Abstract
Epigenetic aberrations and a CpG island methylator phenotype are associated with poor outcome in children with neuroblastoma (NB). Previously, we demonstrated that valproic acid (VPA), a histone deacetylase (HDAC) inhibitor, has antitumor effects in an NB xenograft model. However, the underlying antitumor molecular mechanisms are largely unknown. In this study, we investigated the role of HDAC in cell proliferation, cell cycle progression, gene expression patterns, and epigenome in neuroblastoma. Cell proliferation, cell cycle progression, caspase activity, RNA and protein expression, quantitative methylation, and global DNA methylation were examined in NBL-W-N and LA1-55n NB cell lines. Our studies demonstrated that Inhibition of HDAC decreased NB proliferation and induced G1 growth arrest. Expression patterns of cancer-related genes were modulated by VPA. THBS1, CASP8, SPARC, CDKN1A, HIC1, CDKN1B, and HIN1 expression was upregulated, and MYCN and TIG1 were downregulated. HDAC inhibition decreased methylation levels of THBS1 and RASSF1A promoters. Inhibition of HDAC increased acetylation of histone 4 and global DNA methylation levels. Our studies demonstrated that inhibition of HDAC blocked cell proliferation and cell cycle progression in relation to alteration of cancer related genes, increasing global DNA methylation and decreasing methylation of tumor suppressor genes. Further studies investigating the anti-tumor effects of VPA in NB are warranted.
Keywords: Histone deacetylase, histone acetylation, cell proliferation, cell cycle, valproic acid, neuroblastoma, DNA methylation
Introduction
Neuroblastoma (NB) is the most common pediatric extracranial solid tumor. The cancer is characterized by its broad spectrum of clinical behavior [1]. Although some patients have high cure rates, approximately 45% of the patients present with widely disseminated, high-risk disease which remains difficult to cure [2]. NB originates from neural crest precursor cells as the result of genetic and epigenetic alterations that disrupt the normal developmental program. It has been reported that a CpG island methylator phenotype (CIMP) was a powerful prognostic factor, independent of age and stage in NB [3]. In preclinical studies, NB tumor growth is impaired with agents that inhibit DNA methyltransferase agents, demonstrating the important role the epigenome plays in NB tumor growth [4, 5].
Histone acetylation and DNA methylation are epigenetic modifications whose patterns can be regarded as heritable marks that ensure accurate transmission of the chromatin states and gene expression profiles over many cell generations [6, 7]. Importantly, patterns and levels of DNA methylation and histone acetylation are profoundly changed in human cancers. It has been shown that an epigenetic interplay between DNA methylation and histone acetylation may be involved in the alteration of gene transcription and aberrant gene silencing in a variety of cancers. We have previously showed that 5-aza-2′-deoxycytidine (5-aza-dC), an inhibitor of DNA methyltransferases (DNMTs), is able to increase thrombospondin-1 (THBS1) expression by alteration of histone codes in the promoter region, leading to an accessible chromatin structure [8].
Valproic acid (VPA), a histone deacetylase (HDAC) inhibitor, is a short-chained fatty acid with a broad spectrum of antiepileptic activities. It is also used in migraine prophylaxis, the treatment of bipolar disorders, and neuropathic pain [9, 10]. In addition, VPA has a wide range of therapeutic applications in many cancer types including breast cancer, glioma, acute myelogenous leukemia, thyroid cancer, endometrial carcinoma, and NB [11, 12, 13]. However, little is known regarding the molecular mechanism underlying its antitumor activity. In this study, we investigated the effects of VPA on NB cell proliferation, expression levels of 9 cancer-related genes, promoter methylation of tumor suppressor genes THBS1 and RASSF1A, and global DNA methylation.
Methods
NB cell culture
Two MYCN amplified NB cell lines, LA1-55n and NBL-W-N, were used as previously described [8, 14]. LA1-55n and NBL-W-N cells were grown at 5% CO2 in RPMI 1640 (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen), L-Glutamine, and antibiotics.
VPA treatment
VPA and trichostatin A (TSA) were purchased from Sigma, St. Louis, MO. Cells were treated with the HDAC inhibitor VPA at concentrations of 0.25, 0.5, 1.0, 2.5, and 5.0 mM for 1, 2, and 3 days for cell proliferation assays. For flow cytometry and gene expression experiments, cells were treated with 2.5 mM and 1 mM VPA for 2 days and 7 days respectively. Cells were treated for 2 days with 2.5 mM VPA and 7 days with 1.0 mM VPA or 10 nM TSA for Methylation Sensitive High Resolution Melting analysis and liquid chromatography-mass spectroscopy was performed in NB cells treated with 1 mM VPA or 10 nM TSA for 7 days respectively.
Cell proliferation assay
Cell proliferation was performed by MTS colorimetric assay and BrdU incorporation assay. For MTS colorimetric assays, LA1-55n and NBL-W-N cells were seeded into 96-well plates at a density of 5 × 103 cells/well. After 24 h, VPA was added at various concentrations to each quadruplicate well. After 1, 2, or 3 days of treatment, MTS labeling mixture (Promega, Madison, WI) was added, and cells were further incubated for 3 h. The absorbance of the samples was measured using a Synergy 2 Microplate Reader (BioTek Instruments, Winooski, VT). For BrdU incorporation assay, BrdU label solution (Millipore, Billerica, MA) was added to each well 15–18 h prior to analysis. Denaturing solution was added to each well for 30 min at room temperature after removing the contents of wells. Then, anti-BrdU antibody was added to each well and incubated for 1 h and peroxidase goat anti-mouse IgG HRP conjugate was added to the well for 30 min at room temperature. The absorbance was read at 450–540 nm on a Glomax Multiple Detection System (Promega).
Flow cytometry for analysis of cell cycle
NBL-W-N and LA1-55n cells were harvested at the completion of the respective VPA treatments and washed with phosphate buffered saline (PBS, pH 7.4) twice and then fixed with 70% ethyl alcohol for 15 min on ice. Subsequently, the cells were centrifuged at a 2000 rpm to obtain pellets and residual alcohol was aspirated. Cells were then digested with DNase-free RNase A (2 mg/ml) for 30 min at 37°C. Before flow cytometric analysis, cells were resuspended in 1 ml of 10 mg/ml propidium iodide (PI) (Sigma) for staining cellular DNA, as previously described [15]. Cellular DNA content was then analyzed using an Epics XL-MCL Flow Cytometer (Beckman Coulter, Fullerton, CA).
Caspase assays
Caspase 3/7 activities in LA1-55n and NBL-W-N cells were measured using Caspase-Glo assay kit (Promega). Briefly, the cells were plated in white-walled 96-well plate (Corning Incorporated, Lowell, MA), and incubated for 24 h. The VPA was added into the medium, and incubated for 24h. The proluminescent substrate containing DEVD, which is cleaved by caspase 3 or 7, was added to each well, and incubated at room temperature for 1 h. The luminescence of each sample was measured in a luminometer as directed by the manufacturer (Promega).
cDNA synthesis and SYBR green real-time PCR
RNA was isolated from untreated and VPA-treated LA1-55n and NBL-W-N cells using Trizol reagent (Invitrogen). Reverse transcription was performed using SuperScript III (Invitrogen) and 50 µM oligo(dT)20 at 50°C for 50 min. SYBR green real-time PCR reactions were set up containing 1X Power SYBR Green Master Mix (Applied Biosystems, Foster City, CA), 250 nM forward and reverse primers in a 20 µl reaction. All assays were carried out in a 96-well format. Real-time fluorescent detection of PCR products was performed with an 7500 Fast Real-Time PCR System (Applied Biosystems) using the following thermocycling conditions: 1 cycle of 50°C for 2 min and 95°C for 20 s; 40 cycles of 95°C for 30 s, and 60°C for 1 min. The primers were used and reactions were performed as previously described [8]. For data analysis, the comparative method (∇∇Ct) was used to calculate relative quantities of a nucleic acid sequence. Non-treated LA1-55n and NBL-W-N cells were used as the calibrator sample, and GAPDH was used as an endogenous control.
Western blot analysis
Total protein from NBL-W-N and LA1-55n NB cells were extracted after lysing the cells in RIPA buffer (20mM Tris-HCl–pH 7.5, 150mM NaCl, 1mM EDTA, 1mM EGTA, 1% IGEPAL, 2.5mM sodium pyrophosphate, 1mM β-glycerophosphate) containing protease and phosphatase inhibitor cocktails (Sigma), and protein concentration was determined using a Bradford protein assay kit (Bio-Rad, Hercules, CA). Equal amounts of total protein (25 µg) from cells were subjected to SDS-PAGE and proteins were transferred to nitrocellulose membrane for 90 min at 100 V. Membranes were blocked for 1 h at room temperature in Tris-buffered saline (TBS) containing 5% nonfat powdered milk and probed with primary antibody (anti-AcH4, Abcam, Cambridge, MA) in TBS with 2.5% nonfat powdered milk with 1:1000 dilution and pre-incubated overnight. A secondary antibody labeled with horseradish peroxidase (GE Healthcare, Piscataway, NJ) was used at 1:10000 dilution for 1 h at room temperature, and immuno-reactive bands were detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL) and recorded on photosensitive film. Tubulin (Sigma) was used as a loading control.
DNA isolation and bisulfite modification
Total genomic DNA was extracted from the NB cell lines using the Puregene Core Kit A (Qiagen, Valencia, CA) and modified by sodium bisulfite using the EZ DNA Methylation-Direct kit (Zymo Research, Irvine, CA) according to manufacturer’s protocol.
DNA methylation analysis
Methylation Sensitive High Resolution Melting (MS-HRM) technology [16] was used to determine the methylation degree along the promoter regions of THBS-1 and RASSF1A. PCR amplification and high resolution melting analysis (HRM) were performed on a 7500 Fast Real-Time PCR System. The 97 bp amplicon of THBS1 with 10 CpG dinucleotides was amplified with sense primer 5’-GGTCGGAGGAATTTTTAGGAATG-3’, and antisense primer 5’-CCTAAACTCGCAAACCAACT-3’. MS-HRM for RASSF1A was performed as previously described [17]. PCR Reactions were performed in triplicate in a 20 µl volume containing 1× buffer, 2U Hotstart Taq DNA polymerase (Qiagen), 250 nM of each primer, 1.5 µM SYTO-9, and 10 ng bisulfite-treated DNA template, with 2 mM final MgCl2. The cycling conditions were as follows: 1 cycle at 95°C for 10 minutes, 40 cycles at 95°C for 15 seconds, 60°C for 1 minute; followed by an HRM step of 95°C for 10 seconds, 60°C for 1 minute, and continuous acquisition to 95°C at 1 acquisition per 0.3°C, and 60°C for 15 seconds. A standard curve with known methylation ratios was included in each assay and was used to deduce the methylation ratio of treated and untreated cells. Universal methylated and unmethylated DNAs were purchased from Millipore. HRM data were analyzed using the High Resolution Melting Software (Applied Biosystems).
Liquid chromatography-mass spectroscopy (LC/MS)
Analysis of total cytosine methylation was performed by LC/MS as described previously [18]. Briefly, genomic DNA from LA1-55n and NBL-W-N was hydrolyzed to nucleosides by adding 5U nuclease P1 (Sigma) at 37°C for 2 hrs, 0.002 units of venom phosphodiesterase I (Sigma) at 37°C for 2 hrs, 0.5 units of alkaline phosphatase at 37°C for 1 h. Stock solutions of 2’-deoxycytidine and 5-methyl-2’-deoxycytidine were prepared in water. An eight-point stock mixture of a standard was carefully prepared to give an exact known concentration ratio of 2’-deoxycytidine to 5-methyl-2’-deoxycytidine. The concentration of 2’-deoxycytidine and 5-methyl-2’-deoxycytidine in each sample was calculated from the standard curve. Each DNA sample was analyzed in triplicate. 25 µl (80ng) of sample was injected into the LC and run through an Atlantis DC18 silica column (Waters Corporation, Milford, MA), and identification of 2’-deoxycytidine and 5-methyl-2’-deoxycytidine was obtained by mass spectra of chromatographic peaks.
Statistical analyses
Statistical analyses were performed using a two-tailed Student's t test. A p value of < 0.05 was considered statistically significant.
Results
Inhibition of histone deacetylase significantly decreased NB cell proliferation in vitro
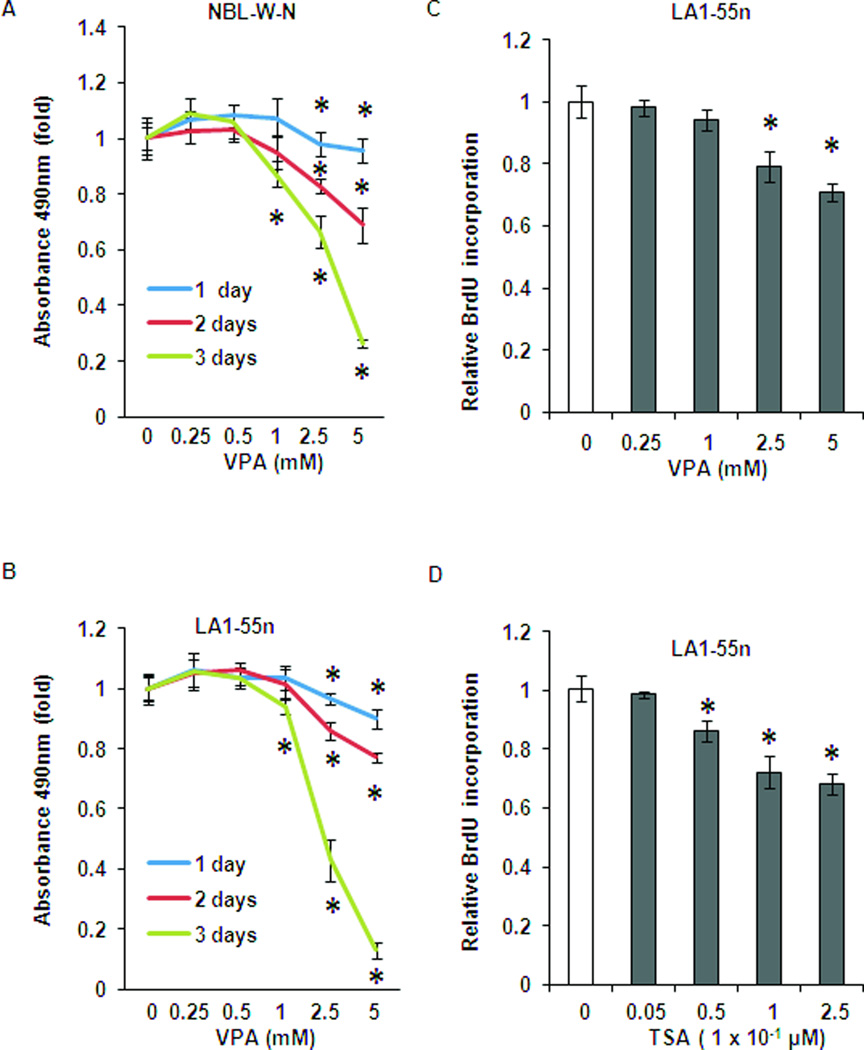
MTS colorimetric assays were performed to determine the effect of VPA on NB cell growth. As shown in Figure 1A, VPA inhibited cell proliferation of NBL-W-N in a dose- and time-dependent manner. After 1 day of treatment with VPA, cell proliferation was decreased with concentrations of 2.5 mM and 5 mM (p<0.01). There was no inhibitory effect of cell growth at a concentration of 1 mM or below. Cell growth was further inhibited with 2 days of treatment at concentrations of 2.5 mM and 5 mM. With 3 days of treatment with VPA, cell growth was further inhibited at a concentration of 1 mM (p<0.01). A similar inhibitory effect by VPA was also seen in LA1-55n NB cells as shown in Figure 1B. After 3 days of treatment with VPA, cell proliferation of LA1-55n cells was decreased by ~ 90% compared with ~75% decrease in NBL-W-N cells.
Figure 1.
Effect of HDAC inhibition on NB growth and cell cycle in vitro. (A) In vitro cell proliferation was examined by MTS colorimetric assays. NBL-W-N cells were grown for 1–3 days in the presence or absence of VPA with varying concentrations as indicated. (B) In vitro cell proliferation was examined by MTS colorimetric assays. LA1-55n cells were grown for 1–3 days in the presence or absence of VPA with varying concentrations as indicated. (C) BrdU incorporation assay was performed in LA1-55n cells treated with various concentrations of VPA for 1 day. (D) BrdU incorporation assay was performed in LA1-55n cells treated with various concentrations of TSA for 1 day. * p<0.05 compared to control
To further determine the effect of VPA on cell proliferation, BrdU incorporation assay was performed in LA1-55n NB cells in addition to the metabolic activity assays above. As shown in Figure 1 C, VPA markedly decreased DNA synthesis in a dose-dependent manner in LA1-55n cells. TSA is a well-known potent inhibitor of histone deacetylase. To determine if TSA has a similar inhibitory effect of cell proliferation in NB, LA1-55n cells were treated with varied concentrations of TSA and subjected to BrdU incorporation assay. As shown in Figure 1D, TSA exhibited an anti-proliferation effect in a dose-dependent manner.
Inhibition of histone deacetylase blocked cell cycle progression
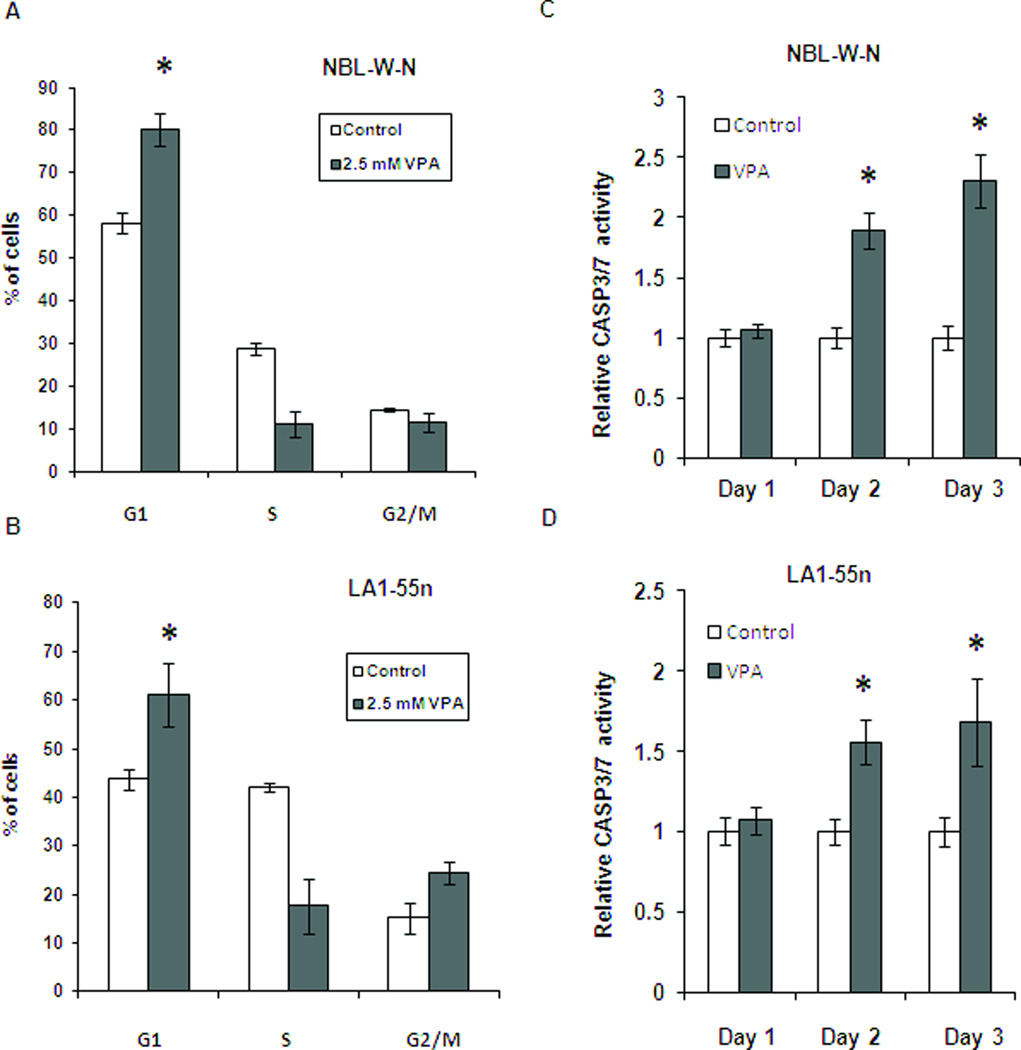
Flow cytometry analysis was performed to investigate if VPA induced cell cycle arrest. As shown in Figures 2A and 2B, cell cycle arrest at G1 was seen following treatment of NBL-W-N and LA1-55n cells with VPA at a concentration of 2.5 mM for 2 days (p<0.01).
Figure 2.
Effect of HDAC inhibition on cell cycle and apoptosis. (A) NBL-W-N cell line was treated with either 2.5 mM of VPA or vehicle control for 2 days and analyzed by flow cytometry. Each column represents the mean of three independent experiments. (B) LA1-55n cell line was treated with either 2.5 mM of VPA or vehicle control for 2 days and analyzed by flow cytometry. (C) NBL-W-N cells were treated with 2.5 mM VPA for 1 day and subjected to caspase 3/7 activity assay. (D) LA1-55n cells were treated with 2.5 mM VPA for 1 day and caspase 3/7 activity between control and VPA-treated samples were measured. * p<0.05 compared to control
It has been reported that concentrations of VPA above 2 mM are cytotoxic to NB cell lines[13]. To determine if inhibition of histone deacetylase attenuated cell proliferation through the apoptotic pathway, caspase 3/7 activity was measured. As shown in Figure 2C, 2D a trend toward increase of caspase 3/7 activity was seen in both NBL-W-N and LA1-55n cells after treatment with VPA at 2.5mM for 1 day, However, no significant difference of caspase 3 activity between VPA and vehicle treated NB cells was observed.
Inhibition of histone deacetylase modulated expression pattern of cancer related genes
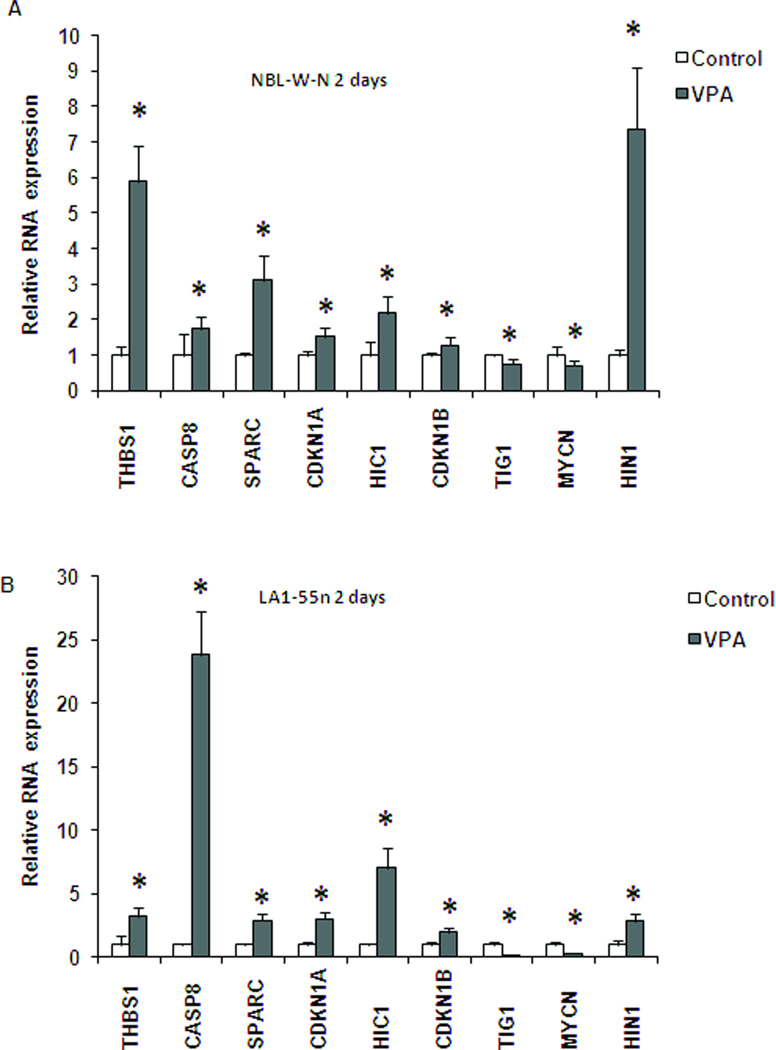
Expression studies revealed changes in the levels of nine cancer-related genes following treatment with VPA. As shown in Figure 3A, the level of expression of seven genes (THBS1, CASP8, SPARC, CDKN1A, HIC1, CDKN1B, and HIN1) was significantly upregulated in the VPA-treated NBL-W-N cells. HIN-1 was especially sensitive in NBL-W-N cells and the level of transcript expression was increased by 7 fold following 2 days of treatment. In contrast, TIG1 and MYCN were downregulated in NBL-W-N cells treated with VPA (Figure 3A, p<0.01).
Figure 3.
Relative RNA expression of control and VPA-treated NB cell lines. Each cell line was treated with either 2.5 mM of VPA or vehicle control for 2 days. Cells were then harvested and RNA isolated from each group. SYBR green real-time PCR reactions were performed to study alterations of gene expression in cells treated with VPA. Each column represents the mean of two independent experiments. (A) NBL-W-N; (B) LA1-55n. .* p<0.05 compared to control
A similar expression pattern was seen in LA1-55n cells treated with 2.5 mM VPA for 2 days. The seven genes whose expression was upregulated in NBL-W-N cells treated with VPA were shown to have increased gene expression in LA1-55n cells as well (Figure 3B). The expression of CASP8 was increased by over 20 folds. In addition, similar downregulation of MYCN and TIG1 was seen in LA1-55n cells. We also examined the expression of these 9 genes in both cell lines treated with VPA for 1 day and 3 days respectively. A similar expression pattern in both cell lines was seen as with 2 days of treatment (data not shown).
Inhibition of histone deacetylase increased acetylation of histone H4
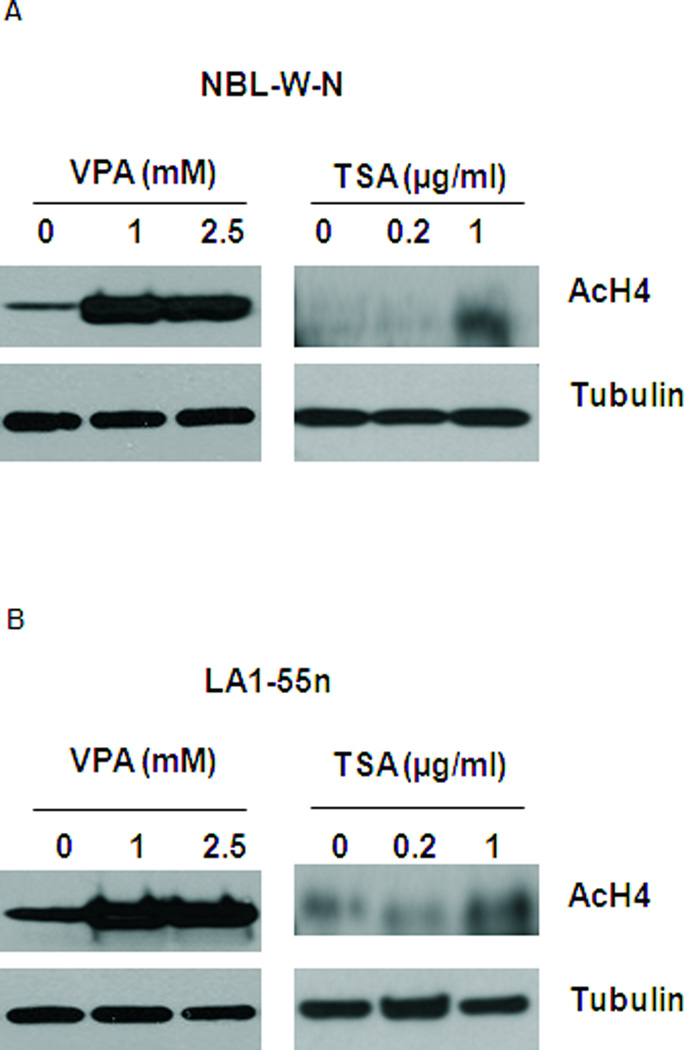
To determine if VPA mediated inhibitory effect of cell proliferation in NB was through histone modification, acetylation level of histone H4 was examined by western blot analysis using antibody against acetyl histone H4. As shown in Figure 4A, the level of acetylation of histone 4 was markedly increased in NBL-W-N cells after treatment with VPA at concentrations of 1 mM and 2.5 mM for 1 day. As a positive control, TSA at concentration of 1 µg/ml upregulated the acetylation level of histone H4. Similar results were seen in LA1-55n NB cells after treatment with VPA and TSA respectively as shown in Figure 4B.
Figure 4.
Effect of HDAC inhibition on the level of acetyl histone 4. (A) Total lysates from vehicle- or VPA-treated NBL-W-N cells were subjected to western blot analysis using antibody against acetyl histone 4. NBL-W-N cells were treated with 1 and 2.5 mM VPA for 1 day. Tubulin was used as a loading control. (B) Total lysates from vehicle- or VPA treated LA1-55n cells were subjected to western blot analysis using antibody against acetyl histone 4. LA1-55n cells were treated with 1 and 2.5 mM VPA for 1 day. Tubulin was used as a loading control. TSA treated samples were used as a positive control of acetyl histone 4. * p<0.05 compared to control
Time course gene specific methylation modulated by Inhibition of histone deacetylase
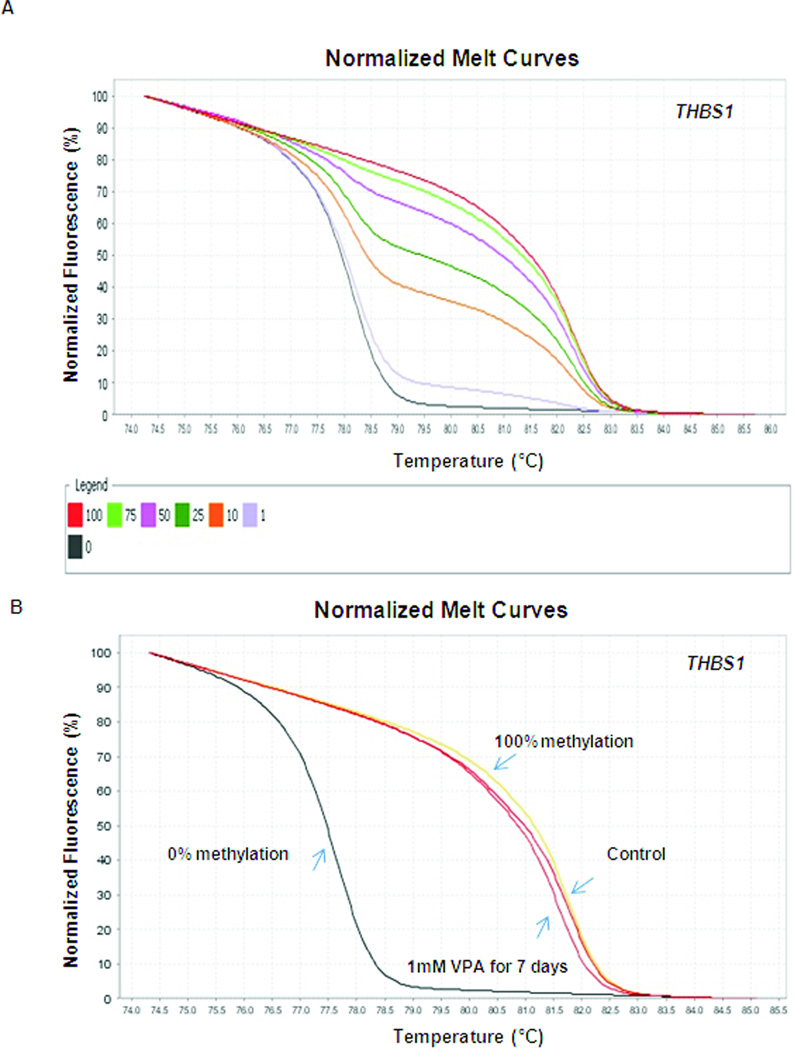
To determine if inhibition of histone deacetylase mediates gene-specific methylation changes, MS-HRM analysis was performed, and the levels of THBS1 and RASSF1A promoter methylation were analyzed in LA1-55n cells treated with VPA for 2 days and 7 days respectively. THBS1 was selected, since we previously demonstrated that its expression is regulated by both DNA methylation and histone modification of the promoter region [5, 8]. We also selected RASSF1A as a target, since we have previously demonstrated that methylation of RASSF1A was associated with poor outcome in NB [19]. As shown in Figure 5A, the sensitivity of the THBS1 HRM analysis was examined by using dilutions of fully methylated DNA with unmethylated DNA and subsequent amplification with THBS1 primers. The HRM standard melting curve was derived from seven samples with the content of methylated DNA (0%, 1%, 10%, 25%, 50% 75%, and 100%). At the annealing temperature of 60°C, such methylation levels can be easily distinguished, and as low as 1% of methylation can be detected. As shown in Figure 5B, we detected a distinct shift in melting profile, which demonstrates that VPA treatment for 7 days resulted in a reduced methylation level of the THBS1 promoter in LA1-55n NB cells.
Figure 5.
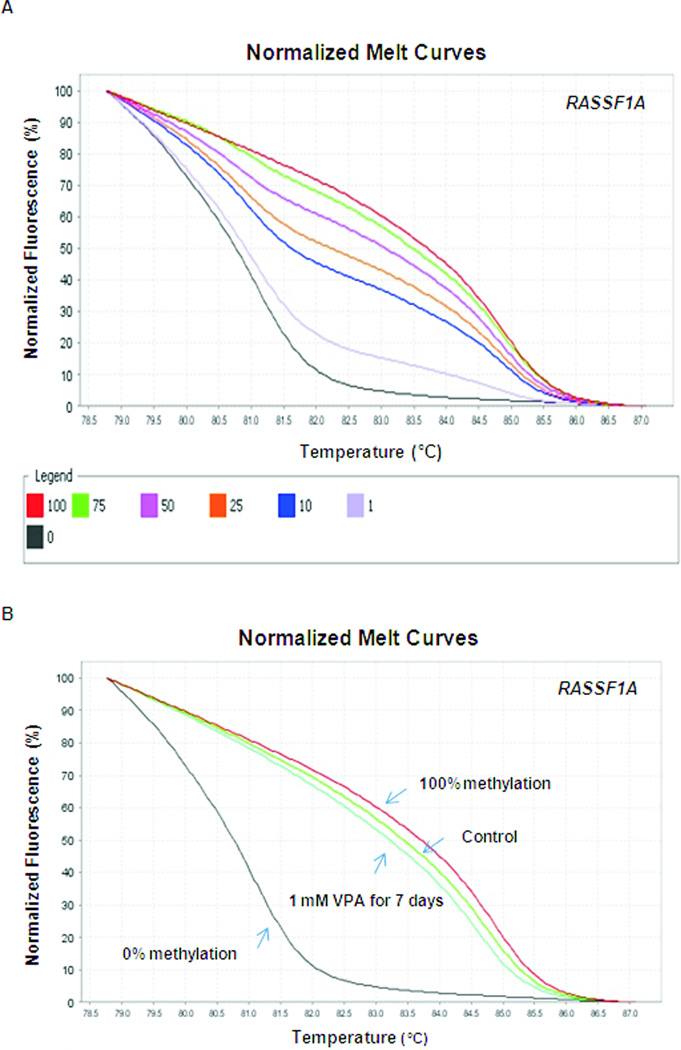
Quantitative RASSF1A methylation detection by high-resolution melt curve analysis. (A) Normalized HRM standard curve of RASSF1A was generated. (B) HRM analysis of DNA methylation status of RASSF1A was performed in LA1-55n cells treated with vehicle and 1 mM VPA for 7 days.
The sensitivity of the RASSF1A HRM analysis was also examined using RASSF1A primers. As shown in Figure 6A, the methylation levels of RASSF1A can also be easily distinguished and 1% of methylation can be detected. VPA treatment for 7 days resulted in reduced methylation level of RASSF1A promoter in LA1-55N NB cells as evidenced by the left shift in melting curve (Figure 6B).
Figure 6.
Quantitative THBS1 methylation detection by high-resolution melt curve analysis. (A) Normalized HRM standard curve of THBS1 was generated. (B) HRM analysis of DNA methylation status of THBS1 was performed in LA1-55n cells treated with vehicle and 1 mM VPA for 7 days. .
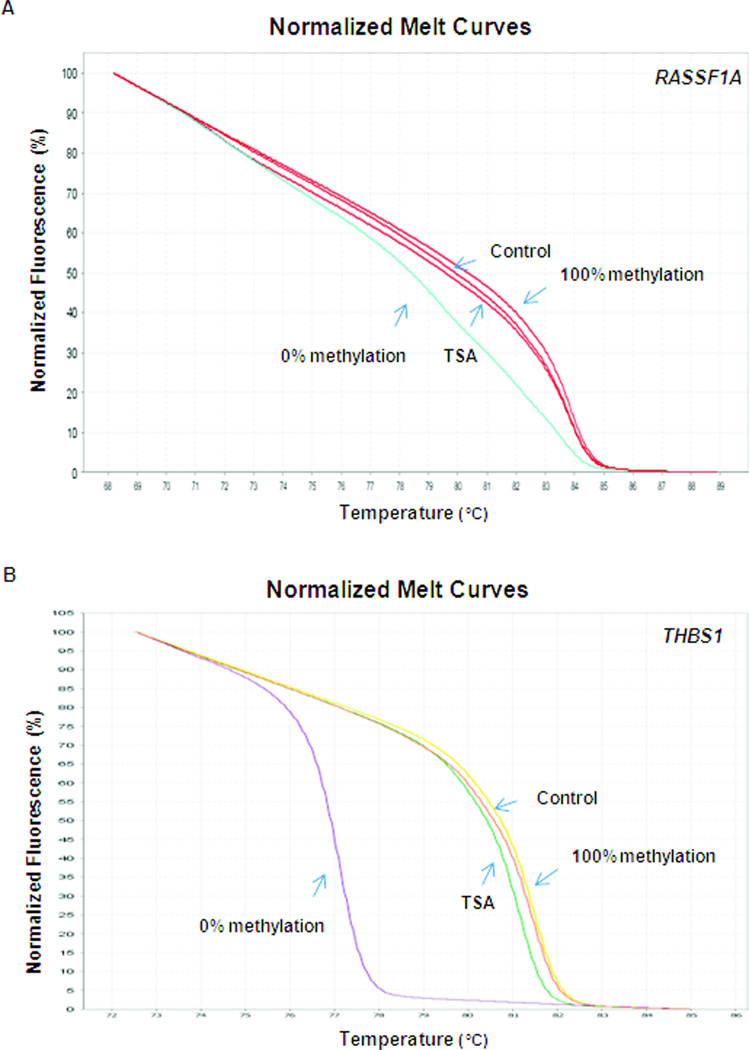
To determine if VPA altered the DNA methylation level around THBS1 and RASSF1A promoter regions within a relative short period time of treatment, HRM analysis was performed in LA1-55n cells after treatment with VPA for 2 days. There is no difference of methylation level around THBS1 and RASSF1A promoter regions (data not shown).
Long-term effect of VPA treatment on gene expression
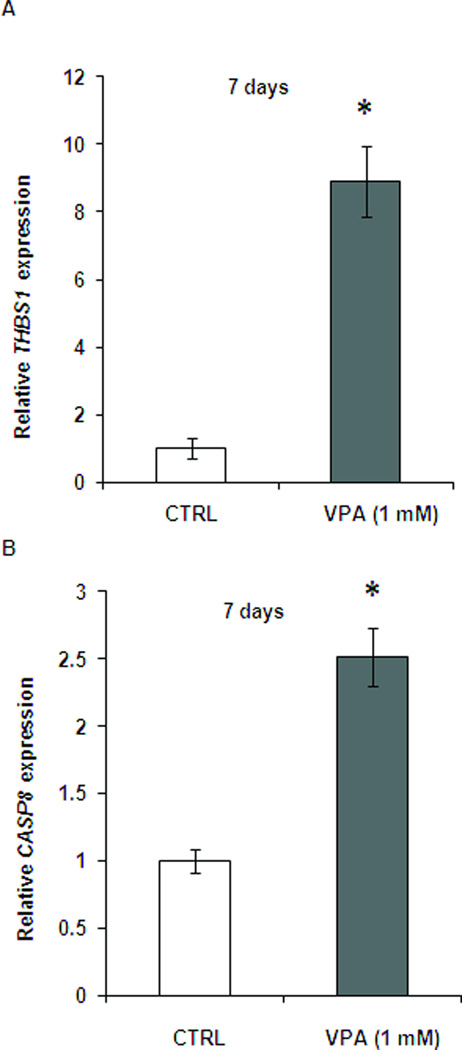
To determine if relative short (2 days) and long (7days) treatments of VPA exhibited distinct effects on gene expression, we measured the gene expression level of THBS1 and CASP8 in LA1-55n cells treated with 1 mM VPA for 7 days. As shown in Figure 7A, the expression of THBS1 was further increased compared with expression in LA1-55n cells treated with VPA for 2 days (Figure 3). The expression of CASP8 in LA1-55n cells treated with VPA for 7 days was also higher compare to control. However the increase of CASP8 in 7 days treatment is not as high as 2 days treatment (Figure 7B).
Figure 7.
Relative RNA expression of vehicle- and VPA-treated LA1-55n cells. (A) The RNA expression of THBS1 was examined in LA1-55n cells treated with vehicle or 1 mM VPA for 7 days. After total RNA isolation and cDNA synthesis, THBS1 expression was determined by SYBR green real-time PCR. (B) The RNA expression of Casp8 was examined by SYBR green real-time PCR in LA1-55n cells treated with vehicle or 1 mM VPA for 7 days. * p<0.05 compared to control
Inhibition of histone deacetylase altered global DNA methylation
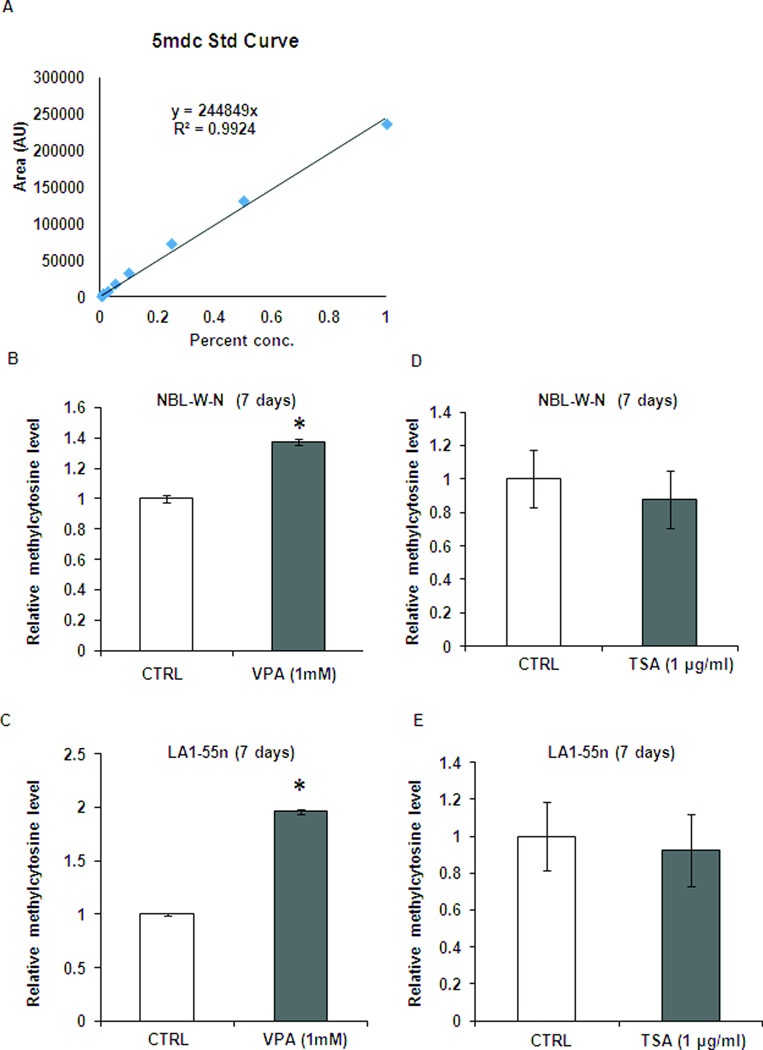
To investigate if inhibition of histone deacetylase affects genome-wide epigenetic changes, we compared the global DNA methylation levels in the NBL-W-N and LA1-55n cells with and without VPA treatment. Using a standard curve average of 5-methylcytosine (Figure 8A), the level of 5-methylcytosine was increased significantly (p<0.01) by 1.4 and 2.0 folds in VPA-treated NBL-W-N and LA1-55n cells compared with controls (Figure 8B, 8C), suggesting that inhibition of histone deacetylase by VPA is able to modulate the epigenome.
Figure 8.
Global methylation level was altered in NB cell lines after treatment with VPA. (A) Stock solutions of 2’-deoxycytidine and 5-methyl-2’-deoxycytidine were prepared in water. An eight-point stock mixture of a standard was carefully prepared to give an exact known concentration ratio of 2’-deoxycytidine and 5-methyl-2’-deoxycytidine. (B) DNA methylation was determined by LC/MS. Genomic DNA was isolated from control and VPA-treated NBL-W-N cells. DNA was then subjected to digestion with nuclease P1, venom phosphodiesterase I, and alkaline phosphatase respectively. The concentration of 2’-deoxycytidine and 5-methyl-2’-deoxycytidine in each sample was calculated from the standard curve. Each DNA sample was analyzed in triplicate. (C) Total DNA methylation was determined by LC/MS in control and VPA-treated LA1-55n cells. * p<0.05 compared to control
Discussion
Epigenetic changes play an important role in the pathogenesis of cancer, and a CpG island methylator phenotype has been shown to be predictive of poor outcome [3, 20, 21]. The effect of HDAC inhibition on NB cells has been intensively investigated by several groups [12, 22–24]. In this study, we examined the role of histone deacetylase in cell proliferation, cell cycle progression, gene expression, gene specific methylation and global methylation status in two MYCN amplified tumorigenic NB cell lines (LA1-55n and NBL-W-N). We show that inhibition of histone deacetylase by VPA significantly decreased NB cell proliferation in vitro in a dose- and time-dependent manner, and induced G1 growth arrest in LA1-55n and NBL-W-N NB cell lines. Consistent with the mechanism of VPA action, these changes were associated with an increase in global methylation and corresponding decrease in the expression level of oncogene MYCN. In contrast, the expression levels of genes which suppress tumor progression were increased. Gene-specific analysis demonstrated that despite the increase in global methylation, promoters of tumor suppressor genes THBS1 and RASSF1A were hypomethylated after VPA treatment, indicating a redistribution of the methylation pattern which is mediated by VPA.
Epigenetics has an established role in promoting pathological cell proliferation in cancers due to decreased expression of tumor suppressor genes. Our present study indicates that VPA is able to rescue the expression of tumor suppressor genes, and tumorigenic expression profiles in NB can be modulated to a benign expression pattern by inhibition of HDAC. For instance, the expression of both CDKN1A and CDKN1B were upregulated in VPA-treated NB cells. CDKN1A and CDKN1B genes encode potent cyclin-dependent kinase inhibitors, which are silenced in several malignancies [25, 26]. The encoded proteins for CDKN1A and CDKN1B have been reported to function as regulators of cell cycle progression at G1 [27]. The mechanism of both CDKN1A and CDKN1B regulation and its consequences parallel those recently discovered in human cancers [27, 28]. The inverse correlation between expression of CDKN1A and CDKN1B and cell proliferation in NB cells treated with VPA was seen in our study. In our previous preclinical model, tumor cell proliferation was inhibited by VPA suggested that VPA may target NB tumor cells [12]. Our present study suggested that modulation of the epigenome is a critical link between expression of cell cycle related genes and regulation of cell proliferation.
In the pediatric cancer NB, clinically aggressive disease is associated with increased levels of angiogenesis stimulators and high vascular index. THBS1 and SPARC encode proteins which function as anti-angiogenesis factors in NB. THBS1 is a well-known natural inhibitor of angiogenesis. Regulation of THBS1 plays an important role in the angiogenic switch in many tumor types [29, 30] and we have shown that ABT510, a THBS1 peptide, inhibited the growth of NB in preclinical models [12]. Further, THBS1 expression is upregulated, angiogenesis is inhibited, and NB tumor growth is impaired following treatment with the demethylating agent, 5-Aza-dC [5]. Here we show that VPA also modifies the pattern of DNA methylation in the THBS1 promoter and induces expression of the gene. Previously we have shown that SPARC is a key anti-angiogenesis factor in NB [31], and modulation of SPARC expression is mediated by the differentiation agent retinoic acid as well as 5-aza-dC, an inhibitor of DNMT [8,31]. In this study, VPA induced THBS1 and SPARC expression in NB cells indicating that histone modification is involved in the regulation of THBS1 and SPARC. The present study supports a key role for THBS1 and SPARC as the link between the angiogenic and epigenetic switches.
CASP8 encodes a member of the cysteine-aspartic acid protease (caspase) family. Sequential activation of caspases plays a central role in the execution-phase of cell apoptosis. The methylation of CASP8 was initially identified in NB patients with amplification of MYCN [32]. Because of the remarkable up-regulation of CASP8 expression that was seen in the LA1-55n cells following treatment with VPA, it is quite possible that VPA mediates decreased cell growth not only due to cell cycle arrest but also cell apoptosis. Our data indicated that although a significant difference in caspase3 activity between vehicle- and VPA-treated cells was not reached, the trend towards an increase of caspase activity was seen in VPA-treated LA1-55n cells. It was recently reported that VPA induced apoptosis and affected acetylation status of p53 in prostate cancer cells [30]. Trichostatin A, another potent inhibitor of HDAC, showed anti-proliferation effects through induction of cell apoptosis in several cancer cell types [33, 34].
HIC1 is a molecule controlling cell growth [35]. We have previously shown that expression of HIC1 is upregulated in NB cells treated with 5-aza-dC. In this study, expression of HIC1 was increased following treatment with VPA suggesting that histone modification/acetylation level is involved in the regulation of HIC1 expression. HIN1 is a putative cytokine with growth inhibitory activities [36]. HIN1 was initially found to be significantly down regulated in human breast carcinomas and in preinvasive lesions. Although the evidence linking HIN1 levels to NB phenotype is unknown, our previous study indicated that methylation of HIN1 is associated with poor outcome in NB [37]. In this study we demonstrated that the expression of HIN1 was upregulated by VPA and is associated with modulation of NB phenotype.
MYCN and TIG1 are the two genes whose expression is down regulated by VPA. MYCN is considered to be a potential specific target for therapy of NB. Our study demonstrated that VPA resulted in a decrease in the expression of MYCN which correlated with inhibited cell proliferation and cell cycle arrest. The effect of down regulation of MYCN by VPA in NBL-W-N and LA1-55n NB cells is consistent with previous observations [12, 13] suggesting that a decrease in MYCN expression by VPA in NB cell lines is a common phenomena. Our data is also in good agreement with previous studies demonstrating that reduction of MYCN mRNA by the use of antisense MYCN can inhibit proliferation and/or induce differentiation in NB cell lines [38, 39]. Although there are studies showing MYCN mediated signal pathways, leading to alteration of NB phenotype, the molecular mechanism underlying regulation of MYCN in NB is largely unknown. There are some other studies showing that expression of MYCN can be regulated by differentiation agents. Treating MYCN amplified NB cells with retinoic acid has been shown to cause NB cells to undergo G1 arrest and differentiation along with down regulation of MYCN [40]. Importantly, over-expression of MYCN counteracts retinoic acid induced G1 arrest and differentiation [41]. TIG1 expression was also down regulated by VPA in this study, and not paralleled with a change of TIG1 by 5-aza-dC. an inhibitor of DNMTs, as we previously described [8]. It is possible that besides a cell specific effect, expression of TIG1 is regulated due to its promoter hypermethylation.
Cancer cells are characterized by a generalized disruption of the DNA methylation pattern involving an overall decrease in the level of 5-methylcytosine along with regional hypermethylation of particular CpG islands. During the development of neoplasms, the degree of hypomethylation of genomic DNA increases as the lesion progresses from a benign proliferation of cells to an invasive cancer [42, 43]. The decrease of DNA methylation is mainly due to the hypomethylation of repetitive DNA sequences and demethylation of some coding regions and introns. The distribution of DNA is altered in a variety of normal cellular processes, including generation of chromosomal instability, reactivation of transposable elements, and loss of imprinting, leading to an increased risk of cancer [44]. Since VPA is a HDAC inhibitor, one may expect that VPA initially targets histone modification and later modulates the DNA methylation. We determined that the acetylation level of histone 4 was modulated by VPA. After 1 day treatment with VPA, we were able to see an increased level of acetylation of histone 4. However VPA-mediated decrease of promoter hypermethylation of THBS1 and RASSF1A was not observed within 2 days treatment with VPA, but the alteration of promoter hypermethylation of THBS1 and RASSF1A was seen after 7 days of VPA treatment, indicating that reprogramming of methylation in certain promoter regions is time-course dependent upon VPA treatment. More importantly, an inverse relationship between THSB1 expression and the extent of DNA methylation was seen in the LA1-55n cells treated with VPA for 2 days and 7 days respectively. A VPA mediated-demethylation study was previously reported [45] which showed that RELN and GAD (67) promoter demethylation was dramatically accelerated after administration of VPA to mice for 48–72 h. VPA effectively increased the binding of acetyl histone-3 to RELN and GAD (67) promoters, suggesting that histone-3 covalent modifications modulate DNA demethylation. Our study supports the view that HDAC inhibitors facilitate DNA demethylation. In addition, quantitative analysis of global 5-methylcytosine levels by LC/MS indicated that inhibition of HDAC resulted in rescuing global DNA methylation levels in both NB cell lines. According to our gene specific and global methylation analysis, inhibition of HDAC is capable of altering the distribution of DNA methylation, which is associated with changes of NB phenotype. Our studies indicated that VPA, as an HDAC inhibitor, is not only able to modulate histone modification but also alter genome wide DNA methylation. According to our knowledge, this is the first report to show that VPA mediates changes in global DNA methylation. That is also consistent with our previous studies showing that an interplay between histone methylation and DNA methylation occurred in NB [8]. The extent of both DNA hypomethylation and hypermethylation in NB cells is likely to reflect distinctive biological and clinical characteristics.
Taken together, our results indicate that the inhibition of HDAC is capable of modulation NB phenotype by changing the expression pattern and the epigenome of NB. Understanding the role of the interaction between histone modification and DNA methylation, and how they function at the molecular level may shed light on the mechanisms involved in the development and progression of NB.
Conclusions
Our data indicated that inhibition of HDAC blocked NB cell proliferation and induced cell cycle arrest. We also showed that inhibition of HDAC decreased the level of promoter methylation in two tumor suppressor genes (THBS1 and RASSFIA), increased global DNA methylation, and modified the level of expression of cancer-related genes towards to non-tumorigenic pattern. These studies indicate that the histone modifier (VPA) may exert anti-tumor effects through alteration of DNA methylation status and patterns. Further studies investigating the anti-tumor effects of VPA in high-risk NB are warranted.
Figure 9.
Acknowledgments
Sources of founding: This work was supported in part by Children’s Neuroblastoma Cancer Foundation (QY), Comer Kids Classic Grant of University of Chicago (QY), National Institutes of Health grants R01 HL075187 (JUR) and R01 HL059435 (JUR).
Footnotes
Conflicts of interest: The authors declare that they have no competing interests.
Contributor Information
Song Gu, Email: gusong@shsmu.edu.cn.
Yufeng Tian, Email: ytian@uchicago.edu.
Alexandre Chlenski, Email: achlensk@uchicago.edu.
Helen R. Salwen, Email: hsalwen@uchicago.edu.
Ziyan Lu, Email: ziyanlu@uic.edu.
J. Usha Raj, Email: usharaj@uic.edu.
References
- 1.Cohn SL, Andrew JP, Wendy BL, Tom M, Peter FA, Garrett MB, et al. The International Neuroblastoma Risk Group (INRG) Classification System: An INRG Task Force Report. J Clin Oncol. 2009;27:289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tom M, Garrett MB, Peter FA, Hervé JB, Giovanni C, Keith H, et al. The International Neuroblastoma Risk Group (INRG) Staging System: An INRG Task Force Report. J Clin Oncol. 2009;27:298–303. doi: 10.1200/JCO.2008.16.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abe M, Ohira M, Kaneda A, Yagi Y, Yamamoto S, Kitano Y, et al. CpG island methylator phenotype is a strong determinant of poor prognosis in neuroblastomas. Cancer Res. 2005;65:828–834. [PubMed] [Google Scholar]
- 4.Yang Q, Liu S, Tian Y, Hasan C, Kersey D, Salwen HR, et al. Methylation-associated silencing of the heat shock protein 47 gene in human neuroblastoma. Cancer Res. 2004;64:4531–4538. doi: 10.1158/0008-5472.CAN-04-0956. [DOI] [PubMed] [Google Scholar]
- 5.Yang QW, Liu S, Tian Y, Salwen HR, Chlenski A, Weinstein J, et al. Methylation-associated silencing of the thrombospondin-1 gene in human neuroblastoma. Cancer Res. 2003;63:6299–6310. [PubMed] [Google Scholar]
- 6.Mossman D, Scott RJ. Long term transcriptional reactivation of epigenetically silenced genes in colorectal cancer cells requires DNA hypomethylation and histone acetylation. PLoS One. 2011;6:e23127. doi: 10.1371/journal.pone.0023127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Georgiou E, Kouidou S. Epigenetically-targeted therapies for the treatment of hematological malignancies. Curr Med Chem. 2011;18:1757–1764. doi: 10.2174/092986711795496791. [DOI] [PubMed] [Google Scholar]
- 8.Yang Q, Tian Y, Ostler KR, Chlenski A, Guerrero LJ, Salwen HR, et al. Epigenetic alterations differ in phenotypically distinct human neuroblastoma cell lines. BMC Cancer. 2010;10:286. doi: 10.1186/1471-2407-10-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bidabadi E, Mashouf M. A randomized trial of propranolol versus sodium valproate for the prophylaxis of migraine in pediatric patients. Paediatr Drugs. 2010;12:269–275. doi: 10.2165/11316270-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 10.Lovell BV, Marmura MJ. Valproate semisodium ER for migraine and cluster headache prophylaxis. Expert Opin Drug Metab Toxicol. 2010;6:495–504. doi: 10.1517/17425251003693547. [DOI] [PubMed] [Google Scholar]
- 11.Venkataramani V, Rossner C, Iffland L, Schweyer S, Tamboli IY, Walter J, et al. Histone deacetylase inhibitor valproic acid inhibits cancer cell proliferation via down-regulation of the alzheimer amyloid precursor protein. J Biol Chem. 2010;285:10678–10689. doi: 10.1074/jbc.M109.057836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Q, Tian Y, Liu S, Zeine R, Chlenski A, Salwen HR, et al. Thrombospondin-1 peptide ABT-510 combined with valproic acid is an effective antiangiogenesis strategy in neuroblastoma. Cancer Res. 2007;67:1716–1724. doi: 10.1158/0008-5472.CAN-06-2595. [DOI] [PubMed] [Google Scholar]
- 13.Cinatl J, Jr, Cinatl J, Scholz M, Driever PH, Henrich D, Kabickova H, et al. Antitumor activity of sodium valproate in cultures of human neuroblastoma cells. Anticancer Drugs. 1996;7:766–773. doi: 10.1097/00001813-199609000-00008. [DOI] [PubMed] [Google Scholar]
- 14.Yang Q, Kiernan CM, Tian Y, Salwen HR, Chlenski A, Brumback BA, et al. Methylation of CASP8, DCR2, and HIN-1 in neuroblastoma is associated with poor outcome. Clin Cancer Res. 2007;13:3191–3197. doi: 10.1158/1078-0432.CCR-06-2846. [DOI] [PubMed] [Google Scholar]
- 15.Janardhanan R, Butler JT, Banik NL, Ray SK. N-(4-hydroxyphenyl) retinamide potentiated paclitaxel for cell cycle arrest and apoptosis in glioblastoma C6 and RG2 cells. Brain Res. 2009;1268:142–153. doi: 10.1016/j.brainres.2009.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wojdacz TK, Hansen LL, Dobrovic A. A new approach to primer design for the control of PCR bias in methylation studies. BMC Research Notes. 2008;1:54. doi: 10.1186/1756-0500-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wojdacz TK, Borgbo T, Hansen LL. Primer design versus PCR bias in methylation independent PCR amplifications. Epigenetics. 2009;4:231–234. doi: 10.4161/epi.9020. [DOI] [PubMed] [Google Scholar]
- 18.Shah MY, Vasanthakumar A, Barnes NY, Figueroa ME, Kamp A, Hendrick C, et al. DNMT3B7, a truncated DNMT3B isoform expressed in human tumors, disrupts embryonic development and accelerates lymphomagenesis. Cancer Res. 2010;70:5840–5850. doi: 10.1158/0008-5472.CAN-10-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Q, Zage P, Kagan D, Tian Y, Seshadri R, Salwen HR, et al. Association of epigenetic inactivation of RASSF1A with poor outcome in human neuroblastoma. Clin Cancer Res. 2004;10:8493–8500. doi: 10.1158/1078-0432.CCR-04-1331. [DOI] [PubMed] [Google Scholar]
- 20.Chan TA, Glockner S, Yi JM, Chen W, Van Neste L, Cope L, et al. Convergence of mutation and epigenetic alterations identifies common genes in cancer that predict for poor prognosis. PLoS Med. 2008;5:e114. doi: 10.1371/journal.pmed.0050114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramos EA, Camargo AA, Braun K, Slowik R, Cavalli IJ, Ribeiro EM, et al. Simultaneous CXCL12 and ESR1 CpG island hypermethylation correlates with poor prognosis in sporadic breast cancer. BMC Cancer. 2010;10:23. doi: 10.1186/1471-2407-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Witt O, Deubzer HE, Lodrini M, Milde T, Oehme I. Targeting histone deacetylases in neuroblastoma. Curr Pharm Des. 2009;15:436–447. doi: 10.2174/138161209787315774. [DOI] [PubMed] [Google Scholar]
- 23.Michaelis M, Suhan T, Cinatl J, Driever PH, Cinatl J., Jr Valproic acid and interferon-alpha synergistically inhibit neuroblastoma cell growth in vitro and in vivo. Int J Oncol. 2004;25:1795–1799. [PubMed] [Google Scholar]
- 24.Rocchi P, Tonelli R, Camerin C, Purgato S, Fronza R, Bianucci F, et al. p21Waf1/Cip1 is a common target induced by short-chain fatty acid HDAC inhibitors (valproic acid, tributyrin and sodium butyrate) in neuroblastoma cells. Oncol Rep. 2005;13:1139–1144. [PubMed] [Google Scholar]
- 25.Liu NA, Jiang H, Ben-Shlomo A, Wawrowsky K, Fan XM, Lin S, et al. Targeting zebrafish and murine pituitary corticotroph tumors with a cyclin-dependent kinase (CDK) inhibitor. Proc Natl Acad Sci U S A. 2011;108:8414–8419. doi: 10.1073/pnas.1018091108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho YJ, Kim JH, Yoon J, Cho SJ, Ko YS, Park JW, et al. Constitutive activation of glycogen synthase kinase-3beta correlates with better prognosis and cyclin-dependent kinase inhibitors in human gastric cancer. BMC Gastroenterol. 2010;10:91. doi: 10.1186/1471-230X-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agarwal C, Singh RP, Dhanalakshmi S, Tyagi AK, Tecklenburg M, Sclafani RA, et al. Silibinin upregulates the expression of cyclin-dependent kinase inhibitors and causes cell cycle arrest and apoptosis in human colon carcinoma HT-29 cells. Oncogene. 2003;22:8271–8282. doi: 10.1038/sj.onc.1207158. [DOI] [PubMed] [Google Scholar]
- 28.Cabello CM, Bair WB, 3rd, Ley S, Lamore SD, Azimian S, Wondrak GT. The experimental chemotherapeutic N6-furfuryladenosine (kinetin-riboside) induces rapid ATP depletion, genotoxic stress, and CDKN1A(p21) upregulation in human cancer cell lines. Biochem Pharmacol. 2009;77:1125–1138. doi: 10.1016/j.bcp.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weinstat-Saslow DL, Zabrenetzky VS, VanHoutte K, Frazier WA, Roberts DD, Steeg PS. Transfection of thrombospondin 1 complementary DNA into a human breast carcinoma cell line reduces primary tumor growth, metastatic potential, and angiogenesis. Cancer Res. 1994;54:6504–6511. [PubMed] [Google Scholar]
- 30.Yao M, Roberts DD, Isenberg JS. Thrombospondin-1 inhibition of vascular smooth muscle cell responses occurs via modulation of both cAMP and cGMP. Pharmacol Res. 2011;63:13–22. doi: 10.1016/j.phrs.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chlenski A, Liu S, Crawford SE, Volpert OV, DeVries GH, Evangelista A, et al. SPARC is a key Schwannian-derived inhibitor controlling neuroblastoma tumor angiogenesis. Cancer Res. 2002;62:7357–7363. [PubMed] [Google Scholar]
- 32.Teitz T, Wei T, Valentine MB, Vanin EF, Grenet J, Valentine VA, et al. Caspase 8 is deleted or silenced preferentially in childhood neuroblastomas with amplification of MYCN. Nat Med. 2000;6:529–535. doi: 10.1038/75007. [DOI] [PubMed] [Google Scholar]
- 33.Fortson WS, Kayarthodi S, Fujimura Y, Xu H, Matthews R, Grizzle WE, et al. Histone deacetylase inhibitors, valproic acid and trichostatin-A induce apoptosis and affect acetylation status of p53 in ERG-positive prostate cancer cells. Int J Oncol. 2011;39:111–119. doi: 10.3892/ijo.2011.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu YF, Sheu JR, Hsiao G, Lin CH, Chang TH, Chiu PT, et al. p53 in trichostatin A induced C6 glioma cell death. Biochim Biophys Acta. 2011;1810:504–513. doi: 10.1016/j.bbagen.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 35.Zhang W, Zeng X, Briggs KJ, Beaty R, Simons B, Yen R-W, et al. A potential tumor suppressor role for Hic1 in breast cancer through transcriptional repression of ephrin-A1. Oncogene. 2010;29:2467–2476. doi: 10.1038/onc.2010.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krop I, Parker MT, Bloushtain-Qimron N, Porter D, Gelman R, Sasaki H, et al. HIN-1, an Inhibitor of Cell Growth, Invasion, and AKT Activation. Cancer Res. 2005;65:9659–9669. doi: 10.1158/0008-5472.CAN-05-1663. [DOI] [PubMed] [Google Scholar]
- 37.Yang Q, Kiernan CM, Tian Y, Salwen HR, Chlenski A, Brumback BA, et al. Methylation of CASP8, DCR2, and HIN-1 in neuroblastoma is associated with poor outcome. Clin Cancer Res. 2007;13:3191–3197. doi: 10.1158/1078-0432.CCR-06-2846. [DOI] [PubMed] [Google Scholar]
- 38.Schwab M, Varmus HE, Bishop JM. Human N-myc gene contributes to neoplastic transformation of mammalian cells in culture. Nature. 1985;316:160–162. doi: 10.1038/316160a0. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt ML, Salwen HR, Manohar CF, Ikegaki N, Cohn SL. The biological effects of antisense N-myc expression in human neuroblastoma. Cell Growth Differ. 1994;5:171–178. [PubMed] [Google Scholar]
- 40.Thiele CJ, Reynolds CP, Israel MA. Decreased expression of N-myc precedes retinoic acid-induced morphological differentiation of human neuroblastoma. Nature. 1985;313:404–406. doi: 10.1038/313404a0. [DOI] [PubMed] [Google Scholar]
- 41.Peverali FA, Orioli D, Tonon L, Ciana P, Bunone G, Negri M, et al. Retinoic acid-induced growth arrest and differentiation of neuroblastoma cells are counteracted by N-myc and enhanced by max overexpressions. Oncogene. 1996;12:457–462. [PubMed] [Google Scholar]
- 42.Frigola J, Solé X, Paz MF, Moreno V, Esteller M, Capellà G, et al. Differential DNA hypermethylation and hypomethylation signatures in colorectal cancer. Hum Mol Genet. 2005;14:319–326. doi: 10.1093/hmg/ddi028. [DOI] [PubMed] [Google Scholar]
- 43.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 44.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 45.Dong E, Guidotti A, Grayson DR, Costa E. Histone hyperacetylation induces demethylation of reelin and 67-kDa glutamic acid decarboxylase promoters. Proc Natl Acad Sci U S A. 2007;104:4676–4681. doi: 10.1073/pnas.0700529104. [DOI] [PMC free article] [PubMed] [Google Scholar]