Abstract
The bacterial Hfq protein is a versatile modulator of RNA function and is particularly important for regulation mediated by small non-coding RNAs. Hfq is a bacterial Sm protein but bears more similarity to the eukaryotic Sm-like (Lsm) family of proteins than the prototypical Sm proteins. Hfq and Lsm proteins share the ability to chaperone RNA-RNA and RNA/protein interactions and an interesting penchant for protecting the 3′ end of a transcript from exonucleolytic decay while encouraging degradation through other pathways. Our view of Lsm function in eukaryotes has historically been informed by studies of Hfq structure and function but mutational analyses and structural studies of Lsm sub-complexes have given important insights as well. Here, we aim to compare and contrast the roles of these evolutionarily related complexes and to highlight areas for future investigation.
Keywords: Sm-like, mRNA decay, RNA chaperone, splicing, oligouridylation, polyadenylation, exoribonuclease
Introduction
Sm proteins exist in all three domains of life and are defined by the ability to adopt the Sm fold, which is comprised of an N-terminal α-helix and five anti-parallel β-strands (Fig. 1).1 While bacteria generally have just one Sm family member known as Hfq, eukaryotes have evolved over 20 of these proteins.2 The eukaryotic proteins can be divided into Sm and Sm-like (Lsm) sub-families and form a variety of multimeric complexes (Table 1). Seven canonical Sm proteins are the founding members of the family and were discovered as the antigens targeted by auto-antibodies in a patient who suffered from systemic lupus erythematosus.3 Seven Sm proteins (predominantly SmB/B’, SmD1, SmD2, SmD3, SmE, SmF and SmG in vertebrates) are assembled as a heptameric ring around a conserved core sequence at the center of the spliceosomal snRNAs.4 Both Hfq and the Lsm proteins can also adopt multimeric ring structures. Hfq forms a hexamer in solution;5 while Lsm proteins typically form heteromers or homomers of six or seven subunits.6 We note though that other oligomeric states of Lsm proteins have been observed, including octamers.7 Complexes containing both Sm and Lsm proteins have also been found, such as the heptamer comprising SmD3, SmB, Lsm10, Lsm11, SmF, SmE and SmG, which assembles on the U7 snRNP involved in vertebrate histone mRNA processing.8 Notably, some of the more recently described Lsm proteins do not appear to form multimers and have other protein domains in addition to the Sm fold. These include EDC3,9 Ataxin210 and RAP55/LSM14,11 all of which have been implicated in mRNA decay and/or translation. In addition, two other proteins with Sm folds, Gemin6 and Gemin7, form a heterodimer and are part of the SMN complex that chaperones association of the Sm proteins with the U snRNAs.12
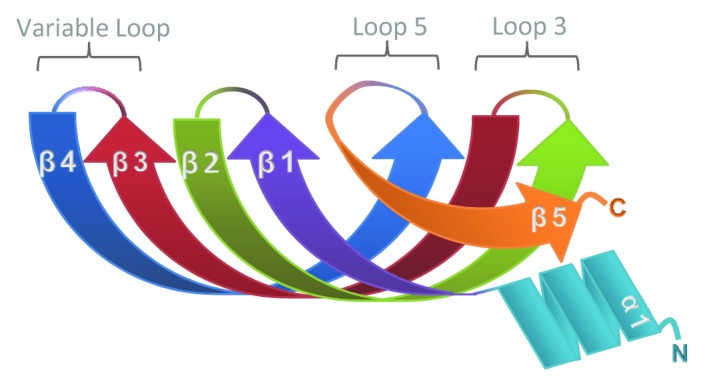
Figure 1. Schematic showing the structure of the Sm fold.1 The five anti-parallel β-strands and single α-helix are depicted in different colors. Loops 3 and 5 contain residues important for RNA-binding. Loop 4 varies in length and amino acid composition.
Table 1. Composition and function of eukaryotic Lsm and Sm complexes.
| Complexes | RNA partners | Cellular functions | Assembly | |
|---|---|---|---|---|
| Sm |
Sm Complex |
U1, U2, U4 and U5 snRNAs3 |
mRNA splicing |
Requires RNA103 In vertebrates, Sm complex association with snRNAs is chaperoned by SMN14 |
| U11, U12 and U4atac snRNAs104 |
mRNA splicing by the minor spliceosome105 |
|||
| TLC1 (S.cerevisiae) TER1 (S.pombe) |
Biogenesis and stability of telomerase RNA subunit106,107 |
|||
| SmB, SmB’, SmD |
hTR scaRNAs |
Associate with telomerase RNA and scaRNAs in human cells, but role is unclear108 |
Unknown but the Sm complex chaperone SMN is also associated with telomerase109 |
|
| Sm/Lsm |
U7 Sm/Lsm complex |
U7 snRNA |
Essential for 3′ processing of histone mRNAs Lsm11 interacts with FLASH to recruit cleavage factors95 |
Requires SMN complex for assembly8 |
| Lsm |
Lsm2–8 complex |
U6 snRNA |
Stabilizes U6 snRNA and chaperone its interactions with U4, the pre-mRNA and Prp24/p11013,27,99 |
Spontaneous assembly in the absence of RNA110 |
| TER1 RNA (S.pombe) |
Promotes association of telomerase catalytic subunit and protects the 3′ end of mature TER1 RNA from exonucleases107 |
|||
|
Lsm1–7 complex |
Oligoadenylated cytoplasmic mRNAs |
Prevents 3′ end trimming by exonucleases39 Recruits the decapping machinery40 |
||
| Oligouridylated histone mRNAs (in vertebrates) |
Recruits the decapping machinery and Eri1 exonuclease to initiate degradation57,59,60 |
|||
| Lsm2–7 complex |
snR528 pre-RNase P20 |
Biological role is not clear |
||
| Lsm2–4/6–8 complex | U8 snoRNA29 | Biological role is not clear | ||
Importantly, Hfq and the canonical eukaryotic Lsm complexes (Lsm1-7 and Lsm2-8) differ fundamentally from the seven spliceosomal Sm proteins in that they spontaneously form stable multimeric rings in the absence of RNA.13 In contrast, Sm complexes assemble only in the presence of RNA in a process that, at least in vertebrates, requires considerable chaperoning from other protein factors.14 In the interests of brevity, we will be focusing mainly on the multimeric complexes formed by Lsm and Hfq proteins in this review.
Functions of Lsm and Hfq proteins
The cellular functions of eukaryotic Lsm proteins and their bacterial counterparts are intimately connected with RNA processing and degradation. In E. coli, the Hfq protein binds small non-coding RNAs (sRNAs) to facilitate sRNA/mRNA interactions that, in turn, modulate translation efficiency and mRNA decay.15 In addition, Hfq can also influence mRNA decay by associating with the 3′ end of the transcript and promoting polyadenylation while also inhibiting 3′-5′ exonuclease activities.16,17 These activities are important for normal stress response and pathogenicity in E.coli and other bacterial pathogens.18
Eukaryotic Lsm complexes have similar functions to those ascribed to Hfq in bacteria in that they chaperone mRNAs and non-coding RNAs through various steps in metabolism.19 In eukaryotes ranging from S. cerevisiae to humans there are multiple complexes of Lsm proteins, but the two best-characterized are Lsm1-7 and Lsm2-8, which are restricted to the cytoplasm and nucleus, respectively.20-24 Lsm1-7 is a major regulator of mRNA decay through its interactions with the decapping machinery,19,22,23 whereas Lsm2-8 is involved primarily in stabilizing the U6 snRNA and chaperoning it through the splicing process.21,25-27 Other minor Lsm complexes include Lsm2-7, which associates with the snR5 snoRNA in yeast28 and also with pre-RNase P20 while Lsm 2-4/6-8 binds the U8 snoRNA.29
At first glance, it seems that both the structure (the Sm fold) and functions (in RNA decay and processing) of Lsm proteins are conserved from bacteria through to eukaryotes. However, as we delve deeper into the structure and RNA-binding activities of these factors, several differences will become evident. During evolution, the Lsm family has shown a high propensity to adapt to different substrates and mechanisms.
Hfq and Lsm associate with RNA 3′ ends
Although Hfq and Lsm proteins are both able to recognize internal RNA sequence elements, in both cases, their interactions with 3′ ends of RNA substrates are best-characterized and arguably most important. Both complexes share affinity for 3′ uridine or adenosine tracts.13,30,31 Such elements can be template encoded, like the U-rich tracts which are formed at the 3′ end of bacterial sRNAs as a result of Rho-independent termination.32 Alternatively, such sequences can be added post-transcriptionally by nucleotidyltransferases such as TUTases and poly(A) polymerases (PAPs).33 Regardless of their provenance, such single-stranded 3′ elements are important determinants of RNA stability as they are vulnerable to 3′ exonucleolytic attack. Hfq and Lsm proteins share the ability to protect these 3′ ends but they also use them as a platform to orchestrate recruitment of other RNA elements and proteins.
3′ polyadenylation
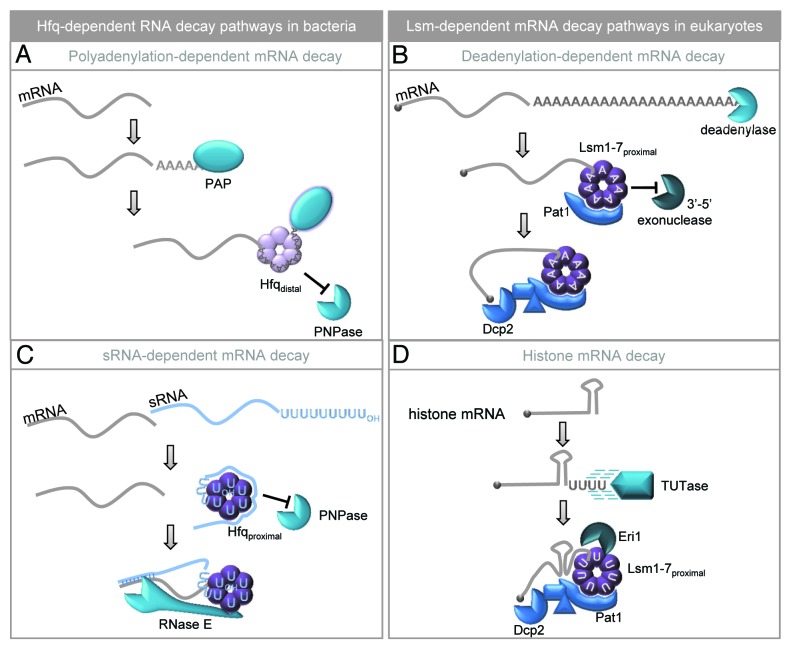
In bacteria, just as in higher organisms, there are multiple pathways to induce decay of mRNAs.16 One of the best studied mechanisms in both eukaryotes and prokaryotes involves a 3′ poly(A) tail. In bacterial cells, polyadenylation mediated by PAP enhances mRNA degradation as it provides a toehold for exonucleases such as polynucleotide phosphorylase (PNPase).17,34 PAP initially acts distributively, adding only a few adenosines; however, Hfq associates avidly with these short poly(A) tails and stimulates PAP such that it becomes more processive (Fig. 2A).35-37 The long poly(A) tails that result from this mechanism remain bound to Hfq and are protected from PNPase action.
Figure 2. Hfq and Lsm proteins bind single-stranded 3′ ends and activate RNA decay. (A) Poly(A) polymerase (PAP) acts distributively to add adenosine residues until the poly(A) tail reaches sufficient length to recruit Hfq. Hfq binds poly(A) through its distal surface and stimulates PAP activity to extend the poly(A) tail. This interaction also prevents the 3′-5′ exonuclease, polynucleotide phosphorylase (PNPase), from attacking the 3′ end of the transcript.35-37 (B) In eukaryotic cells, poly(A) tails are shortened by deadenylase enzymes prior to decay.38 Once the tail is short enough (~10–12 residues), Lsm1-7 associates using the proximal binding surface and Pat1 is also recruited.78 The Lsm-Pat1 complex prevents further shortening from the 3′ end while activating decapping at the 5′ end by interacting with a number of accessory factors to engage the decapping enzyme (Dcp2).40 (C) Bacterial sRNAs terminate in a tract of seven to nine uridines generated through Rho-independent termination. This 3′ U-tract is recognized by Hfq through its proximal binding surface resulting in protection of the 3′OH group from attack by PNPase. Additional regions of the sRNA can bind the lateral surface of Hfq while the distal surface recognizes mRNA targets. Once the mRNA and sRNA anneal, RNase E joins the complex and cleaves both the mRNA and sRNA to initiate their degradation.42,44,45 (D) Vertebrate histone mRNAs are degraded through a pathway that requires 3′ oligouridylation by a terminal uridyltransferase (TUTase).57,58 This provides a platform to recruit the Lsm1–7/Pat1 complex which, in turn, initiates mRNA decapping.57,59 Oligouridylation of histone mRNAs also promotes 3′-5′ exonucleolytic decay as Lsm1-7 recruits the Eri1 exonuclease.60
In eukaryotic cells, the poly(A) tail is added co-transcriptionally and expedites many required steps of mRNA metabolism such as splicing, export and translation. However, it is also intimately involved in mRNA decay. Deadenylation, mediated by dedicated deadenylases such as CCR4/NOT or PARN, is the first rate-limiting step in decay of most mRNAs and generates mRNAs with short poly(A) tails of 10 or less residues, much like those generated by PAP during decay of bacterial mRNAs.38 These oligoadenylated mRNAs are specifically recognized by the Lsm1-7 complex (Fig. 2B).30 Although the function of Lsm in mRNA decay is not fully understood, there are two clear consequences to Lsm1-7 binding. First, the 3′ end of the mRNA is protected from exonucleases; yeast Lsm1 mutants accumulate mRNAs with 3′ ends that are trimmed by 20–30 nt.39 Second, Lsm1-7 recruits factors required for decapping at the 5′ end of the transcript; loss of Lsm1-7 function results in accumulation of deadenylated intermediates in yeast and mRNAs are stabilized.40 There are several parallels with Hfq function that can be highlighted here. The most obvious of these is that both Hfq and Lsm1-7 associate with the 3′ ends of oligoadenylated mRNAs that are destined for degradation. In addition, both complexes protect the 3′ end from exoribonucleases; Hfq blocks PNPase while Lsm prevents attack by the exosome (which interestingly contains six subunits with homology to PNPase41). There is also at least one surprising difference: Hfq promotes extension of the mRNA poly(A) tail by PAP and, in fact, associates more intensely with longer poly(A).37 In contrast, Lsm1-7 has a distinct preference for oligoadenylated mRNAs over those with longer poly(A) tails.30
3′ oligouridylation
Many bacterial sRNAs end in 3′ oligo(U) tracts of seven to nine residues which are acquired through Rho-independent termination. These regions provide a platform for recruitment of Hfq to stabilize sRNAs against 3′ exonucleolytic decay. Notably, the U-tracts appear to be precisely sized to favor Hfq binding as an sRNA with a 3′-U6 tract shows significantly reduced association and one with just four 3′ uridines fails to bind Hfq at all.42 Importantly, Hfq binding is also essential for annealing of the sRNA to its mRNA target, most commonly at sites within the 5′UTR. Annealing of the sRNA can modulate translation initiation by occluding or exposing the ribosome-binding site on the mRNA.43 In addition, an sRNA/mRNA/Hfq complex can be recognized by the endonuclease RNase E resulting in rapid decay of both the mRNA and sRNA (Fig. 2C).44,45 In this case, Hfq protects the sRNA 3′ end in order to facilitate its decay through an alternative pathway. This is surprisingly reminiscent of the role of Lsm1-7 in mRNA degradation; it associates with 3′ oligoadenylated mRNAs and protects them from 3′-5′ decay while simultaneously promoting decapping and decay through the 5′-3′ pathway (Fig. 2B).39
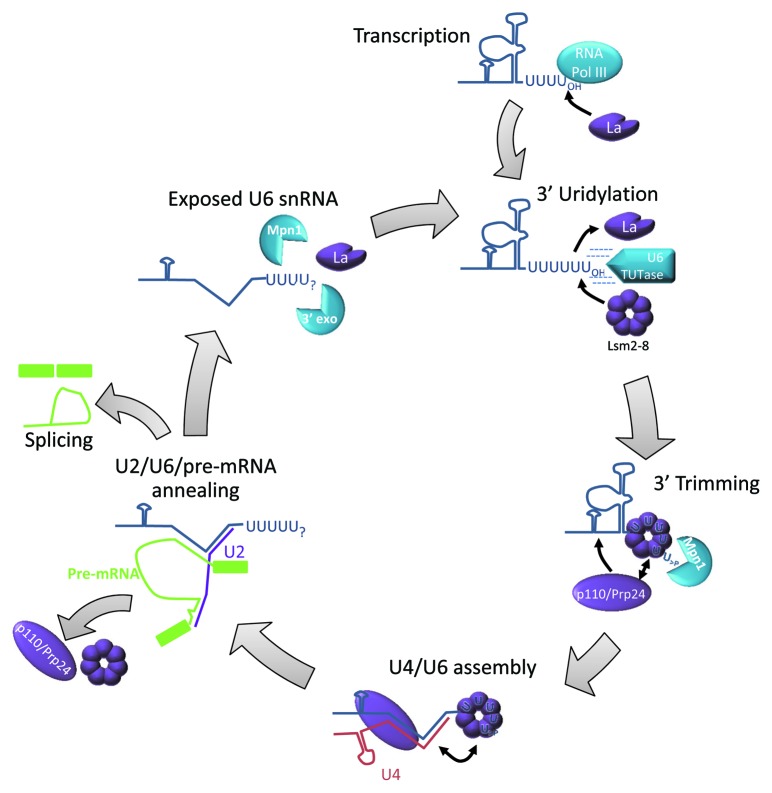
Short 3′ uridine tracts are important binding sites for eukaryotic Lsm proteins as well. For example, the nuclear Lsm2-8 complex binds a U5 tract at the 3′ end of U6 snRNA and influences stability of the transcript.13,46 As Lsm2-8 is essential for almost all functions of U6 in the splicing reaction (Fig. 3), the U-tract also represents a vital platform for Lsm recruitment. Interestingly, the length of the U-tract appears to be important in Lsm binding. Nascent U6 snRNA has just four uridines at its 3′ end, which allow association with the La protein.47,48 The 3′ U-tract is subsequently extended by a TUTase49 and then trimmed by an exonuclease known as MPN1.50,51 During processing, Lsm displaces La and protects U6 against excessive 3′ shortening.52 It is possible that cycles of oligouridylation and trimming serve to ensure U6 snRNA is maintained intact following each splicing event.
Figure 3. U6 snRNP metabolism requires Lsm2-8. U6 snRNA is transcribed in the nucleus by RNA polymerase III and terminates in four uridine residues, which recruit the La RNA-binding protein.48 The U tract is extended by U6 TUTase leading to exchange of La for Lsm2-8.49,52 The U-tract is then trimmed to five residues by Mpn1 and consequently terminates in a 2’,3′ cyclic phosphate.50,51 Lsm binding allows association of Prp24/p110 to generate mature U6 snRNA.99 Lsm facilitates annealing of U4 and U6 and the complex enters the spliceosome. Subsequent rearrangements allow U6 to interact with U2 and the pre-mRNA and Lsm2-8 and p110 are then ejected prior to the first step of splicing.88 After splicing, U6 is released in a form that is accessible to various RNA-modifying enzymes. U6 snRNP must then be reassembled prior to being re-used. Lsm2-8 and the U6 TUTase may participate in a quality control mechanism to ensure the U-tract remains intact.
Recent studies have uncovered novel roles for Lsm1-7 and oligouridylation in cytoplasmic mRNA decay pathways. The vast majority of cytoplasmic S. cerevisiae mRNAs degrade primarily through the deadenylation-dependent pathway described above, and this pathway is also conserved in higher organisms.38 However, there is compelling evidence for the existence of additional decay mechanisms in other fungi and in plants. Moreover, these alternate pathways also involve Lsm1-7 interactions at the 3′ end. In S. pombe, a significant fraction of mRNAs decay through a deadenylation-independent pathway that involves the action of a terminal uridyl transferase known as Cid1.53 Cid1 is responsible for addition of one or two non-templated uridines at the 3′ end of the poly(A) tail of certain transcripts including act1, urg1 and others. Interestingly, this terminal modification induces decapping without prior deadenylation, almost certainly through the recruitment of Lsm1-7 and its associated decapping factors. A similar pathway has also been uncovered in the filamentous fungus Aspergillus nidulans, where mRNAs obtain a 3′CUCU modification following shortening of the poly(A) tail to around 15 adenosine residues.54 This modification is performed by two nucleotidyltransferases known as CutA and CutB, both of which are related to the S.pombe Cid1 enzyme.54,55 Although Lsm proteins are essentially uncharacterized in Aspergillus, it seems likely that they will be involved in this decay mechanism. Similar events have also been detected in Arabidopsis where one or two pyrimidines, generally uridines, can be found at the 3′ end of oligoadenylated mRNAs.55 It still remains unclear whether this decay pathway for polyadenylated mRNAs is conserved in mammals or other eukaryotes but, as described below, a very similar mechanism is employed for degradation of the non-adenylated histone mRNAs in mammalian cells.
In higher eukaryotes, the replication-dependent histones lack poly(A) tails and instead undergo unique 3′ end processing that results in a stem-loop structure at their 3′ end. This stem-loop associates with a specific set of factors, including the U7 snRNP and stem loop binding protein (SLBP) among others.56 These factors work together to ensure that histone production is tightly restricted to S phase and facilitate the rapid degradation of histone mRNAs as S phase comes to a close, or when DNA replication is inhibited. Importantly, 3′ oligouridylation is the initiating step in turnover of histone mRNA species57 and is mediated by the ZCCHC11 TUTase58 (which is again related to Cid1). This 3′ toehold allows Lsm1, most likely as part of the Lsm1-7 complex, to be recruited to the 3′ end of histone transcripts.57 Binding of Lsm1 induces histone mRNA turnover by decapping and 5′-3′degradation,57-59 and also enhances 3′-5′ decay by recruiting an exonuclease known as Eri1 (Fig. 2D).60 We note that although histone mRNAs are polyadenylated in yeast, Lsm1 still is essential for their controlled turnover as Lsm1 mutants have defects in genomic stability caused by excess accumulation of histone mRNAs.61
At this point we should note that a large number of eukaryotic non-coding RNAs, such as miRNAs and piRNAs, also experience post-transcriptional 3′ uridylation and/or adenylation.62,63 It is too early to say what role, if any, Lsm complexes play in the metabolism of these extended small RNAs but it is conceivable that Lsm modulates stability or even annealing of these regulatory RNAs with their targets much as Hfq influences bacterial sRNA functions.
The structural basis of 3′ end recognition
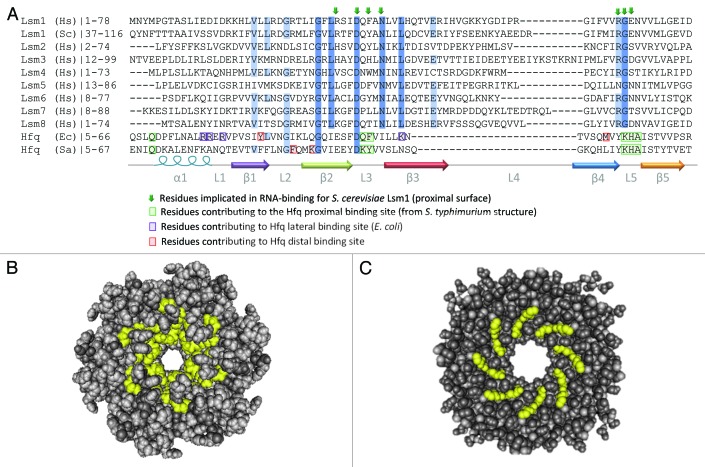
Several studies have investigated exactly how Hfq and Lsm complexes interact with RNA, particularly with A-rich and U-rich oligonucleotides. A preponderance of our current knowledge derives from structural studies on Hfq from the bacteria S.aureus,64,65 E.coli,66-68 S. typhimurium,69 P. aeruginosa,70,71 B. subtilis72 and the cyanobacteria Anabaena and Synechocystis,73 but additional insights have been gleaned from mutational analyses of Lsm1,74 structural studies using Lsm sub-complexes75,76 and binding assays performed with recombinant Lsm complexes.77,78 One over-arching similarity between Lsm and Hfq complexes is that both recognize oligo(U) through a surface that lies close to the central pore on the proximal side of the toroid. For Hfq, conserved residues important for RNA-binding lie within Loop 3 and Loop 5 of each subunit and form a propeller-like arrangement that comprises six individual binding sites, each of which can interact with a single uridine (Fig. 1 and 4).64,69 The residues required for RNA-binding in Lsm1 were predicted based on the structure of Hfq and subsequent mutational analysis supported their requirement for both mRNA decay and 3′ end protection.74 There is no high-resolution structure available for either Lsm1-7 or Lsm2-8 complexes. In an Lsm3 octamer, the putative RNA-binding residues are arranged close to the central pore, as in Hfq (Fig. 4).76 Despite the very similar overall structures of Hfq and Lsm complexes, the primary sequence conservation is insufficient to allow prediction of the exact contacts between RNA and each Lsm subunit.
Figure 4. Conservation of RNA-binding residues in Lsm and Hfq proteins. (A) Alignment of primary sequence of Sm domains for human Lsm1-8 proteins, S. cerevisiae Lsm1 and Hfq from E. coli and S. aureus. C-terminal domains and some N-terminal sequences are excluded from this alignment for clarity. The alignment was generated using CLUSTAL-W100 and edited in JalView.101 Secondary structure is indicated below the alignment and colored as in Figure 1. Residues shown to be important for RNA-binding are highlighted (Hfq) or indicated by arrows (Lsm proteins). Conserved residues are indicated by blue shading with darker blue denoting more conservation. The accession numbers for the protein sequences used in the alignment are: H.s. Lsm1 NP_055277.1, S.c. Lsm1 NP_012411.1, H.s. Lsm2 NP_067000.1, H.s. Lsm3 NP_055278.1, H.s. Lsm4 NP_036453.1, H.s. Lsm5 NP_036454.1, H.s. Lsm6 NP_009011.1, H.s. Lsm7 NP_057283.1, H.s. Lsm8 NP_057284.1, E.c.Hfq ACE63256.1, S.a.Hfq AEW65270.1. (B) Space-filling structure of proximal surface of S. Hfq (PDB ID:2YLB). Residues involved in RNA-binding are highlighted in yellow. (C) Space-filling structure of proximal surface of an S. cerevisiae Lsm3 octamer (PDB ID: 3BW1). Residues likely to be involved in RNA recognition are highlighted in yellow. The images in (B and C) were generated using Cn3D Structure Viewer Ver 4.3.102
While both Hfq and Lsm proteins show clear affinity for the 3′ end of RNA, S.typhimurium Hfq has a unique interaction with the 3′ end that allows it to distinguish and protect nascent RNAs, which bear a 3′ hydroxyl group, while ignoring decay intermediates that terminate with a 3′ phosphate.69 This interaction relies on a hydrogen bond between the histidine in Loop 5 and the 3′OH. The histidine residue is conserved in other bacteria, but Loop 5 is very different in Lsm proteins (Fig. 4), suggesting that they may have distinct interactions with the 3′ end. Indeed, while Hfq strongly prefers a 3′OH over a 2’,3′ cyclic phosphate (3′>P),69 the Lsm2-8 complex demonstrates a clear preference for 3′>P over 3′OH.52 This correlates well with the fact that 3′>P is the predominant terminal moiety of the mature U6 snRNA that is complexed with Lsm2-8 in vertebrates.52
There is quite a dramatic divergence between Hfq and Lsm in their mechanisms for binding to oligo(A). Hfq associates with oligo(A) and other A-rich sequence elements using a binding surface on the distal face.65,67,68,79 This surface varies in charge distribution between Hfq complexes from different bacteria and there is no evidence to date to suggest that its function is conserved in eukaryotes. Lsm appears to utilize the same proximal binding site to recognize 3′ oligo(A) as it does to bind 3′ oligo(U). Notably, in contrast to Hfq, which prefers long poly(A), the Lsm1-7 complex has evolved significant specificity for short A tails and binds less well to longer tails.30,78 Residues within the proximal RNA-binding pocket of Lsm1 confer this specificity. This observation is very consistent with the fact that the two complexes recognize oligo(A) through completely different regions.
Additional RNA-binding surfaces
Both Hfq and Lsm complexes certainly have additional means of interacting with RNA as the ability to simultaneously contact RNA in multiple places, or to simultaneously bind multiple RNAs, is implicit for their chaperone function. A third binding site with generally positive charge has been detected on the lateral surface of E. coli Hfq, but this charge distribution is not well-conserved, even in other bacteria.80 The C-terminal extension has also been implicated in RNA-binding; C-terminally deleted E. coli Hfq fails to associate with mRNAs81 and the C-terminal region of the P.aerophilum SmAP3 protein bears some structural resemblance to domains that associate with single-stranded nucleic acids.82 Indeed, the C-terminal regions of this family vary widely in both length and composition, so it remains very possible that they contribute to RNA-binding in eukaryotic cells, either through direct interaction with RNA, or by influencing access of the RNA to other binding sites. In support of this, yeast Lsm1 mutants lacking from 28‒55 amino acids of the C-terminal region have reduced affinity for RNA, although they still maintain the ability to distinguish short and long poly(A) tails.77
RNA chaperones
To this point we have focused predominantly on the interactions of Lsm and Hfq proteins with RNA, but it is equally important to discuss the central role of these complexes in chaperoning RNA-RNA and RNA-protein interactions.27,83,84 As mentioned above, Hfq has many RNA-binding surfaces that allow it to interface with multiple transcripts simultaneously. This property is crucial in the course of sRNA/mRNA annealing. Recent studies support that in addition to binding the sRNA 3′ end through the proximal binding site, Hfq interacts with additional sRNA sequences through its lateral-binding surface.80 These other interactions likely involve the internal stem-loop and short U-rich region that lie between the 3′ oligo(U) tract and the region that base-pairs with the mRNA.85 Recruitment of an mRNA target to the distal-binding site then results in rearrangements that favor annealing of the mRNA to the sRNA. Annealing can lead to translation inhibition and decay of both the mRNA and sRNA through RNase E cleavage (Fig. 2C),44,45 but translation can also be enhanced in some cases.86
Although the binding surfaces on Lsm proteins are less well-characterized, the interactions of Lsm2-8 with U6 snRNA may well involve multiple surfaces as well as other RNAs. First, although Lsm2-8 is clearly initially engaged through binding to the 3′ U tract, it can also be cross-linked to other regions of the U6 snRNA, suggesting that it may bind other U6 sequences as well.87 Second, the splicing process itself involves several changes in U6 snRNA conformation (Fig. 3): U6 snRNP is a transient entity which is readily incorporated into the U4/U6 di-snRNP and then the U4/U6.U5 tri-snRNP, before entering the spliceosome where U4 is ejected. Within the spliceosome, U6 base-pairs with U2 as well as interacting with the pre-mRNA but the Lsm complex is dismissed prior to the first step of splicing. U6 is released following completion of the splicing event.88 Lsm is known to enhance U4/U6 interactions, but it seems likely that it may also be involved in the subsequent remodeling events that occur as the U6 snRNA unwinds to interact with U2. It would be interesting to examine whether novel Lsm-binding surfaces are implicated in the remodeling of U6 snRNAs.
Interactions of Lsm and Hfq with other proteins
As well as binding RNA, both Hfq and Lsm complexes interact directly with other proteins. Of note, both Hfq and Lsm recruit decay/processing enzymes to their substrates. Following binding to sRNAs and annealing of the sRNA to its mRNA target, Hfq recruits the endonuclease RNase E to cleave and initiate decay of both mRNA and sRNA.44,45 There appears to be a direct interaction between RNase E and Hfq in this case,89,90 although the region of Hfq that binds RNase E has not been defined. In addition, Hfq is reported to co-purify with both PAP and PNPase, which presumably assists Hfq-mediated regulation of polyadenylation-induced mRNA decay.35
The cytoplasmic Lsm1-7 complex associates with multiple proteins required for mRNA decapping. Most notably, in S. cerevisiae, Pat1p readily co-purifies with Lsm1-7.91 Pat proteins are essentially scaffold factors that enable recruitment of other factors needed for decapping.92 In particular, Pat1 binds the decapping enzyme and enhances its activity.93,94 Pat1 recruitment also inhibits translation.92 A major portion of Lsm function in mRNA decay is likely mediated through its interaction with Pat1 and it has been suggested that Pat1 is required for association of Lsm with mRNAs.93 The region(s) of Lsm1-7 that interact with Pat1 have not been defined to date.
The U7 snRNP required for vertebrate histone mRNA 3′ end processing contains two Lsm proteins, Lsm10 and Lsm11, as well as five Sm proteins.8 Interestingly, the ~170 amino-acid N-terminal extension of Lsm11 plays a vital role in recruiting other factors to the U7 snRNP and eventually to histone pre-mRNAs. The N-terminal region of Lsm11 interacts directly with a large protein called FLASH to form a platform that recruits mRNA 3′ end processing factors including the endonuclease CPSF73.95
Hfq also appears to rely on protein-protein interactions for its assembly. Specifically, the RelA protein encourages assembly of Hfq monomers into the hexameric form. In E. coli RelA mutants, active Hfq becomes limiting and many of its activities in sRNA-mediated regulation are lost.96 It is not clear whether there are similar factors that promote assembly of the canonical Lsm1-7 and Lsm2-8 heptamers in higher organisms. We do note though that the Sm complex employs a number of factors including the SMN complex and PRMT5 to favor its association into snRNPs.14 Indeed, many of the same factors are involved in assembly of the U7 snRNP, which contains a mixture of Sm and Lsm factors.8 Moreover, SMN itself has been reported to associate with Lsm4 and Lsm6 in vitro.97
Conclusions and Perspectives
Over the last few years, structural studies of Hfq bound to RNA have brought invaluable insights into the mechanisms by which this RNA chaperone functions. Although such experiments are much more challenging in eukaryotes due to the heteromeric nature of Lsm complexes, such studies are essential to allow better definition of the interactions between Lsm and its RNA substrates. There are numerous interesting questions on the horizon for Lsm proteins and the complexes that they generate in eukaryotic cells. Given the explosion in the numbers of non-coding RNA regulators, the roles of Lsm complexes in the biogenesis and function of these transcripts should be investigated. Furthermore, unlike Hfq, which forms homomeric complexes, all Lsm proteins are not created equal; each has a unique C-terminal region as well as significant divergence within the Sm domain. It is already clear that Lsm1 and Lsm8 are unique to the cytoplasm and nucleus, respectively, and have distinct roles, but the individual contributions of Lsm2-7 remain to be elucidated. Hints as to the properties of individual Lsm proteins are currently available through recent structural studies on subunits and intersubunit interactions.75,76 These data should stimulate the development of testable hypotheses for the functions of individual Lsm subunits. Finally, the observation that both Hfq and Lsm proteins have been usurped by evolutionarily diverse viruses to promote viral gene expression and replication98 underscores the versatility of these protein complexes in RNA biology. Studies of these interactions will undoubtedly give further insights into the cellular roles of Lsm proteins.
Acknowledgments
We are grateful to the anonymous reviewers for their diligent reading of the manuscript and constructive criticism. Research into mechanisms of mRNA decay in the Wilusz laboratories is supported by R01 awards from the National Institutes of Health (NIAID-GM072481 to J.W. and NIAMS-AR059247 to C.J.W.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We apologize to those whose work we were unable to cite due to lack of space.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/23695
References
- 1.Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, Raker VA, et al. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–87. doi: 10.1016/S0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- 2.Veretnik S, Wills C, Youkharibache P, Valas RE, Bourne PE. Sm/Lsm genes provide a glimpse into the early evolution of the spliceosome. PLoS Comput Biol. 2009;5:e1000315. doi: 10.1371/journal.pcbi.1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lerner MR, Steitz JA. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci USA. 1979;76:5495–9. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walke S, Bragado-Nilsson E, Séraphin B, Nagai K. Stoichiometry of the Sm proteins in yeast spliceosomal snRNPs supports the heptamer ring model of the core domain. J Mol Biol. 2001;308:49–58. doi: 10.1006/jmbi.2001.4549. [DOI] [PubMed] [Google Scholar]
- 5.Franze de Fernandez MT, Hayward WS, August JT. Bacterial proteins required for replication of phage Q ribonucleic acid. Purification and properties of host factor I, a ribonucleic acid-binding protein. J Biol Chem. 1972;247:824–31. [PubMed] [Google Scholar]
- 6.Khusial P, Plaag R, Zieve GW. LSm proteins form heptameric rings that bind to RNA via repeating motifs. Trends Biochem Sci. 2005;30:522–8. doi: 10.1016/j.tibs.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Wu D, Jiang S, Bowler MW, Song H. Crystal structures of Lsm3, Lsm4 and Lsm5/6/7 from Schizosaccharomyces pombe. PLoS One. 2012;7:e36768. doi: 10.1371/journal.pone.0036768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pillai RS, Grimmler M, Meister G, Will CL, Lührmann R, Fischer U, et al. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 2003;17:2321–33. doi: 10.1101/gad.274403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tritschler F, Eulalio A, Truffault V, Hartmann MD, Helms S, Schmidt S, et al. A divergent Sm fold in EDC3 proteins mediates DCP1 binding and P-body targeting. Mol Cell Biol. 2007;27:8600–11. doi: 10.1128/MCB.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albrecht M, Golatta M, Wüllner U, Lengauer T. Structural and functional analysis of ataxin-2 and ataxin-3. Eur J Biochem. 2004;271:3155–70. doi: 10.1111/j.1432-1033.2004.04245.x. [DOI] [PubMed] [Google Scholar]
- 11.Marnef A, Sommerville J, Ladomery MR. RAP55: insights into an evolutionarily conserved protein family. Int J Biochem Cell Biol. 2009;41:977–81. doi: 10.1016/j.biocel.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 12.Ma Y, Dostie J, Dreyfuss G, Van Duyne GD. The Gemin6-Gemin7 heterodimer from the survival of motor neurons complex has an Sm protein-like structure. Structure. 2005;13:883–92. doi: 10.1016/j.str.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 13.Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Lührmann R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18:5789–802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer U, Englbrecht C, Chari A. Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdiscip Rev RNA. 2011;2:718–31. doi: 10.1002/wrna.87. [DOI] [PubMed] [Google Scholar]
- 15.Wagner G. Function of Hfq in sRNA-mediated regulation. RNA Biol. 2013 In press. [Google Scholar]
- 16.Silva IJ, Saramago M, Dressaire C, Domingues S, Viegas SC, Arraiano CM. Importance and key events of prokaryotic RNA decay: the ultimate fate of an RNA molecule. Wiley Interdiscip Rev RNA. 2011;2:818–36. doi: 10.1002/wrna.94. [DOI] [PubMed] [Google Scholar]
- 17.Régnier P, Hajnsdorf E. Role of Hfq in polyadenylation-dependent mRNA decay. RNA Biol. 2013 In press. [Google Scholar]
- 18.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–89. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tharun S. Roles of eukaryotic Lsm proteins in the regulation of mRNA function. Int Rev Cell Mol Biol. 2009;272:149–89. doi: 10.1016/S1937-6448(08)01604-3. [DOI] [PubMed] [Google Scholar]
- 20.Salgado-Garrido J, Bragado-Nilsson E, Kandels-Lewis S, Séraphin B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18:3451–62. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayes AE, Verdone L, Legrain P, Beggs JD. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 1999;18:4321–31. doi: 10.1093/emboj/18.15.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bouveret E, Rigaut G, Shevchenko A, Wilm M, Séraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–71. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boeck R, Lapeyre B, Brown CE, Sachs AB. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol Cell Biol. 1998;18:5062–72. doi: 10.1128/mcb.18.9.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ingelfinger D, Arndt-Jovin DJ, Lührmann R, Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–501. [PMC free article] [PubMed] [Google Scholar]
- 25.Séraphin B. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J. 1995;14:2089–98. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pannone BK, Xue D, Wolin SL. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998;17:7442–53. doi: 10.1093/emboj/17.24.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verdone L, Galardi S, Page D, Beggs JD. Lsm proteins promote regeneration of pre-mRNA splicing activity. Curr Biol. 2004;14:1487–91. doi: 10.1016/j.cub.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez CF, Pannone BK, Chen X, Fuchs G, Wolin SL. An Lsm2-Lsm7 complex in Saccharomyces cerevisiae associates with the small nucleolar RNA snR5. Mol Biol Cell. 2004;15:2842–52. doi: 10.1091/mbc.E04-02-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomasevic N, Peculis BA. Xenopus LSm proteins bind U8 snoRNA via an internal evolutionarily conserved octamer sequence. Mol Cell Biol. 2002;22:4101–12. doi: 10.1128/MCB.22.12.4101-4112.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chowdhury A, Mukhopadhyay J, Tharun S. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA. 2007;13:998–1016. doi: 10.1261/rna.502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sauer E. The RNA binding properties of Hfq. RNA Biol. 2013 doi: 10.4161/rna.24201. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naville M, Gautheret D. Transcription attenuation in bacteria: theme and variations. Brief Funct Genomics. 2010;9:178–89. doi: 10.1093/bfgp/elq008. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt M-J, Norbury CJ. Polyadenylation and beyond: emerging roles for noncanonical poly(A) polymerases. Wiley Interdiscip Rev RNA. 2010;1:142–51. doi: 10.1002/wrna.16. [DOI] [PubMed] [Google Scholar]
- 34.Lisitsky I, Schuster G. Preferential degradation of polyadenylated and polyuridinylated RNAs by the bacterial exoribonuclease polynucleotide phosphorylase. Eur J Biochem. 1999;261:468–74. doi: 10.1046/j.1432-1327.1999.00285.x. [DOI] [PubMed] [Google Scholar]
- 35.Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol Microbiol. 2004;54:905–20. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- 36.Hajnsdorf E, Régnier P. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc Natl Acad Sci USA. 2000;97:1501–5. doi: 10.1073/pnas.040549897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Folichon M, Allemand F, Régnier P, Hajnsdorf E. Stimulation of poly(A) synthesis by Escherichia coli poly(A)polymerase I is correlated with Hfq binding to poly(A) tails. FEBS J. 2005;272:454–63. doi: 10.1111/j.1742-4658.2004.04485.x. [DOI] [PubMed] [Google Scholar]
- 38.Chen C-YA, Shyu A-B. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip Rev RNA. 2011;2:167–83. doi: 10.1002/wrna.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He W, Parker R. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics. 2001;158:1445–55. doi: 10.1093/genetics/158.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–8. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 41.Januszyk K, Lima CD. Structural components and architectures of RNA exosomes. Adv Exp Med Biol. 2011;702:9–28. doi: 10.1007/978-1-4419-7841-7_2. [DOI] [PubMed] [Google Scholar]
- 42.Otaka H, Ishikawa H, Morita T, Aiba H. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc Natl Acad Sci USA. 2011;108:13059–64. doi: 10.1073/pnas.1107050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–91. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morita T, Aiba H. RNase E action at a distance: degradation of target mRNAs mediated by an Hfq-binding small RNA in bacteria. Genes Dev. 2011;25:294–8. doi: 10.1101/gad.2030311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bandyra KJ, Said N, Pfeiffer V, Górna MW, Vogel J, Luisi BF. The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol Cell. 2012;47:943–53. doi: 10.1016/j.molcel.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Achsel T, Stark H, Lührmann R. The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc Natl Acad Sci USA. 2001;98:3685–9. doi: 10.1073/pnas.071033998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stefano JE. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell. 1984;36:145–54. doi: 10.1016/0092-8674(84)90083-7. [DOI] [PubMed] [Google Scholar]
- 48.Rinke J, Steitz JA. Association of the lupus antigen La with a subset of U6 snRNA molecules. Nucleic Acids Res. 1985;13:2617–29. doi: 10.1093/nar/13.7.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trippe R, Sandrock B, Benecke BJ. A highly specific terminal uridylyl transferase modifies the 3′-end of U6 small nuclear RNA. Nucleic Acids Res. 1998;26:3119–26. doi: 10.1093/nar/26.13.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mroczek S, Krwawicz J, Kutner J, Lazniewski M, Kuciński I, Ginalski K, et al. C16orf57, a gene mutated in poikiloderma with neutropenia, encodes a putative phosphodiesterase responsible for the U6 snRNA 3′ end modification. Genes Dev. 2012;26:1911–25. doi: 10.1101/gad.193169.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shchepachev V, Wischnewski H, Missiaglia E, Soneson C, Azzalin CM. Mpn1, mutated in poikiloderma with neutropenia protein 1, is a conserved 3′-to-5′ RNA exonuclease processing U6 small nuclear RNA. Cell Rep. 2012;2:855–65. doi: 10.1016/j.celrep.2012.08.031. [DOI] [PubMed] [Google Scholar]
- 52.Licht K, Medenbach J, Lührmann R, Kambach C, Bindereif A. 3′-cyclic phosphorylation of U6 snRNA leads to recruitment of recycling factor p110 through LSm proteins. RNA. 2008;14:1532–8. doi: 10.1261/rna.1129608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rissland OS, Norbury CJ. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol. 2009;16:616–23. doi: 10.1038/nsmb.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morozov IY, Jones MG, Razak AA, Rigden DJ, Caddick MX. CUCU modification of mRNA promotes decapping and transcript degradation in Aspergillus nidulans. Mol Cell Biol. 2010;30:460–9. doi: 10.1128/MCB.00997-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morozov IY, Jones MG, Gould PD, Crome V, Wilson JB, Hall AJ, et al. mRNA 3′ tagging is induced by nonsense-mediated decay and promotes ribosome dissociation. Mol Cell Biol. 2012;32:2585–95. doi: 10.1128/MCB.00316-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA: getting closer to the end. Gene. 2007;396:373–90. doi: 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidt M-J, West S, Norbury CJ. The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA. 2011;17:39–44. doi: 10.1261/rna.2252511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Su W, Slepenkov SV, Slevin MK, Lyons SM, Ziemniak M, Kowalska J, et al. mRNAs containing the histone 3′ stem-loop are degraded primarily by decapping mediated by oligouridylation of the 3′ end. RNA. 2013;19:1–16. doi: 10.1261/rna.034470.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoefig KP, Rath N, Heinz GA, Wolf C, Dameris J, Schepers A, et al. Eri1 degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nat Struct Mol Biol. 2013;20:73–81. doi: 10.1038/nsmb.2450. [DOI] [PubMed] [Google Scholar]
- 61.Herrero AB, Moreno S. Lsm1 promotes genomic stability by controlling histone mRNA decay. EMBO J. 2011;30:2008–18. doi: 10.1038/emboj.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ji L, Chen X. Regulation of small RNA stability: methylation and beyond. Cell Res. 2012;22:624–36. doi: 10.1038/cr.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burroughs AM, Ando Y, de Hoon MJL, Tomaru Y, Nishibu T, Ukekawa R, et al. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 2010;20:1398–410. doi: 10.1101/gr.106054.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schumacher MA, Pearson RF, Møller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–56. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Horstmann N, Orans J, Valentin-Hansen P, Shelburne SA, 3rd, Brennan RG. Structural mechanism of Staphylococcus aureus Hfq binding to an RNA A-tract. Nucleic Acids Res. 2012;40:11023–35. doi: 10.1093/nar/gks809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sauter C, Basquin J, Suck D. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 2003;31:4091–8. doi: 10.1093/nar/gkg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci USA. 2009;106:19292–7. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun X, Wartell RM. Escherichia coli Hfq binds A18 and DsrA domain II with similar 2:1 Hfq6/RNA stoichiometry using different surface sites. Biochemistry. 2006;45:4875–87. doi: 10.1021/bi0523613. [DOI] [PubMed] [Google Scholar]
- 69.Sauer E, Weichenrieder O. Structural basis for RNA 3′-end recognition by Hfq. Proc Natl Acad Sci USA. 2011;108:13065–70. doi: 10.1073/pnas.1103420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moskaleva O, Melnik B, Gabdulkhakov A, Garber M, Nikonov S, Stolboushkina E, et al. The structures of mutant forms of Hfq from Pseudomonas aeruginosa reveal the importance of the conserved His57 for the protein hexamer organization. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:760–4. doi: 10.1107/S1744309110017331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nikulin A, Stolboushkina E, Perederina A, Vassilieva I, Blaesi U, Moll I, et al. Structure of Pseudomonas aeruginosa Hfq protein. Acta Crystallogr D Biol Crystallogr. 2005;61:141–6. doi: 10.1107/S0907444904030008. [DOI] [PubMed] [Google Scholar]
- 72.Someya T, Baba S, Fujimoto M, Kawai G, Kumasaka T, Nakamura K. Crystal structure of Hfq from Bacillus subtilis in complex with SELEX-derived RNA aptamer: insight into RNA-binding properties of bacterial Hfq. Nucleic Acids Res. 2012;40:1856–67. doi: 10.1093/nar/gkr892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bøggild A, Overgaard M, Valentin-Hansen P, Brodersen DE. Cyanobacteria contain a structural homologue of the Hfq protein with altered RNA-binding properties. FEBS J. 2009;276:3904–15. doi: 10.1111/j.1742-4658.2009.07104.x. [DOI] [PubMed] [Google Scholar]
- 74.Tharun S, Muhlrad D, Chowdhury A, Parker R. Mutations in the Saccharomyces cerevisiae LSM1 gene that affect mRNA decapping and 3′ end protection. Genetics. 2005;170:33–46. doi: 10.1534/genetics.104.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mund M, Neu A, Ullmann J, Neu U, Sprangers R. Structure of the LSm657 complex: an assembly intermediate of the LSm1-7 and LSm2-8 rings. J Mol Biol. 2011;414:165–76. doi: 10.1016/j.jmb.2011.09.051. [DOI] [PubMed] [Google Scholar]
- 76.Naidoo N, Harrop SJ, Sobti M, Haynes PA, Szymczyna BR, Williamson JR, et al. Crystal structure of Lsm3 octamer from Saccharomyces cerevisiae: implications for Lsm ring organisation and recruitment. J Mol Biol. 2008;377:1357–71. doi: 10.1016/j.jmb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 77.Chowdhury A, Raju KK, Kalurupalle S, Tharun S. Both Sm-domain and C-terminal extension of Lsm1 are important for the RNA-binding activity of the Lsm1-7-Pat1 complex. RNA. 2012;18:936–44. doi: 10.1261/rna.029876.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chowdhury A, Tharun S. lsm1 mutations impairing the ability of the Lsm1p-7p-Pat1p complex to preferentially bind to oligoadenylated RNA affect mRNA decay in vivo. RNA. 2008;14:2149–58. doi: 10.1261/rna.1094208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol. 2004;11:1206–14. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sauer E, Schmidt S, Weichenrieder O. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc Natl Acad Sci USA. 2012;109:9396–401. doi: 10.1073/pnas.1202521109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Večerek B, Rajkowitsch L, Sonnleitner E, Schroeder R, Bläsi U. The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res. 2008;36:133–43. doi: 10.1093/nar/gkm985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mura C, Phillips M, Kozhukhovsky A, Eisenberg D. Structure and assembly of an augmented Sm-like archaeal protein 14-mer. Proc Natl Acad Sci USA. 2003;100:4539–44. doi: 10.1073/pnas.0538042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol Cell. 2002;9:11–22. doi: 10.1016/S1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 84.Møller T, Franch T, Højrup P, Keene DR, Bächinger HP, Brennan RG, et al. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell. 2002;9:23–30. doi: 10.1016/S1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 85.Ishikawa H, Otaka H, Maki K, Morita T, Aiba H. The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3′ poly(U) tail. RNA. 2012;18:1062–74. doi: 10.1261/rna.031575.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr Opin Microbiol. 2004;7:140–4. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 87.Karaduman R, Dube P, Stark H, Fabrizio P, Kastner B, Lührmann R. Structure of yeast U6 snRNPs: arrangement of Prp24p and the LSm complex as revealed by electron microscopy. RNA. 2008;14:2528–37. doi: 10.1261/rna.1369808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Valadkhan S. snRNAs as the catalysts of pre-mRNA splicing. Curr Opin Chem Biol. 2005;9:603–8. doi: 10.1016/j.cbpa.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 89.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–86. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ikeda Y, Yagi M, Morita T, Aiba H. Hfq binding at RhlB-recognition region of RNase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol Microbiol. 2011;79:419–32. doi: 10.1111/j.1365-2958.2010.07454.x. [DOI] [PubMed] [Google Scholar]
- 91.Tharun S. Purification and analysis of the decapping activator Lsm1p-7p-Pat1p complex from yeast. Methods Enzymol. 2008;448:41–55. doi: 10.1016/S0076-6879(08)02603-7. [DOI] [PubMed] [Google Scholar]
- 92.Marnef A, Standart N. Pat1 proteins: a life in translation, translation repression and mRNA decay. Biochem Soc Trans. 2010;38:1602–7. doi: 10.1042/BST0381602. [DOI] [PubMed] [Google Scholar]
- 93.Pilkington GR, Parker R. Pat1 contains distinct functional domains that promote P-body assembly and activation of decapping. Mol Cell Biol. 2008;28:1298–312. doi: 10.1128/MCB.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nissan T, Rajyaguru P, She M, Song H, Parker R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell. 2010;39:773–83. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang X-C, Sabath I, Debski J, Kaus-Drobek M, Dadlez M, Marzluff WF, et al. A complex containing the CPSF73 endonuclease and other polyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3′ end processing. Mol Cell Biol. 2013;33:28–37. doi: 10.1128/MCB.00653-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Argaman L, Elgrably-Weiss M, Hershko T, Vogel J, Altuvia S. RelA protein stimulates the activity of RyhB small RNA by acting on RNA-binding protein Hfq. Proc Natl Acad Sci USA. 2012;109:4621–6. doi: 10.1073/pnas.1113113109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Friesen WJ, Dreyfuss G. Specific sequences of the Sm and Sm-like (Lsm) proteins mediate their interaction with the spinal muscular atrophy disease gene product (SMN) J Biol Chem. 2000;275:26370–5. doi: 10.1074/jbc.M003299200. [DOI] [PubMed] [Google Scholar]
- 98.Li Z, Nagy PD. Diverse roles of host RNA binding proteins in RNA virus replication. RNA Biol. 2011;8:305–15. doi: 10.4161/rna.8.2.15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ryan DE, Stevens SW, Abelson J. The 5′ and 3′ domains of yeast U6 snRNA: Lsm proteins facilitate binding of Prp24 protein to the U6 telestem region. RNA. 2002;8:1011–33. doi: 10.1017/S1355838202026092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 101.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–91. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH. Cn3D: sequence and structure views for Entrez. Trends Biochem Sci. 2000;25:300–2. doi: 10.1016/S0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]
- 103.Raker VA, Plessel G, Lührmann R. The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J. 1996;15:2256–69. [PMC free article] [PubMed] [Google Scholar]
- 104.Montzka KA, Steitz JA. Additional low-abundance human small nuclear ribonucleoproteins: U11, U12, etc. Proc Natl Acad Sci USA. 1988;85:8885–9. doi: 10.1073/pnas.85.23.8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Turunen JJ, Niemelä EH, Verma B, Frilander MJ. The significant other: splicing by the minor spliceosome. Wiley Interdiscip Rev RNA. 2013; 4:61-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–80. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 107.Tang W, Kannan R, Blanchette M, Baumann P. Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature. 2012;484:260–4. doi: 10.1038/nature10924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fu D, Collins K. Human telomerase and Cajal body ribonucleoproteins share a unique specificity of Sm protein association. Genes Dev. 2006;20:531–6. doi: 10.1101/gad.1390306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bachand F, Boisvert F-M, Côté J, Richard S, Autexier C. The product of the survival of motor neuron (SMN) gene is a human telomerase-associated protein. Mol Biol Cell. 2002;13:3192–202. doi: 10.1091/mbc.E02-04-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zaric B, Chami M, Rémigy H, Engel A, Ballmer-Hofer K, Winkler FK, et al. Reconstitution of two recombinant LSm protein complexes reveals aspects of their architecture, assembly, and function. J Biol Chem. 2005;280:16066–75. doi: 10.1074/jbc.M414481200. [DOI] [PubMed] [Google Scholar]