Abstract
African swine fever virus (ASFV) is a large double-stranded DNA virus that replicates in discrete areas in the cytosol of infected cells called viral factories. Recent studies have shown that assembling virions acquire their internal envelopes through enwrapment by membranes derived from the endoplasmic reticulum (ER). However, the mechanisms that underlie the formation of viral factories and progenitor viral membranes are as yet unclear. Analysis of the published genome of the virus revealed a conserved multigene family that encodes proteins with hydrophobic signal sequences, indicating possible translocation into the ER lumen. Strikingly, two of these genes, XP124L and Y118L, encoded proteins with KDEL-like ER retention motifs. Analysis of XP124L and Y118L gene product by biochemical and immunofluorescence techniques showed that the proteins were localized to pre-Golgi compartments and that the KEDL motif at the C terminus of pXP124L was functional. XP124L expression, in the absence of other ASFV genes, had a dramatic effect on the contents of the ER that was dependent precisely on the C-terminal sequence KEDL. The normal subcellular distribution of a number of proteins resident to this important, cellular organelle was drastically altered in cells expressing wild-type XP124L gene product. PXP124L formed unusual perinuclear structures that contained resident ER proteins, as well as proteins of the ER-Golgi intermediate compartment. The data presented here hint at a role for MGF110 gene product in preparing the ER for its role in viral morphogenesis; this and other potential functions are discussed.
African swine fever virus (ASFV) is a large double-stranded DNA virus of the family Asfarviridae that infects members of the family Suidae, in particular domestic swine, the European wild boar (Sus scrofa ferus), warthogs (Phacochoerus aethiopicus), and bushpigs (Potamochoerus porcus). ASFV is also the only known DNA arbovirus since the virus infects soft ticks of the family Ornithodoros, and these may act as a vector for the virus in areas where the disease is endemic (8, 22, 40, 55). ASFV can induce a range of symptoms in the domestic pig varying from fatal hemorrhagic disease (33, 37, 41) to chronic and subclinical infections (7, 37, 41). Interestingly, ASFV infection is essentially asymptomatic in bushpigs and warthogs (33, 41).
The sequencing of the genome of ASFV represented a major step forward in allowing the complicated life cycle of ASFV to be understood in molecular terms (15, 61). The viral genome of the Badajoz 1971 Vero-adapted strain (Ba71v) of ASFV is 170,101 bp in length and expresses at least 150 open reading frames, several of which share homology with cellular proteins and have the potential to modulate host signal transduction and immunoregulatory pathways. These include proteins able to modulate IκB and calcineurin phosphatase, inhibitors of apoptosis, and a CD2 homolog (1, 6, 9, 10, 31, 42, 44, 46, 52). An intriguing facet of ASFV is the presence of five multigene families (MGFs), located in the left- and right-hand variable regions of the genome. These are named after the average number of codons in each gene and are referred to as MGF100, -110, -300, -360, and -505/530 (2, 18, 47, 59, 61, 63). A link between MGF proteins and the complex life cycle of ASFV is provided by the observation that a number of genes expressed in the MGF360 and MGF530 loci determine host range (64) and virulence (35) and that MGF110 family proteins are lost when the virus is passaged through tissue culture (2, 13, 39). Sequencing and comparative restriction mapping of the left-hand region of several ASFV genomes have suggested that, on adaptation to tissue culture, the Lisbon 60 strain loses two MGF110 genes, whereas the Badajoz 1971 strain loses seven (2, 13, 39). Restriction mapping of isolates from soft ticks shows little genetic diversity in the left-hand variable region of the genome expressing MGF genes (14), suggesting that maintenance of MGFs may be important for the propagation of ASFV in the tick vector.
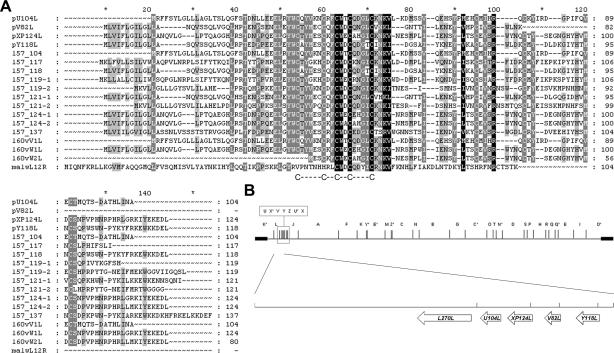
The cellular functions of most MGF proteins are largely unknown and difficult to predict because the proteins show limited homology to host proteins. For MGF110 proteins, however, sequence analysis does provide some clues to function. All of the MGF110 encoded proteins have potential signal peptides, suggesting they enter the secretory pathway. The MGF110 proteins also have a highly conserved central cysteine-rich domains [C-(X)5-C-(X)2-C-(X)2-C-(X)4-C] (Fig. 1), suggesting that they function in an oxidizing environment as found in the lumen of the endoplasmic reticulum (ER) or outside the cell. The MGF110 cysteine motif shares homology to a bovine posterior pituitary peptide (43), suggesting that MGF110 proteins form disulfide bonds in a similar way. However, the identification of a member of MGF110 from the Malawi Lil20/1 isolate of ASFV (59) with a histidine residue substituted for one cysteine suggests that this motif may be a metal-binding structure. Interestingly, the two C-(X)2-C sequences in the MGF110 cysteine domain also resemble the thioredoxin motifs that are present in the lumenal ER thioreductase enzymes: ERp57 and protein disulfide isomerase (PDI). In addition, the central C-(X)2-C-(X)2-C sequence of the MGF110 motif is found in the lumenal ER protein Ero1-Lα (ER oxoreductin), a protein involved in the maintenance of disulfide bonding in the ER (56). This motif is characteristic of some iron cluster proteins (4, 23) and is a structural determinant of Ero1-Lα (5). An ER location is also strongly suggested by the presence of ER retention motifs (38) at the C terminus of MGF110 family proteins. PY118L, for example, contains the classical ER retention sequence, KDEL, whereas pXP124L contains a homologous motif, KEDL, in which the D and E residues of the motif have been reversed. This motif is highly unusual and has only been previously described in the calreticulin gene of Trypanosoma cruzi and Leishmania donovani. Mutagenesis studies have shown that the KEDL sequence introduced artificially into ER proteins functions as a retention signal (20, 25); however, the KEDL sequence has not been described as a functional ER retention motif in a biological system.
FIG. 1.
MGF110 proteins are conserved between different ASFV isolates. (A) Soluble MGF110 protein sequences (2, 13, 15, 39) from four ASFV isolates were aligned by using the PILEUP program of GCG10.1. Protein sequences from the Ba71v isolate have a “p” prefix, sequences from the LIS57 isolate have an “l57” prefix, sequences from the Lisbon 60 Vero cell-adapted isolate have an “l60v” prefix, and a sequence from the Malawi Lil20/1 isolate is indicated by a “malw” prefix. Shading of the alignment was carried out with the Genedoc program and indicates the degree of similarity between residues, with black, dark gray shading, and light gray shading indicating 100, 80, and 60%, respectively. The position of the MGF110 cysteine motif is also highlighted. (B) EcoRI restriction map of the Ba71v isolate of ASFV, with the positions of MGF110 genes shown in an enlarged section below.
Retention of MGF110 proteins in the ER could serve several functions for ASFV. ASFV, for example, gains two internal envelopes by being wrapped by ER membrane cisternae (3, 11, 48), and MGF110 proteins within the lumen of the ER may facilitate recruitment of ER cisternae into virus assembly sites. Indeed, pXP124L has been shown to localize to assembling virions and membranous material within cytoplasmic virus factories by immunogold electron microscopy (48). ASFV infection also induces the loss of the trans-Golgi network (TGN) (30), and this may be mediated by an effect of MGF110 proteins on the trafficking of resident TGN proteins from the ER. Furthermore, ASFV infects mononuclear phagocytic cells (12, 45), and an ability to modulate the ER may offer the virus opportunities to regulate the trafficking of immunoregulatory or anticoagulant proteins to the surface of these cells. Taken together, such effects mediated within the ER could have profound consequences for host range and virulence.
As a first step to understanding the function of MGF110 in ASFV infection, we have studied the subcellular localization of the two members of MGF110 that encode different potential ER retention motifs. Surprisingly, even though both of these proteins had similar sequences, they had different cellular distributions, suggesting that they functioned in different cellular compartments. As anticipated from the C-terminal KDEL sequence, pY118L was localized to the ER; however, pXP124L with the KEDL sequence localized to post-ER-pre-Golgi structures. Intracellular retention of the XP124L protein in the post-ER compartment was dependent on the unusual ER retention sequence KEDL. Strikingly, expression of XP124L alone in cells produced an enlarged pre-Golgi membrane compartment that sequestered lumenal ER proteins. This redistribution was dependent on the sequence KEDL and may involve interaction of the KEDL sequence with the KDEL receptor that controls the distribution of lumenal ER proteins within pre-Golgi membrane compartments.
MATERIALS AND METHODS
Cells and viruses.
Vero cells (ECACC 84113001) were grown at 37°C in HEPES-buffered Dulbecco modified Eagle medium containing 10% (vol/vol) fetal calf serum, 100 U of streptomycin/ml, 100U of penicillin/ml, and 20 mM l-glutamine. Porcine aortic endothelial cells (PAECs) have been described previously (57) and were cultured on gelatinized flasks and coverslips at 37°C in HEPES-buffered Roswell Park Memorial Institute 1640 medium containing 10% (vol/vol) fetal calf serum, 100 U of streptomycin/ml, 100 U of penicillin/ml, and 20 mM l-glutamine. Porcine bone marrow cells were cultured at 37°C in Earle's saline medium containing 13% (vol/vol) heat-inactivated pig serum, 100 U of streptomycin/ml, and 100 U of penicillin/ml. The Lisbon 1957 (LIS57) and Ba71v strains of ASFV have been described previously (16, 27). Modified vaccinia virus Ankara-T7 polymerase (MVA-T7) is an attenuated, host-range-restricted, vaccinia virus strain expressing the bacteriophage T7 RNA polymerase gene (51) and was a generous gift from Gerd Sutter (Instit für Molekulare Virologie, Oberschleiβrheim, Germany).
Antibodies.
Rabbit antibody R29 and mouse anti-pXP124L were raised to the synthetic peptide SNPVPHNRPHRLGRKIYEK that corresponds to amino acids 102 to 121 of pXP124L (48). Rabbit antibody R30 was raised to the synthetic peptide QLVGQLRPTEDPPEEELEYWC that corresponds to amino acids 19 to 40 of pY118L. Antibody TW34, which recognizes p34, has been described previously (21). Monoclonal antibody C18 recognizes the early ASFV protein p30 and was a generous gift from Dan Rock (Plum Island Animal Disease Center). Mouse antibody to ASFV capsid protein p73 (17LD3) was purchased from Ingenasa. Mouse (1D3) and rabbit antibodies to PDI have been described previously (58, 62). Mouse anti-γ tubulin (GTU-88) was purchased from Sigma. Mouse antibodies to hsp47 (M16.10A1) and the KDEL receptor (KR10) were purchased from Stressgen Biotechnology Corp. Antibody to ERp57 has been described previously (48), and antibody recognizing calnexin was a generous gift of Ineke Braakman (Academic Medical Centre, University of Amsterdam). Antibodies to the mammalian homologues of Sec31 and Sec13 were a kind gift of Wanjin Hong (Institute of Molecular and Cell Biology, National University of Singapore).
Plasmids.
pDsRed2-ER was purchased from BD Biosciences Clontech, and p58-YFP (60) was kindly supplied by Nihal Alton (National Institutes of Health). XP124L was amplified from the HindIII-C fragment of the Ba71v genome (kindly provided by the late Eladio Viñuela) by PCR with the primers 5′-CTAGAGTTCATCTTTTTTCCA and 5′-GGATCCATGTTGGTGATCTTTTTGGGA. Y118L was amplified from the HindIII-C fragment of the Ba71v genome by PCR with the primers 5′-CCCGGGCCAGTATCTTAGCTCCTCTTCCAGG and 5′- GGATCCTTCAGCCAGCCGTAGCAATG. XP124LΔKEDL, XP124L-KDEL and Y118LΔKDEL were amplified by PCR from the full-length clones of their respective genes. XP124LΔKEDL was amplified with the primers 5′-TCATTCATAAATTTTCCTTCCCAA and 5′-GGATCCTTCAGCCAGCCGTAGCAATG, XP124L-KDEL was amplified with the primers 5′-TCACAGTTCATCCTTTTCATA and 5′-GGATCCTTCAGCCAGCCGTAGCAATG, and Y118LΔKDEL was amplified with the primers 5′-CTATTTCCATTCCTTCCTGAAGTA and 5′-GGATCCATGTTGGTGATCTTTTTGGGA. PCR fragments were cloned into pT7blue T-vector (Novagen and R&D Systems, Inc.) and then subcloned into pcDNA3.1zeo(+) (Invitrogen). Transfection quality DNA was subsequently maxiprepped by using Qiagen columns (Qiagen).
MVA-T7 infection and transfections.
Vero cells were grown to 70% confluency and then infected for 1 h with MVA-T7. Cells were then transfected with plasmid DNA by using Transfast (Promega) for 1 h at 37°C and incubated overnight in Dulbecco modified Eagle medium-HEPES (with 2% [vol/vol] fetal calf serum). Experiments were performed on transfected cells the following day. p58-YFP and DsRed2-ER were transfected as described above but in the absence of MVA-T7.
Indirect immunofluorescence and digital deconvolution.
Cells were fixed to coverslips by incubation for 30 min at room temperature with either 4% (wt/vol) paraformaldehyde in phosphate-buffered saline (PBS), 2% paraformaldehyde (wt/vol) containing 0.05% glutaraldehyde (vol/vol) in PBS, or methanol. Coverslips were then incubated for 15 min at room temperature with shaking in blocking buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 0.2% [wt/vol] gelatin, 1% [vol/vol] Nonidet P-40, 30% [vol/vol] normal goat serum). Primary antibodies were diluted in blocking buffer, and cells were stained with shaking at room temperature for 1 h. Primary antibodies were visualized by the addition of appropriate secondary antibodies conjugated to Alexa-488 or Alexa-594 (Molecular Probes). These were diluted 1:500 in blocking buffer, and cells were stained as for primary antibodies in the dark. Cells were washed with PBS between all stages. Next, cells were stained with 500 ng of DAPI (4′,6′-diamidino-2-phenylindole/ml; Sigma) in PBS to visualize nuclear and extranuclear viral DNA and then finally rinsed in distilled water before being mounted on Superfrost microscope slides (BDH) with Fluoromount G (Southern Biotechnology). Cells were viewed at ×60/1.4-numerical-aperture or ×100/1.3-numerical-aperture with a Nikon E800 microscope. The images were captured with a Hamamatsu C-4746A charge-coupled device camera and were deconvolved by using Improvision Openlab 2.1.3 or 3.1 software. Optical sections 0.2 μm thick were analyzed.
Metabolic labeling and immunoprecipitation.
Cells were starved in methionine-cysteine-free Eagle medium for 20 min and then labeled with 0.75 MBq of 35S-Promix (Amersham Life Sciences) per ml in the same medium. Cells were then chased by replacing the labeling medium with normal culture medium. At appropriate times after incubation at 37°C, cells were washed with PBS and lysed at 4°C in immunoprecipitation buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 1%[vol/vol] Brij 33, 10 mM iodoacetimide, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mg of leupeptin/ml, 1 mg of pepstatin/ml, 1 mg of chymostatin/ml, 1 mg of antipain/ml). To immunoprecipitate extracellular antigen, the medium was removed before the cells were washed; Brij 33 and leupeptin, pepstatin, chymostatin, and antipain were then added to the same concentrations as for the immunoprecipitation buffer. Unlysed cells were removed by centrifugation (10,000 × g, 4°C, 10 min), and the lysates were then precleared by incubating them at 4°C for 1 h with insoluble protein A (Sigma). Insoluble protein A was removed by centrifugation (10,000 × g, 4°C, 15 min), and antigens were immunoprecipitated from the samples by overnight incubation with antibodies immobilized with protein A-Sepharose B beads. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and detected by autofluorography.
Digestion with endo-H.
Immunoprecipitates were denatured by boiling for 3 min in 10 μl of 1% SDS. Digestions were performed at 37°C overnight in 50 μl of 50 mM sodium phosphate (pH 6.0) containing 1 mU of endo-H (endo-β-N-acetylglucosaminidase H; Boehringer Mannheim).
Western blotting.
Proteins were resolved by SDS-PAGE and transferred onto Protan BA85 cellulose nitrate membrane (Schleicher & Schuell, Dassel, Germany). The membrane was blocked overnight at 4°C with 10% (vol/vol) normal goat serum-5%(wt/vol) skimmed milk powder in PBS; the membrane was then washed in PBS prior to incubation with primary antibody. Primary antibody was removed, and the membrane was washed three times with changes in PBS before incubation with secondary antibody conjugated to horseradish peroxidase (Promega). After three further washes with changes in PBS, bands were revealed by enhanced chemiluminescence (Amersham Life Sciences).
SDS-PAGE.
Unless specified otherwise, proteins were resolved on 12.5% acrylamide gels by using the Tricine system (50). Gradient gels (10 to 20%) were purchased from Sigma.
RESULTS
MGF110 protein pY118L, with a C-terminal KDEL sequence, is localized to the ER.
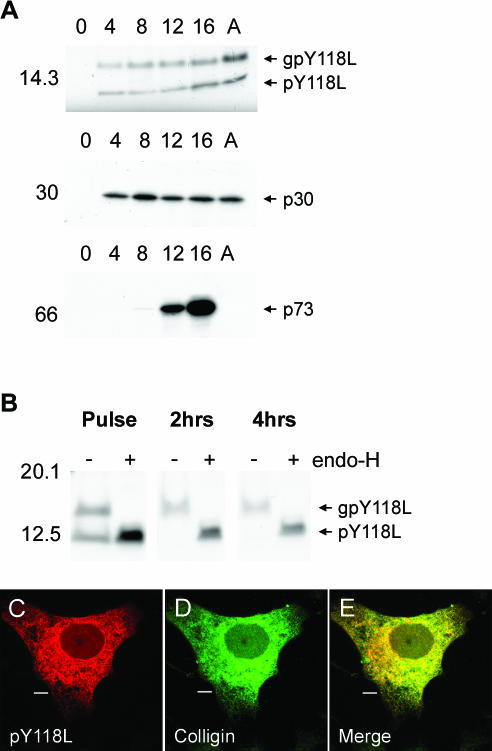
Analysis of the gene encoding pY118L predicts a 12-kDa protein with a hydrophobic N-terminal signal sequence, a single N-linked glycosylation site, and a C-terminal KDEL ER retention motif. To monitor the onset of synthesis of pY118L, cells infected with ASFV were pulse-labeled for 30 min at increasing times postinfection (Fig. 2A). Lysates were immunoprecipitated with an anti-peptide antibody specific for pY118L and with antibodies binding p30 and p73 to determine the onset of early and late gene expression, respectively. The anti-peptide antibody immunoprecipitated proteins of 14 and 12 kDa 4 h after infection, and the levels of these proteins increased modestly during the time course. The results suggested that pY118L was an early gene product, and this was supported by the observation that expression matched that of the early ASFV proteins p30 and was not inhibited when viral DNA replication was blocked by cytosine βD-arabinoside (Fig. 2A, lanes A). The two bands immunoprecipitated by the anti-peptide antibody could represent different glycoforms of pY118L or inefficient cleavage of pY118L by signal peptidase, or they could be two different proteins. The presence of N-linked oligosaccharides attached to pY118L was tested by digestion of immunoprecipitates with endo-H (Fig. 2B). Endo-H digestion converted the 14-kDa protein to a 12-kDa protein, indicating that the molecular mass difference was due to the presence of N-linked oligosaccharides. Interestingly, the pY118L protein remained sensitive to endo-H 4 h into the chase, suggesting that pY118L was retained in the ER. This was confirmed by indirect immunofluorescence microscopy where pY118L colocalized with the lumenal ER protein colligin (Fig. 2C and D). The two antibodies produced similar, reticular staining patterns that spread out to the periphery of the cell, suggesting that pY118L was localized to the ER; this finding was confirmed in Fig. 2E.
FIG. 2.
pY118L is an early ASFV protein localized to the ER. (A) Control Vero cells (lanes 0), cells infected with Ba71v for increasing times (in hours) as indicated, or infected for 16 h in the presence of cytosine β-d-arabinofuranoside (lanes A) were pulsed for 30 min with [35S]methionine and [35S]cysteine, lysed, and immunoprecipitated with antibodies specific for pY118L (R30), p30 (C18), or p73 (17LD3). (B) Vero cells were infected for 12 h and then pulse-labeled as described above before being chased in complete medium for the indicated lengths of time. PY118L was immunoprecipitated with R30 and subjected to endo-H digestion (lanes +). Proteins were resolved by SDS-PAGE and subjected to autofluorography. Sizes of molecular mass markers are indicated in kilodaltons on the left. (C to E) Vero cells infected for 8 h with Ba71v were fixed with 4% paraformaldehyde and probed with antibody R30 (pY118L) (C) and anti-colligin (D). Primary antibody was visualized with appropriate goat antibodies conjugated to Alexa-488 (D) or Alexa-594 (C). Panel E is a digitally merged image of panels C and D. Digital sections (0.2 μm) were captured at ×100 magnification and digitally deconvolved by using Openlab 2.1.3. Bars, 10 μm.
MGF110 protein pXP124L, with a C-terminal KEDL sequence, is localized to post-ER compartments.
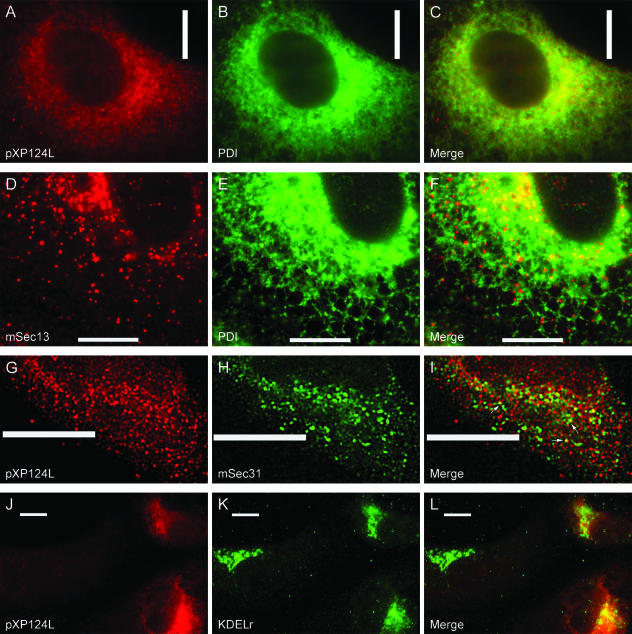
The sequence of pXP124L, the second member of MGF110 under study, suggested a protein very similar to pY118L. The protein was predicted to be a 14-kDa glycoprotein with a N-terminal signal sequence and a possible ER retention motif. Unlike pY118L, however, the motif contained the sequence KEDL rather than the classical ER retention motif KDEL. Database searches suggested that the KEDL sequence was unusual since the sequence has not been described for higher eukaroytes. The subcellular distribution of the pXP124L protein in infected cells was therefore studied. An antibody raised against pXP124L showed weak staining throughout the infected cell (Fig. 3A), but higher levels were concentrated next to the nucleus. This distribution was similar but not identical to that of the lumenal ER protein PDI (Fig. 3B), and this can be seen more clearly in the merged image shown in Fig. 3C. Although pXP124L staining was perinuclear and peripheral, the pattern was more vesicular than the reticular stain seen for PDI, and this vesicular distribution was seen in more detail by analyzing cells at a higher magnification (Fig. 3G).
FIG. 3.
pXP124L localizes to pre-Golgi compartments. Vero cells infected for 8 h with Ba71v were fixed with either glutaraldehyde solution (A to C) or 4% paraformaldehyde (D to L). Cells were then probed with either antibodies R29 (pXP124L in A, G, and J) or 1D3 (PDI in B and E) and antibodies recognizing either mSec13 (D), mSec31 (H), or the KDEL receptor (K). Primary antibody was visualized with appropriate goat antibodies conjugated to Alexa-488 (B, E, H, and K) or Alexa-594 (A, D, G, and J). Panels C, F, I, and L are digitally merged images of panels A and B, D and E, G and H, and J and K, respectively. Digital sections (0.2 μm) were visualized at ×60 (A to F and J to L) or ×100 (G to I) magnification and digitally deconvolved by using Openlab 2.1.3. Bars, 10 μm.
It was possible that pXP124L, like the KDEL receptor responsible for the retention of lumenal ER proteins, may recycle constitutively through ER-Golgi intermediate membrane compartments (ERGIC). This would explain the vesicular nature of the pXP124L stain and the observation that the viral protein predominantly colocalized with ER proteins in perinuclear regions of the cell, which contain pre-Golgi compartments, and not at the periphery. If pXP124L were recycling, then some of the protein would be expected to colocalize with ERGIC proteins and possibly ER exit sites. Markers for the latter subcellular compartment include Sec13 (53) and Sec31 (54), which are components of the COPII coats formed at ER exit sites. Antibodies recognizing the mammalian homologue of Sec13 (mSec13) and PDI were used to identify ER exit sites in Vero cells (Fig. 3D and E). The merged image in Fig. 3F shows successful labeling of exit sites superimposed on the reticular ER stain provided by the antibody recognizing PDI. This pattern for mSec13 was similar to that published for HeLa cells (19) and, interestingly, was also similar to the stain seen for pXP124L. The distributions of pXP124L and ER exit sites were therefore compared in infected cells (Fig. 3G and H). Both anti-mSec31 and the antibody to pXP124L produced punctate vesicular stains; however, the merged image in Fig. 3I demonstrates that the distributions of the proteins were not identical. There were colocalization in perinuclear regions and some colocalization in the cell periphery (arrows in Fig. 3I); however, most of the signal was separate. The results suggested that pXP124L may pass through ER exit sites, but at steady state most of the viral protein is in vesicles that do not stain for mSec31 or ER markers. To test for colocalization between pXP124L and an ERGIC marker, infected cells were double stained for pXP124L and the KDEL receptor. Figure 3J, K, and L indicate that pXP124L codistributed with the KDEL receptor at perinuclear sites but that the degree of colocalization varied from cell to cell.
The immunofluorescence results described above suggested that pXP124L was located in several post-ER membrane compartments. To test the extent of trafficking of pXP124L along the secretory pathway to the Golgi, the processing of N-linked oligosaccharides attached to pXP124L was analyzed. Cells infected with Ba71v were pulse-labeled for 30 min and then chased for up to 4 h. The protein was immunoprecipitated at the indicated times and digested with endo-H. The results in Fig. 4A show that the levels of pXP124L in cells remained constant during the chase. The protein was not therefore secreted from cells. The protein also remained sensitive to endo-H during the chase and did not therefore reach Golgi membrane compartments. Given that the fluorescence results failed to find the protein in the ER, the results suggest that pXP124L localizes to pre-Golgi/ERGIC compartments in ASFV-infected cells.
FIG. 4.
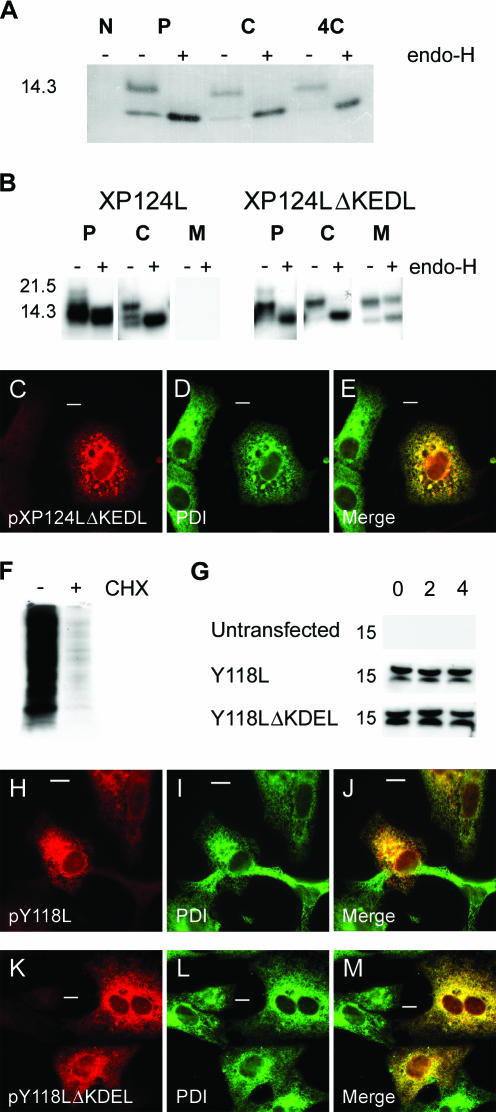
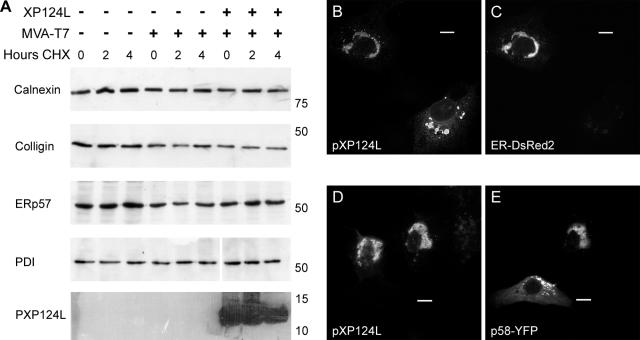
Role of the C-terminal sequences in the cellular localization of pXP124L and pY118L. (A) Uninfected (N) and Vero cells infected for 12 h with ASFV were pulse-labeled for 30 min with [35S]methionine and [35S]cysteine (P) and then chased in complete medium for 2 (C) or 4 (4C) h. Cells were lysed and then immunoprecipitated with antibody R29 and digested with endo-H (lanes +). (B) Wild-type XP124L and XP124LΔKEDL were expressed in Vero cells by using MVA-T7. Cells were pulse-labeled for 30 min (P) and then chased for 2 h (C). Cell lysates (P and C) and the medium from the chase (M) were immunoprecipitated with antibody R29 and digested with endo-H (“+” lanes). Proteins were resolved by SDS-PAGE and subjected to autofluorography. Sizes of molecular mass markers are indicated in kilodaltons on the left. (C to E and H to M) Vero cells expressing XP124LΔKEDL (C to E), Y118L (H to J), or Y118LΔKDEL (K to M) were fixed with 4% paraformaldehyde and then probed with antibody R29 (C) or R30 (H and K) and antibody 1D3 (PDI in panels D, I, and L). Primary antibody was visualized with appropriate goat antibodies conjugated to Alexa-488 (D, I, and L) or Alexa-594 (C, H, and K). Images were captured at ×60 magnification and processed as described in the legend to Fig. 2. Bars, 10 μm. (F) Cells infected with MVA-T7 were pulse-labeled for 4 h in the presence (lane “+”) or absence (lane “−”) of 200 μg of cycloheximide/ml (lane CHX). Cell lysates were resolved by SDS-PAGE and subjected to autofluorography. (G) Control cells or cells expressing Y118L or Y118LΔKDEL were incubated in the presence of cycloheximide (200 μg/ml) for the indicated lengths of time (in hours), and the levels of pY118L and pY118LΔKDEL were determined by Western blotting with antibody R30.
The KEDL sequence localizes pXP124L to pre-Golgi membrane compartments.
To test whether the KEDL motif of pXP124L determined the pre-Golgi retention of the viral protein, the KEDL sequence was removed, and this deletion mutant, along with the wild-type gene, was expressed in Vero cells by using a recombinant vaccinia virus expressing the T7 RNA polymerase gene (51). Cells expressing XP124L and XP124LΔKEDL were pulse-labeled and then chased for 2 h. Viral proteins were immunoprecipitated from cell lysates and culture media and then digested with endo-H (Fig. 4B). Wild-type pXP124L was not secreted into the medium during the 2-h chase and remained sensitive to endo-H; however, deletion of the KEDL motif from pXP124L resulted in the appearance of endo-H-resistant protein in culture supernatants. This showed that removal of the KEDL sequence allowed transit through the Golgi, where the sugar processing occurred, and then secretion into the extracellular medium. Approximately two-thirds of the labeled protein, however, remained in cells after the chase and was still endo-H sensitive. Taken together, the results showed that removal of KEDL motif allowed secretion of some of the newly synthesized protein; however, transport from the ER to the Golgi and hence to the extracellular medium was slow.
The effects of the KEDL sequence on the subcellular distribution of pXP124L were studied by using immunofluorescence. Figure 4C shows that when the KEDL motif was removed the protein showed a reticular stain spreading through the cytosol. There were also bright cytosolic spots of fluorescence in continuity with the reticular stain. The reticular stain seen for the deletion mutant suggested an ER location, and cells were also labeled with antibody specific for PDI (Fig. 4D). The viral protein lacking the KEDL sequence colocalized with PDI (Fig. 4E), suggesting that the reticular stain and the bright cytosolic spots were ER.
Taken together, the results showed that pXP124L was localized to pre-Golgi membrane compartments by a C-terminal KEDL motif. The results were, however, complex. Removal of the KEDL motif allowed secretion of approximately one-third of the protein over 2 h, but the remainder was retained in the ER. The results were unexpected since they implied that amino acids other than those at the C terminus conferred ER retention. In other words, the unusual KEDL sequence targets the protein to post-ER and pre-Golgi compartments, but in the absence of the motif the protein is retained in the ER. Given the overall sequence similarity between members of the MGF110 family (Fig. 1), it was possible this may be true for the other MGF110 proteins. To test this, the C-terminal KDEL sequence was removed from pY118L. The antibody raised against pY118L cross-reacted with a vaccinia virus protein of similar size, making interpretation of immunoprecipitations of the protein expressed from vaccinia vectors difficult. Fortunately, no cross-reactivity was noticed on immunoblots of lysates from MVA-T7-infected cells. Therefore, the trafficking of pY118L was studied by using a cycloheximide chase experiment to compare the cellular levels of pY118L and pY118LΔKDEL over time. Figure 4F shows the successful inhibition of protein synthesis by cycloheximide. In Fig. 4G, immunoblotting of lysates from cells expressing Y118L and Y118LΔKDEL revealed two bands corresponding to the predicted molecular masses of glycosylated and nonglycosylated pY118L. There was no decrease in the steady-state levels of either protein over time. The Y118LΔKDEL protein was not therefore secreted, suggesting that the KDEL sequence alone did not therefore determine ER retention of pY118L. This was confirmed when immunostaining of cells expressing Y118L and Y118LΔKDEL revealed that both proteins colocalized with the lumenal ER protein PDI (Fig. 4H to M). MGF110 proteins may, therefore, have two domains determining cellular distribution. The C-terminal KEDL motif of pXP124L retains the protein in pre-Golgi compartments, but surprisingly, the KDEL motif in pY118L is redundant since when the C-terminal motif was removed the protein was retained in the ER.
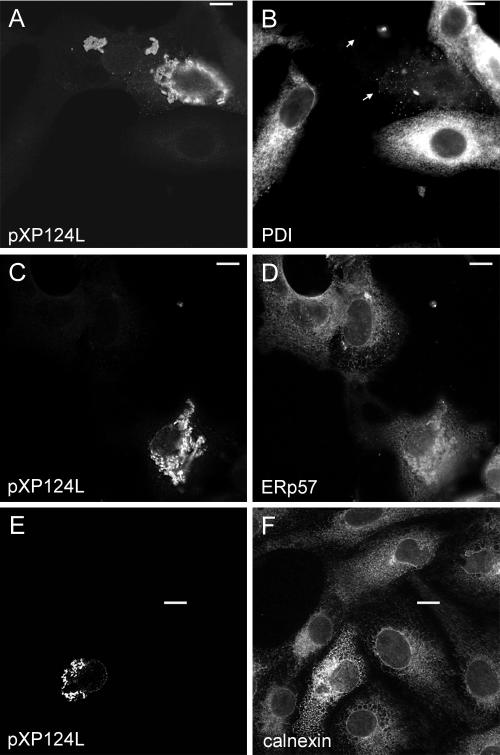
Expression of pXP124L alone in cells results in profound rearrangement of lumenal ER proteins.
The KDEL retrieval pathway is responsible for the retention of resident lumenal protein in the ER. The presence of the unusual KEDL sequence present at the C terminus of pXP124L prompted us to follow the effects of pXP124L on the distribution of other proteins normally found in pre-Golgi compartments. Figure 5A showed that when XP124L was expressed in the absence of other ASFV gene products, the protein produced strong labeling of punctate perinuclear structures, with some labeling of the nuclear envelope. The punctate stain suggested that, as seen in infected cells, the protein did not locate to the ER but was found in post-ER compartments. To test this, the distribution of PDI was examined (Fig. 5B). Surprisingly, expression of pXP124L greatly diminished the immunofluorescence signal for PDI. The integrity of the ER in cells expressing XP124L was further examined by using antibodies against calnexin, an abundant integral ER membrane protein, and two further ER lumenal proteins, ERp57 and colligin. The distribution of calnexin was not greatly affected by XP124L expression. There was a subtle enhanced perinuclear distribution of calnexin in areas staining for pXP124L (Fig. 5F); importantly, however, the reticular, peripheral ER stain of calnexin and staining of the nuclear envelope were not affected by pXP124L. The viral protein did not therefore cause a loss of the ER from cells. In the cell expressing XP124L, ERp57 maintained a peripheral reticular stain (Fig. 5D) but, as seen for calnexin, ERp57 was redistributed in the perinuclear regions containing pXP124L. Interestingly, as seen for PDI, the pXP124L protein led to complete loss of the signal for colligin (data not shown). Taken together, these results show that the reticular structure of the ER remains intact in cells expressing the viral protein but that there is selective redistribution of some lumenal ER proteins into regions enriched for pXP124L and, in the case of PDI and colligin, a complete loss of signal.
FIG. 5.
Effect of pXP124L on the early secretory pathway. Vero cells expressing XP124L were fixed with methanol (C to F) or 4% paraformaldehyde (A and B). Cells were then probed with antibody R29 (pXP124L in panel A) or mouse anti-pX124L (pXP124L in panels C and E) and antibody 1D3 (PDI in panel B), anti-ERp57 (D), or anti-calnexin (F). Primary antibody was visualized with appropriate goat antibodies conjugated to Alexa-488 (B, D, and F) or Alexa-594 (A, C, and E), and images were captured at ×60 magnification and processed as described in the legend to Fig. 2. Bars, 10 μm.
The signals for PDI and colligin could have been lost because of enhanced degradation or secretion of the protein from cells. Alternatively, the immunofluorescence signals may have been masked in some way. PXP124L may, for example, block the epitopes recognized by the antibodies. If XP124L expression induced the secretion or degradation of PDI or other resident ER proteins, then steady-state levels would be predicted to decrease over time in the absence of protein synthesis. Cells expressing XP124L were therefore incubated with cycloheximide for increasing times, and the levels of the lumenal ER proteins colligin, ERp57, and PDI, as well as the integral membrane protein calnexin, were analyzed by Western blotting. Figure 6A shows that there was no obvious change in the levels of any of these proteins when cells were incubated with cycloheximide for 2 or 4 h. These results suggested that pXP124L did not cause degradation or secretion of PDI or colligin but blocked recognition of the proteins by antibodies. This could easily explain the lack of PDI or colligin staining of punctate pXP124L-positive structures but did not explain the loss of reticular PDI or colligin stain from areas of the ER negative for pXP124L. One possibility was that pXP124L induced redistribution of PDI and colligin from the ER into the structures that stained brightly for the viral protein. To test this, the ER was labeled with the fluorescent protein ER-DsRed2. This protein contains ER targeting sequences derived from calreticulin and is naturally fluorescent, avoiding the need for antibody staining. Cells expressing ER-DsRed2 and pXP124L are shown in Fig. 6B and C. In the absence of pXP124L the DsRed2 localized to a reticular structure indicative of the ER (data not shown). Figure 6B shows the bright perinuclear staining for pXP124L. Two cells expressed XP124L, but only one was coexpressing ER-DsRed2. In this cell the localization of the ER-DsRed2 was identical to that of pXP124L. The bright inclusions induced by pXP124L therefore contain lumenal ER proteins, and the lack of peripheral reticular stain for DsRed2 showed that pXP124L redistributed the fluorescent lumenal ER marker protein into areas rich in pXP124L.
FIG. 6.
Effect of pXP124L on distribution of resident ER proteins. (A) Control Vero cells, cells infected with MVA-T7, or cells infected with MVA-T7 and transfected with XP124L were incubated in the presence of cycloheximide (200 μg/ml) for the indicated lengths of time (in hours). The levels of ER proteins and pXP124L were determined by Western blotting. Sizes of molecular weight markers are indicated on the right of each panel in kDa. (B to E) Vero cells transiently expressing ER-DsRed2 (B and C) or p58-YF (D and E) were infected with MVA-T7 and then transfected with a construct encoding wild-type XP124L. Cells were fixed with 4% paraformaldehyde and then probed with antibody R29 (pXP124L in panels B and D). Primary antibody was visualized with goat anti-rabbit antibody conjugated to Alexa-488 (B) or Alexa-594 (D). YFP and DsRed2 were observed through their natural fluorescence. Digital sections (0.2 μm) were captured at ×60 magnification and digitally deconvolved by using Openlab 2.1.3. Bars, 10 μm.
The observation that pXP124L located to post-ER-pre-Golgi compartments in infected cells raised the possibility that the punctate structures seen when pXP124L was expressed alone in cells represented an enlarged ERGIC compartment. This possibility was tested by coexpressing pXP124L with a fluorescently tagged ERGIC protein p58-YFP. Representative images are shown in Fig. 6D and E. The cell in the middle of the images is expressing both proteins, and it is clear that p58 and pXP124L reside in the same compartment. Taken together, the results suggest that pXP124L causes the redistribution of lumenal ER protein to an enlarged ERGIC compartment.
The KEDL sequence of pXP124L is necessary to cause redistribution of PDI to ERGIC membrane compartments.
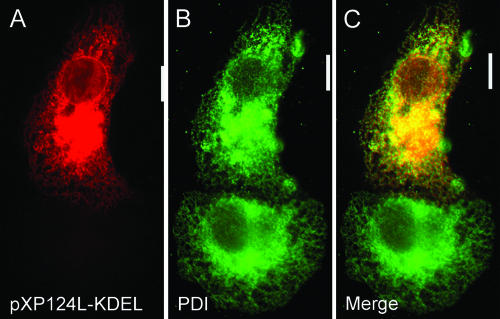
The redistribution of ER proteins caused by pXP124L could be nonspecific, resulting from overexpression of the protein in the ER, or it could be a specific effect mediated by the C-terminal KEDL sequence. The specific role played by the KEDL sequence was therefore tested by changing the KEDL sequence to the well-characterized KDEL ER retention motif. Figure 7 shows that this reversion of amino acids to KDEL changed the distribution of the viral protein. The perinuclear bright spots of fluorescence seen in Fig. 5A, C, and E were lost, and the viral protein was distributed to reticular structures throughout the cell (Fig. 7A) that colocalized with PDI (Fig. 7B and C). Overexpression of the protein in the ER did not therefore lead to redistribution of PDI. The bright pre-Golgi structures formed by pXP124L and the redistribution of lumenal ER proteins into these structures were dependent, precisely, on the reversal of the two central amino acids in the KDEL ER retention motif.
FIG. 7.
Expression of XP124L-KDEL. Cells expressing XP124L-KDEL were fixed with 4% paraformaldehyde and then stained with R29 (A) and antibody 1D3 (PDI) (B). Primary antibody was visualized with appropriate goat antibodies conjugated to Alexa-488 (B) or Alexa-594 (A). Digital sections (0.2 μm) were captured at ×60 magnification and digitally deconvolved by using Openlab 2.1.3. Bars, 10 μm.
Redistribution of ER proteins and expression of MGF110 in primary cells.
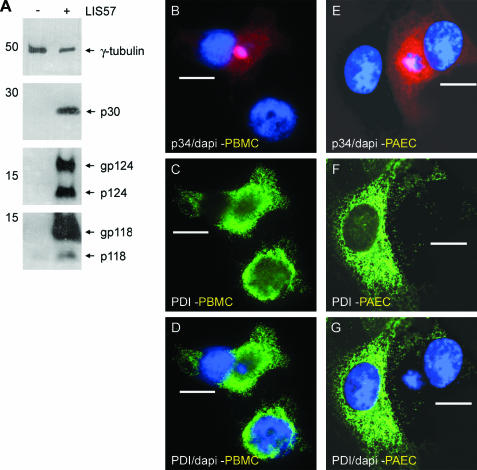
Interestingly, previous work has reported exclusion of PDI from virus assembly sites in monkey Vero cells infected with Ba71v, a tissue culture-adapted strain of ASFV (3). In the next experiment this ability of ASFV to modify the ER was tested in a more biological context by studying primary porcine macrophages and endothelial cells infected with LIS57 virulent isolate of ASFV. The LIS57 strain was chosen because sequence analysis had shown the presence of MFG110 Y118L and XP124L equivalents (13), and Western blot analysis demonstrated the expression of gene product in bone marrow macrophages infected with virus (Fig. 8A). Infected macrophages or endothelial cells were located by using an antibody for the viral matrix protein p34, and virus assembly sites were located by extranuclear DAPI staining of viral DNA (Fig. 8B and E). Inspection of Fig. 8C and D and of Fig. 8F and G shows that the area containing viral DNA was essentially free of PDI; in both macrophages and endothelial cells, similar results were obtained with antibody to ERp57, a second lumenal ER protein (data not shown). Significantly, as seen above for expression of XP124L alone in Vero cells (Fig. 5), infection of PAECs with LIS57 greatly diminished the immunofluorescence signal for PDI (Fig. 8F).
FIG. 8.
Expression of MGF110 proteins and redistribution of ER proteins in LIS57-infected cells. (A) Uninfected or LIS57-infected bone marrow cells were lysed and subjected to SDS-PAGE. Protein was transferred to nitrocellulose membranes, and proteins were detected by Western blotting. The sizes of molecular mass markers are indicated on the left of each panel in kilodaltons. (B to G) Porcine bone marrow cells (PBMC) (B to D) and PAECs (E to G) infected overnight with LIS57 were fixed with 4% paraformaldehyde and then stained with TW34 (B and E), 1D3 (C and F), and DAPI (B, D, E, and G). Primary antibody was visualized by using appropriate goat antibodies conjugated to Alexa-488 (C, D, F, and G) or Alexa-594 (B and E). Digital sections (0.2 μm) were captured at ×100 magnification and digitally deconvolved by using Openlab 3.1. Merged images of TW34 and DAPI staining (B and E) or 1D3 and DAPI staining (D and G) were created digitally. Bars, 10 μm.
DISCUSSION
We investigated the subcellular location of two ASFV MGF110 proteins that differ in the nature of a C-terminal ER retention motif. PY118L has the KDEL motif used by many cellular lumenal ER proteins, whereas pXP124L has a C-terminal KEDL sequence that has not been reported for ER proteins for higher eukaryotes. Metabolic labeling coupled with endo-H digestion showed that pY118L was a 14-kDa glycoprotein that was retained in the ER. This was confirmed by immunofluorescence analysis showing that pY118L colocalized with the resident lumenal ER protein colligin in a reticular network spreading throughout the cell. Surprisingly, the pXP124L protein with the KEDL sequence did not localize to reticular structures indicative of the ER but was found in heterogeneous vesicular structures concentrated to one side of the nucleus. Analysis of the N-linked sugars of pXP124L indicated a lack of Golgi processing, suggesting that pXP124L was located in pre-Golgi structures. Colocalization experiments with several ER and pre-Golgi markers showed pXP124L in a range of structures, including the ER, ER exit sites (mSec31), and ERGIC compartments containing the KDEL receptor. These results suggest that in infected cells pXP124L may recycle through ER and pre-Golgi compartments but is primarily localized in the ERGIC compartments rather than in the ER itself.
When pY118L or pXP124L was expressed alone in cells, the results were broadly similar to those seen in infected cells. PY118L localized to the ER and colocalized with PDI, whereas pXP124L was located in perinuclear vesicular structures. pXP124L remained sensitive to endo-H, again indicating retention in structures intermediate between the ER and the Golgi. Surprisingly, pXP124L produced a complete loss of immunofluorescence signal for PDI and colligin. The protein did not cause ER loss because a reticular ER network containing calnexin was retained in cells expressing XP124L. Instead, pXP124L caused a selective redistribution of lumenal ER proteins into an expanded perinuclear ERGIC compartment. Interestingly, the ability of pXP124L to redistribute ER proteins varied. The effect was most pronounced for PDI and colligin and much less evident for ERp57 and the integral membrane protein calnexin. The immunofluorescence signal for ERp57 and calnexin increased in perinuclear areas containing pXP124L, suggesting a degree of redistribution, but they were still present in a reticulum throughout the cell characteristic of the ER. The difference in the observed redistribution of ERp57 and PDI caused by pXP124L could be due to different C-terminal retention or retrieval sequences used by these proteins. ERp57 has a QEDL motif at its C terminus, whereas PDI and colligin have KDEL and RDEL sequences, respectively (38). Furthermore, retention of calnexin is independent of the KDEL receptor pathway and is mediated by COPI coats recruited to a dilysine motif in the cytoplasmic tail of the protein (26). The large structures formed by MVA-T7-mediated expression of XP124L may have been due to the large amount of protein produced when XP124L expression is driven by the MVA-encoded T7 polymerase system. This leads to an expansion of post-ER-pre-Golgi compartments and enhanced redistribution of PDI and colligin into these structures.
The ability of ASFV to cause redistribution of lumenal ER proteins in infected cells was tested by infecting primary PAECs and porcine macrophages with the virulent LIS57 strain of ASFV. A large-scale loss of PDI staining was seen in infected endothelial cells, and PDI was excluded from sites of replication in porcine macrophages. The left-hand region of LIS57 strain of ASFV encodes one Y118L and two XP124L homologues (Fig. 1), which were detected in infected cells by Western blotting. It is possible, therefore, that these MGF proteins cause the ER rearrangements seen in infected cells. Confirmation will, however, require the generation of recombinant viruses lacking these MGF proteins.
To ascertain whether the localization of the MGF110 proteins was dependent on the C-terminal motifs, the sequences were removed and the subcellular location of the proteins was tested. Surprisingly, removal of the KDEL sequence did not change the distribution of pY118L and both wild type, and the deletion mutant were retained in cells where they colocalized with lumenal ER proteins. These observations suggest pY118L is retained in the cell by domains of the protein other than the C-terminal KDEL sequence. Removal of the KEDL sequence from pXP124L, on the other hand, had a profound effect on the protein, leading to a loss of ERGIC targeting, secretion of approximately one-third of the protein, and redistribution of the remaining protein to the ER. The KEDL sequence therefore appears to exert a dominant role in targeting pXP124L to the ERGIC. This was further confirmed by converting the KEDL sequence to KDEL, whereupon pXP124L-KDEL protein failed to reach the ERGIC and was retained in the ER. Similar functions have been ascribed to two phenylalanine residues of ERGIC53, a protein that cycles between the ER and Golgi. The phenylalanine residues serve to export the protein from the ER, whereas two lysines confer ER retention (24). This combination of an export and retention signal are most likely responsible for the localization of ERGIC53 to the ERGIC at steady state.
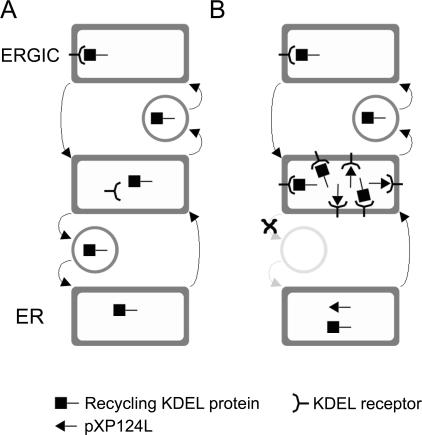
In light of the great variety of cellular proteins with C-terminal ER retention motifs (38), it is quite striking that so few use the KEDL motif. It is possible that the sequence is in some way incompatible with the KDEL receptor and that ASFV exploits this to modify the function of the early secretory pathway. If the KEDL sequence were to bind strongly or irreversibly to the KDEL receptor, XP124L expression could induce the redistribution of cellular ER proteins by perturbing the KDEL receptor recycling pathway (Fig. 9). It is possible that proteins that are more rapidly recycled through the ERGIC are affected more than those that are more actively retained within the ER. This would explain why lumenal ER proteins such as PDI and colligin are sequestered in an ERGIC compartment in cells expressing XP124L. Perhaps KEDL dissociates more slowly from the KDEL receptor than other retention motifs. This may trap the receptor and pXP124L, as well as resident ER proteins such as PDI and colligin, at the retrograde cargo-sorting stage in the ERGIC (29, 36, 49).
FIG. 9.
Possible mechanism for pXP124L induced redistribution of resident ER proteins. (A) In resting cells, escaped KDEL proteins are bound in the ERGIC by the KDEL receptor. They are then moved to areas specialized for concentrating retrograde cargo, where the receptor dissociates, and then returned to the ER in COPI-coated vesicles. (B) In cells overexpressing XP124L, KDEL proteins leave the ER normally, bind the KDEL receptor, but then become caught while returning to the ER, potentially by slow release of the KDEL bearing pXP124L from the KDEL receptor. The high levels of the viral protein may therefore inhibit the recycling of cellular proteins, and this would explain the colocalization of pXP124L with resident ER proteins in perinuclear regions of the cell.
How may exploitation of the KDEL recyling pathway be advantageous to ASFV? Although speculative, it is possible that the redistribution of resident ER proteins induced by XP124L expression plays a role in the envelopment of ASFV. Staining of bone marrow cells and endothelial cells showed that ASFV moves ER proteins away from areas of viral replication, and similar results have been obtained in Vero cells infected with Ba71v (3, 34). The redistribution of lumenal ER proteins by XP124L expression may implicate the protein in this part of the viral life cycle by preparing ER membranes for a role in virus envelopment. The number of MGF110 proteins encoded by different ASFV isolates varies, and adaptation to tissue culture results in attenuation of the virus and a reduction in the total number of MGF110 open reading frames in a given viral genome. Recent work has shown a dose-dependent effect linking MGF360 gene numbers to the ability of ASFV isolates to grow in porcine macrophages (64). There may be a similar link between expression of MGF110 family members and host range and virulence. Again speculative but nonetheless interesting is the possibility that the redistribution of resident ER proteins caused by these proteins could impair the ability of the ER to fold important cytokines and/or antigen presentation molecules, both of which are common targets for viral immune evasion (17, 32). There is no direct correlation at present between MGF110 copy number and host range or virulence. The LIS57 isolate, for example, has 12 MGF110 genes and Lisbon 60 has 5 (13), and both are virulent viruses (27, 28). Interestingly, the available sequence data show that there is always an MGF110 protein containing a KEDL motif present in the genome, even if the copy number of genes is variable. Indeed, of the three MGF110 genes identified in the Lisbon 60 Vero cell-adapted strain, two contain KEDL motifs, despite one of these (protein w2L) lacking a hydrophobic N terminus. It is possible, therefore, that the KDEL ER retention sequence at the C terminus of pY118L is a mutation of the viral KEDL-ERGIC targeting sequence rather than the other way round.
Acknowledgments
This study was supported by The Biotechnology and Biological Research Council and Department of Environment, Food, and Rural Affairs grant SE1509.
We thank Geoff Pero, Sheila Wilsden, Jenny Willgoss, Penny Powell, and Rebecca Herbert for providing tissue culture resources.
REFERENCES
- 1.Afonso, C. L., J. G. Neilan, G. F. Kutish, and D. L. Rock. 1996. An African swine fever virus bcl-2 homolog, 5-HL, suppresses apoptotic cell death. J. Virol. 70:4858-4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almendral, J. M., F. Almazán, R. Blasco, and E. Viñuela. 1990. Multigene families in African swine fever virus: family 110. J. Virol. 64:2064-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrés, G., R. García-Escudero, C. Simón-Mateo, and E. Viñuela. 1998. African swine fever virus is enveloped by a two-membraned collapsed cisterna derived from the endoplasmic reticulum. J. Virol. 72:8988-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beinert, H., R. H. Holm, and E. Münck. 1997. Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277:653-659. [DOI] [PubMed] [Google Scholar]
- 5.Benham, A. M., A. Cabibbo, A. Fassio, N. Bulleid, R. Sitia, and I. Braakman. 2000. The CXXCXXC motif determines the folding, structure and stability of human Ero1-Lα. EMBO J. 19:4493-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borca, M. V., G. F. Kutish, C. L. Afonso, P. Irusta, C. Carrillo, A. Brun, M. Sussman, and D. L. Rock. 1994. An African swine fever virus gene with similarity to the T-lymphocyte surface antigen CD2 mediates hemadsorption. Virology 199:463-468. [DOI] [PubMed] [Google Scholar]
- 7.Botija, C. S. 1962. Estudios sobre la peste porcina. Africana España Bull. Off. Int. Epiz. 58:707-727. [Google Scholar]
- 8.Botija, C. S. 1963. Reservorios del virus de la peste porcina Africana. Bull. Off. Int. Epiz. 60:890-895. [Google Scholar]
- 9.Brun, A., C. Rivas, M. Esteban, J. M. Escribano, and C. Alonso. 1996. African swine fever virus gene A179L, a viral homologue of bcl-2, protects cells from programmed cell death. Virology 225:227-230. [DOI] [PubMed] [Google Scholar]
- 10.Chacón, M. R., F. Almazán, M. L. Nogal, E. Viñuela, and J. F. Rodríguez. 1995. The African swine fever virus IAP homolog is a late structural polypeptide. Virology 214:670-674. [DOI] [PubMed] [Google Scholar]
- 11.Cobbold, C., J. T. Whittle, and T. Wileman. 1996. Involvement of the endoplasmic reticulum in the assembly and envelopment of African swine fever virus. J. Virol. 70:8382-8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colgrove, G. S., E. O. Haelterman, and L. Coggins. 1969. Pathogenesis of African swine fever in young pigs. Am. J. Vet. Res. 30:1343-1359. [PubMed] [Google Scholar]
- 13.de la Vega, I., E. Viñuela, and R. Blasco. 1990. Genetic variation and multigene families in African swine fever virus. Virology 179:234-246. [DOI] [PubMed] [Google Scholar]
- 14.Dixon, L. K., and P. J. Wilkinson. 1988. Genetic diversity of African swine fever virus isolates from soft ticks (Ornithodoros moubata) inhabiting warthog burrows in Zambia. J. Gen. Virol. 69:2981-2993. [DOI] [PubMed] [Google Scholar]
- 15.Dixon, L. K., S. R. F. Twigg, S. A. Baylis, S. Vydelingum, C. Bristow, J. M. Hammond, and G. L. Smith. 1994. Nucleotide sequence of a 55-kbp region from the right end of the genome of a pathogenic African swine fever virus isolate (Malawi LIL20/1). 75:1655-1684. [DOI] [PubMed]
- 16.Enjuanes, L., A. L. Carrascosa, M. A. Moreno, and E. Viñuela. 1976. Titration of African swine fever (ASF) virus. J. Gen. Virol. 32:471-477. [DOI] [PubMed] [Google Scholar]
- 17.Früh, K., A. Gruhler, R. M. Krishna, and G. J. Schoenhals. 1999. A comparison of viral immune escape strategies targeting the MHC class I assembly pathway. Immunol. Rev. 168:157-166. [DOI] [PubMed] [Google Scholar]
- 18.González, A., V. Calvo, F. Almazán, J. M. Almendral, J. C. Ramírez, I. de la Vega, R. Blasco, and E. Viñuela. 1990. Multigene families in African swine fever virus: family 360. J. Virol. 64:2073-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammond, A. T., and B. S. Glick. 2000. Dynamics of transitional endoplasmic reticulum sites in vertebrate cells. Mol. Biol. Cell 11:3013-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haugejorden, S. M., M. Srinivasan, and M. Green. 1991. Analysis of the retention signals of two resident lumenal endoplasmic reticulum proteins by in vitro mutagenesis. J. Biol. Chem. 266:6015-6018. [PubMed] [Google Scholar]
- 21.Heath, C. M., M. Windsor, and T. Wileman. 2001. Aggresomes resemble sites specialized for virus assembly. J. Cell Biol. 153:449-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hess, W. R. 1987. African swine fever virus in nature, p. 5-9. In Y. Becker (ed.), African swine fever. Martinus Nijhoff Publishing, Boston, Mass.
- 23.Johnson, M. K. 1998. Iron-sulfur proteins: new roles for old clusters. Curr. Opin. Chem. Biol. 2:173-181. [DOI] [PubMed] [Google Scholar]
- 24.Kappeler, F., D. R. C. Klopfenstein, M. Foguet, J.-P. Paccaud, and H.-P. Hauri. 1997. The recycling of ERGIC-53 in the early secretory pathway. J. Biol. Chem. 272:31801-31808. [DOI] [PubMed] [Google Scholar]
- 25.Kreitman, R. J., and I. Pastan. 1995. Importance of the glutamate residue of KDEL in increasing the cytotoxicity of Pseudomonas exotoxin derivatives and for increased binding to the KDEL receptor. Biochem. J. 307:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letourner, F., E. C. Gaynor, S. Hennecke, C. Démollière, R. Duden, S. D. Emr, H. Reizman, and P. Cosson. 1994. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell 79:1199-1207. [DOI] [PubMed] [Google Scholar]
- 27.Manso Ribeiro, J., J. A. Rosa Azevedo, M. J. O. Teixeira, M. C. Braço Forte, A. M. Rodrigues Ribeiro, F. Oliveira, E. Noronha, C. Grave Pereira, and J. Dias Vigario. 1958. Peste porcine provoquée par une souche différente (souche L) de la souche classique. Bull. Off. Int. Epiz. 50:516-534.
- 28.Manso Ribeiro, J., and J. Rosa Azevedo. 1961. Réapparition de la peste porcine Africaine (P.P.A.) au Portugal. Bull. Off. Int. Epiz. 55:88-106.
- 29.Martínez-Menárguez, J. A., H. J. Gueze, J. W. Slot, and J. Klumperman. 1999. Vesicular tubular clusters between the ER and Golgi mediate concentration of soluble secretory proteins by exclusion from COPI-coated vesicles. Cell 98:81-90. [DOI] [PubMed] [Google Scholar]
- 30.McCrossan, M., M. Windsor, S. Ponnambalam, J. Armstrong, and T. Wileman. 2001. The trans-Golgi network is lost from cells infected with African swine fever virus. J. Virol. 75:11755-11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miskin, J. E., C. C. Abrams, L. C. Goatley, and L. K. Dixon. 1998. A viral mechanism for inhibition of the cellular phosphatase calcineurin. Science 281:562-565. [DOI] [PubMed] [Google Scholar]
- 32.Mocarksi, E. S., Jr. 2002. Immunomodulation by cytomegaloviruses: manipulative strategies beyond evasion. Trends Microbiol. 10:332-339. [DOI] [PubMed] [Google Scholar]
- 33.Montgomery, R. E. 1921. On a form of swine fever occurring in British East Africa (Kenya colony). J. Comp. Pathol. Ther. 34:159-191.
- 34.Moura Nunes, J. F., J. D. Vigário, and A. M. Terrinha. 1975. Ultrastructural study of African swine fever virus replication in cultures of swine bone marrow cells. Arch. Virol. 49:59-66. [DOI] [PubMed] [Google Scholar]
- 35.Neilan, J. G., L. Zsak, Z. Lu, G. F. Kutish, C. L. Afonso, and D. L. Rock. 2002. Novel swine virulence determinant in the left variable region of the African swine fever virus genome. J. Virol. 76:3095-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orci, L., M. Stamnes, M. Ravazzola, M. Amherdt, A. Perrelet, T. H. Söllner, and J. E. Rothman. 1997. Bidirectional transport by distinct populations of COPI-coated vesicles. Cell 90:335-349. [DOI] [PubMed] [Google Scholar]
- 37.Ordas-Alvarez, A., and M. A. Marcotegui. 1987. African swine fever: clinical aspects, p. 11-20. In Y. Becker (ed.), African swine fever. Martinus Nijhoff Publishing, Boston, Mass.
- 38.Pelham, H. R. B. 1990. The retention signal for soluble proteins of the endoplasmic reticulum. Trends Biochem. Sci. 15:483-486. [DOI] [PubMed] [Google Scholar]
- 39.Pires, S., G. Riberio, and J. V. Costa. 1997. Sequence and organization of the left multigene family 110 region of the Vero-adapted L60V strain of African swine fever virus. Virus Genes 15:271-274. [DOI] [PubMed] [Google Scholar]
- 40.Plowright, W., J. Parker, and M. A. Pierce. 1969. The epizootiology of African swine fever virus in Africa. Vet. Rec. 85:668-675. [PubMed] [Google Scholar]
- 41.Plowright, W., G. R. Thomson, and J. A. Neser. 1994. African swine fever, p. 568-599. In J. A. W. Coetzer, G. R. Thomson, R. C. Tustin, and N. P. J. Kriek (ed.), Infectious diseases of livestock, with special reference to Southern Africa, vol. 1. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 42.Powell, P. P., L. K. Dixon, and R. M. E. Parkhouse. 1996. An IκB homolog encoded by African swine fever virus provides a novel mechanism for downregulation of proinflammatory cytokine responses in host macrophages. J. Virol. 70:8527-8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Preddie, E. C., and M. Saffran. 1965. Isolation of a large polypeptide from bovine posterior pituitary powder. J. Biol. Chem. 240:4189-4193. [PubMed] [Google Scholar]
- 44.Revilla, Y., A. Cebrián, E. Baixerás, C. Martínez-A., E. Viñuela, and M. L. Salas. 1997. Inhibition of apoptosis by the African swine fever virus Bcl-2 homologue: role of the BH1 domains. Virology 228:400-404. [DOI] [PubMed] [Google Scholar]
- 45.Rodríguez, F., A. Fernández, J. Pérez, J. Martín de las Mulas, M. A. Sierra, and A. Jover. 1996. African swine fever: morphopathology of a viral haemorrhagic disease. Vet. Rec. 139:249-254. [DOI] [PubMed] [Google Scholar]
- 46.Rodríguez, J. M., R. J. Yáñez, F. Almazán, E. Viñuela, and J. F. Rodriguez. 1993. African swine fever virus encodes a CD2 homolog responsible for the adhesion of erythrocytes to infected cells. J. Virol. 67:5312-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodríguez, J. M., R. J. Yáñez, R. Pan, J. F. Rodriguez, M. L. Salas, and E. Viñuela. 1994. Multigene families in African swine fever virus: family 505. J. Virol. 68:2746-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rouiller, I., S. M. Brookes, A. D. Hyatt, M. Windsor, and T. Wileman. 1998. African swine fever virus is wrapped by the endoplasmic reticulum. J. Virol. 72:2373-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scales, S. J., R. Pepperkok, and T. E. Kries. 1997. Visualization of ER-to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell 90:1137-1148. [DOI] [PubMed] [Google Scholar]
- 50.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 51.Sutter, G., M. Ohlmann, and V. Erfle. 1995. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 371:9-12. [DOI] [PubMed] [Google Scholar]
- 52.Tait, S. W. G., E. B. Reid, D. R. Greaves, T. E. Wileman, and P. P. Powell. 2000. Mechanism of inactivation of NF-κB by a viral homologue of IκBα. J. Biol. Chem. 275:34656-34664. [DOI] [PubMed] [Google Scholar]
- 53.Tang, B. L., F. Peter, J. Krijnse-Locker, S. H. Low, G. Griffiths, and W. Hong. 1997. The mammalian homolog of yeast sec13p is enriched in the intermediate compartment and is essential for protein transport from the endoplasmic reticulum to the Golgi apparatus. Mol. Cell. Biol. 17:256-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang, B. L., T. Zhang, D. Y. H. Low, E. T. Wong, H. Horstmann, and W. Hong. 2000. Mammalian homologues of yeast sec31p. J. Biol. Chem. 275:13597-13604. [DOI] [PubMed] [Google Scholar]
- 55.Thomson, G., M. Gainaru, A. Lewis, H. Biggs, E. Nevill, H. van der Pypekamp, L. Gerber, J. Esterhuysen, R. Bengis, D. Bezuidenhout, and J. Condy. 1983. The relationship between African swine fever virus, the warthog and Ornithodoros species in southern Africa, p. 85-100. In P. J. Wilkinson (ed.), Proceedings of CEC/FAO Research Seminar. Commission of the European Communities, Luxembourg, Belgium.
- 56.Tu, B. P., S. C. Ho-Schleyer, K. J. Travers, and J. S. Weissman. 2000. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science 290:1571-1574. [DOI] [PubMed] [Google Scholar]
- 57.Vallée, I., S. W. G. Tait, and P. P. Powell. 2001. African swine fever virus infection of porcine aortic endothelial cells leads to inhibition of inflammatory responses, activation of the thrombotic state, and apoptosis. J. Virol. 75:10372-10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaux, D., J. Tooze, and S. Fuller. 1990. Identification by anti-idiotype antibodies of an intracellular membrane protein that recognizes a mammalian endoplasmic reticulum retention signal. Nature 345:495-502. [DOI] [PubMed] [Google Scholar]
- 59.Vydelingum, S., S. A. Baylis, C. Bristow, G. L. Smith, and L. K. Dixon. 1993. Duplicated genes within the variable right end of the genome of a pathogenic isolate of African swine fever virus. J. Gen. Virol. 74:2125-2130. [DOI] [PubMed] [Google Scholar]
- 60.Ward, T. H., R. S. Polishchuk, S. Caplan, K. Hirschberg, and J. Lippincott-Schwartz. 2001. Maintenance of Golgi structure and function depends on the integrity of ER export. J. Cell Biol. 155:557-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yáñez, R. J., J. M. Rodríguez, M. L. Nogal, L. Yuste, C. Enríquez, J. F. Rodríguez, and E. Viñuela. 1995. Analysis of the complete nucleotide sequence of African swine fever virus. Virology 208:259-278. [DOI] [PubMed] [Google Scholar]
- 62.Young, J., L. P. Kane, M. Exley, and T. Wileman. 1993. Regulation of selective protein degradation in the endoplasmic reticulum by redox potential. J. Biol. Chem. 268:19810-19818. [PubMed] [Google Scholar]
- 63.Yowza, T., G. F. Kutish, C. L. Afonso, Z. Lu, and D. L. Rock. 1994. Two novel multigene families, 530 and 300, in the terminal variable regions of African swine fever virus genome. Virology 202:997-1002. [DOI] [PubMed] [Google Scholar]
- 64.Zsak, L., Z. Lu, T. G. Burrage, J. G. Neilan, G. F. Kutish, D. M. Moore, and D. L. Rock. 2001. African swine fever virus multigene family 360 and 530 genes are novel macrophage host range determinants. J. Virol. 75:3066-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]