Abstract
The lipid raft membrane has been shown to be the site of hepatitis C virus (HCV) RNA replication. The mechanism of formation of the replication complex is not clear. We show here that the formation of the HCV RNA replication complex on lipid raft (detergent-resistant membranes) requires interactions among the HCV nonstructural (NS) proteins and may be initiated by the precursor of NS4B, which has the intrinsic property of anchoring to lipid raft membrane. In hepatocyte cell lines containing an HCV RNA replicon, most of the other NS proteins, including NS5A, NS5B, and NS3, were also localized to the detergent-resistant membranes. However, when individually expressed, only NS4B was associated exclusively with lipid raft. In contrast, NS5B and NS3 were localized to detergent-sensitive membrane and cytosolic fractions, respectively. NS5A was localized to both detergent-sensitive and -resistant membrane fractions. Furthermore, we show that a cellular vesicle membrane transport protein named hVAP-33 (the human homologue of the 33-kDa vesicle-associated membrane protein-associated protein), which binds to both NS5A and NS5B, plays a critical role in the formation of HCV replication complex. The hVAP-33 protein is partially associated with the detergent-resistant membrane fraction. The expression of dominant-negative mutants and small interfering RNA of hVAP-33 in HCV replicon cells resulted in the relocation of NS5B from detergent-resistant to detergent-sensitive membranes. Correspondingly, the amounts of both HCV RNA and proteins in the cells were reduced, indicating that hVAP-33 is critical for the formation of HCV replication complex and RNA replication. These results indicate that protein-protein interactions among the various HCV NS proteins and hVAP-33 are important for the formation of HCV replication complex.
Detergent-insoluble cholesterol- and sphingolipid-rich microdomains, known as lipid rafts, of cellular membranes play important roles in signal transduction (29), protein sorting (12), and the budding and assembly of enveloped viruses (1, 19, 21, 24). Human immunodeficiency virus type 1 (HIV-1) particles contain a high concentration of cholesterol and sphingomyelin (21, 33). Influenza virus also contains a large amount of detergent-insoluble complex and selects ordered lipid domains during its budding from the plasma membrane (24). Influenza virus hemagglutinin (HA) protein interacts with lipid raft directly via its transmembrane domain (25). Our recent studies indicate that lipid raft is also involved in the formation of hepatitis C virus (HCV) RNA replication complex (27).
HCV is the causative pathogen of non-A, non-B hepatitis. The positive-sense, single-stranded 9.6-kb RNA genome encodes a polypeptide of 3,010 to 3,030 amino acids that is processed by host and viral proteases into 10 structural and nonstructural (NS) proteins (8, 18). Most of the NS proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) of HCV were associated with the endoplasmic reticulum (ER) or other subcellular membranes when these proteins were expressed individually or as a polyprotein (10, 11, 20, 31). However, the newly synthesized HCV RNA and most of the NS proteins in the cells supporting HCV replicons were also localized to distinct speckle-like structures, which consist of detergent-resistant membrane resembling lipid raft (27). It is not clear how the NS proteins are assembled from the ER membranes to form a complex on these lipid raft membranes.
Previously, we have shown that HCV NS5B and NS5A proteins bind to two different domains of a vesicle membrane protein hVAP-33 (31), which shares characteristics with proteins of the syntaxin family (14). It is an integral membrane protein with an immunoglobulin G-like domain at the N terminus, a coiled-coil domain, and a hydrophobic transmembrane domain at the C terminus. A wide range of proteins involved in membrane fusion, including VAMP1 and VAMP2 (34, 35), interact with hVAP-33. Recent studies have shown that at least several soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor (SNARE) proteins, such as syntaxin 1A, syntaxin 3, and VAMP2, are lipid raft associated (4, 12, 13). hVAP-33 also interacts with the oxysterol-binding protein (36), which binds to phosphatidylinositol and oxysterol ligands and mediates the effects of oxysterols on cholesterol homeostasis (30). Therefore, it is highly likely that hVAP-33 may play a role in the assembly of the HCV replication complex on cholesterol-rich lipid raft.
Here we analyzed the mechanism of lipid raft association of HCV NS proteins. Membrane flotation analysis demonstrated that although the individual NS proteins were localized in the different membrane or cytosolic fractions, the NS proteins in the context of the whole complement of NS proteins (NS3-NS5) or in the HCV replicon cells were localized mostly on detergent-resistant membranes. Dominant-negative mutants or small interfering RNA (siRNA) of hVAP-33 could inhibit these associations and correspondingly inhibit HCV RNA replication. These studies therefore identified a critical vesicle membrane protein in HCV replication and suggested a possible mechanism of the formation of HCV RNA replication complex.
MATERIALS AND METHODS
Cell lines, constructs, and transfection.
An HCV subgenomic replicon derived from plasmid 1bneo/delS (9) was kindly provided by C. Seeger of Fox Chase Cancer Center, Philadelphia, Pa. A stable cell line Huh7-replicon, in which active HCV RNA replication takes place, was selected from G418-resistant Huh7 cell colonies after electroporation with the RNA transcribed from plasmid 1bneo/delS. Cells were maintained in Dulbecco modified Eagle medium with 10% fetal bovine serum and 0.5 mg of G418/ml.
An adenovirus vector with deletions in the E1 and E3 regions was used to generate the construct expressing the HCV NS3 to NS5 proteins by using the published procedure (17). The HCV cDNA encoding NS3, NS4A, NS4B, NS5A, and NS5B proteins was cloned from a genotype 1b isolate (pCV-J4L6S [37]) by amplifying the entire NS3 to 3′-untranslated region (UTR) region by using the Expand High-Fidelity System (Roche) and cloning it into a NotI- and XbaI-digested pAdTrack-CMV shuttle vector (Q. Biogene). A control adenovirus was made from the empty pAdTrack-CMV. The resultant constructs were linearized with PmeI and cotransformed with the adenovirus backbone vector pAdeasy-1 (Q. Biogene) into Escherichia coli BJ5183 by electroporation to achieve recombination. The adenovirus recombinants were amplified in E. coli DH10B and transfected into human embryonic kidney (HEK-293) cells by using Lipofectamine (Invitrogen). Recombinant adenoviruses were harvested from cells after three cycles of freezing and thawing at day 7 posttransfection and further propagated by serial passages in HEK-293 cells.
Plasmids pcDNA3.1-NS5B and pcDNA3-NS5A were used to express HCV NS5B and NS5A proteins in Huh7 cells, as previously described (31). The PCR-generated NS4B fragment (genotype 1a, isolate H77) with or without a Flag sequence at the N terminus was cloned into BamHI site of pcDNA3.1 (Invitrogen). The PCR-generated NS3 fragment (genotype 1b) was cloned into XhoI site of a eukaryotic vector pCAGGS (22). Plasmid pcDNA3.1-Flag-hVAP-33 was used to express Flag-tagged hVAP-33 as previously described (31). The PCR-generated truncated hVAP-33 fragments were cloned into BamHI and EcoRI sites of pcDNA3.1.
Huh7 cells on six-well plates were transfected with 2 μg of DNA by using FuGENE 6 transfection reagent (Roche). The siRNA against hVAP-33, 5′-AAAGTGAAGACTACAGCACCT-3′, and the scrambled siRNA were purchased from Dharmacon Research, Inc. (Lafayette, Colo.), and transfected with Lipofectamine2000 (Invitrogen) according to the manufacturer's protocol. Huh7-replicon cells were incubated in Dulbecco modified Eagle medium with 10% fetal bovine serum at 1 day posttransfection.
Antibodies, immunoprecipitation, and immunoblotting.
The primary antibodies used in the present study included a monoclonal anti-NS5B antibody as described previously (6), a monoclonal anti-NS5A antibody (Biodesign International, Saco, Maine), a monoclonal anti-NS3 antibody (clone MMM33; Novocastra Laboratories), a monoclonal anti-tubulin antibody (Sigma), a monoclonal anti-caveolin-2 antibody (New England Biolabs), a monoclonal anti-GRP78 antibody (Stressgen), and polyclonal rat sera against hVAP-33 (31). An HCV patient serum (31) was also used to detect NS5A, NS5B, and NS4B proteins. Secondary antibodies were respective peroxidase-conjugated antibodies (American Qualex). For immunoprecipitation studies, the transfected cells were harvested at 48 h posttransfection and washed with cold phosphate-buffered saline (PBS). The samples were then collected in 1 ml of radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 12 mM deoxycholate sodium salt, 0.1% sodium dodecyl sulfate [SDS], 1% Triton X-100, 0.2 mM sodium vanadate, 1 mM phenylmethylsulfonyl fluoride). After 10 passages of the cells through a 25-gauge needle, the cell lysates were centrifuged at maximum speed in a microfuge for 5 min at 4°C, and the supernatant was collected. Immunoprecipitation was performed at 4°C in TM10 buffer (50 mM Tris-HCl [pH 7.9], 100 mM KCl, 12.5 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride). Cell lysates (50 μl) were incubated with 3 μl of anti-Flag M2-agarose affinity gel (Sigma) in a total volume of 300 μl of reaction buffer. After incubation overnight, beads were washed four times with TM10 buffer. The precipitates were then boiled for 5 min in Laemmli sample buffer and run on a 10% polyacrylamide gel. Proteins were transferred to a nitrocellulose membrane (Hybond-ECL plus) and analyzed by an enhanced chemiluminescence detection method (ECL Plus; Amersham).
Immunofluorescence staining.
Transfected cells were fixed with 4% formaldehyde in PBS and permeabilized by 0.1% Triton X-100. The cells were then incubated with the primary antibody and TRITC (tetramethyl rhodamine isothiocyanate)-conjugated secondary antibody.
Membrane flotation assay.
The membrane flotation assay was performed as previously described (28). The cells were first lysed in 0.5 ml of hypotonic buffer (10 mM Tris-HCl [pH 7.5], 10 mM KCl, 5 mM MgCl2) and passed through a 25-gauge needle 20 times. The nuclei and unbroken cells were removed by centrifugation at 1,000 × g for 5 min in a microfuge at 4°C. The cell lysates were then mixed with 3 ml of 72% sucrose in low-salt buffer (LSB; 50 mM Tris-HCl [pH 7.5], 25 mM KCl, 5 mM MgCl2) and overlaid with 4 ml of 55% sucrose in LSB, followed by 1.5 ml of 10% sucrose in LSB. To identify detergent-resistant membrane, the cell lysates were treated with 1% NP-40 on ice or 1% Triton X-100 at 37°C for 1 h before being loaded onto a sucrose gradient. The sucrose gradient was centrifuged at 38,000 rpm in a Beckman SW41Ti rotor for 14 h at 4°C. One-milliliter fractions were taken from the top of the gradient, and each was concentrated by passage through a Centricon YM-30 filter unit (Millipore, Bedford, Mass.). The sample was resuspended in SDS sample buffer and analyzed on a 4 to 15% gradient polyacrylamide gel containing SDS (Bio-Rad Laboratories, Hercules, Calif.).
Northern blot analysis.
Total cellular RNA was extracted with TRIzol reagent (Gibco-BRL). Ten micrograms of total RNA was denatured by glyoxal-dimethyl sulfoxide treatment, electrophoresed through a 1% agarose gel in 10 mM sodium phosphate (pH 7.0), transferred to a nylon membrane, and immobilized by UV cross-linking. RNA was glyoxalated before being electrophoresed on the gel. An in vitro-transcribed, 32P-labeled negative-strand 3′-UTR RNA was used as a probe to detect positive-strand HCV replicon RNA. A plasmid containing the GAPDH (glyceraldehyde-3-phosphate dehydrogenase)-coding sequence was used to generate 32P-labeled negative-strand GAPDH RNA by in vitro transcription. The membrane was hybridized in Rapid-Hyb buffer (Amersham Pharmacia Biotech) for 2 h at 70°C, washed once in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS for 20 min at room temperature, twice in 1× SSC-0.1% SDS, and then twice in 0.1× SSC-0.1% SDS for 20 min at 65°C. The dried membrane was then exposed to X-ray film.
RESULTS
Conditional association of HCV NS proteins with detergent-resistant membrane fractions.
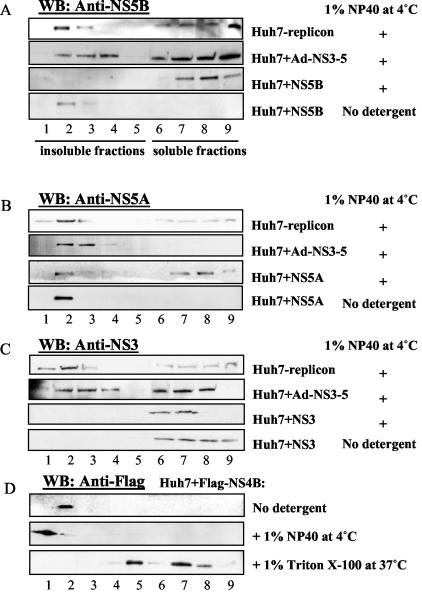
To examine the distribution of the components of HCV replication complex, we performed membrane flotation analysis of a human hepatocyte cell line harboring an HCV RNA replicon. The cellular lysates were separated on 62-55-10% discontinuous sucrose gradients after solubilization of cells with cold 1% NP-40, a mild nonionic detergent. Under this condition, insoluble membrane proteins and lipid floated to the interface between the 55 and 10% sucrose layers, whereas cytosolic proteins and solubilized membrane proteins remained in the 62% sucrose layer. Similar to the previous report (27), a substantial proportion of NS5B and NS5A proteins was found to be associated with detergent-resistant membrane fractions (Fig. 1A and B, Huh7-replicon, fractions 1 to 5), whereas an ER marker protein, GRP78, was released to the cytosol under the same condition (28) (data not shown). The majority of NS3 protein (Fig. 1C, Huh7-replicon) was also recovered with insoluble membrane fractions (fractions 1 to 3). A similar distribution of NS proteins was observed when NS3 to NS5B proteins were expressed as polyproteins from a recombinant adenovirus (Fig. 1A to C, Huh7+Ad-NS3-5), except that the relative proportions of the various NS proteins in the detergent-resistant membranes were lower than those in the Huh7-replicon cells. In contrast, when NS proteins were expressed individually in Huh7 cells, the distributions of the individual NS proteins were significantly different. All of the NS3 was in the cytosolic fractions even without the NP-40 treatment (Fig. 1C, Huh7+NS3, fractions 6 to 9). NS5B was localized exclusively in the membrane fractions that were solubilized by the detergent (Fig. 1A, Huh7+NS5B); NS5A was present in both the detergent-resistant and the detergent-sensitive membrane fractions (Fig. 1B, Huh7+NS5A). In contrast, when a Flag-tagged NS4B was transfected alone in Huh7 cells, NS4B was completely recovered in the detergent-resistant membrane fractions (fractions 1 and 2 [Fig. 1D]). Characteristic of all of the lipid raft proteins, NS4B was almost completely solubilized by incubation with the detergent TX-100 at 37°C (Fig. 1D). These data together indicated that NS4B is the only HCV NS protein that has the intrinsic ability to associate with lipid raft. All of the other NS proteins become associated with the detergent-resistant membranes only in the presence of the complete complement of NS proteins.
FIG. 1.
Association of HCV NS proteins with detergent-resistant membrane fractions. Huh7-replicon cells were harvested at 48 h after seeding. Huh7 cells were harvested 48 h after transfection with plasmids expressing individual NS proteins or infection with a recombinant adenovirus expressing NS3-NS5. Cell lysates were treated with 1% NP-40 for 1 h on ice or with 1% Triton X-100 for 1 h at 37°C and then fractionated by discontinuous sucrose gradient centrifugation as described in Materials and Methods. One-milliliter fractions were collected from top to bottom of the sucrose gradient, concentrated in YM-30 Centricons, and analyzed on polyacrylamide gels. The proteins were detected by immunoblotting with monoclonal antibodies to NS5A, NS5B, NS3, or Flag.
NS4B formed a complex with NS5A, NS5B, and hVAP-33.
Since NS4B is the only NS protein that, by itself, is localized to the detergent-resistant membrane fraction, it is likely to be the key protein that forms the core of the HCV replication complex. Thus, we investigated how NS4B interacted with the other NS proteins and cellular proteins.
We first studied the possible interaction between NS4B and NS5A. E. coli-expressed glutathione S-transferase (GST)-NS4B fusion protein was incubated with in vitro-translated, [35S]methionine-labeled NS5A and then pulled down with glutathione beads. The result showed that NS5A was pulled down together with GST-NS4B (Fig. 2A, lane 3) under the in vitro binding conditions. In contrast, NS5A did not bind GST protein under the same conditions (Fig. 2A, lane 4). As previously shown (31), NS5A interacted with GST-hVAP-33 (lane 2). We further confirmed the interaction between NS4B and NS5A by in vivo coimmunoprecipitation. NS5A was precipitated by anti-Flag antibody when it was coexpressed with Flag-tagged NS4B (Fig. 2B, lanes 1 and 2) but not in its absence (Fig. 2B, lanes 3 and 4). Furthermore, when NS5B and NS5A were cotransfected with Flag-tagged NS4B, both NS5A and NS5B were precipitated by anti-Flag antibody (lanes 5 and 6). In contrast, when NS4B and Flag-tagged NS5B were coexpressed, NS4B could not be coimmunoprecipitated by an anti-Flag antibody (Fig. 2B, lanes 7 and 8). These results combined suggest that there was no direct interaction between NS4B and NS5B; however, NS4B and NS5B could form a complex through direct or indirect NS4B-NS5A-NS5B interactions in vivo.
FIG. 2.
In vitro and in vivo coimmunoprecipitation of various NS proteins and hVAP-33. (A) In vitro GST pull-down assay. In vitro-translated [35S]methionine-labeled NS5A was mixed with various GST fusion proteins. The precipitated NS5A protein was detected by autoradiography. (B and C) Various Flag-tagged and untagged constructs were cotransfected into Huh7 cells. At 48 h posttransfection, cell lysates were immunoprecipitated with anti-Flag-antibody cross-linked Sepharose-4B beads. An HCV patient serum was used to detect NS5A, NS5B, and NS4B by immunoblotting. Lanes 1, 3, 5, and 7 represent 10% of lysates prior to immunoprecipitation.
We next examined whether other components were necessary for the formation of the NS4B-NS5A-NS5B complex. Since NS5B binds to hVAP-33 (31), which, in turn, binds to NS5A (31) (Fig. 2A), we examined whether hVAP-33 was involved in the formation of such a complex. First, NS4B was cotransfected with Flag-hVAP-33 into Huh7 cells; the lysates were precipitated with anti-Flag antibody. The precipitates were then detected by an HCV patient's serum that recognized most of the HCV NS proteins. NS4B could not be precipitated by anti-Flag antibody, indicating that there is no direct interaction between NS4B and hVAP-33 (Fig. 2C, lanes 1 and 2). However, when Flag-tagged hVAP-33, NS5A, and NS4B were coexpressed, NS4B was coimmunoprecipitated by Flag-tagged hVAP-33 (Fig. 2C, lanes 3 and 4). The patient serum used in this immunoblot also recognized several background bands that might represent partially processed NS proteins or their degradation products (e.g., from NS5A). In contrast, when NS5B, NS4B, and Flag-hVAP-33 were coexpressed, NS4B could not be coprecipitated with Flag-hVAP-33 (lanes 5 and 6). These results combined indicate that NS5B did not interact directly with NS4B and that NS4B did not interact directly with hVAP-33. However, NS5B, NS5A, and NS4B could form a complex with hVAP-33 through a series of protein-protein interactions. This conclusion is consistent with our previous finding that hVAP-33 interacts with both NS5A and NS5B (31).
hVAP-33 is partially associated with detergent-resistant membrane fractions.
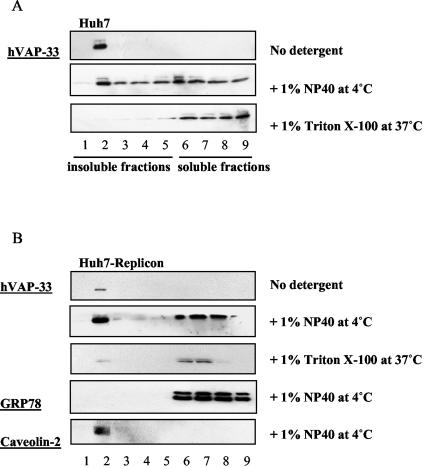
The results shown above suggested that hVAP-33 may be a component of the HCV replication complex. We therefore examined whether hVAP-33 was also associated with detergent-resistant membranes. Figure 3A shows that in Huh7 cells, in the absence of detergent treatment, hVAP-33 was recovered in membrane fraction 2 only. When the cell lysates were preincubated with cold 1% NP-40 at 4°C for an hour, about half of hVAP-33 remained in membrane fractions (fractions 2 to 5), whereas the other half became soluble and retained in fractions 6 to 9. All of the hVAP-33 became soluble after treatment with Triton X-100 at 37°C. These results indicate that part of hVAP-33 is associated with lipid raft membrane. Similar distribution of hVAP-33 was seen in Huh7-replicon cells (Fig. 3B). An ER marker, GRP78, which is known not to associate with lipid raft, was completely solubilized by 1% NP-40 at 4°C. In contrast, caveolin-2, a lipid raft membrane protein (27, 29), was completely resistant to the detergent treatment at 4°C. These data suggest that hVAP-33 is present in both detergent-resistant and detergent-sensitive membranes and that its distribution is not affected by the HCV proteins. Combined with the observations that hVAP-33 binds to NS5A and NS5B, these findings strongly suggest that hVAP-33 is a component of the HCV replication complex on lipid raft membrane.
FIG. 3.
Association of hVAP-33 protein with detergent-resistant membrane fractions. (A) Huh7 cells and (B) Huh7-replicon cells were harvested at 48 h after seeding. Cell lysates were subjected to various treatments as indicated and fractionated by discontinuous sucrose gradient centrifugation as described in Materials and Methods. Endogenous hVAP-33 was detected by immunoblotting with a polyclonal antibody.
siRNA of hVAP-33 inhibited the association of HCV NS proteins with detergent-resistant membrane fractions.
To establish the importance of hVAP-33 in the formation of the HCV replication complex, we first examined the effects of an siRNA against hVAP-33 in Huh7-replicon cells. The protein expression level was determined by Western blotting at 3 days posttransfection. The expression level of hVAP-33 was significantly reduced by the siRNA against hVAP-33 but not by the scrambled siRNA (Fig. 4A). As a control, the expression level of tubulin was not affected. Significantly, the level of NS5A was reduced, suggesting that HCV replication was reduced in the presence of the siRNA against hVAP-33 (Fig. 4A). To rule out the possibility that the siRNA might have induced nonspecific cytotoxic effects, we studied the growth rate of the transfected cells in the absence of G418. No differences in growth rate or percentage of cell death between the cells transfected with hVAP-33 siRNA and with scrambled siRNA were observed (data not shown), indicating that the siRNAs did not exert nonspecific cytotoxic effects. We then examined whether the distribution of NS proteins was affected by the hVAP-33-specific siRNA. As shown in Fig. 4A, the overall levels of HCV NS proteins were lower in the cells transfected with hVAP-33 siRNA. Therefore, the films for the samples that expressed hVAP-33 siRNA were exposed longer than their control samples. Figure 4B shows that most of NS5B was recovered in detergent-soluble fractions 6 to 9 in the cells expressing the hVAP-33-specific siRNA. In contrast, the distributions of NS5A and NS3 were not significantly affected (Fig. 4B). These results indicate that hVAP-33 is important for the association of NS5B with lipid raft membrane, whereas NS3 and NS5A rely on other factors for this association.
FIG. 4.
Effects of siRNA of hVAP-33 on the association of HCV NS proteins with detergent-resistant membrane fractions. (A) Immunoblotting analysis of protein expression level after transfection of siRNA against hVAP-33 or control siRNA in Huh7-replicon cells at 3 days posttransfection. Thirty micrograms of protein was loaded into each lane. Tubulin was detected as a loading control. Lanes: (−), untransfected; H2O, mock transfected. (B) Membrane flotation analysis. Huh7-replicon cells transfected with siRNA of hVAP-33 or control siRNA were harvested at 3 days posttransfection. Cell lysates were treated with 1% NP-40 for 1 h on ice and then fractionated by discontinuous sucrose gradient centrifugation, as detailed in Materials and Methods. NS proteins were analyzed by immunoblotting with individual monoclonal antibodies.
Dominant-negative mutants of hVAP-33 inhibited the association of HCV NS proteins with insoluble membrane fractions and reduced the expression level of NS5A protein and HCV RNA in Huh7-replicon cells.
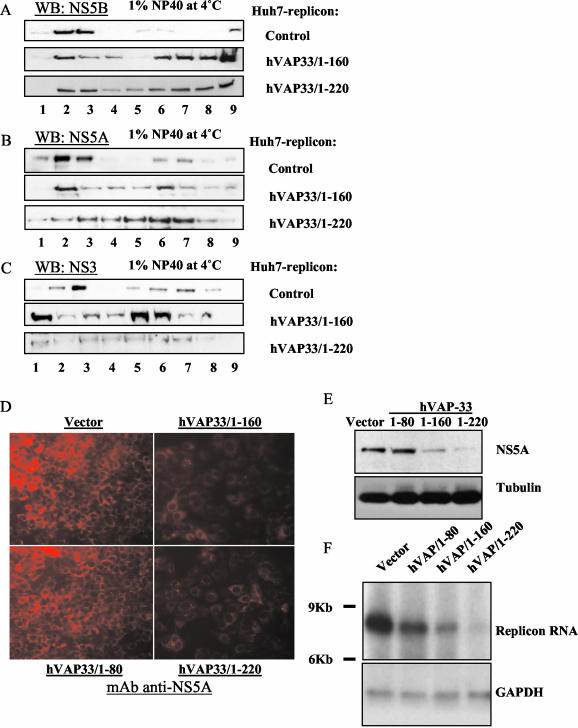
We further confirmed the importance of hVAP-33 in the formation of the HCV replication complex by examining the effects of the truncated mutants of hVAP-33 (as potential dominant-negative mutants) on the distributions of NS proteins. Plasmid pcDNA3.1-hVAP/1-220 encodes a truncated hVAP-33 that lacks the C-terminal 22 amino acids, which constitutes the transmembrane domain (31). Another truncated mutant hVAP-33/1-160 lacks both the coiled-coil region and the transmembrane domain. In the Huh7-replicon cells transfected with either one of the truncated hVAP-33 mutants, the relative proportions of NS5B, NS5A, and NS3 in detergent-soluble membrane fractions increased to various degrees (Fig. 5A to C). Thus, these truncated mutants served as dominant-negative mutants and inhibited the association of NS proteins with the detergent-resistant membrane, probably because they cannot integrate into vesicle membranes but retain the ability to interact with other cellular proteins associated with lipid rafts (31). The effects were particularly evident for NS5B and NS3, but less so for NS5A. Interestingly, hVAP-33/1-160, which did not bind NS5A (31), did not cause significant alteration of the association of NS5A with detergent-resistant membrane fractions, whereas it disrupted that of NS5B and NS3. Thus, the interaction between NS5A and hVAP-33 is important for the dominant-negative effects of these mutants. The overexpression of the full-length hVAP-33 did not have any effect on the distributions of NS proteins (data not shown). These results are generally consistent with the siRNA experiments (Fig. 4), except that the expression of the dominant-negative mutants of hVAP-33 had moderate effects on NS3 and NS5A, whereas the siRNA of hVAP-33 did not. Nevertheless, these data together suggested that hVAP-33 plays a role in the formation of the HCV NS protein complex.
FIG. 5.
Effects of tuncation mutants of hVAP-33 on association of HCV NS proteins with detergent-resistant membrane fractions. (A to C) Membrane flotation analysis. Huh7-replicon cells transfected with hVAP-33/1-160 or hVAP-33/1-220 were harvested at 3 days posttransfection. Cell lysates were treated with 1% NP-40 for 1 h on ice and then fractionated by discontinuous sucrose gradient centrifugation. (D) Immunofluorescence staining of NS5A in Huh7-replicon cells at 4 days posttransfection. Cells seeded on chamber slides were fixed in 4% formaldehyde-PBS, permeabilized by 0.1% Triton X-100-PBS, and immunostained with monoclonal antibodies to NS5A, followed by the addition of secondary antibodies conjugated with TRITC. Images were taken under fluorescence microscopy with the Zeiss program for the same length of exposure time for all panels. (E) Western blot analysis of protein expression in transfected Huh7-replicon cells at 4 days posttransfection. Thirty micrograms of postnuclear cell lysate from each transfection was separated by SDS-PAGE, transferred to nitrocellulose membrane, and probed with monoclonal antibody to NS5A. The same membrane was reprobed with anti-tubulin antibody as loading control. (F) Northern blot analysis of total RNA extracted from Huh7-replicon cells transfected with the various plasmids expressing truncated hVAP-33 or vector at 3 days posttransfection. Ten micrograms of total RNA from each plate was used. A specific RNA probe complementary to the HCV 5′-UTR RNA was used in hybridization. GADPH served as a loading control.
To further confirm that the association of NS proteins with detergent-resistant membranes was important for HCV replication, we monitored the expression levels of both NS proteins and viral RNA in Huh7-replicon cells transfected with hVAP-33/1-160 or hVAP-33/1-220. The expression of the hVAP-33 truncation mutants did not affect cell growth or cell death in the absence of G418 (data not shown), which was used to maintain HCV replicon. However, when the cells transfected with these mutants were grown in the presence of 0.5 mg of G418/ml, these cells grew more slowly than the control cells without hVAP-33 transfection, suggesting a reduced production of neomycin phosphotransferase that was encoded from the HCV replicon RNA. Figure 5D shows that immunofluorescence staining of NS5A protein in the Huh7-replicon cells transfected with hVAP-33/1-160 or hVAP-33/1-220 was reduced compared to the cells transfected with hVAP-33/1-80, which expressed only the N-terminal 80 amino acids of hVAP-33 and did not interact with NS5A pr NS5B (31), or untransfected cells. The reduction of NS5A protein in these cells was probably a secondary effect of the reduced viral RNA replication, since it was not apparent until 3 to 4 days posttransfection. The hVAP-33/1-80 lacks the NS5A-interacting domain, which is localized within amino acids 81 to 160 (31); correspondingly, it did not affect the NS5A level. This result was confirmed by immunoblotting the NS5A protein (Fig. 5E). The level of tubulin served as a loading control.
To determine whether the reduced protein level correlated with the reduction of viral RNA replication, we performed Northern blot analysis of HCV RNA in these replicon cells at 3 days posttransfection. Figure 5F shows that the HCV RNA level was significantly lower in cells transfected with hVAP-33/1-160 and hVAP-33/1-220 than that in the cells transfected with the vector plasmid or hVAP/1-80. GAPDH RNA was measured as a loading control. These results confirmed that the disruption of the association of HCV NS proteins with detergent-resistant membrane correlated with the reduced viral RNA and NS protein synthesis.
DISCUSSION
In the present study we characterized the mechanism of formation of HCV replication complex on lipid raft (detergent-resistant) membrane. We showed the critical role of the interactions between the various viral NS proteins and between the NS proteins and a cellular membrane protein hVAP-33 in the formation of the HCV replication complex on detergent-resistant membranes. Lipid raft is typified by the plasma membrane caveolae, which contain caveolin-1 and caveolin-2. Caveolin-2 is a cofactor for caveola formation but is also localized on Golgi and could function in separate pathways (3). Considering that caveolae are absent in Huh7 cells (32) and that some caveolin-2 protein cofractionated with HCV NS proteins in the detergent-resistant membrane fraction (27) (Fig. 3B), these detergent-resistant membranes are most likely intracellular lipid raft, derived from the ER, Golgi, or trans-Golgi network. In addition to their association with the lipid raft, some of the NS proteins, including NS3, NS5A, and NS5B, appeared to be localized on the detergent-soluble membrane in the HCV replicon cells. Indeed, several previous reports suggested that the viral NS proteins are associated with the ER membranes (20, 23). However, these reports did not examine the detergent sensitivity of the membrane association of these proteins. Since many viral NS proteins, such as NS5A and NS3, perform multiple functions in viral replication and pathogenesis, they may be recruited to lipid raft only when participating in RNA replication. Indeed, the proportion of the NS proteins associated with detergent-resistant membrane was higher in the cells that have more active RNA replication (unpublished data). The NS proteins present in the detergent-soluble membranes (e.g., the ER) or elsewhere in the cells may be involved in other processes of viral replication, such as translation. Viral RNA molecules or the replicating RNA may also help spur the formation of the replication complex, since a higher proportion of NS proteins was associated with lipid raft in HCV replicon cells than in the cells in the absence of RNA replication (Fig. 1).
Some of the HCV NS proteins, such as NS5B and NS3, could not associate with lipid raft by themselves. Individually expressed NS3 remained in cytosolic fractions (Fig. 1C), whereas individually expressed NS5B was localized in detergent-soluble membranes. Only when they were expressed as a polyprotein did they become associated with detergent-resistant membrane, suggesting that the interactions between the HCV NS proteins are important for their lipid raft association. In the HCV replication cycle, the entire NS proteins are expressed as a polypeptide, which is then proteolytically processed by NS3-NS4A protease into individual proteins through a series of processing intermediates. The NS3-NS4A and NS5A-NS5B sites are rapidly processed, generating a more stable intermediate NS4AB-NS5A (23). Since NS4B by itself has the ability to associate with lipid raft (Fig. 1), the NS4AB-NS5A peptide is likely the peptide that initiates the formation of the replication complex on lipid raft membrane. This finding is consistent with the previous observation that NS4B by itself can induce a membrane web structure (5), which colocalized with the HCV replication complex (7). Since NS4A is a cofactor of NS3 (15, 16), NS4AB-NS5A may then recruit NS3 to lipid raft. Since NS5B is separated from NS4AB-NS5A relatively early during the polyprotein processing, the entry of NS5B into the replication complex will require additional factors. Our study shows that the cellular membrane protein hVAP-33, which interacts with both NS5A and NS5B, may perform such a role. This conclusion is supported by the finding that the relative distribution of NS5B between detergent-sensitive and detergent-resistant membranes was altered by the dominant-negative mutants and the siRNA of hVAP-33. hVAP-33 appears to play a minor role in the recruitment of NS3 and NS5A. In any case, the role of hVAP-33 in the formation of the HCV replication complex on lipid raft and in RNA replication was substantiated by the experiments with siRNA and dominant-negative mutants of hVAP-33 presented here.
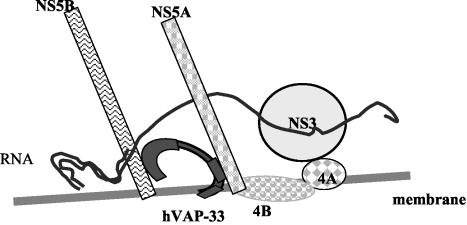
The model in Fig. 6 demonstrates the presumptive steps in the formation of HCV replication complex on lipid raft. We hypothesize that soon after the HCV polypeptide is translated, NS3 and NS5B are cleaved from the polypeptide rapidly; the uncleaved NS4A-NS4B-NS5A is recruited to lipid raft by the intrinsic lipid raft-anchoring property of NS4B. NS3 then becomes associated with lipid raft by binding to its cofactor NS4A. Both NS5A and NS5B bind to hVAP-33, which is present in both the detergent-resistant and the detergent-soluble membranes, probably at the site of their synthesis on the ER. Since hVAP-33 could be localized on, and potentially shuffled between, various types of membranes in the cell, NS5B and hVAP-33 are then recruited to join NS4A-NS4B-NS5A and NS3 on the lipid raft. Since the distribution of the various NS proteins in the replicon cells was slightly different from that in the cells expressing NS3-NS5 proteins only, a replication-competent HCV RNA may also play a role in forming this complex. This diagram is consistent with the known membrane topology of the NS proteins. NS5A is a type II membrane protein, with its N terminus as a membrane anchor (2). In contrast, NS5B is a type I membrane protein (26). The C terminus of hVAP-33 binds to the N terminus of NS5A, whereas the N terminus of hVAP-33 binds to the C terminus of NS5B (unpublished). Thus, the NS5A- and NS5B-interacting domains of hVAP-33 are likely close to the membrane. NS proteins likely are enclosed by lipid raft membrane as lipid rafts coalesce. This interpretation is consistent with the finding that the NS proteins and viral RNAs in the replication complex are resistant to protease and RNase, respectively (unpublished data).
FIG. 6.
Model for the mechanism of formation of HCV replication complex on lipid raft. After NS3 and NS5B are proteolytically cleaved from the polyprotein, the uncleaved NS4A-4B-5A is recruited to lipid raft by the intrinsic membrane-anchoring character of NS4B. NS3 becomes associated with lipid raft by binding to its cofactor NS4A. NS5B binds to hVAP-33 and moves to lipid raft and form a complex with NS4A-4B-5A via the interaction between NS5B and NS5A. The mechanism of entry of RNA into this complex is not known.
Finally, we reported here the association of hVAP-33 with both detergent-resistant and detergent-sensitive membranes. Several SNARE proteins, including the potential binding partners of hVAP-33, have been found to associate with lipid raft (4, 13, 14). The SNARE-associated lipid raft may play a role in organizing exocytotic pathways. Our finding provided an additional piece of evidence that hVAP-33 is present in specific membranes where its binding partners are localized.
Acknowledgments
We thank C. Seeger (Fox Chase Cancer Center) for providing HCV replicon1bneo/delS construct. We thank T.-Y. Hsieh for providing an NS4B construct.
This study was partially supported by NIH grants R01AI47348 and U19AI40038.
REFERENCES
- 1.Bavari, S., C. M. Bosio, E. Wiegand, G. Ruthel, A. B. Will, T. W. Geisbert, M. Hevey, C. Schmaljohn, A. Schmaljohn, and M. J. Aman. 2002. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 195:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. [DOI] [PubMed] [Google Scholar]
- 3.Breuza, L., S. Corby, J. P. Arsanto, M. H. Delgrossi, P. Scheiffele, and A. Le Bivic. 2002. The scaffolding domain of caveolin 2 is responsible for its Golgi localization in Caco-2 cells. J. Cell Sci. 115:4457-4467. [DOI] [PubMed] [Google Scholar]
- 4.Chamberlain, L. H., R. D. Burgoyne, and G. W. Gould. 2001. SNARE proteins are highly enriched in lipid rafts in PC12 cells: implications for the spatial control of exocytosis. Proc. Natl. Acad. Sci. USA 98:5619-5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao, L., H. Tu, S. T. Shi, K. J. Lee, M. Asanaka, S. B. Hwang, and M. M. Lai. 2003. Interaction with a ubiquitin-like protein enhances the ubiquitination and degradation of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 77:4149-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hijikata, M., H. Mizushima, Y. Tanji, Y. Komoda, Y. Hirowatari, T. Akagi, N. Kato, K. Kimura, and K. Shimotohno. 1993. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc. Natl. Acad. Sci. USA 90:10773-10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang, S. B., K. J. Park, Y. S. Kim, Y. C. Sung, and M. M. Lai. 1997. Hepatitis C virus NS5B protein is a membrane-associated phosphoprotein with a predominantly perinuclear localization. Virology 227:439-446. [DOI] [PubMed] [Google Scholar]
- 12.Lafont, F., P. Verkade, T. Galli, C. Wimmer, D. Louvard, and K. Simons. 1999. Raft association of SNAP receptors acting in apical trafficking in Madin-Darby canine kidney cells. Proc. Natl. Acad. Sci. USA 96:3734-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lang, T., D. Bruns, D. Wenzel, D. Riedel, P. Holroyd, C. Thiele, and R. Jahn. 2001. SNAREs are concentrated in cholesterol-dependent clusters that define docking and fusion sites for exocytosis. EMBO J. 20:2202-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lapierre, L. A., P. L. Tuma, J. Navarre, J. R. Goldenring, and J. M. Anderson. 1999. VAP-33 localizes to both an intracellular vesicle population and with occludin at the tight junction. J. Cell Sci. 112(Pt. 21):3723-3732. [DOI] [PubMed] [Google Scholar]
- 15.Lin, C., and C. M. Rice. 1995. The hepatitis C virus NS3 serine proteinase and NS4A cofactor: establishment of a cell-free trans-processing assay. Proc. Natl. Acad. Sci. USA 92:7622-7626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, C., J. A. Thomson, and C. M. Rice. 1995. A central region in the hepatitis C virus NS4A protein allows formation of an active NS3-NS4A serine proteinase complex in vivo and in vitro. J. Virol. 69:4373-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, Z. X., H. Nishida, J. W. He, M. M. Lai, N. Feng, and G. Dennert. 2002. Hepatitis C virus genotype 1b core protein does not exert immunomodulatory effects on virus-induced cellular immunity. J. Virol. 76:990-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohmann, V., J. O. Koch, and R. Bartenschlager. 1996. Processing pathways of the hepatitis C virus proteins. J. Hepatol. 24:11-19. [PubMed] [Google Scholar]
- 19.Manie, S. N., S. Debreyne, S. Vincent, and D. Gerlier. 2000. Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J. Virol. 74:305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mottola, G., G. Cardinali, A. Ceccacci, C. Trozzi, L. Bartholomew, M. R. Torrisi, E. Pedrazzini, S. Bonatti, and G. Migliaccio. 2002. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology 293:31-43. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen, D. H., and J. E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 23.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheiffele, P., A. Rietveld, T. Wilk, and K. Simons. 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274:2038-2044. [DOI] [PubMed] [Google Scholar]
- 25.Scheiffele, P., M. G. Roth, and K. Simons. 1997. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 16:5501-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt-Mende, J., E. Bieck, T. Hugle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276:44052-44063. [DOI] [PubMed] [Google Scholar]
- 27.Shi, S. T., K. J. Lee, H. Aizaki, S. B. Hwang, and M. M. Lai. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 77:4160-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi, S. T., S. J. Polyak, H. Tu, D. R. Taylor, D. R. Gretch, and M. M. Lai. 2002. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology 292:198-210. [DOI] [PubMed] [Google Scholar]
- 29.Smart, E. J., G. A. Graf, M. A. McNiven, W. C. Sessa, J. A. Engelman, P. E. Scherer, T. Okamoto, and M. P. Lisanti. 1999. Caveolins, liquid-ordered domains, and signal transduction. Mol. Cell. Biol. 19:7289-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor, F. R., S. E. Saucier, E. P. Shown, E. J. Parish, and A. A. Kandutsch. 1984. Correlation between oxysterol binding to a cytosolic binding protein and potency in the repression of hydroxymethylglutaryl coenzyme A reductase. J. Biol. Chem. 259:12382-12387. [PubMed] [Google Scholar]
- 31.Tu, H., L. Gao, S. T. Shi, D. R. Taylor, T. Yang, A. K. Mircheff, Y. Wen, A. E. Gorbalenya, S. B. Hwang, and M. M. Lai. 1999. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology 263:30-41. [DOI] [PubMed] [Google Scholar]
- 32.Vainio, S., S. Heino, J. E. Mansson, P. Fredman, E. Kuismanen, O. Vaarala, and E. Ikonen. 2002. Dynamic association of human insulin receptor with lipid rafts in cells lacking caveolae. EMBO Rep. 3:95-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, J. K., E. Kiyokawa, E. Verdin, and D. Trono. 2000. The Nef protein of HIV-1 associates with rafts and primes T cells for activation. Proc. Natl. Acad. Sci. USA 97:394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weir, M. L., A. Klip, and W. S. Trimble. 1998. Identification of a human homologue of the vesicle-associated membrane protein (VAMP)-associated protein of 33 kDa (VAP-33): a broadly expressed protein that binds to VAMP. Biochem. J. 333(Pt. 2):247-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weir, M. L., H. Xie, A. Klip, and W. S. Trimble. 2001. VAP-A binds promiscuously to both v- and tSNAREs. Biochem. Biophys. Res. Commun. 286:616-621. [DOI] [PubMed] [Google Scholar]
- 36.Wyles, J. P., C. R. McMaster, and N. D. Ridgway. 2002. Vesicle-associated membrane protein-associated protein-A (VAP-A) interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum. J. Biol. Chem. 277:29908-29918. [DOI] [PubMed] [Google Scholar]
- 37.Yanagi, M., M. St Claire, M. Shapiro, S. U. Emerson, R. H. Purcell, and J. Bukh. 1998. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology 244:161-172. [DOI] [PubMed] [Google Scholar]