binding proteins [also known as the RNA-binding domain (RBD) or ribonucleoprotein domain (RNP)]. It possesses three RRMs, which share high-sequence homology with TIA-1 (RRM1: 79%, RRM2: 92%, RRM3: 91%) and a glutamine-rich carboxyl terminal region, which shares 51% homology with TIA-1 (Fig. 1A). Longer isoforms (TIARa and TIA-1a) created by alternative splicing also exist for both TIAR and TIA-132 and it has been shown that the relative expression of TIAR and TIA-1 isoforms varies in different tissues and cell lines suggesting distinct functional properties and regulatory mechanisms underlying the isoform expression.33 SELEX studies by Dember and colleagues reported that TIAR preferentially binds to U-rich sequences.34 They showed that the three RRMs of TIAR confer high affinity binding to U-rich RNA sequences with each contributing variously to the interaction (overall KD ~20 nM), while the C-terminal 90-amino acid residue glutamine-rich sequence is essential for stress-granule formation.13,15,34,35 Since then, it has been recognized that TIAR also forms stable interactions with AU-rich sequences.31
Figure 1. TIA protein sequence alignment and domain structure. (A) Sequence alignment of TIA protein isoforms highlighting secondary structural elements within the RRMs as derived from structural information (in white font) and positions of RNP motifs (boxed). (B) Cartoon representations of TIAR RRM domains determined by NMR (PDB IDs: 2CQI; 2DH7, 1X4G) showing labeled secondary structural elements. (C) Schematic showing the bounds of TIAR constructs used in the current and a previous study.31
The RRM is a very commonly used motif in the cell (with 0.5‒1% of all human genes utilizing an RRM) and its general structure is well described.36,37 The RRM is 70–90 amino acids long and consists of a four-stranded β-sheet packed against two α-helices with βαββαβ topology (Fig. 1B). It has been observed that most RRMs specifically recognize between two and eight single-stranded nucleotides against the surface of the four-stranded β-sheet. In particular, RRMs contain two ribonucleoprotein (RNP) consensus domains (known as RNP1 and RNP2) that contain three conserved aromatic residues. The RNPs accommodate two bound nucleotides via ring-stacking interactions and a network of specific hydrogen bonds that dictate molecular specificity.36,37 The RNA-binding is not always limited to the RNPs, however. Recent studies reveal deviations from the classic mode of interaction where the linker region between RRMs, the loops between secondary structure components or the C-terminal extension of the RRM also contribute significantly toward RNA-binding specificity and affinity.38-41
TIAR and TIA-1 RRMs have been subject to both structural and biophysical studies to better understand their interactions with oligonucleotide. No structure of a TIA protein RRM in complex with RNA is yet reported, though structures of the individual TIAR RRMs have been elucidated using NMR (PDB IDs: 1X4G, 2DH7, 2CQI). The three RRMs all share the canonical RRM fold with the three conserved aromatics in the RNPs. The structure of the TIA-1 RRM2 has also been solved, by both NMR and X-ray crystallography, and its RNA interaction surface has been investigated using NMR.40,42 Chemical shift perturbation analyses with pentamer U-rich RNA (5′-UUUUU-3′) revealed that most of the RRM2 β-sheet surface, including residues within or adjacent to conserved RNP consensus motifs and the β4 C-terminal extension, is involved in the RNA binding.
Despite their seeming similarity in structural features for the accommodation of oligonucleotide, the three RRMs of TIA proteins do not contribute equally to oligonucleotide binding. The early work by Dember et al.,34 using nitrocellulose filter binding assays, showed that RRM2 is both sufficient and necessary for binding to U-rich elements and that RRM3 can bind to RNA but may have other specificities besides U-rich elements. This is in agreement with our work using surface plasmon resonance (SPR) to examine TIAR binding to U-rich and AU-rich sequences.31 There was no increased affinity of a TIAR construct comprising all three RRMs (RRM123) compared with TIAR comprising only the first two RRMs (RRM12) to a U-rich sequence, and just a small enhancement in binding to an AU-rich sequence—in agreement with there being only a minor role for RRM3 binding to such sequences. The early Dember study also detected no binding to U-rich RNA by RRM1 alone, though a slightly higher affinity by RRM12 (KD = 40 nM) than RRM2 (KD = 50 nM) suggested that RRM1 does contribute a little toward binding to U-rich sequences.34 Later studies reporting affinity measurements using a UV-cross-linking method by Suswam et al.10 revealed that while TIAR RRM1 interactions with U-rich RNA were not detectible, interactions with T-rich ssDNA occurred with significant affinity (KD = 43 nM), suggesting that the primary role of RRM1 is to interact with DNA. In a slightly different approach, Bauer et al.43 tested U-rich RNA binding by TIA-1 RRM mutants using ITC. Their work was consistent with all three RRMs being involved in binding to U-rich RNA, but with unequal contributions.
Having established that TIAR RRM3 contributes negligibly to the overall binding affinity to U-rich sequences as detected by SPR, we turned our attention to the first two RRM domains of TIAR. Here, we investigate both the affinity and kinetics of different TIAR RRM domains binding to a 17-nt U-rich RNA sequence to fully explore the roles of separate TIAR domains in achieving the high affinity (nanomolar) interactions observed. Kinetics studies provide additional insight into the basis for complex affinity and specificity. Since it has been shown that TIAR RRM123 interacts strongly with ssDNA as well as with RNA,10,31 we further characterized the roles of the first two RRMs of TIAR in T-rich DNA binding compared with U-rich RNA binding, showing the involvement of both domains in DNA interactions. We also report the unexpected enhancement by the C-terminal extension of TIAR RRM2 toward binding to substrate U-rich RNA and T-rich DNA. The unstructured region outside or between the classic RRM motifs has previously been noted for its ability to interact with target oligonucleotide.39-41,44,45 Thus, it was of interest to discover how it affects RNA and DNA binding by TIAR. Finally, NMR spectroscopy was employed to obtain further insight into the involvement of the specific TIAR RRM2 residues as well as those in the C-terminal extension in binding RNA in solution.
Results
Different TIAR domains have distinctive roles in RNA binding
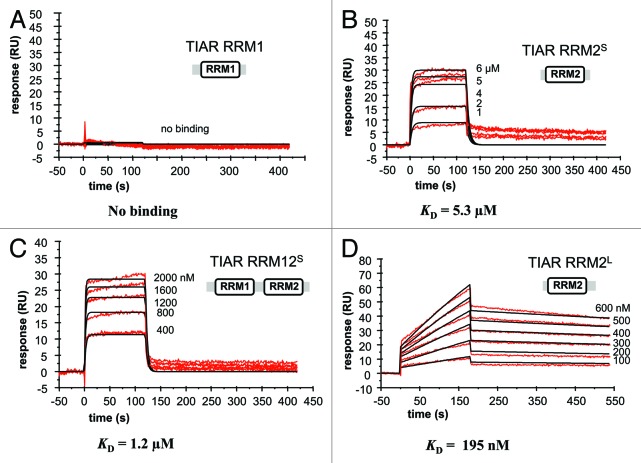
Proteins representing one and two RRM domains of TIAR isoform b (RRM1, RRM2 and RRM12) were prepared in order to characterize their binding to U-rich RNA and T-rich DNA sequences using SPR. It was noted that the original RRM12 and RRM2 constructs (from Dember and colleagues)34 extend 36 amino acids C-terminal to RRM2 (to amino acid residue 208). Shorter constructs were therefore cloned that extend only three residues C-terminal to RRM2 domain as defined by the end of the determined secondary structure (to amino acid residue 175; PDB ID: 2DH7). These are referred to as “RRM12 short” (RRM12S) and “RRM2 short” (RRM2S) (Fig. 1C). It became apparent, however, in the course of our studies, that the original RRM12 and RRM2 are susceptible to specific proteolytic cleavage at residue 181 (as determined by the mass spec-determined molecular weight of the cleaved RRM12 and RRM2 and subsequently confirmed by the full sequential assignments of backbone resonances for RRM2 by NMR). For the purpose of this study, these constructs are referred to as “RRM12 long” (RRM12L) and “RRM2 long” (RRM2L), respectively, for ease of comparison with the shorter constructs. The four sensorgrams (Fig. 2) show the binding of a range of concentrations of RRM1, RRM2S, RRM2L and RRM12S when injected across the U-rich RNA-coated chip. The association rate constants (ka), dissociation rate constants (kd) and overall equilibrium dissociation constants (KD) for each protein, as approximated by a simple 1:1 Langmuir binding model, are shown in Table 1 and the residual plots and statistics (χ2) for the fitting are supplied in Figure S1.
Figure 2. Kinetic analysis of the interactions of different TIAR constructs with U-rich RNA using SPR. The binding of (A) TIAR1, (B) TIAR2S, (C) TIAR12S and (D) TIAR2L to a U-rich RNA is shown. Biotinylated RNA was captured on SA-coated sensor chip and increasing concentrations of protein were injected over the surface. Injections were performed for 120 sec (association phase), followed by a 300 sec flow of running buffer to assess dissociation. The experiments were conducted in duplicate and showed good overlap. The red line represents the binding responses for injections of protein analyte at specified concentration over the RNA surface. The kinetic data were fit by a 1:1 Langmuir binding model. Mass transport effects were not evident. The black curves superimposed on top of the sensor grams represent the model fitted curves. The rate constants ka and kd were determined simultaneously as global fitting parameters from which the KD was determined. The resulting parameter values are given in Table 1.
Table 1. Kinetic and affinity constants for the interactions of different TIAR RRMs with U-rich RNA.
| Protein | RNA | ka (1/Ms) | kd (1/s) | KD (kd/ka1 nM) |
|---|---|---|---|---|
| TIAR RRM1 |
U-rich |
N/A |
N/A |
no binding |
| TIAR RRM2S |
U-rich |
(3.49 ± 0.09) × 104 |
(1.84 ± 0.03) × 10−1 |
5,300 ± 200 |
| TIAR RRM12S |
U-rich |
(2.30 ± 0.02) × 105 |
(2.77 ± 0.02) × 10−1 |
1,200 ± 20 |
| TIAR RRM2L | U-rich | (1.84 ± 0.01) × 103 | (3.60 ± 0.1) × 10−4 | 200 ± 6 |
The association and dissociation rate constants (ka and kd) were determined as global fitting parameters for a 1:1 binding model. The error shown is derived from the fit of the calculated curves to the data. The equilibrium dissociation constant KD was determined as kd/ka.
Our previous studies showed that RRM12L and RRM123 both bind U-rich RNA with nM affinities (KD ~1 nM) and bind with similar association and dissociation rate constants, demonstrating that RRM3 does not contribute detectably to, or impact on, the overall binding affinity to a U-rich RNA sequence.30,31 RRM1 showed no detectable binding to the U-rich RNA (Fig. 2A). Since strong binding was observed for the TIAR RRM12L construct, this confirms RRM2 as the major U-rich RNA-binding domain as consistent with the report by Dember et al.34 We therefore examined the binding of RRM2 alone (RRM2S) with U-rich RNA anticipating strong nanomolar binding. But unexpectedly, we observed a much lower binding affinity of KD of ~5 μM with a fast off-rate evident in the rapid return of the sensorgams to zero at the end of the injection period (Fig. 2B). From this observation, we concluded that either there is a contribution to binding from RRM1 N-terminal to RRM2, and/or that the six extra residues at the C terminus of TIAR RRM12L were also contributing to binding. Thus, we examined the binding of RRM12S and RRM2L to U-rich RNA to be able to distinguish between the role of RRM1 and the C-terminal extension in contributing to RRM2 binding respectively (Fig. 2C and D). Interestingly, RRM12S bound with a 5-fold higher affinity (KD ~1 μM) than RRM2S, though a fast off-rate was still observed, and RRM2L bound with > 25-fold higher affinity than RRM2S (KD = 195 nM), reflecting the dramatically slower off-rate kinetics. This suggests that not only is the RRM2 domain responsible for U-rich RNA binding, but that some or all of TIAR N-terminal to RRM2, and especially the C-terminal extension beyond the classically structured RRM2 motif, contribute significantly toward U-rich RNA binding.
The detailed kinetic analyses of separate TIAR RRMs binding U-rich RNA is shown in Table 1, and it reveals some very interesting “hidden” information about their binding. The presence of six extension residues (KPPAPK) at the C terminus of RRM2 has a remarkable effect on the dissociation rate constant, but not so much on the association rate constants of binding. RRM2L as well as RRM12L from our previous studies31 both have much slower dissociation rate constants of ~10−3 to 10−4s−1 compared with ~10−1 s−1 for those without the extension (RRM2S and RRM12S) (Table 1). However, the association rate constants are closer in value for both RRM2S and RRM2L (~104 to 103 M−1s−1) as well as for RRM12S and RRM12L (~105 to 106 M−1s−1) (Table 1; ref. 31). This suggests that the C-terminal extension residues are more likely to be involved in the stability of the TIAR-RNA interactions, but not so much in the initial formation of interactions.
In contrast, the effect of the presence of RRM1 is on the association rate constant. A comparison of RRM12L and RRM2L reveals that they both have dissociation rate constants that are close in value (~10−3 to 10−4s−1) but a very much increased association rate constant when RRM1 is present (~103 for RRM2L compared with 106 M−1s−1 for RRM12L) (Table 1; ref. 31). Comparing RRM12S vs RRM2S also shows that they have similar dissociation rate constants (~10−1s−1) but that the association rate constant increases when RRM1 is present (~104 for RRM2S compared with ~105 M−1s−1 for RRM12S). These results suggest that the RRM1 and/or the linker region between RRM1 and 2 could potentially be involved in the initial interaction with the U-rich RNA, giving rise to the faster association rate constants, but not likely to be involved in the stability of the TIAR-RNA complex.
Different TIAR domains also have distinctive roles in DNA binding
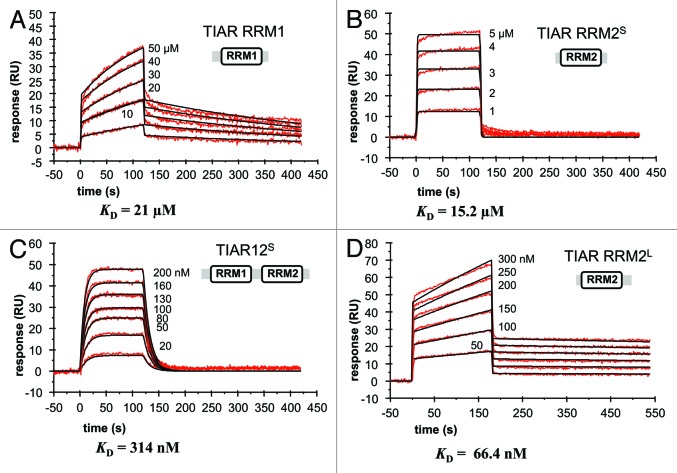
Since TIAR has also been identified by ourselves and others as interacting with T-rich DNA via its first two RRM domains,10,31 it was of interest to also perform a detailed binding study with the same TIAR constructs (RRM1, RRM2S, RRM2L and RRM12S). Their binding to a T-rich DNA sequence was therefore characterized using SPR (Fig. 3). The four sensorgrams show the binding of a range of concentrations of RRM1, RRM2S, RRM2L and RRM12S when injected across the T-rich DNA-coated chip. The association and dissociation rate constants (ka and kd) and overall equilibrium dissociation constants (KD) for each binding were estimated by a 1:1 Langmuir binding model (Table 2) and the residual plots and statistics (χ2) for the fitting are supplied in Figure S1.
Figure 3. Kinetic analysis of the interactions of different TIAR constructs with T-rich DNA using SPR. The binding of (A) TIAR1, (B) TIAR2S, (C) TIAR12S and (D) TIAR2L to a T-rich DNA is shown. Biotinylated DNA was captured on SA-coated sensor chip and increasing concentrations of protein were injected over the surface. Injections were performed for 120 sec (association phase), followed by a 300 sec flow of running buffer to assess dissociation. The experiments were conducted in duplicate and showed good overlap. The red line represents the binding responses for injections of protein analyte at specified concentration over the DNA surface. The kinetic data were fit by a 1:1 Langmuir binding model. Mass transport effects were not evident. The black curves superimposed on top of the sensor grams represent the model fitted curves. The rate constants ka and kd were determined simultaneously as global fitting parameters from which the KD was determined. The resulting parameter values are given in Table 2.
Table 2. Kinetic and affinity constants for the interactions of different TIAR RRMs with T-rich DNA.
| Protein | DNA | ka (1Ms) | kd (1/s) | KD (kd/ka1 nM) |
|---|---|---|---|---|
| TIAR RRM1 |
T-rich |
(1.12 ± 0.02) × 102 |
(2.34 ± 0.02) × 10−3 |
20,900 ± 550 |
| TIAR RRM2S |
T-rich |
(7.13 ± 0.1) × 104 |
1.09 ± 0.01 |
15,200 ± 350 |
| TIAR RRM12s |
T-rich |
(2.54 ± 0.01) × 105 |
(7.98 ± 0.02) × 10−2 |
314 ± 2 |
| TIAR RRM2L | T-rich | (3.36 ± 0.06) × 103 | (2.23 ± 0.05) × 10−4 | 66 ± 3 |
The association and dissociation rate constants (ka and kd) were determined as global fitting parameters for a 1:1 binding model. The error shown is derived from the fit of the calculated curves to the data. The equilibrium dissociation constant KD was determined as kd/ka.
Figure 3A shows that RRM1 binds DNA with micromolar affinity (21 μM) indicating a dramatic preference for binding DNA over RNA. RRM12L from our previous study however showed strong binding to the T-rich DNA with low nanomolar affinity (200-fold stronger binding), confirming RRM2 as the major DNA-binding domain.31 We therefore examined the binding of RRM2 alone (RRM2S) to DNA again anticipating strong nanomolar affinity binding. But just as we observed with the RNA binding, we measured a relatively low affinity of KD ~15 μM (Fig. 3B). This suggested that again, there is a contribution to binding from RRM1 N-terminal to RRM2, and/or that the six extra residues at the C terminus of TIAR RRM12L also contribute to binding. Thus, we examined the binding of RRM12S and RRM2L to T-rich DNA to be able to distinguish between the role of RRM1 and the C-terminal extension residues in contributing to RRM2 binding to DNA, respectively. Interestingly, RRM12S bound with ~50-fold higher affinity (~300 nM) than RRM2S and RRM2L binds with > 100-fold higher affinity than RRM2S (Fig. 3C and D). These results demonstrate that the first two RRM domains of TIAR both participate in T-rich DNA binding. TIAR RRM2 is still the major binding domain and the six C-terminal extension residues of RRM2 is heavily involved in binding again. RRM1, however, shows μM binding to T-rich DNA whereas it did not show any binding to U-rich RNA on its own.
The detailed kinetic analysis of separate TIAR RRMs binding T-rich DNA is shown in Table 2, and again it provides us with insight into TIAR binding. As was observed for RNA binding, the presence of the C-terminal extension mainly impacts on the dissociation rate constant. RRM2L has a much slower dissociation rate constant (~10−4s−1) compared with RRM2S (~1 sec−1), whereas the association rate constants are relatively similar (~104 to 105 M−1s−1). This suggests that the C-terminal extension of RRM2 plays an important role stabilizing complex formation with DNA, but does not contribute to the initial formation of the complex.
The presence of RRM1, however, impacts on both the association and dissociation rate constants in a way that promotes TIAR binding to DNA. RRM12S has a slower dissociation rate constant (10−2 s−1) than RRM2S (~1 sec−1) and a slightly faster association rate constant (~105 M−1s−1) compared with that of RRM2S (~104 M−1s−1) (Table 2). RRM1 then appears to contribute to the initial interaction with DNA and also to the stability of the TIAR-DNA complex.
Key residues in TIAR RRM2 and its C-terminal linker region involved in binding U-rich RNA
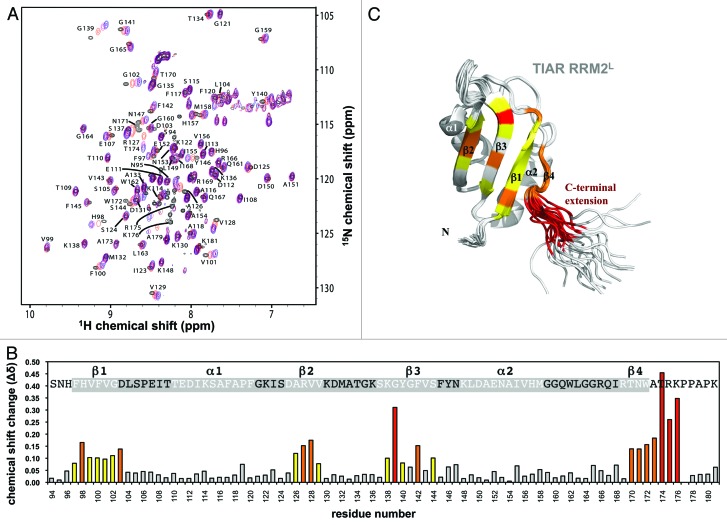
Having confirmed TIAR RRM2 with its short C-terminal extension of six amino acid residues (KPPAPK) as the key RNA (and DNA) binding domain, it was of interest to identify the specific regions (residues) involved in binding U-rich RNA, especially in the extension region. Thus, NMR spectroscopy was employed to examine their interactions in solution. The involvement of linker regions in binding to U-rich RNA has been reported previously in other RRM proteins such as TIA-1 and HuR/D,39,40,44,45 but identification of specific residues within the region and their direct impact on binding affinity and kinetics to U-rich targets has not been fully explored. Double-labeled (15N,13C)-RRM2L was prepared from which the sequence-specific resonance assignments of H, HN, C’, Cα and Cβ were generated for RRM2L (94–181), both in the apo state and in complex with AU-rich RNA (5′ UUAUUU 3′). The assignments of the complex were essentially complete with the exception of the resonances for P177. Figure 4A shows (15N,1H)-HSQC spectra of RRM2L alone and in complex with different concentrations (protein:RNA molar ratio of 1:0, 1:0.4, 1:0.8 and 1:1.2) of AU-rich RNA (5′ UUAUUU 3′). The chemical shift perturbation values of backbone amide, Cα and Cβ resonances upon binding of RNA were quantified from the HNCACB spectra acquired in the apo state and at a protein:RNA ratio of 1:1.2. The length of the 6-nt AU-rich RNA was chosen to maximize the interaction with the protein while minimizing the possibility of observing multiple modes of binding, and its sequence was designed to provide a more stable complex with TIAR than U-rich sequence based on our previous findings from the kinetic studies of TIAR binding U- vs. AU-rich RNA.31
Figure 4. Perturbations of TIAR2L chemical shifts upon RNA binding. (A) An overlay of the assigned (15N,1H)-HSQC spectra of TIAR2L alone and in complex with different concentrations of 6-nt AU-rich RNA (5′-UUAUUU-3′). The molar ratio of TIAR2L to RNA is 1:0 (black), 1:0.4 (red), 1:0.8 (pink) and 1:1.2 (blue). (B) Chemical shift perturbation values for each residue were calculated at the TIAR2L to RNA 1:1.2 ratio as Δδ = {[0.154 (δ15N)2+ 0.25 (δ13Cα)2+ 0.25 (δ13Cβ)2+ (δ1H)2]/4}1/2 and plotted against the amino acid sequence of TIAR2L. The 20 residues showing the largest perturbations upon RNA-binding are colored (red > 0.20 ppm; orange > 0.14 ppm; yellow > 0.08 ppm). (C) Cartoon representation of the TIAR RRM2 structure (PDB ID: 2DH7) with residues affected by RNA-binding colored as in (B) with residues that showed lesser chemical shift perturbations upon RNA binding colored gray.
Upon addition of the RNA, progressive chemical shift perturbations of specific RRM2L amide resonances were observed with increasing RNA concentrations, indicating that these RRM2L resonances are in fast chemical exchange between their apo- and RNA-bound states compared with the NMR time-scale. Overall, the cross-peaks strongly affected by the binding to AU-rich RNA are located on the β-sheet side of RRM2L and include β1 (F97-D103), β2 (A126-V129), β3 (K138, G139, G141, F142 and S144), β4 (T170-A173) and the C-terminal extension residues T174, R175 and K176 (Fig. 4). Interestingly, the C-terminal extension residues, T174, R175 and K176, were among the most strongly affected by RNA binding. These results strongly support our SPR binding data as they are consistent with both RRM2 and the C-terminal extension region interacting with the RNA target underlying the observed nanomolar affinity. Moreover, residues P178, A179, P180 and K181 show only small chemical shift perturbation on RNA binding suggesting that these residues are not involved.
Discussion
The roles of TIAR domains in the formation of stable complexes with RNA and DNA
TIA proteins play critical roles in mRNA splicing, translational repression and sequestration of mRNA into stress granules. Previous studies have shown that TIA-1 and TIAR strongly interact with both RNA and DNA targets with nanomolar affinities.10,30,31,34 They do so via direct interactions with U-rich RNA and with T-rich DNA, respectively, primarily through RRM domains 1 and 2. In the current study, we further explore and compare the unique roles of the first two RRM domains of TIAR binding by studying the kinetics and affinity of their interactions with U-rich RNA and T-rich DNA using SPR. Four constructs were prepared that encompassed either TIAR RRM1 alone (RRM1) or in tandem with a “short” version of RRM2 (RRM12S), a “short” version of RRM2 alone (RRM2S) and a “longer” form of RRM2 with six extra residues at the C-terminal end of the construct (RRM2L). Between these four constructs we could tease out the contributions of the various domains to RNA and DNA binding affinity and kinetics.
Interactions with U-rich RNA are driven by RRM2, with no binding to RNA observed for RRM1 alone. In the case of T-rich DNA binding, both RRM1 and RRM2S individually bind with low micromolar affinity, thus making equivalent and synergistic contributions to binding. The presence of RRM1, in both cases enhances the rate of association with oligonucleotide. Even though binding by RRM1 alone to U-rich RNA could not be observed, its presence could enhance the probability of an initial interaction between the protein and RNA. Only in the case of T-rich DNA binding did RRM1 also contribute to the stability of the complex, as reflected in the slower disassociation constant. Binding of RRM1 to DNA has previously been reported and suggested to facilitate a shuttling between DNA and RNA molecules at the site of active transcription.10 The current data support this model, as RRM1 and RRM2 together confer good affinity and stability to T-rich DNA. Upon the close proximity of U-rich RNA, however, RRM2 could preferentially bind to RNA, weakening the overall affinity to DNA and facilitating a transfer to the RNA upon active transcription of the DNA strand.
The important roles of RRM1 and RRM2 have bearing on the potential differences between TIA protein isoform activities. The TIA protein isoforms (Fig. 1A) are expressed to different extents in different tissues, consistent with their possessing slightly different activities. TIA-1b, the variant more similar to TIAR, exhibits enhanced splicing regulatory activity compared with TIA-1a.46 TIA-1a has an extra 11 residues inserted between RRM1 and RRM2. This suggests that the cooperative behavior of RRMs 1 and 2 may be compromised by this insert. Whether the two TIAR isoforms have different activities is not yet known. Here, we have investigated the shorter of the two isoforms, TIARb. The longer TIARa isoform contains an extra 17 residues in the loop between β2- and β3-strands of RRM1. This extended loop may impact on oligonucleotide binding and awaits further investigation.
An unexpected feature of TIAR’s interactions with oligonucleotide was discovered due to our use of two different length RRM2 constructs. The shorter constructs (RRM2S and RRM12S) were cloned to facilitate structural studies, but upon testing their binding to RNA and DNA, we determined that their binding to oligonucleotide was greatly compromised. In a direct comparison of RRM2S and RRM2L we found that the extra 6 C-terminal residues in the longer construct contributed 25-fold and 100-fold to binding affinities of U-rich RNA and T-rich DNA respectively. This was not due to a big change in the association rate constant of the interactions but, rather, to a dramatic reduction in the dissociation rate constant. Thus, it is this C-terminal extension to RRM2 that is required for the nanomolar affinities reported for TIAR binding to RNA and DNA.10,31
The importance of linker regions in RBP interactions with target RNA has also been shown in studies involving Hu-proteins.39,44 It was shown that the linker region between RRM2 and RRM3 of HuD protein affects the stability of its interaction with substrate RNAs, as the dissociation rate constant increased significantly in its absence whereas the association rate constant remained relatively similar.44 In the case of HuR, the linker region between RRM2 and RRM3 again contributed significantly to binding AU-rich RNA in a length-dependent manner by increasing the RNA-binding affinity (> 30-fold).39 The specific residue(s) within the hinge region of HuR/D proteins responsible for affecting the kinetics and affinity of their AU-rich sequences are yet to be identified.
TIAR RRM2 as well as its C-terminal extension residues are significantly involved in ARE-binding
The above findings clearly indicate that RRM2 and its C-terminal extension residues are the key contributors to the nanomolar affinity interactions observed between TIAR and its substrate RNAs. Thus, we employed NMR spectroscopy to obtain further insight into the involvement of specific residues of TIAR RRM2 and its C-terminal extension in binding target RNA sequences in solution. Chemical shift perturbation mapping revealed involvement of specific residues of RRM2L in forming complex with the 6-nt AU-rich RNA target (5′-UUAUUU-3′). These include residues in RNP1 and 2, as well as the residues on β-sheets and β-α loop. These are classic RNA target-binding sites on the RRM as shown in many previous studies involving the RRM family of proteins and their RNA targets.36,37
However, chemical shift mapping also implicates three residues C-terminal to RRM2: T174, R175 and K176. This finding supports our SPR data and provides direct evidence for the involvement of these residues in the stable TIAR-RNA complex formation. The proline C-terminal to these residues, P177 might also be involved in the interaction but since it precedes another proline, P178 its resonances cannot be assigned using HNCACB/HN(CO)CACB spectra acquired in the current study. However, P178, A179, P180 and K181 were not significantly perturbed, suggesting that the interaction does not extend to this part of the linker region. More interestingly, of the three residues at the C terminus of RRM2 involved in binding, two (T174 and R175) are included in all our TIAR constructs containing RRM2. K176 however is only included in the longer RRM2 constructs used in our SPR experiments (RRM2L as well as RRM12L from our previous studies),30,31 which showed higher affinity interactions with U-rich RNA as a result of their slower dissociation rate constants relative to the shorter constructs. Thus, K176 is likely to be a key contact residue and play an important role in stabilizing the TIAR-RNA complex. This finding highlights the involvement of not only the residues within the classically structured TIAR RRM2, but also the residues outside the structured motif in strong binding to U-rich RNA elements.
The involvement of the unstructured region outside the RRM in RNA-binding has also been documented in previous studies involving other RBPs.41 TIA-1 RRM2, which shares 92% sequence homology and an identical C-terminal extension with TIAR RRM2, has been investigated for its mode of interaction with a target RNA.40 Chemical shift perturbation analyses of TIA-1 RRM2 with 5-nt U-rich RNA revealed that amino acids perturbed by RNA-binding include residues across the expected β-sheet RNA-binding site as well as the C-terminal extension (A173, R175 and K176).40 Their data are in agreement with our results for TIAR RRM2 interactions with 6-nt AU-rich RNA suggesting that they have similar modes of interaction. Some differences were observed, however. V101 (RNP2), Y140 and (RNP1), and Y146 and N147 (β3-α2 loop) in RRM2L were significantly affected by the AU-rich RNA binding, but not by the U-rich RNA binding in TIA-1 RRM2, possibly due to their sequence specificity (6-nt AU- vs. 5-nt U-rich RNA). More importantly, however, K176 at C-terminal extension of RRM2 was strongly affected by both U- and AU-rich RNA binding in TIA-1 and TIAR proteins, respectively. Therefore, this positively charged residue at the conserved C-terminal extension of TIA-proteins is likely to make a significant contribution toward high affinity interactions between TIA-proteins and their target RNA sequences.
The structural basis for the C-terminal interaction will not be known until a TIA RRM2 domain is characterized in complex with target RNA or DNA. There are several instances, however, where the C-terminal extension region of an RRM has been seen to adopt a defined structure upon oligonucleotide binding and to form additional contacts with the oligonucleotide.36,47-50 In the current study, analysis of the NMR data using TALOS+ (Fig. S2) suggest that the C-terminal extension in the RRM2L construct is extended in both the apo and RNA-bound states. Furthermore, there is no evidence for a major change in the secondary structure in this region of the protein upon RNA binding. Thus, it appears likely that additional electrostatic or hydrogen bond interactions from sidechains of T174, R175 and K176 to the oligonucleotide may underlie the remarkable extra stability and, thus, high affinity that is demonstrated in the current study.
Materials and Methods
Plasmid construction and protein purification
Constructs for TIAR isoform b (NCBI Refseq: NP_003243) RRM1 (residues 1–94), RRM2S (residues 96–175), RRM2L (residues 94–181) and RRM12S (residues 8–175) (Fig. 1) were expressed in E. coli strain BL21 (DE3). The culture was grown till an OD600 of ~0.8 was reached, induced with IPTG (0.5 mM) for 2.5 ~3 h and the cells were harvested by centrifugation. The cell pellets were stored at -80°C overnight, thawed on ice and resuspended in lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 5% Glycerol, 1 mM DTT, 0.1 mM PMSF). Cells were lysed by gentle sonication and debris was removed by centrifugation at 15,000 × g for 1 h. Supernatant was then incubated with washed glutathione (GSH)-Sepharose beads (GE Healthcare) with gentle mixing at 4°C for 4~16 h. After the beads were washed with wash buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% Triton X-100, 5% Glycerol) for four times or more, the TIAR proteins were cleaved from the GSH beads by incubating them with thrombin at 4°C for 16–20 h and cleavage confirmed by SDS-PAGE analysis. The proteins were further purified by size-exclusion and cation-exchange chromatography. The concentration of each protein was determined using the Bradford assay (BioRad) and by A280 measurements using theoretical molar extinction coefficients (ProtParam). The extinction coefficients were validated for folded protein; A280 measurements were within 10% of measurements made in 6.0 M guanidinium hydrochloride. The purity of each protein was confirmed by SDS-PAGE.
Biosensor experiments
The dynamics of RNA/DNA-protein interactions were characterized by surface plasmon resonance (SPR) using a BIACORE T100 instrument (GE Healthcare). The oligonucleotides used in the analyses were: The U-rich RNA [containing poly (U) stretches; 5′-GGGGGGUUUUUUUUUUUUUUUUUGGGGG-3′] and T-rich DNA (5′-TTTTTTTTTTTTTTTTTTTT-3′). The oligonucleotides were chemically synthesized carrying a 5′-biotin tag and supplied (as PAGE purified oligonucleotides; Dharmacon Research, now part of Thermo Scientific) to allow immobilization of the RNA/DNA onto streptavidin-coated sensor chips (Series S Sensor Chip SA, GE Healthcare). The RNA was deprotected (according to the manufacturer’s instructions for propriety 2'-ACE deprotection), and both RNA and DNA diluted to a final concentration of 1 µM in HBS buffer (10 mM HEPES, pH 7.4, 150 mM NaCl) followed by heating at 80°C for 10 min and cooling to room temperature. The sample was then diluted 500-fold in running buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM DTT, 0.025% surfactant P20; GE Healthcare) and injected over the sensor chip surface at 10 µl/min at 25°C to generate a ~50 response unit (RU) RNA/DNA surface (for a low-density surface). Proteins were serially diluted in running buffer at a range of concentrations and injected at 25°C at a flow rate of 50 μl/min for 2–3 min. Surface regeneration to remove any protein that remained bound after 3–6 min of dissociation was achieved using a 1-min injection of 2 M NaCl at 50 μl/min. Analyses of protein concentrations were done in duplicates and any background signal from a streptavidin-only reference flow cell was subtracted from every data set. Data were analyzed using a simple 1:1 Langmuir interaction model using the Biacore T100 evaluation software (Biacore Inc.) to determine the kinetics (association/dissociation rate constants; ka/kd) as well as the affinities (KD) of the protein-RNA/DNA interactions.
Preparation of RRM2L (15N,13C)
Uniformly labeled RRM2L (15N,13C) for NMR experiments was expressed in E. coli BL21 (DE3) in M9 salt-based minimal media supplemented with 15N NH4Cl (1 g/L) and 13C glucose (3 g/L) (Cambridge Isotope Laboratory). The culture was grown till the desired OD600 of 0.8~1.0 was reached, induced with IPTG (0.5 mM) for 3 h and the cells were harvested by centrifugation. The cell pellets were stored at -80°C overnight, thawed on ice and resuspended in lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 5% Glycerol, 1 mM DTT, 0.1 mM PMSF) and purified as a GST-fusion protein in the same way as the unlabeled TIAR RRM2L. The purified protein was then dialyzed into the phosphate buffer (50 mM phosphate buffer pH6, 100 mM NaCl) and concentrated to a final concentration of 300 μM with 10% D2O.
NMR spectroscopy
Experiments were performed on a Varian Unity AS600 MHz spectrometer at 25°C equipped with a cryogenically cooled triple resonance gradient probe. Standard triple resonance experiments as well as 2D (15N,1H)-HSQC were run to obtain the H, HN, C’ Cα and Cβ assignments of RRM2L. These include HNCA, HNCACB, HNCACO, HNCO and CBCA(CO)NH. (15N,1H)-HSQC spectra were recorded to monitor the chemical shifts of RRM2L alone and with the increasing concentrations of RNA. For RNA-titration experiments, 6-nt AU-rich RNA (5′-UUAUUU-3′) (Dharmacon) was added to RRM2L at protein:RNA molar ratios of 1:0, 1:0.4, 1:0.8 and 1:1.2. Triple resonance HNCA, HNCACB, HNCO and CBCA(CO)NH spectra were acquired for RRM2L at the final protein:RNA ratio of 1:1.2. Data were processed using NMRPipe51 and analyzed with SPARKY (Goddard and Kneller, UCSF). Assignments are deposited at the Biological Magnetic Resonance Bank for apo- and RNA-bound TIAR RRM2L (BMRB accession numbers 19063 and 19064).
Conclusion
TIAR is a multi-functional protein shuttling between nucleus and cytoplasm and plays an important role in the regulation of gene expression, interacting strongly with both RNA and DNA targets. This study reports the unique roles of individual domains of TIAR as well as the linker region between them in binding RNA and DNA targets by examining detailed kinetics and affinity of their interactions. TIAR RRM2 is the major binding domain for both RNA and DNA targets but the strong low nanomolar affinity observed between TIAR and its targets is only achieved in the presence of its C-terminal extension as well as RRM1. Kinetic analysis reveals that the RRM2 extension residues may play an important role in the stability of the TIAR-RNA/DNA complexes. RRM1, in contrast, contributes toward the stability of TIAR-DNA complexes. The specific structural role of the residues in the linker region C-terminal to RRM2 in RNA/DNA binding remains to be determined.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Australian Research Council (DP110102056 awarded to M.C.J.W., J.A.W. and M.G.). M.C.J.W. is a fellow of the National Health and Medical Research Council of Australia. M.G. was supported by the National Institute on Aging-Intramural Research Program, National Institutes of Health.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/24341
References
- 1.Förch P, Valcárcel J. Molecular mechanisms of gene expression regulation by the apoptosis-promoting protein TIA-1. Apoptosis. 2001;6:463–8. doi: 10.1023/A:1012441824719. [DOI] [PubMed] [Google Scholar]
- 2.Anderson P, Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones. 2002;7:213–21. doi: 10.1379/1466-1268(2002)007<0213:VSTROE>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aznarez I, Barash Y, Shai O, He D, Zielenski J, Tsui LC, et al. A systematic analysis of intronic sequences downstream of 5′ splice sites reveals a widespread role for U-rich motifs and TIA1/TIAL1 proteins in alternative splicing regulation. Genome Res. 2008;18:1247–58. doi: 10.1101/gr.073155.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Förch P, Puig O, Kedersha N, Martínez C, Granneman S, Séraphin B, et al. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol Cell. 2000;6:1089–98. doi: 10.1016/S1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 5.Förch P, Puig O, Martínez C, Séraphin B, Valcárcel J. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 2002;21:6882–92. doi: 10.1093/emboj/cdf668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Izquierdo JM, Valcárcel J. Fas-activated serine/threonine kinase (FAST K) synergizes with TIA-1/TIAR proteins to regulate Fas alternative splicing. J Biol Chem. 2007;282:1539–43. doi: 10.1074/jbc.C600198200. [DOI] [PubMed] [Google Scholar]
- 7.Le Guiner C, Lejeune F, Galiana D, Kister L, Breathnach R, Stévenin J, et al. TIA-1 and TIAR activate splicing of alternative exons with weak 5′ splice sites followed by a U-rich stretch on their own pre-mRNAs. J Biol Chem. 2001;276:40638–46. doi: 10.1074/jbc.M105642200. [DOI] [PubMed] [Google Scholar]
- 8.Shukla S, Dirksen WP, Joyce KM, Le Guiner-Blanvillain C, Breathnach R, Fisher SA. TIA proteins are necessary but not sufficient for the tissue-specific splicing of the myosin phosphatase targeting subunit 1. J Biol Chem. 2004;279:13668–76. doi: 10.1074/jbc.M314138200. [DOI] [PubMed] [Google Scholar]
- 9.Zhu H, Hasman RA, Young KM, Kedersha NL, Lou H. U1 snRNP-dependent function of TIAR in the regulation of alternative RNA processing of the human calcitonin/CGRP pre-mRNA. Mol Cell Biol. 2003;23:5959–71. doi: 10.1128/MCB.23.17.5959-5971.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suswam EA, Li YY, Mahtani H, King PH. Novel DNA-binding properties of the RNA-binding protein TIAR. Nucleic Acids Res. 2005;33:4507–18. doi: 10.1093/nar/gki763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson P, Kedersha N. Stressful initiations. J Cell Sci. 2002;115:3227–34. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- 12.Mazan-Mamczarz K, Lal A, Martindale JL, Kawai T, Gorospe M. Translational repression by RNA-binding protein TIAR. Mol Cell Biol. 2006;26:2716–27. doi: 10.1128/MCB.26.7.2716-2727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.López de Silanes I, Galbán S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, et al. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol. 2005;25:9520–31. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–84. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–42. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–50. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA. 1986;83:1670–4. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–70. doi: 10.1016/S0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 19.Chen CY, Xu N, Shyu AB. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol Cell Biol. 1995;15:5777–88. doi: 10.1128/mcb.15.10.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damgaard CK, Lykke-Andersen J. Translational coregulation of 5’TOP mRNAs by TIA-1 and TIAR. Genes Dev. 2011;25:2057–68. doi: 10.1101/gad.17355911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30:963–9. doi: 10.1042/BST0300963. [DOI] [PubMed] [Google Scholar]
- 22.Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, et al. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–68. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, et al. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–64. doi: 10.1016/S0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 24.Loflin P, Chen CY, Shyu AB. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 1999;13:1884–97. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, DeMaria CT, Crater D, et al. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–65. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168–81. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–77. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–8. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips K, Kedersha N, Shen L, Blackshear PJ, Anderson P. Arthritis suppressor genes TIA-1 and TTP dampen the expression of tumor necrosis factor alpha, cyclooxygenase 2, and inflammatory arthritis. Proc Natl Acad Sci USA. 2004;101:2011–6. doi: 10.1073/pnas.0400148101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HS, Kuwano Y, Zhan M, Pullmann R, Jr., Mazan-Mamczarz K, Li H, et al. Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR. Mol Cell Biol. 2007;27:6806–17. doi: 10.1128/MCB.01036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim HS, Wilce MCJ, Yoga YMK, Pendini NR, Gunzburg MJ, Cowieson NP, et al. Different modes of interaction by TIAR and HuR with target RNA and DNA. Nucleic Acids Res. 2011;39:1–14. doi: 10.1093/nar/gkq837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beck AR, Medley QG, O’Brien S, Anderson P, Streuli M. Structure, tissue distribution and genomic organization of the murine RRM-type RNA binding proteins TIA-1 and TIAR. Nucleic Acids Res. 1996;24:3829–35. doi: 10.1093/nar/24.19.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Izquierdo JM, Valcárcel J. Two isoforms of the T-cell intracellular antigen 1 (TIA-1) splicing factor display distinct splicing regulation activities. Control of TIA-1 isoform ratio by TIA-1-related protein. J Biol Chem. 2007;282:19410–7. doi: 10.1074/jbc.M700688200. [DOI] [PubMed] [Google Scholar]
- 34.Dember LM, Kim ND, Liu KQ, Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J Biol Chem. 1996;271:2783–8. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- 35.Tian Q, Streuli M, Saito H, Schlossman SF, Anderson P. A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell. 1991;67:629–39. doi: 10.1016/0092-8674(91)90536-8. [DOI] [PubMed] [Google Scholar]
- 36.Cléry A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol. 2008;18:290–8. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Auweter SD, Oberstrass FC, Allain FH. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 2006;34:4943–59. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dominguez C, Fisette JF, Chabot B, Allain FH. Structural basis of G-tract recognition and encaging by hnRNP F quasi-RRMs. Nat Struct Mol Biol. 2010;17:853–61. doi: 10.1038/nsmb.1814. [DOI] [PubMed] [Google Scholar]
- 39.Fialcowitz-White EJ, Brewer BY, Ballin JD, Willis CD, Toth EA, Wilson GM. Specific protein domains mediate cooperative assembly of HuR oligomers on AU-rich mRNA-destabilizing sequences. J Biol Chem. 2007;282:20948–59. doi: 10.1074/jbc.M701751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuwasako K, Takahashi M, Tochio N, Abe C, Tsuda K, Inoue M, et al. Solution structure of the second RNA recognition motif (RRM) domain of murine T cell intracellular antigen-1 (TIA-1) and its RNA recognition mode. Biochemistry. 2008;47:6437–50. doi: 10.1021/bi7024723. [DOI] [PubMed] [Google Scholar]
- 41.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–31. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 42.Kumar AO, Swenson MC, Benning MM, Kielkopf CL. Structure of the central RNA recognition motif of human TIA-1 at 1.95A resolution. Biochem Biophys Res Commun. 2008;367:813–9. doi: 10.1016/j.bbrc.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bauer WJ, Heath J, Jenkins JL, Kielkopf CL. Three RNA recognition motifs participate in RNA recognition and structural organization by the pro-apoptotic factor TIA-1. J Mol Biol. 2012;415:727–40. doi: 10.1016/j.jmb.2011.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park S, Myszka DG, Yu M, Littler SJ, Laird-Offringa IA. HuD RNA recognition motifs play distinct roles in the formation of a stable complex with AU-rich RNA. Mol Cell Biol. 2000;20:4765–72. doi: 10.1128/MCB.20.13.4765-4772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park-Lee S, Kim S, Laird-Offringa IA. Characterization of the interaction between neuronal RNA-binding protein HuD and AU-rich RNA. J Biol Chem. 2003;278:39801–8. doi: 10.1074/jbc.M307105200. [DOI] [PubMed] [Google Scholar]
- 46.Izquierdo JM, Valcárcel J. Two isoforms of the T-cell intracellular antigen 1 (TIA-1) splicing factor display distinct splicing regulation activities. Control of TIA-1 isoform ratio by TIA-1-related protein. J Biol Chem. 2007;282:19410–7. doi: 10.1074/jbc.M700688200. [DOI] [PubMed] [Google Scholar]
- 47.Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–45. doi: 10.1016/S0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 48.Ding J, Hayashi MK, Zhang Y, Manche L, Krainer AR, Xu RM. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–15. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazza C, Segref A, Mattaj IW, Cusack S. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 2002;21:5548–57. doi: 10.1093/emboj/cdf538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cléry A, Jayne S, Benderska N, Dominguez C, Stamm S, Allain FH. Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-β1. Nat Struct Mol Biol. 2011;18:443–50. doi: 10.1038/nsmb.2001. [DOI] [PubMed] [Google Scholar]
- 51.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.