Abstract
RNA enables the material interpretation of genetic information through time and in space. The creation, destruction and activity of RNA must be well controlled and tightly synchronized with numerous cellular processes. We discuss here the pathways and mechanism of bacterial RNA turnover, and describe how RNA itself modulates these processes as part of decision-making networks. The central roles of RNA decay and other aspects of RNA metabolism in cellular control are also suggested by their vulnerability to sabotage by phages; nonetheless, RNA can be used in defense against phage infection, and these processes are described here. Salient aspects of RNA turnover are drawn together to suggest how it could affect complex effects such as phenotypic diversity in populations and responses that persist for multiple generations.
Keywords: RNA turnover, riboregulation, post-transcriptional network, ribonuclease, messenger RNA, small regulatory RNA
Introduction
While transcripts are transient entities in the lifetime of a cell, their limited lifespans serve as a means of manipulating the strength and duration of gene expression. The adjustment of transcript turnover sustains homeostasis and is a key aspect of the rapid adaptation to environmental changes.1,2 In Escherichia coli, protein-encoding RNAs have on average overall chemical half-lives of 7–8 min,3,4 but individual transcripts each have their own characteristic lifetime that can be modulated according to growth stage and stress response. The modulation of transcript lifetime is affected by protective binding partners and the accessibility to the machinery of RNA decay that include ribonucleases, their accessory proteins and helper RNAs that catalyze RNA turnover.
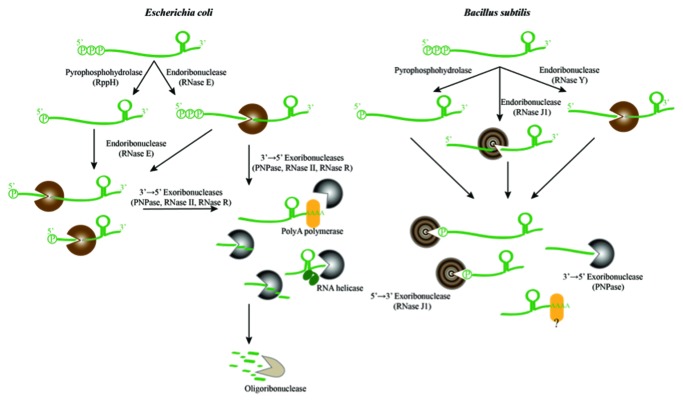
The general scheme for degradation of RNA and the key participating enzymes for the Gram-negative Escherichia coli and the Gram-positive Bacillus subtilis are shown in the schematic in Figure 1. These bacterial species are thought to have diverged at least 1 billion years ago,5 and each is considered representative in genomic character for many of the currently known bacteria. In both E. coli and B. subtilis, the principal degradation pathways begin with endoribonuclease cleavage that destines the transcript to a fate of complete destruction. The cleavage(s) made by an endoribonuclease can be followed by degradation catalyzed by exoribonucleases.
Figure 1. A billion years of bacterial RNA degradation. The general scheme of RNA degradation pathways in Escherichia coli (left panel) and Bacillus subtilis (right panel). The key enzymes involved in the different steps are listed in parentheses, and represented in brown (endonucleases) or gray (exoribonucleases). RNase J has both endo- and exo-activities, and it is depicted with the concentric gray and brown circles. “Decapping” of the 5′ terminal triphosphate to leave a 5′ monophosphate may convert some transcripts into preferred substrates for RNase E in E. coli and RNase J in B. subtilis. At the 3′ end of transcripts, poly(A) tails are added by poly(A) polymerase or another enzyme with such activity (yellow ellipse) in B. subtilis, contributing to the processivity of RNA turnover in these bacteria. The stem loop structure on the 3′ end of the transcripts at the top are rho-independent transcription terminators.
Despite the similarity in the overall pathways of E. coli and B. subtilis, surprisingly few of the participating enzymes are common to both species. It seems remarkable that entirely different enzymes, with unrelated structures or evolutionary origins, have evolved with analogous function in the two organisms. This likely reflects the biological importance of the pathway and the power of convergent evolution to meet the cellular demands for RNA metabolism. In E. coli and many other proteobacteria, one of the key enzymes in the decay pathway is the hydrolytic endoribonuclease RNase E,6-9 which is highlighted in the E. coli decay pathway shown in the left panel of Figure 1. In B. subtilis, RNase Y serves a functionally analogous role to E. coli RNase E, as shown in the right panel of Figure 1.10-13
Although RNase E cleaves RNA internally, it often interacts with the 5′ end of its many substrates and prefers those harboring a monophosphate group.14 In bacteria, primary transcripts are transcribed with a 5′ terminal triphosphate group, which can confer protection from RNase E attack. However, a pyrophosphohydrolase (RppH) can remove the pyrophosphate from the 5′ end of the RNA, leaving a monophosphorylated RNA that can be rapidly degraded by RNase E.15,16 The B. subtilis ribonuclease RNase J1 also has a preference for substrates with a 5′ monophosphate.17 A homolog of the RppH pyrophosphohydrolase in B. subtilis appears to play an analogous role to decap the triphosphate group of transcripts.18 It seems likely that 5′ end recognition is also important in the decay pathway B. subtilis although the structural basis for the ribonuclease activation has an entirely different stereochemical origin;19 this parallel seems yet another remarkable case of evolutionary convergence.
Following the initiating endoribonuclease cleavage(s), exoribonuclease activities in E. coli are provided by 3′ to 5′ exoribonucleases such as the phosphorolytic polynucleotide phosphorylase (PNPase), or the hydrolytic enzymes RNase R or RNase II. Exoribonuclease cleavages in B. subtilis are mediated by the hydrolytic RNase J in the 5′ to 3′ direction, and by PNPase, with the opposite polarity.17,20,21 These processes are assisted by other enzymes, such as poly(A) polymerase (PAP), which adds 3′-terminal poly-adenine tails that become sites for processive degradation.22,23 Poly-adenine tails may also help initiate degradation, as polyadenylation affects the lifetime of many transcripts in E. coli22,24,25 and potentially helps control levels of tRNA.26 Moreover, in B. subtilis, poly(A) and heteropolymeric tails seem to mark the RNA for degradation as well, although the enzyme responsible for adding the tails has not yet been identified.27
In considering the degradation pathways of the two representative bacteria, it is interesting to note that the archaea harbor some bacterial-like enzymes such as RNase J and some eukaryotic-like RNA metabolic assemblies, such as the exosome (described further below). The archaeal pathways will not be described here, except to draw occasional parallels between the bacterial components.
Several of the enzymes depicted in Figure 1 play additional functional roles, as they are also involved in processing precursors of structured RNA. For instance, E. coli RNase E also takes part in RNA maturation by cleaving precursors of structured RNAs.8 This activity reveals a more controlled operation mode for the ribonuclease, in which the cleavage products are not marked for destruction but rather processed further to form a functional RNA molecule. How this discrimination is achieved is an intriguing puzzle. The processing activity of RNase E is a dominating aspect of its in vivo activity, and it has been suggested that the enzyme may catalyze as many processing as degradation cleavages during exponential growth stage.8 Processing activities are also likely to be a dominating mode for RNase Y and RNase J in B. subtilis and other species.
A multi-enzyme assembly for RNA degradation
In many bacterial species, a multi-enzyme assembly known as the RNA degradosome acts as a key machinery of RNA decay and processing.28 The components of the degradosome cooperate to degrade RNA transcripts and to process precursors of structured RNAs. In E. coli and several other bacteria, the core of the complex is formed by RNase E, which was mentioned above. The protein composition of the degradosome assemblies differs between evolutionarily divergent bacterial species, and some degradosome assemblies include enzymes such as exoribonucleases and metabolic enzymes, but they often include a DEAD-box helicase that directly associates to an endoribonuclease.29
The exoribonuclease PNPase, mentioned above as an important contributor to prokaryotic RNA degradation, is a canonical component of the E. coli RNA degradosome (Fig. 2). There are interesting structural and functional parallels between PNPase with the archaeal and eukaryotic exosomes. All share a common evolutionary origin with the phosphorolytic enzyme RNase PH and have chamber-like quaternary architecture that forms a central channel containing the active site. In both PNPase and the exosome, the entrance to the central channel narrows to form an aperture wide enough only for single-stranded RNA substrate to enter, suggesting a necessity for structured RNA substrates to be un-wound prior to processing by the degradative machinery.
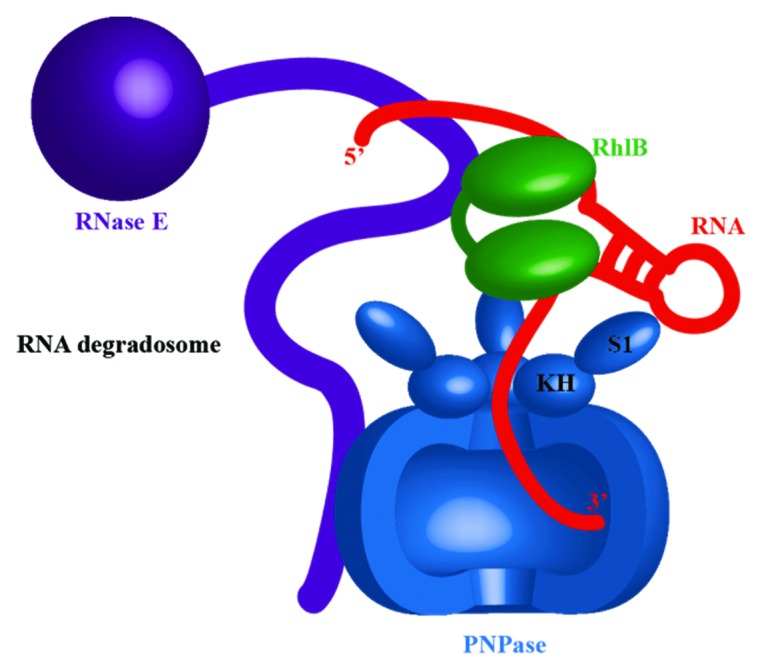
Figure 2. The Escherichia coli RNA degradosome, and facilitated substrate delivery to the exoribonuclease component, polynucleotide phosphorylase (PNPase, blue). A portion of the PNPase ring has been removed to illustrate the central chamber that harbors the active site. An ATP-dependent DEAD box helicase RhlB (green), another degradosome component, is unwinding the RNA stem-loop (red), providing a single-stranded RNA for PNPase. Both enzymes are associated with RNase E (purple). For clarity, other degradosome components, such as enolase, were omitted.
The RNA helicase (RhlB) as part of the E. coli RNA degradosome aides the processing of RNA substrates by the assembly.30-32 The unwinding or remodeling activities of Rh1B are required to improve the efficiency of mRNA degradation, since the ribonuclease activities of RNase E and PNPase are specific for single-stranded RNA (Fig. 2). The ATPase activity of RhlB is required to facilitate the degradation by PNPase of structured RNA transcripts with a repetitive element that forms stable stem-loop structures.30-33 The helicase binds to a site in RNase E that is flanked by RNA-binding domains, and these perhaps help to capture substrates for the helicase, as does also an arginine-rich tail on the C terminus of RhlB.29
Riboregulation: RNA turnover mediated by small regulatory RNAs
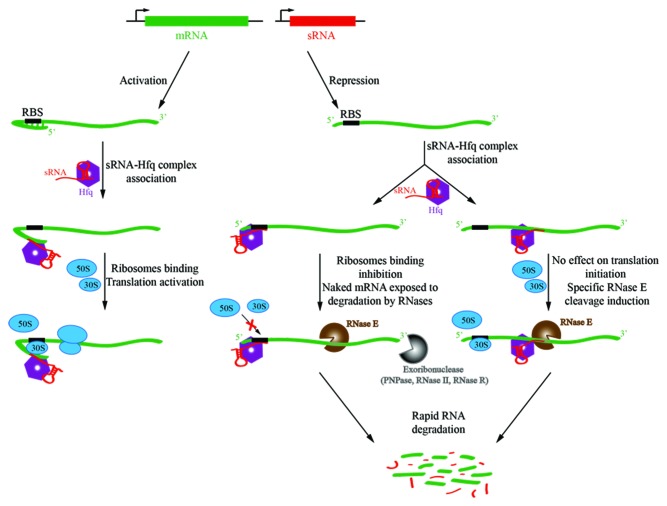
Small regulatory RNAs (sRNAs) are an important class of trans-acting regulators of post-transcriptional gene regulation that can modulate transcript lifetimes, rapidly and with specificity (Fig. 3). They appear to be ubiquitous throughout the bacterial lineages, and mediate numerous processes, including membrane stress response, carbon regulation, virulence effects, biofilm formation and countless other complex behaviors.34,35 Many sRNAs act by impeding translation and increasing likelihood for encounters with a ribonuclease with subsequent turnover, but some also have entirely the opposite effect, and act by exposing the ribosome-binding site to favor translation initiation;36 this presumably protects the RNA against ribonuclease activity.
Figure 3. Schematic of small RNA (sRNA)-mediated regulation of gene expression. In the top panel, the sRNAs (red) are transcribed in trans to the target gene (green). A cognate seed region that pairs to a recognition site in the target RNA can relieve repression of translation by secondary structure that masks the ribosome-binding site (RBS, or Shine Dalgarno sequence). In this way, sRNAs can activate gene expression (left panel). Conversely, they can mask the RBS (ribosome binding site), thereby preventing translation initiation and making the naked RNA vulnerable to ribonuclease attack, as shown in the middle panel. sRNA can also bind within the coding sequence of the target mRNA and the sRNA 5′ end can allosterically activate RNase E to cleave target mRNA (right panel).
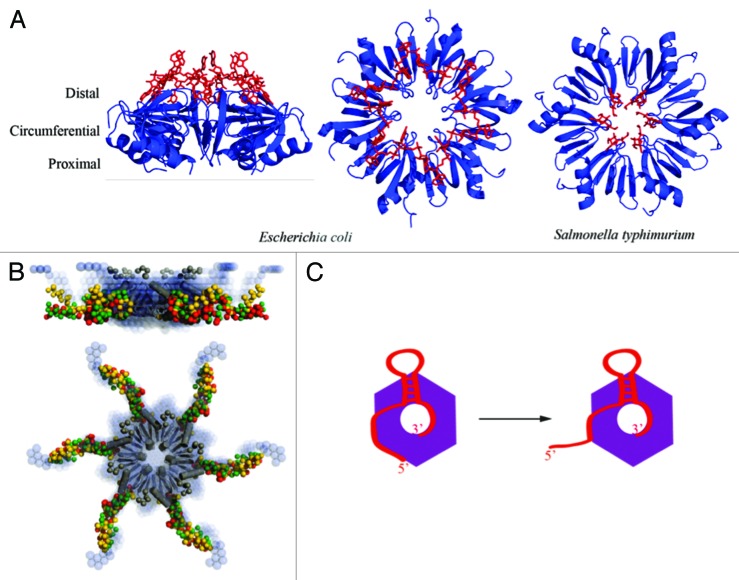
sRNAs can target multiple transcripts, and some transcripts are the targets for several different sRNAs.34 They emulate elements of complex logic circuits, and help to build post-transcriptional networks that are as rich in complexity as the well-characterized transcriptional regulator networks.37 In many bacteria, a key facilitator of sRNA function in these regulatory networks is the RNA chaperone protein Hfq.38 The protein belongs to the extensive Sm-family, characterized by a conserved fold and propensity to assemble into a ring-like oligomeric architecture (Fig. 4). Hfq can interact with sRNA and target transcripts through three different binding surfaces on this ring: proximal, distal and circumferential. These interactions help to mediate pairing of sRNAs to targets. The limited cellular quantity of the protein means that RNAs must compete for binding Hfq in sRNA-mediated riboregulation, with implications for inter-connecting different regulatory networks.39
Figure 4. The structure and proposed RNA-binding modes of the RNA chaperone Hfq. (A) Crystal structures revealing the ring-like architecture of the hexameric Hfq protein and RNA binding to the distal (left and central panels, E. coli)98 and proximal faces (right panel, Salmonella typhimurium)99 of the ring. (B) C-terminal tails emanate from the core, based on small angle X-ray solution scattering. These tails have some weak conservation in families and are likely to be flexible.100,101 They may help to capture RNA and accommodate species with complex folds38 or serve for interaction with other proteins.102 (C) A proposed binding model for sRNA (red) in which the 3′ poly(U) tail is engaged on the proximal side of the Hfq hexamer (purple), and the body of the sRNA is engaged on the circumferential rim of the ring.99,103 The mRNA recognition region, here located on the 5′ end of the sRNA, is proposed to be able to peel off to mediate cognate base pairing with the target mRNA.
In E. coli, RNase E appears to be the dominant enzyme responsible for sRNA-mediated response and for degradation of the sRNAs.40 Another enzyme that was shown to influence sRNAs that are not associated with Hfq is PNPase, and this exoribonuclease is responsible for their degradation in stationary phase.41 However, PNPase has been identified in a genetic screen to be linked to Hfq-mediated sRNA activity in E. coli and, surprisingly, it affects the stability of some sRNAs in vivo by protecting them from degradation by other ribonucleases.42 It seems paradoxical that a ribonuclease might confer a protective effect on an sRNA in vivo, but there is precedent for example in RNase II playing a protective role for other RNAs by removing 3′-poly(A) tails.43 As PNPase is a processive exonuclease, it might be expected to drive the degradation of the substrate once it has loaded, but perhaps in the case of structured sRNAs it might trap them into protective complexes. This mechanism awaits experimental confirmation.
sRNAs are also present in B. subtilis, however the study of the networks involving these regulatory molecules in this organism are still at an early stage, and the roles of sRNAs are not yet as well defined as in E. coli. However, it has already been shown in B. subtilis that a small RNA and a variety of potential RNA chaperones are involved in iron sensing.44 The B. subtilis RNase Y may also be involved in the turnover of cis-acting regulatory RNA, such as riboswitches.12 Studies of Staphylococcus aureus (which is gram positive but not in the firmicute group of B. subtilis) show clear differences from sRNA regulation in E. coli, such as the apparent lack of importance of Hfq.45
Substrate recognition and cues for initiating degradation
As mentioned above, the 5′ end of a transcript can boost RNase E and RNase J activity. The 5′ monophosphate group is engaged in the RNase E 5′ binding pocket, localized in the N-terminal catalytic domain of the enzyme and triggers domain closure at the active site.46,47 In RNase J, a pocket near the hydrolytic active site has been identified which recognizes the 5′ monophosphate group.19
The 5′ monophosphate group necessary for stimulated cleavage of an RNA transcript need not be provided by the target RNA itself, as it has been shown that an sRNA harboring a 5′ monophosphate group can act in trans to target triphosphorylated RNAs to RNase E and in this way act as an allosteric activator of ribonuclease activity.48 A recently discovered class of sRNA are liberated from mRNAs by processing the 3′ UTRs (untranslated regions),49 and it seems likely these also could have scope to play roles in trans-activating RNase E. Interesting in this regard to note that mRNA fragments may play regulatory roles in eukaryotes.50
The cleavage of some RNA substrates in E. coli is not significantly influenced by the status of their 5′ end, these are recognized and cleaved probably by RNase E through an internal entry pathway, which does not require enzyme activation by the monophosphate bound in the 5′ binding pocket.51-55 The processes are assisted by the C-terminal half of RNase E, which is a mostly unstructured region that contains two RNA-binding domains important for the substrate recognition and alignment on the enzyme.51,53-57,60
The 3′ end of a transcript is also important for affecting its lifetime. In E. coli, the decay of many mRNAs is affected by polyadenylation,59-62 and the half-lives of certain mRNAs are found to be boosted in the absence of polyadenylation.61 The poly(A) tails are added by poly(A) polymerase (PAPI), and the levels of poly(A) tails increase in a mutant lacking PNPase.61 It has been proposed that polyadenylation of tRNA may serve as a means of detecting defective species and directing them to degradation.61,63,64
In principle, polyadenylation may help to recruit ribonucleases. Taken together, several studies provide evidence in vitro and in vivo for a decay pathway in the turnover of structured mRNA degradation intermediates that is dependent on polyadenylation and involves RNase R and PAP I.31,65,66 PNPase is also likely to be involved in some cases, as the polyadenylation of the anti-sense RNA I in vivo may recruit PNPase and lead to decay of that RNA. Polyadenylation and linked degradation in bacteria has an interesting parallel with surveillance in eukaryotes, where the helicase-bearing TRAMP complex recognizes aberrant transcripts, certain non-coding RNA and snoRNAs, tagging them with poly(A) tails and directing them to the exosome.67 Perhaps a somewhat similar process occurs in bacteria, with the poly(A) polymerase marking RNAs with the poly(A) tails, the RNA unwinding role being fulfilled by the DEAD box helicase RhlB and cleavage is associated with RNase E together with PNPase in the RNA degradosome assembly.
RNA turnover and energy economy
Under starvation conditions, E. coli enters the stationary phase, where growth is slow, metabolism is less active overall and there is a general downregulation of both transcription and translation.68 Counter intuitively, translation of some metabolic enzymes must increase as cells enter stationary phase: this enables the bacterium to use a broader range of carbon and nitrogen sources. The starvation responsive genes are part of a stress regulon controlled by the sigma factor RpoS. In turn, RpoS levels are regulated by proteolytic degradation, and its transcript levels are also controlled by sRNAs. Interestingly, the protease adaptor SprE (also known as RssB) that affects RpoS stability also affects polyadenylation and the control of mRNA stability.69 In exponentially growing E. coli, SprE stimulates polyadenylation to boost decay of specific mRNAs, and it also affects the localization of PAPI to the cytoplasmic membrane. Polyadenylation also affects the recovery from stationary phase.69 The SprE protein thus plays two distinct roles in stationary phase, by regulating the stability of RpoS and by affecting the stability of a subset of mRNAs by stimulating their polyadenylation and directing them to destruction.
Turnover of RNA can proceed either through hydrolysis or phosphorolysis. The latter is more useful for shunting into DNA precursors for cell division, and would be expected to occur close to the onset of DNA replication. It is interesting to note that levels of the hydrolytic exoribonuclease RNase R decreases in stationary phase,70,71 but they increase in stress conditions.72 This switch is affected by acetylation of a specific lysine in RNase R. Metabolic enzymes in Salmonella are also regulated by lysine acetylation,73 suggesting that this modification could be the downstream effect of a signaling pathway. We speculate that there is a likely link of cell division and ribonuclease pathway, sensitive to the metabolic state of the cell.74 The speculative link is supported by the functionally important role of ribonucleases for controling certain cell division genes.75-77 Perhaps in spore-forming species such as Streptomyces and Bacillus, this proposed linkage might be important for energy conservation for cells as they enter the vegetative storage stage.
RNA is manipulated by phage in infection, but also used in host defense
Given the role of RNA metabolism in the regulation of complex genetic networks, it might seem unsurprising that this pathway can be targeted by phage during infection in numerous ways. For instance, RNase E is directly manipulated through phosphorylation by a kinase encoded by T7 phage, and the modification changes the ribonuclease activity to selectively stabilize phage-encoded transcripts.8,78 Recently, bacteriophage T4 polynucleotide kinase has been found to destabilize host mRNAs.21
Small RNAs are used in host defense against phage infection. The Type III toxin-antitoxin system comprises a toxic endoribonuclease and a non-coding RNA that inhibits the enzyme.79-82 In the ToxIN system of Pectobacterium atrosepticum, the RNA forms a pseudoknot structure from a repetitive element that is processed by the ToxN ribonuclease. Thus, the ToxI antitoxins inhibit their own parent-processing enzymes and consequently protect the cell from a harmful general ribonuclease. This system confers immunity to a population of bacteria by triggering death of infected cells. It is not presently known what triggers toxin activation—whether foreign metabolite, RNA or abnormal state. Other types of toxin-antitoxin systems contribute to the phenomenon of persistence, in which sub-populations in an isogenic population are more resistant to the action of antibiotic.83
Parallels can be drawn between ToxN proteins and Type I Cas6 ribonucleases of the anti-viral CRISPR/Cas systems, which provide adaptive immunity against phage infection (CRISPR, clustered regularly interspaced short palindromic repeats; Cas, CRISPR-associated proteins). Like the ToxN protein, the Cas6 enzyme also remains associated with its own catalytic product following cleavage of the repetitive CRISPR transcript substrates into crRNAs.84-86 Binding of crRNAs to their Cas6-processing enzymes, which do not cleave other RNAs, leads to formation of the Cascade ribonucleoprotein complex, and its highly specific recognition of invading nucleic acids. In the archeon Sulfolobus solfataricus, a complex composed of seven CAS proteins forms a CMR complex that carries targeting crRNAs and cleaves target RNA and the guide RNA by endoribonuclease activity.87 The CMR has some functional parallels with the DICER silencing assembly of eukaryotes and the pairing of RNase E and sRNA in E. coli, where sRNA, in complex with Hfq and the mRNA, guides the enzyme to destroy the target.
Gaining substrate access, and the problems of finding a cognate partner quickly and accurately in riboregulation
It was widely assumed that translation and transcription are tightly coupled in bacteria, so that nascent transcripts would only rarely be found outside large assemblies that could potentially protect them from ribonuclease activity. Indeed, the efficiency of translation impacts on mRNA decay rates,53,88,89 and the 5′ end phosphorylation status of the RNA can influence the decay of the transcripts that are poorly bound by the ribosomes.90 However, recent evidence shows that certain transcripts in E. coli may in fact be transcribed at some distance from the site of translation,91 and in these cases there may be other means of protecting the RNA against ribonuclease attack. RNase E and the degradosome are compartmentalized to the cytoplasmic membrane.92 This spatial separation from transcription sites has been proposed to provide a time delay for transcripts to be recognized and tagged by sRNAs before they reach the degradosome for rapid destruction.8
Within its generation time, each E. coli cell synthesizes an estimated 430,000 transcripts. How do sRNAs find their targets in such a thicket? The number of potential pairwise interactions is astronomical—taking by analogy the potential interactions in the protein interactome, for a sRNA to find a target by exhaustive search would require improbable rates of discriminating encounters.93 We suggest that the problem is more acute in RNA compared with protein, because of the propensity of even random RNA to fold into semi-compact states.
The solution to the puzzle might rest in part with compartmentalization. It is becoming apparent that RNA encodes patterns of expression in space, with the consequence that transcripts can be targeted for particular compartments. There is spatial organization of the chromosome for temporal patterns of expression,94 and it is conceivable that a similar order might help to compartmentalize sRNAs with their targets. Also, the specificity and speed in riboregulation may be aided by the activities of helicases to unwind duplexes formed with sRNAs, or to help remodel protein-RNA interactions to expose potential pairing sites for the sRNAs. Driving the system out of equilibrium in an energy-dependent way has some parallels with kinetic proofreading to enhance fidelity of molecular recognition processes.
Summary and perspective
When the transcriptional inhibitor rifampicin is added to cultures, many transcripts in E. coli are found to decay with exponential timecourse.4 Does this decay arise from stochastic access to the transcripts, or from a defined process that is precise and accurate? The published decay rates suggest that the processes may be multi-exponential, and the deviation of decay rates from single-exponential imply that the process of RNA turnover is not entirely random, and can arise from numerous processes that affect gene expression—for instance, the hypothetical possibility of counting translation rounds for each transcript, or a synchronized delay in the onset of turnover. Another key question concerning transcript decay is the extent of variation in the lifetime for a particular type of transcript. It seems likely that the detailed analysis of the key degradation parameters may be very illuminating for understanding the underlying processes.
RNA metabolism and sRNA-mediated regulation can give rise to complex phenotypes. sRNAs have multiple targets, and some target transcripts have multiple sRNA regulators. This results in networks of sRNAs forming a complement to the networks controlling transcription and mediated by transcription factors.37 These networks have, for instance, multi-output feedforward loops that can act to reduce leaky expression and maintain repression of target genes with changing nutrient conditions.37 One striking feature of sRNA-mediated regulation is the sensitivity of the response curve to the ratio of regulatory RNA to targets. As the ratio approaches unitary stoichiometry, sRNA effects are maximized, and in the case of small numbers of transcripts, this could result in tremendous cell-to-cell variations in response to sRNAs.37 It therefore seems likely that the processes of riboregulation can affect phenotypic diversity in populations. There is precedent for such a process in eukaryotes, where antagonist long non-coding RNAs affect heterogeneity in isogenic populations of yeast (Saccharomyces cerevisiae).95,96
The use of RNA to distinguish self from non-self is also an important mechanism. The CRISPR system represents a prokaryotic equivalent to a system that retains some memory of self vs. non-self RNA. Intriguingly, there are eukaryotic regulatory RNAs (piRNA) that are generated in response to non-self RNA expression that can be propagated through multiple generations through the germ line in the nematode, Caenorhabditis elegans.97 It is conceivable, but presently untested, that analogous complex multi-generation processes could occur in prokaryotes, for instance, mediated by long-lived sRNAs whose half-lives exceed the average cellular generation time. The impact of post-transcriptional control of gene expression is strongly dependent on numerous parameters, including the lifetime of transcripts and sRNAs under different growth conditions, their dwell times on the Hfq chaperone and the number of times an individual transcript is translated. These are not presently known in depth, and these details might explain some of the more mysterious puzzles of how gene expression is controlled, and how targets are recognized with specificity and speed.
Genomes seem to bear more information than is encoded as binary bits, as the information acts back on itself. That is, the act of generating a transcript affects the subsequent generation of that transcript itself—and this can be achieved either directly or indirectly. This makes the system self-referential and, accordingly, the information is much richer than the sequence pattern itself. The use of sRNAs in riboregulation enriches and expands this capacity for self-information. Genomic information is thus not only in the digital composition, but also in its base-pairing and fold of cognate matches to RNA. Establishing the full information content of a system like the genome remains a challenging but important problem for experiment and theory, and the mechanisms of RNA metabolism and riboregulation are likely to be a key aspect to address this challenge in the future.
Acknowledgments
The authors are supported by the Wellcome Trust, and K.J.B. is the recipient of a BBSRC studentship. We thank Steven Hardwick and A.J. Carpousis for helpful comments and stimulating discussions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/24393
References
- 1.Grunberg-Manago M. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu Rev Genet. 1999;33:193–227. doi: 10.1146/annurev.genet.33.1.193. [DOI] [PubMed] [Google Scholar]
- 2.Miller C, Schwalb B, Maier K, Schulz D, Dümcke S, Zacher B, et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol Syst Biol. 2011;7:458. doi: 10.1038/msb.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohanty BK, Kushner SR. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol Microbiol. 1999;34:1094–108. doi: 10.1046/j.1365-2958.1999.01673.x. [DOI] [PubMed] [Google Scholar]
- 4.Selinger DW, Saxena RM, Cheung KJ, Church GM, Rosenow C. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 2003;13:216–23. doi: 10.1101/gr.912603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–56. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 6.Carpousis AJ. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- 7.Carpousis AJ, Luisi BF, McDowall KJ. Endonucleolytic initiation of mRNA decay in Escherichia coli. Prog Mol Biol Transl Sci. 2009;85:91–135. doi: 10.1016/S0079-6603(08)00803-9. [DOI] [PubMed] [Google Scholar]
- 8.Mackie GA. RNase E: at the interface of bacterial RNA processing and decay. Nat Rev Microbiol. 2013;11:45–57. doi: 10.1038/nrmicro2930. [DOI] [PubMed] [Google Scholar]
- 9.Stead MB, Marshburn S, Mohanty BK, Mitra J, Pena Castillo L, Ray D, et al. Analysis of Escherichia coli RNase E and RNase III activity in vivo using tiling microarrays. Nucleic Acids Res. 2011;39:3188–203. doi: 10.1093/nar/gkq1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durand S, Gilet L, Bessières P, Nicolas P, Condon C. Three essential ribonucleases-RNase Y, J1, and III-control the abundance of a majority of Bacillus subtilis mRNAs. PLoS Genet. 2012;8:e1002520. doi: 10.1371/journal.pgen.1002520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehnik-Habrink M, Lewis RJ, Mäder U, Stülke J. RNA degradation in Bacillus subtilis: an interplay of essential endo- and exoribonucleases. Mol Microbiol. 2012;84:1005–17. doi: 10.1111/j.1365-2958.2012.08072.x. [DOI] [PubMed] [Google Scholar]
- 12.Shahbabian K, Jamalli A, Zig L, Putzer H. RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J. 2009;28:3523–33. doi: 10.1038/emboj.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao S, Bechhofer DH. Initiation of decay of Bacillus subtilis rpsO mRNA by endoribonuclease RNase Y. J Bacteriol. 2010;192:3279–86. doi: 10.1128/JB.00230-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mackie GA. Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998;395:720–3. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- 15.Celesnik H, Deana A, Belasco JG. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol Cell. 2007;27:79–90. doi: 10.1016/j.molcel.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008;451:355–8. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 17.Mathy N, Bénard L, Pellegrini O, Daou R, Wen T, Condon C. 5′-to-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell. 2007;129:681–92. doi: 10.1016/j.cell.2007.02.051. [DOI] [PubMed] [Google Scholar]
- 18.Richards J, Liu Q, Pellegrini O, Celesnik H, Yao S, Bechhofer DH, et al. An RNA pyrophosphohydrolase triggers 5′-exonucleolytic degradation of mRNA in Bacillus subtilis. Mol Cell. 2011;43:940–9. doi: 10.1016/j.molcel.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li de la Sierra-Gallay I, Zig L, Jamalli A, Putzer H. Structural insights into the dual activity of RNase J. Nat Struct Mol Biol. 2008;15:206–12. doi: 10.1038/nsmb.1376. [DOI] [PubMed] [Google Scholar]
- 20.Condon C. What is the role of RNase J in mRNA turnover? RNA Biol. 2010;7:316–21. doi: 10.4161/rna.7.3.11913. [DOI] [PubMed] [Google Scholar]
- 21.Durand S, Richard G, Bontems F, Uzan M. Bacteriophage T4 polynucleotide kinase triggers degradation of mRNAs. Proc Natl Acad Sci USA. 2012;109:7073–8. doi: 10.1073/pnas.1119802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joanny G, Le Derout J, Bréchemier-Baey D, Labas V, Vinh J, Régnier P, et al. Polyadenylation of a functional mRNA controls gene expression in Escherichia coli. Nucleic Acids Res. 2007;35:2494–502. doi: 10.1093/nar/gkm120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu F, Lin-Chao S, Cohen SN. The Escherichia coli pcnB gene promotes adenylylation of antisense RNAI of ColE1-type plasmids in vivo and degradation of RNAI decay intermediates. Proc Natl Acad Sci USA. 1993;90:6756–60. doi: 10.1073/pnas.90.14.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hajnsdorf E, Braun F, Haugel-Nielsen J, Régnier P. Polyadenylylation destabilizes the rpsO mRNA of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:3973–7. doi: 10.1073/pnas.92.9.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohanty BK, Kushner SR. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 2006;34:5695–704. doi: 10.1093/nar/gkl684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohanty BK, Kushner SR. Deregulation of poly(A) polymerase I in Escherichia coli inhibits protein synthesis and leads to cell death. Nucleic Acids Res. 2013;41:1757–66. doi: 10.1093/nar/gks1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campos-Guillén J, Bralley P, Jones GH, Bechhofer DH, Olmedo-Alvarez G. Addition of poly(A) and heteropolymeric 3′ ends in Bacillus subtilis wild-type and polynucleotide phosphorylase-deficient strains. J Bacteriol. 2005;187:4698–706. doi: 10.1128/JB.187.14.4698-4706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Górna MW, Carpousis AJ, Luisi BF. From conformational chaos to robust regulation: the structure and function of the multi-enzyme RNA degradosome. Q Rev Biophys. 2012;45:105–45. doi: 10.1017/S003358351100014X. [DOI] [PubMed] [Google Scholar]
- 29.Hardwick SW, Luisi BF. Rarely at rest: RNA helicases and their busy contributions to RNA degradation, regulation and quality control. RNA Biol. 2012;10 doi: 10.4161/rna.22270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coburn GA, Miao X, Briant DJ, Mackie GA. Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3′ exonuclease and a DEAD-box RNA helicase. Genes Dev. 1999;13:2594–603. doi: 10.1101/gad.13.19.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khemici V, Carpousis AJ. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol Microbiol. 2004;51:777–90. doi: 10.1046/j.1365-2958.2003.03862.x. [DOI] [PubMed] [Google Scholar]
- 32.Py B, Higgins CF, Krisch HM, Carpousis AJ. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–72. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 33.Chandran V, Poljak L, Vanzo NF, Leroy A, Miguel RN, Fernandez-Recio J, et al. Recognition and cooperation between the ATP-dependent RNA helicase RhlB and ribonuclease RNase E. J Mol Biol. 2007;367:113–32. doi: 10.1016/j.jmb.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–91. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomason MK, Fontaine F, De Lay N, Storz G. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol Microbiol. 2012;84:17–35. doi: 10.1111/j.1365-2958.2012.07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fröhlich KS, Vogel J. Activation of gene expression by small RNA. Curr Opin Microbiol. 2009;12:674–82. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 37.Beisel CL, Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol Rev. 2010;34:866–82. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–89. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moon K, Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol Microbiol. 2011;82:1545–62. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–83. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andrade JM, Pobre V, Matos AM, Arraiano CM. The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq. RNA. 2012;18:844–55. doi: 10.1261/rna.029413.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Lay N, Gottesman S. Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA. 2011;17:1172–89. doi: 10.1261/rna.2531211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marujo PE, Hajnsdorf E, Le Derout J, Andrade R, Arraiano CM, Régnier P. RNase II removes the oligo(A) tails that destabilize the rpsO mRNA of Escherichia coli. RNA. 2000;6:1185–93. doi: 10.1017/S135583820000073X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaballa A, Antelmann H, Aguilar C, Khakh SK, Song KB, Smaldone GT, et al. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc Natl Acad Sci USA. 2008;105:11927–32. doi: 10.1073/pnas.0711752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romilly C, Caldelari I, Parmentier D, Lioliou E, Romby P, Fechter P. Current knowledge on regulatory RNAs and their machineries in Staphylococcus aureus. RNA Biol. 2012;9:402–13. doi: 10.4161/rna.20103. [DOI] [PubMed] [Google Scholar]
- 46.Callaghan AJ, Marcaida MJ, Stead JA, McDowall KJ, Scott WG, Luisi BF. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature. 2005;437:1187–91. doi: 10.1038/nature04084. [DOI] [PubMed] [Google Scholar]
- 47.Garrey SM, Blech M, Riffell JL, Hankins JS, Stickney LM, Diver M, et al. Substrate binding and active site residues in RNases E and G: role of the 5′-sensor. J Biol Chem. 2009;284:31843–50. doi: 10.1074/jbc.M109.063263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bandyra KJ, Said N, Pfeiffer V, Górna MW, Vogel J, Luisi BF. The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol Cell. 2012;47:943–53. doi: 10.1016/j.molcel.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–19. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tuck AC, Tollervey D. RNA in pieces. Trends Genet. 2011;27:422–32. doi: 10.1016/j.tig.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 51.Baker KE, Mackie GA. Ectopic RNase E sites promote bypass of 5′-end-dependent mRNA decay in Escherichia coli. Mol Microbiol. 2003;47:75–88. doi: 10.1046/j.1365-2958.2003.03292.x. [DOI] [PubMed] [Google Scholar]
- 52.Dreyfus M. Killer and protective ribosomes. Prog Mol Biol Transl Sci. 2009;85:423–66. doi: 10.1016/S0079-6603(08)00811-8. [DOI] [PubMed] [Google Scholar]
- 53.Joyce SA, Dreyfus M. In the absence of translation, RNase E can bypass 5′ mRNA stabilizers in Escherichia coli. J Mol Biol. 1998;282:241–54. doi: 10.1006/jmbi.1998.2027. [DOI] [PubMed] [Google Scholar]
- 54.Leroy A, Vanzo NF, Sousa S, Dreyfus M, Carpousis AJ. Function in Escherichia coli of the non-catalytic part of RNase E: role in the degradation of ribosome-free mRNA. Mol Microbiol. 2002;45:1231–43. doi: 10.1046/j.1365-2958.2002.03104.x. [DOI] [PubMed] [Google Scholar]
- 55.Lopez PJ, Marchand I, Joyce SA, Dreyfus M. The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol Microbiol. 1999;33:188–99. doi: 10.1046/j.1365-2958.1999.01465.x. [DOI] [PubMed] [Google Scholar]
- 56.Anupama K, Leela JK, Gowrishankar J. Two pathways for RNase E action in Escherichia coli in vivo and bypass of its essentiality in mutants defective for Rho-dependent transcription termination. Mol Microbiol. 2011;82:1330–48. doi: 10.1111/j.1365-2958.2011.07895.x. [DOI] [PubMed] [Google Scholar]
- 57.Bouvier M, Carpousis AJ. A tale of two mRNA degradation pathways mediated by RNase E. Mol Microbiol. 2011;82:1305–10. doi: 10.1111/j.1365-2958.2011.07894.x. [DOI] [PubMed] [Google Scholar]
- 58.Garrey SM, Mackie GA. Roles of the 5′-phosphate sensor domain in RNase E. Mol Microbiol. 2011;80:1613–24. doi: 10.1111/j.1365-2958.2011.07670.x. [DOI] [PubMed] [Google Scholar]
- 59.Kushner SR. mRNA decay in Escherichia coli comes of age. J Bacteriol. 2002;184:4658–65, discussion 4657. doi: 10.1128/JB.184.17.4658-4665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mohanty BK, Kushner SR. Genomic analysis in Escherichia coli demonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Mol Microbiol. 2003;50:645–58. doi: 10.1046/j.1365-2958.2003.03724.x. [DOI] [PubMed] [Google Scholar]
- 61.Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol Microbiol. 2004;54:905–20. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- 62.O’Hara EB, Chekanova JA, Ingle CA, Kushner ZR, Peters E, Kushner SR. Polyadenylylation helps regulate mRNA decay in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:1807–11. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Z, Reimers S, Pandit S, Deutscher MP. RNA quality control: degradation of defective transfer RNA. EMBO J. 2002;21:1132–8. doi: 10.1093/emboj/21.5.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mohanty BK, Maples VF, Kushner SR. Polyadenylation helps regulate functional tRNA levels in Escherichia coli. Nucleic Acids Res. 2012;40:4589–603. doi: 10.1093/nar/gks006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andrade JM, Hajnsdorf E, Régnier P, Arraiano CM. The poly(A)-dependent degradation pathway of rpsO mRNA is primarily mediated by RNase R. RNA. 2009;15:316–26. doi: 10.1261/rna.1197309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng ZF, Deutscher MP. An important role for RNase R in mRNA decay. Mol Cell. 2005;17:313–8. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 67.Schneider C, Kudla G, Wlotzka W, Tuck A, Tollervey D. Transcriptome-wide analysis of exosome targets. Mol Cell. 2012;48:422–33. doi: 10.1016/j.molcel.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kolter R, Siegele DA, Tormo A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–74. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 69.Carabetta VJ, Mohanty BK, Kushner SR, Silhavy TJ. The response regulator SprE (RssB) modulates polyadenylation and mRNA stability in Escherichia coli. J Bacteriol. 2009;191:6812–21. doi: 10.1128/JB.00870-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liang W, Deutscher MP. Post-translational modification of RNase R is regulated by stress-dependent reduction in the acetylating enzyme Pka (YfiQ) RNA. 2012;18:37–41. doi: 10.1261/rna.030213.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liang W, Malhotra A, Deutscher MP. Acetylation regulates the stability of a bacterial protein: growth stage-dependent modification of RNase R. Mol Cell. 2011;44:160–6. doi: 10.1016/j.molcel.2011.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen C, Deutscher MP. Elevation of RNase R in response to multiple stress conditions. J Biol Chem. 2005;280:34393–6. doi: 10.1074/jbc.C500333200. [DOI] [PubMed] [Google Scholar]
- 73.Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–7. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nurmohamed S, Vincent HA, Titman CM, Chandran V, Pears MR, Du D, et al. Polynucleotide phosphorylase activity may be modulated by metabolites in Escherichia coli. J Biol Chem. 2011;286:14315–23. doi: 10.1074/jbc.M110.200741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tamura M, Lee K, Miller CA, Moore CJ, Shirako Y, Kobayashi M, et al. RNase E maintenance of proper FtsZ/FtsA ratio required for nonfilamentous growth of Escherichia coli cells but not for colony-forming ability. J Bacteriol. 2006;188:5145–52. doi: 10.1128/JB.00367-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cam K, Rome G, Krisch HM, Bouché JP. RNase E processing of essential cell division genes mRNA in Escherichia coli. Nucleic Acids Res. 1996;24:3065–70. doi: 10.1093/nar/24.15.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Faubladier M, Cam K, Bouché JP. Escherichia coli cell division inhibitor DicF-RNA of the dicB operon. Evidence for its generation in vivo by transcription termination and by RNase III and RNase E-dependent processing. J Mol Biol. 1990;212:461–71. doi: 10.1016/0022-2836(90)90325-G. [DOI] [PubMed] [Google Scholar]
- 78.Marchand I, Nicholson AW, Dreyfus M. Bacteriophage T7 protein kinase phosphorylates RNase E and stabilizes mRNAs synthesized by T7 RNA polymerase. Mol Microbiol. 2001;42:767–76. doi: 10.1046/j.1365-2958.2001.02668.x. [DOI] [PubMed] [Google Scholar]
- 79.Blower TR, Pei XY, Short FL, Fineran PC, Humphreys DP, Luisi BF, et al. A processed noncoding RNA regulates an altruistic bacterial antiviral system. Nat Struct Mol Biol. 2011;18:185–90. doi: 10.1038/nsmb.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Blower TR, Short FL, Rao F, Mizuguchi K, Pei XY, Fineran PC, et al. Identification and classification of bacterial Type III toxin-antitoxin systems encoded in chromosomal and plasmid genomes. Nucleic Acids Res. 2012;40:6158–73. doi: 10.1093/nar/gks231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci USA. 2009;106:894–9. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Short FL, Pei XY, Blower TR, Ong SL, Fineran PC, Luisi BF, et al. Selectivity and self-assembly in the control of a bacterial toxin by an antitoxic noncoding RNA pseudoknot. Proc Natl Acad Sci USA. 2013;110:E241–9. doi: 10.1073/pnas.1216039110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gerdes K, Maisonneuve E. Bacterial persistence and toxin-antitoxin loci. Annu Rev Microbiol. 2012;66:103–23. doi: 10.1146/annurev-micro-092611-150159. [DOI] [PubMed] [Google Scholar]
- 84.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haurwitz RE, Sternberg SH, Doudna JA. Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA. EMBO J. 2012;31:2824–32. doi: 10.1038/emboj.2012.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–8. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 87.Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell. 2012;45:303–13. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Deana A, Belasco JG. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 2005;19:2526–33. doi: 10.1101/gad.1348805. [DOI] [PubMed] [Google Scholar]
- 89.Hammarlöf DL, Hughes D. Mutants of the RNA-processing enzyme RNase E reverse the extreme slow-growth phenotype caused by a mutant translation factor EF-Tu. Mol Microbiol. 2008;70:1194–209. doi: 10.1111/j.1365-2958.2008.06472.x. [DOI] [PubMed] [Google Scholar]
- 90.Richards J, Luciano DJ, Belasco JG. Influence of translation on RppH-dependent mRNA degradation in Escherichia coli. Mol Microbiol. 2012;86:1063–72. doi: 10.1111/mmi.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bakshi S, Siryaporn A, Goulian M, Weisshaar JC. Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Mol Microbiol. 2012;85:21–38. doi: 10.1111/j.1365-2958.2012.08081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Khemici V, Poljak L, Luisi BF, Carpousis AJ. The RNase E of Escherichia coli is a membrane-binding protein. Mol Microbiol. 2008;70:799–813. doi: 10.1111/j.1365-2958.2008.06454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tompa P, Rose GD. The Levinthal paradox of the interactome. Protein Sci. 2011;20:2074–9. doi: 10.1002/pro.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sobetzko P, Travers A, Muskhelishvili G. Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc Natl Acad Sci USA. 2012;109:E42–50. doi: 10.1073/pnas.1108229109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bumgarner SL, Neuert G, Voight BF, Symbor-Nagrabska A, Grisafi P, van Oudenaarden A, et al. Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Mol Cell. 2012;45:470–82. doi: 10.1016/j.molcel.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tuck AC, Tollervey D. An RNA reset button. Mol Cell. 2012;45:435–6. doi: 10.1016/j.molcel.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 97.Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Jr., et al. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci U S A. 2009;106:19292–7. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sauer E, Weichenrieder O. Structural basis for RNA 3′-end recognition by Hfq. Proc Natl Acad Sci USA. 2011;108:13065–70. doi: 10.1073/pnas.1103420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vincent HA, Henderson CA, Ragan TJ, Garza-Garcia A, Cary PD, Gowers DM, et al. Characterization of Vibrio cholerae Hfq provides novel insights into the role of the Hfq C-terminal region. J Mol Biol. 2012;420:56–69. doi: 10.1016/j.jmb.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Beich-Frandsen M, Vecerek B, Konarev PV, Sjöblom B, Kloiber K, Hämmerle H, et al. Structural insights into the dynamics and function of the C-terminus of the E. coli RNA chaperone Hfq. Nucleic Acids Res. 2011;39:4900–15. doi: 10.1093/nar/gkq1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vecerek B, Rajkowitsch L, Sonnleitner E, Schroeder R, Bläsi U. The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res. 2008;36:133–43. doi: 10.1093/nar/gkm985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sauer E, Schmidt S, Weichenrieder O. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc Natl Acad Sci USA. 2012;109:9396–401. doi: 10.1073/pnas.1202521109. [DOI] [PMC free article] [PubMed] [Google Scholar]