Abstract
The efficiency and specificity of gene transfer with human adenovirus (hAd)-derived gene transfer vectors would be improved if the native viral tropism could be modified. Here, we demonstrate that the minor capsid protein IX (pIX), which is present in 240 copies in the Ad capsid, can be exploited as an anchor for heterologous polypeptides. Protein IX-deleted hAd5 vectors were propagated in hAd5 helper cells expressing pIX variants, with heterologous carboxyl-terminal extensions of up to 113 amino acids in length. The extensions evaluated consist of alpha-helical spacers up to 75 Å in length and to which peptide ligands were fused. The pIX variants were efficiently incorporated into the capsids of Ad particles. On intact particles, the MYC-tagged-pIX molecules were readily accessible to anti-MYC antibodies, as demonstrated by electron microscopic analyses of immunogold-labeled virus particles. The labeling efficiency improved with increasing spacer length, suggesting that the spacers lift and expose the ligand at the capsid surface. Furthermore, we found that the addition of an integrin-binding RGD motif to the pIX markedly stimulated the transduction of coxsackievirus group B and hAd receptor-deficient endothelioma cells, demonstrating the utility of pIX modification in gene transfer. Our data demonstrate that the minor capsid protein IX can be used as an anchor for the addition of polypeptide ligands to Ad particles.
To date, 51 human adenoviruses (hAds) serotypes classified into six distinct subgroups (or species), A to F, according to their genomic homologies and agglutination properties have been identified (15). The virions are composed of at least 11 different structural proteins and a linear double-stranded DNA genome of ca. 36,000 bp (42). Proteins II (hexon), III (penton base), IIIa, IV (fiber), VI, VIII, and IX form the icosahedral capsid (8, 45-47), while the other four structural proteins (V, VII, μ, and tp) are packaged with the DNA genome within the virus particle (VP) (12, 17, 42, 48). The fiber and penton base proteins bind to cellular receptors and determine the tropism (37). The initial step in infection is the binding of the primary cellular receptor by the globular knob of the fiber. The coxsackievirus group B and hAd receptor (CAR) has been identified as a primary receptor for subgroup A, C, D, E, and F hAd (6, 40). In addition, the major histocompatibility complex class I α2 subunit and sialic acid-containing proteins have been implicated as receptors for hAd5 (subgroup C) and hAd37 (subgroup D), respectively (2). After binding of the primary receptor by the fiber, the penton base recruits αvβ integrins via an RGD motif, thereby facilitating internalization of the virion via receptor-mediated endocytosis (27, 37).
In the previous years, hAds have attracted considerable attention as vectors for in vitro and in vivo delivery of heterologous genes. However, several cell types are relatively refractory to infection by the subgroup C-derived hAd vectors due to the paucity or lack of the cellular hAd receptors on these cells (e.g., CHO, endothelial cells, certain tumor cells) (13). Several strategies have been pursued to improve gene transfer into CAR-negative cells (31, 50), such as the use of vectors that have been derived from non-CAR-binding serotypes (e.g., hAd35), and “fiber-swap” vectors, where (parts of) specific regions of the fiber proteins are replaced by the homologous sequences from non-CAR-binding serotypes (36, 43, 49). Alternatively, recombinant hAds with altered tropism have also been generated by engineering new ligands for cellular receptors into the surface loops of the major capsid components; the C terminus and the HI loop of the fiber knob, the RGD loop of penton base, and the L1 loop of the hexon (4, 5, 29, 51). Although effective, the applicability of this approach can be limited by the restricted tolerance for inserting new ligands at these positions. In another approach, the knob and shaft of the fiber have been replaced by an artificial trimerization domain, which was linked to a receptor-binding ligand (26, 28, 31). However, such viruses appear to be difficult to produce at high titers, thus limiting the applicability of this approach.
A novel strategy for modifying the tropism of hAd vectors relies on fusing polypeptides to the C terminus of the minor capsid protein IX (16). This approach is based on the observation that pIX is dispensable for the virus: Ads lacking the pIX gene can be grown to wild-type titers. However, the pIX-deficient hAds are more heat labile than wt.hAd and do not form the so-called group of nine (GON) hexon capsomers. The GONs form the central part of each of the facets of the icosahedral capsid (9). Hence, pIX has been suggested to function as a molecular cement, which enhances the structural integrity of the particles by stabilizing the interaction between hexon capsomers (21, 22). Cryoelectron microscopy image analyses reveal a continuous density for the clusters of three pIX molecules that reside between three hexon capsomers in the GONs (21, 45, 46). This has led to the hypothesis that pIX forms trimers. The ability of pIX to multimerize has been confirmed by coimmunoprecipitation experiments (41). The main mass of pIX is located in the central cavities of the GONs at ca. 65 Å below the top of the hexon capsomers (21, 45, 46). However, the precise location of pIX within the virus capsid and the pIX-hexon interaction domains are not known. Immune accessibility studies with antibodies directed against the N-terminal and the C-terminal parts of hAd3 pIX suggest that the N terminus is between the hexon capsomer cavities and inaccessible. The C terminus is accessible to immunoglobulins, suggesting that it may be pointing toward the outer surface of the virion (1). These data, together with the data from the cryoelectron microscopy image analysis, suggest that if ligands are to be fused with pIX, a spacer may be required to lift the ligand above the outer surface of the icosahedrical surface formed by the hexon capsomers to ensure its availability for a cellular receptor.
In this present study, we examined whether insertion of alpha-helical spacers between pIX and a fused ligand increases the ligand accessibility. Using pIX-incorporation assays and immunogold-labeling electron microscopy experiments, we demonstrate that pIX variants carrying alpha-helical spacers and peptide ligands at their C termini are well tolerated in the viral capsid and expose their ligands in an accessible manner. From these data we conclude that modification of pIX is a feasible approach for retargeting hAd vectors.
MATERIALS AND METHODS
Cells.
The murine microvascular endothelioma cell line Eoma (38), derived from a mouse hemangioendothelioma, was obtained from R. Auerbach (Laboratory of Developmental Biology, University of Wisconsin, Madison). The primary human foreskin fibroblasts (VH10) were provided by A. G. Jochemsen (Department of Molecular Cell Biology, Leiden University, Leiden, The Netherlands), whereas the U118 human glioma cell line was obtained from B. de Leeuw (Department of Neurology, Erasmus University Medical Centre, Rotterdam, The Netherlands). The CHO cell line was obtained from the American Type Culture Collection. The hAd5 E1-transformed helper cell lines PER.C6 and 911 (18, 19) were used to propagate and titer Ad vectors. All cell lines were maintained at 37°C in a humidified atmosphere of 5% CO2 in Dulbecco modified Eagle medium medium (Gibco-BRL, Breda, The Netherlands) supplemented with 8% fetal bovine serum (Gibco-BRL) and 0.3% glucose (J. T. Baker, Deventer, The Netherlands). Infections of the cells with hAds were carried out in infection medium containing 2% fetal bovine serum.
Construction of pIX-ligand eukaryotic expression plasmids.
For construction of the pIX expressing plasmid pAd5pIX.flag, the 480-bp coding sequence of hAd5 pIX was obtained from plasmid pAd5SalB (7) by PCR, with the protIX forward and reverse primers (Table 1) providing an EcoRI site and a XbaI site upstream from the translation-initiation codon and downstream of the translation-termination codon, respectively. In addition, primer protIX (reverse) adds the codons for a FLAG-tag sequence (TAC AAG CTG GCC GAC, encoding Tyr-Lys-Leu-Ala-Asp) (32) immediately before the translation-termination codon. The resulting PCR product was digested with EcoRI and XbaI and cloned into EcoRI- and XbaI-digested pCDNA3.1, generating plasmid pAd5pIX.flag. To insert the ligands at the C terminus of the pAd5pIX.flag, the translation-stop codon was removed, and two unique sites (XhoI and EcoRV) were introduced by replacing the flag tag sequence by inserting the synthetic oligonucleotide pair flag-XE. This flag-XE fragment (which has a 5′ blunt end and a 3′ sticky XbaI overhang) was cloned between the HpaI and XbaI sites in the pAd5pIX.flag plasmid, generating plasmid pAd5pIX.flagXE. For construction of plasmid pAd5pIX, a stop codon was inserted directly after pIX by site-directed mutagenesis PCR (QuickChange site-directed mutagenesis kit), with the pIXstop primers (Table 1) and the pAd5pIX.flag plasmid as a template. The pAd5pIX.RGD plasmid was obtained by annealing the RGD oligonucleotides (Table 1), cloning the duplex in HpaI and XbaI-digested plasmid pAd5pIX.flag. A similar approach was used for construction of plasmid pAd5pIX.flag.RGD. Here the same RGD DNA duplex was inserted into EcoRV- and XbaI-digested plasmid pAd5pIX.flagXE. The 30-Å spacer DNA duplex was obtained by annealing the 30-Å spacer oligonucleotides (Table 1). The coding polypeptide of this sequence was predicted to form an alpha helix of ∼30 Å (20, 23). Insertion of the 30-Å spacer duplex in XhoI-digested plasmid pAd5pIX.flag.RGD resulted in pAd5pIX.flag-30.RGD. The 45-Å spacer was obtained via PCR by amplifying the longest alpha helix (nucleotides 373 to 472; GenBank entry XM_008844) from ApoE4 cDNA by using the ApoE primers (Table 1). After digestion with SalI and XhoI, the 45-Å spacer fragment was cloned in XhoI-digested plasmid pAd5pIX.flagXE to generate plasmid pAd5pIX.flag-45XE. The RGD4c polypeptide was inserted as described above, generating the pAd5pIX.flag-45.RGD plasmid. For construction of the pAd5pIX.flag-45-30.RGD, the 30.RGD EcoRV-ApaI fragment from plasmid pAd5pIX.flag-30.RGD was inserted into EcoRV-ApaI-digested pAd5pIX.flag-45XE. For construction of the MYC-tagged variants, the RGD codons were replaced by a MYC(3×) tag by three-fragment ligation with two sets of oligonucleotides MYCa and MYCb (Table 1). The DNA duplexes were inserted by ligation into the various pIX constructs digested with EcoRV and ApaI.
TABLE 1.
Primer and oligonucleotide list
| Primer, orientationa | Sequence (5′-3′)b |
|---|---|
| protIX, For | CCGGAATTCGCCATGAGCACCAACTCGTTT |
| protIX, Rev | TGCTCTAGATTAGCCGTCGGCCAGCTTGTAGTCGCTCCCTCCGCCGTTAACCGCATTGGGAGG |
| Flag XE,For | AACGGCGGAGGGAGCGACTACAAGCTGGCCGACGGCCTCGAGGATATCTAGT |
| Flag XE,Rev | CTAGACTAGATATCCTCGAGGCCGTCGGCCAGCTTGTAGTCGCTCCCTCCGCCGTT |
| pIXstop, For | CCTCCCCTCCCAATGCGGTTTAAGGCGGAGGGAGCGACTACAA |
| pIXstop, Rev | TTGTAGTCGCTCCCTCCGCCTTAAACCGCATTGGGAGGGGAGG |
| RGD, For | TGTGATTGTAGAGGCGATTGCTTCTGCGGATGA |
| RGD, Rev | CTAGATCATCCGCAGAAGCAATCGCCTCTACAATCACAGGC |
| 30 spacer, For | TCGAGATCCCAACCTATCTGAGCGAAGATGAACTGAAAGCCGCCGAAGCCGCCTTCAAACGCCACAACCCAACCGCGG |
| 30 spacer, Rev | TCGACCGCGGTTGGGTTGTGGCGTTTGAAGGCGGCTTCGGCGGCTTTCAGTTCATCTTCGCTCAGATAGGTTGGGATC |
| ApoE, For | ACCGGTCGACGAGGAGACGCGGGCACGGCT |
| ApoE, Rev | ATCCTCGAGGGCATCCTCGCCGCGGTACTGC |
| MYCa, For | GGCGCCGAGCAGAAACTCATCTCTGAAGAAGATCTGGAACAAAAGTTGATTTCAGAA |
| MYCa, Rev | CTTCTTCTGAAATCAACTTTTGTTCCAGATCTTCTTCAGAGATGAGTTTCTGCTCGGCGCC |
| MYCb, For | GAAGATCTGGAACAGAAGCTCATCTCTGAGGAAGATCTGCACGTGTAAGGGCC |
| MYCb, Rev | CTTACACGTGCAGATCTTCCTCAGAGATGAGCTTCTGTTCCAGAT |
| pIXScaI, For | GTTTTGCAGCAGCCGCCGCCAGTACTAGCACCAACTCGTTTGATGG |
| pIXScaI, Rev | CCATCAAACGAGTTGGTGCTAGTACTGGCGGCGGCTGCTGCAAAAC |
| pIXSpeI, For | GGTTTCTGCCCTGAAGGCTTACTAGTCTCCCAATGCGGTTTAAAAC |
| pIXSpeI, Rev | GTTTTAAACCGCATTGGGAGACTAGTAAGCCTTCAGGGCAGAAACC |
For, forward; Rev, reverse.
Restriction sites mentioned in Materials and Methods are underscored.
Ad vectors.
The pTrackCMV-GFP is an E1- and E3-deleted hAd5-based shuttle plasmid that carries green fluorescent protein (GFP) transgene each under the control of the human cytomegalovirus (CMV) immediate-early promoter. For construction of ΔpIX pTrackCMV-GFP, two unique restriction sites were introduced, a ScaI site at the start codon and a SpeI site at the C terminus of pIX, by site-directed mutagenesis PCR (primers pIXScaI and pIXSpeI, Table 1). The ΔpIX pTrackCMV-GFP plasmid was constructed by blunt-end self-ligation after ScaI and SpeI digestion and treatment with the Klenow fragment of E. coli DNA polymerase. Replication-incompetent hAd5 vectors hAd5CMV-GFP and hAd5CMV-GFPΔpIX were generated as described elsewhere (25). The hAd5dl313 virus lacks the 2,307 bp of the E1 region, including the 5′ portion of the pIX gene. This mutant does not express detectable levels of pIX (14). Viruses were titrated via standard plaque assay protocol (19).
Incorporation assay.
For incorporation of the pIX-ligand fusion proteins in ΔpIX viruses (hAd5dl313 and hAd5CMV-GFPΔpIX), 70 to 80% confluent layer 911 helper cells were transfected with the appropriated expression plasmids by the calcium phosphate coprecipitation technique (24). For five Cellstar 550-ml flasks (Greiner bio-one, Frickenhausen, Germany), 300 μg of plasmid DNA was used in a total volume of 25 ml of buffered calcium-chloride (Merck, Darmstadt, Germany) solution. At 18 h after transfection the cells were infected with ΔpIX viruses. After 2 days, the virus was harvested and CsCl purified as described before (19).
Immunofluorescence assays.
For immunofluorescence assays, 911 cells were grown on glass coverslips in six-well plates, fixed in methanol, washed with phosphate-buffered saline (PBS) containing 0.05% Tween 20, and incubated with primary antibody, (1:800 diluted in PBS, 3% bovine serum albumin [BSA]) for 60 min at room temperature. The rabbit antiserum recognizing pIX has been described previously (10). The cells were washed and incubated with secondary fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit serum (diluted 1:100 in PBS-3% BSA) for 30 min at room temperature. Nuclei were visualized by using propidium iodide. Subsequently, the cells were washed and mounted on object glasses by using Dabco-glycerol (glycerol, 0.02 M Tris-Cl [pH 8.0], and 2.3% 1,4-diazabicyclo-[2.2.2]-octane). Cells were visualized with a Leica DM-IRBE microscope.
Western analysis.
Cell lysates were made in radioimmunoprecipitation assay lysis buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 0.1% sodium dodecyl sulfate [SDS], 0.5% deoxycholate, and 1% NP-40). Protein concentrations were measured with a protein assay (Bio-Rad Laboratories, Veenendaal, The Netherlands). Samples for Western analyses were prepared by boiling 15 μg of the protein extract with sample buffer (10% glycerol, 2% SDS, 60 mM Tris-Cl [pH 6.7], 2.5% β-mercaptoethanol, and 2.5% bromophenol blue from a 1:20-diluted saturated solution) for 4 min at 100°C. Samples were analyzed on 12% polyacrylamide-SDS gels (30% acrylamide-bisacrylamide [37.5:1]; Bio-Rad) overlaid by a 5% stacking gel (30% acrylamide-bisacrylamide [37.5:1]; Bio-Rad). The proteins were transferred to Immobilon-P (Immobilon-P transfer membrane [polyvinylidene difluoride]; Millipore, Etten-Leur, The Netherlands) and visualized by standard protocols with anti-pIX (1:2000), anti-actin (1:500, clone C4; ICN Biomedicals, Inc., Zoetermeer, The Netherlands), or anti-hexon, a polyclonal serum that recognizes the major capsid proteins (1:500, rat polyclonal serum), as the primary antibody. All antibodies were diluted in TBST (0.2% Tween 20, 150 mM NaCl, and 10 mM Tris) with 5% nonfat dried milk (Protifar Plus; Nutricia BV, Zoetermeer, The Netherlands). Virus lysates were prepared by adding 5 × 1010 virus particles (measured by a standard OD260 protocol (33) directly to Western sample buffer.
Flow cytometry analysis.
For fluorescence-activated cell sorting analyses, cells were treated with trypsin gently, the volume was increased by adding PBS supplemented with 1% fetal bovine serum, and the cells were kept on ice. The samples were analyzed with a FACScan flow cytometer (Becton Dickinson). GFP fluorescence was detected by using a 530±30-nm band-pass filter (FL1 channel) after excitation with an argon ion laser source at 488 nm. Using a forward scatter-side scatter representation of events, a region was defined in order to exclude cellular debris from the analysis. A number-of-events/FL1 (which reflects the fluorescence intensity) histogram was then established according to this region, and percentages of GFP-positive cells were determined in comparison to the negative control (untreated cells). Data analysis was performed by using CellQuest 3.1 software (Becton Dickinson). For each sample, 10,000 events were collected.
Immunoelectron microscopy.
Viruses were brought on copper grids with a carbon-coated Formvar film and washed with PBS (PBS plus 2% BSA), incubated with anti-MYC (diluted 1:200 in PBS-2% BSA, clone 9E10; Upstate Biotechnology, Charlottesville, Va.) for 60 min at room temperature. After a washing step, grids were incubated with rabbit anti-mouse serum (Dako Cytomation A/S, Copenhagen, Denmark) (diluted 1:200 in PBS-2% BSA) for 30 min, washed with PBS-2% BSA, and incubated with gold-labeled protein-A (Amersham Biosciences Europe GmbH, Freiburg, Germany) for 30 min at room temperature. Subsequently, these samples were fixed in 1.5% glutaraldehyde in cacodylate buffer and negative stained with 1% uranyl acetate for 15 min. The viruses were examined by using a Philips EM 410-LS transmission electron microscope at 80 kV.
RESULTS
Ad pIX resides at a deep position (hidden) below the tops of the hexon capsomers (21, 45, 46). Hence, small C-terminal extensions to pIX may be difficult to access from the outside the capsid surface. Although small C-terminal ligands are capable of interacting with cellular structures with some efficiency (16), the position of pIX in the capsid predicts that lifting the peptide ligands toward the top of the hexon cavities may increase ligand accessibility, and thereby, should improve ligand-receptor interaction.
To test whether insertions of alpha-helical spacers between the pIX C terminus and the ligand could improve the accessibility of the ligand, we made fusion constructs with spacers of varied lengths joining the pIX C terminus to MYC and RGD epitopes (Fig. 1). These were used in 911 helper cells to complement pIX deficiency during propagation of hAd5ΔpIX viruses on these cells. During infection, wild-type pIX (wt.pIX) accumulates almost exclusively in the nucleus, where it resides in small bodies called pIX bodies (41). To test whether the pIX fusion proteins used in the present study also localize to pIX bodies, we transfected 911 cells with the corresponding expression plasmids encoding the fusion protein; after 2 days, the cells were stained with anti-pIX and FITC-labeled goat anti-rabbit immunoglobulin and then examined with a fluorescence microscope. As shown in Fig. 2, all pIX variants localized to the nuclei of the 911 cells, as did wt.pIX. Identical distribution patterns were seen with antiserum recognizing the Flag epitope fused to the C terminus of pIX (data not shown).
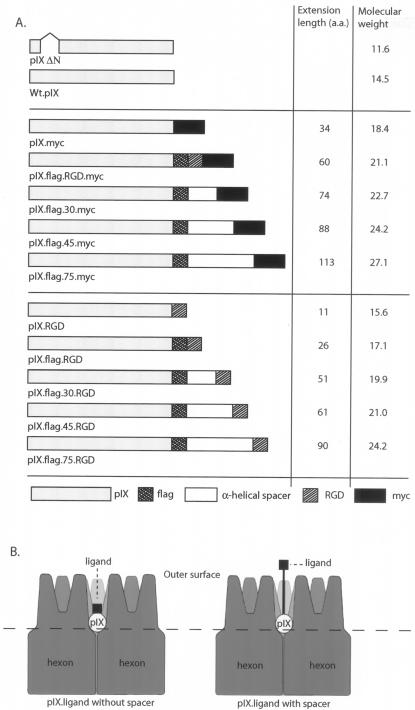
FIG. 1.
(A) Schematic representation of the pIX variants used in the present study. The RGD4c motif and MYC peptide, which are used as ligands, are fused directly with pIX or linked via a spacer. The values 30, 45, and 75 are the predicted lengths of the α-helical spacers in angstroms. (B) Schematic representation of the location of pIX between the hexon capsomers. Electron microscopic image reconstruction suggests that pIX resides between the hexon capsomers, hidden ca. 65 Å below the hexon tops. Introducing spacers between pIX and the ligand may improve the accessibility of ligands fused to the C terminus of pIX.
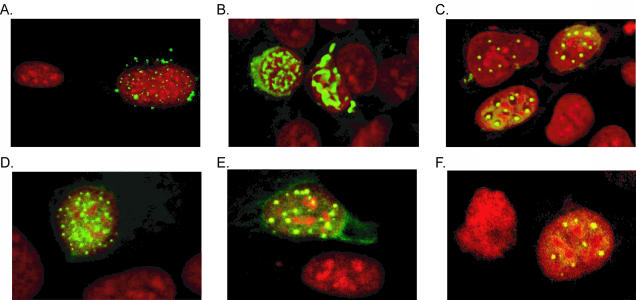
FIG. 2.
Subcellular localization of pIX variants. The subcellular localization of the various pIX proteins was tested by immunofluorescence microscopy. 911 cells grown on coverslips were transfected with the various pIX.RGD expression plasmids. At 2 days postinfection, the localization of pIX was visualized with anti-pIX and FITC-labeled goat anti-rabbit antibodies. The nuclei were stained with propidium iodide. (A) wt.pIX; (B) pIX.RGD; (C) pIX.flag.RGD (D); pIX.flag.30.RGD; (E) pIX.flag.45.RGD; (F) pIX.flag.75.RGD. All pIX variants are localized in nuclear aggregates, as is wt.pIX.
To determine whether these modified pIX molecules could be incorporated into the viral capsid, we used an incorporation assay based on transient expression of the pIX variants in 911 cells. The cells were transfected with the various pIX expression vectors; at 18 h posttransfection, the cells were infected with hAd5dl313, an hAd5 deletion mutant lacking a functional pIX gene. At 48 h postinfection, viruses were harvested and purified by CsCl density gradient centrifugation and subjected to Western blotting with pIX antiserum. All pIX.MYC fusion proteins accumulated to similar levels in 911 helper cells (Fig. 3B) and were efficiently incorporated (Fig. 3C) into virus particles. From these data we conclude that the C-terminal extensions up to 113 amino acids do not impair the incorporation of pIX into adenoviral capsids. In contrast, as previously reported, deletion of the pIX N terminus prevented incorporation into viruses (Fig. 3A).
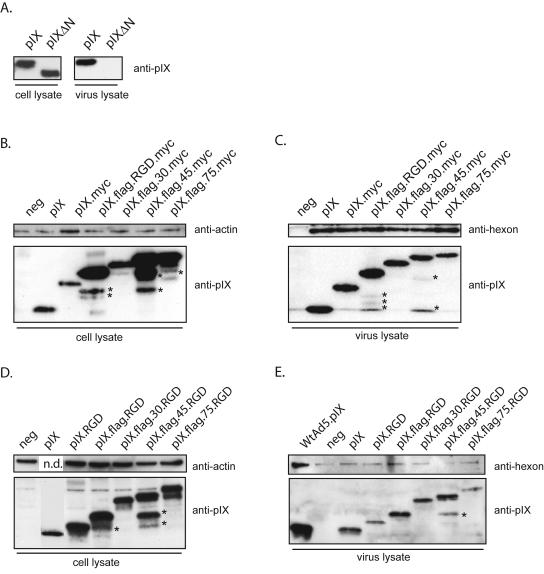
FIG. 3.
Western blot pIX incorporation assays. (A) To test the specificity of the incorporation assay, wt.pIX and pIX-ΔN expression plasmids were transfected into 911 cells. At 18 h posttransfection, the cells were infected with hAd5dl313. The infected cells were lysed 48 h postinfection. Protein extracts were prepared from aliquots of the lysates, whereas the remainder was used for virus isolation. Protein extracts of the cell lysates and of the purified viruses were analyzed by Western blotting with an anti-pIX serum. Although both pIX variants were expressed in 911 cells, only wt.pIX was incorporated into the virion. (B and C) To test the incorporation of the pIX variants, 911 cells were transfected with the various pIX expression plasmids. After overnight incubation, the cells were infected with hAd5dl313 or hAd5CMV-GFPΔpIX viruses. Protein extracts were prepared from the cell lysate and from the purified viruses and analyzed by Western blotting. The blots were probed with anti-pIX, anti-actin, or anti-hexon as described in the text. All pIX MYC constructs were expressed in equivalent amounts by the 911 cells (B), and all were incorporated in equivalent amounts into hAd5dl313 virions (C). (D and E) An identical analysis was performed for the various pIX.RGD constructs. All pIX variants were expressed in 911 cells (D) and incorporated efficiently into hAd5CMV-GFPΔpIX virions (E). As a negative control (neg), 911 cells were mock transfected and infected with hAd5dl313 and hAd5CMV-GFPΔpIX. The extra bands marked with an asterisk are proteolytic degradation products.
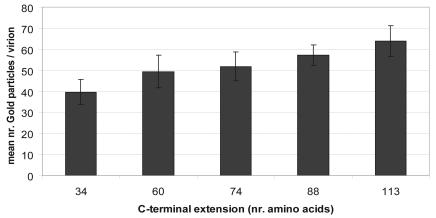
Next, we determined whether the MYC peptides were accessible to immunoglobulins. Viruses were incubated with mouse antiserum recognizing the MYC peptide and then with rabbit anti-mouse antiserum and protein A labeled with 10-nm gold particles. Electron microscopy analysis revealed no gold particles associated with hAd5dl313 viruses or with hAd5dl313 particles loaded with wt.pIX. In contrast, hAd5dl313 incorporating the MYC-tagged pIX constructs were efficiently labeled (Fig. 4). Quantitative analyses of the photomicrographs indicated that particles loaded with any of the spacer constructs labeled more efficiently than those loaded with the pIX.MYC construct that had only the 34-amino-acid MYC tag (P < 0.0001 [unpaired Student t test]) (Fig. 5). Nonlabeled virions were seen occasionally (e.g., Fig. 4D); these most likely resulted from virus propagation in the nontransfected cells in the cultures. Taken together, these data confirm that the pIX-variants carrying C-terminal extensions are efficiently incorporated into the hAd capsid and that the ligands linked to these proteins are accessible on the outer surface of the virion. In addition, these data provide evidence that the ligand accessibility improves when the length of the alpha-helical spacers is increased.
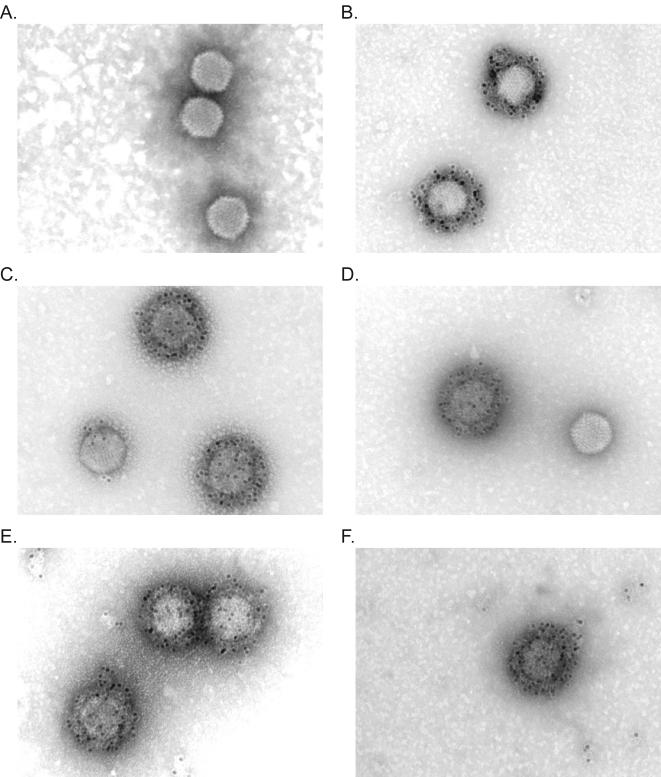
FIG. 4.
Immunoelectron microscopic analysis of ligand accessibility. To test the accessibility of the MYC epitope on hAd5dl313, viruses that were loaded with the various pIX variants were bound on copper grids with a carbon-coated Formvar film. The MYC epitope was detected with anti-MYC antibody, followed by rabbit anti-mouse immunoglobulin and gold-labeled Prot.A. Visualization was done by using a Philips EM 410-LS transmission electron microscope. The viruses were loaded with wt.pIX (A), pIX.MYC (B), pIX.flag.RGD.MYC (C), pIX.flag.30.MYC (D), pIX.flag.45.MYC (E), and pIX.flag.75.MYC (F), respectively.
FIG. 5.
Quantification of the number of gold particles bound to the viruses. The mean number of gold particles bound to the viruses (y axis) is plotted against the length (in amino acids) of the C-terminal extension (x axis). The labeling efficiency of each variant is greater than the pIX.MYC construct (34 amino acids) (P < 0.0001 [unpaired Student t test]).
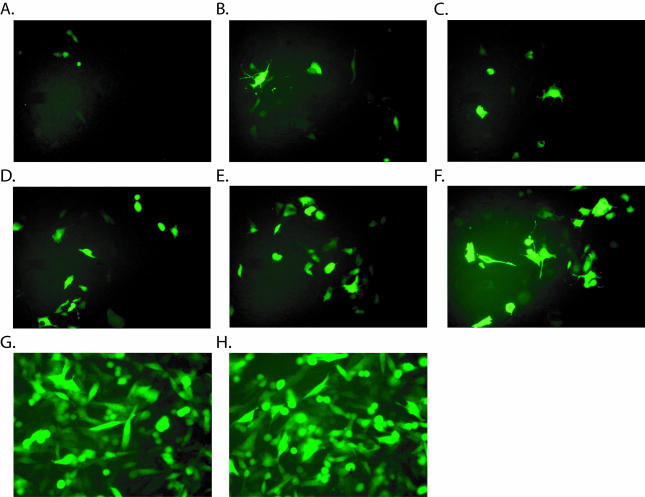
To test whether ligands fused to the C terminus of pIX can bind cellular receptors and whether the recruitment of cellular receptors is improved by addition of a spacer, the MYC peptide was replaced by a small ligand, RGD4c. These pIX.RGD fusion proteins were incorporated into virions encoding GFP. The RGD4c binds efficiently to αvβ3 and αvβ5 integrins (3), which are broadly expressed on many cell types, including endothelial cells (11, 39, 44, 52). To evaluate CAR-independent transduction, we used the mouse hemangioendothelioma cell line Eoma (38) as target. Whereas Eoma cells do not express detectable amounts of CAR receptor on their cellular membrane, αvβ3 and αvβ5 integrins were readily detectable by flow cytometry (J. Vellinga, unpublished data). For these experiments we used the vector hAd5CMV-GFPΔpIX. The vector was propagated on transfected helper cells as described above. All pIX-RGD4c fusion proteins were produced adequately in the 911 helper cells (Fig. 3D). Western blot analysis revealed equivalent incorporation of the different pIX.RGD4c variants into virus particles (Fig. 3E). Where Eoma cells were infected with the GFP vector loaded with wt.pIX, only a few cells were seen to express GFP (Fig. 6A). Cultures infected with vectors loaded with the pIX.RGD variants showed an increased number of GFP-positive cells; the efficiency of transduction appear to increase with spacer lengths (Fig. 6B to F). In contrast, the presence of the RGD motif and the length of the spacers had no effect on transduction efficiency in CAR-positive HeLa cells (data are shown for wt.pIX and pIX.flag.75.RGD in Fig. 6G and H, respectively). To show that the improved infection efficiency was not restricted to the Eoma cells, three other CAR-negative cell lines were infected with GFP vectors incorporating either pIX.RGD or pIX.flag.75.RGD, and number of transduced cells was quantified by flow cytometry analysis (Table 2). Transduction by pIX.flag.75.RGD was five- to sevenfold more efficient on these human (VH10 and U118) and rodent (CHO and Eoma) cell lines than when transduced by the pIX.RGD (which lacks a spacer). In the absence of the RGD peptide, vectors incorporated pIX spacers showed no increased efficiency of transduction. Thus, the improved efficiency seen with pIX.flag.75.RGD reflected the improved accessibility of the RGD motif and not the presence of the spacer itself.
FIG. 6.
Infection assay. To test whether the spacers increase the infection efficiency, hAd5CMV-GFPΔpIX viruses were propagated in 911 cells expressing the pIX.RGD variants. The capacity of the resulting viruses to transduce cells independent of CAR was tested by exposing CAR-negative Eoma cells to the CsCl purified viruses at 1,000 particles/cell. At 48 h postinfection, the cells were evaluated for GFP expression. (A to F) hAd5CMV-GFPΔpIX with wt.pIX (A), pIX.RGD (B), pIX.flag.RGD (C), pIX.flag.30.RGD (D), pIX.flag.45.RGD (E), or pIX.flag.75.RGD (F). To show that hAd5CMV-GFPΔpIX with wt.pIX (G) and hAd5CMV-GFPΔpIX bearing pIX.flag.75.RGD (H) have equivalent infectious particle numbers, HeLa cells were infected with the corresponding viruses.
TABLE 2.
Infection assay
| Cell line | CARa | MOIb | % GFP+ cellsc
|
|||
|---|---|---|---|---|---|---|
| pIX.RGD | pIX.flag.75.RGD | pIX.myc | pIX.flag.75.myc | |||
| HeLa | + | 40 | 30 ± 3 | 32 ± 5 (1.1) | 29 ± 4 | 28 ± 2 |
| CHO | − | 1000 | 1.5 ± 0.5 | 7.8 ± 1.0 (5.2) | 1.1 ± 0.5 | 0.9 ± 0.1 |
| Eoma | − | 1000 | 1.3 ± 0.1 | 9.3 ± 1.1 (7.2) | 0.8 ± 0.4 | 1.1 ± 0.5 |
| VH10 | − | 1000 | 3.1 ± 0.6 | 20.7 ± 2.5 (6.7) | 1.4 ± 0.8 | 1.2 ± 0.2 |
| U118 | − | 1000 | 9.3 ± 0.1 | 28.9 ± 3.1 (3.1) | 7.8 ± 0.8 | 8.1 ± 0.6 |
The data for the CAR expression were obtained from B. de Leeuw.
MOI, multiplicity of infection.
The data represent the mean of three independent experiments. Values in parentheses are the ratio percent GFP-positive cells (pIX.RGD) to percent GFP-positive cells (pIX.flag.75.RGD).
DISCUSSION
In the present study, we show that Ad minor capsid protein pIX variants, carrying alpha-helical spacers and ligands at their C termini, are efficiently incorporated into the Ad capsid. pIX is hidden between hexon capsomers, where it associates with the hexon to stabilize the capsid. Immunoelectron microscopy studies with hAd3 demonstrated that the C terminus of pIX, in contrast to its N terminus, is accessible to immunoglobulins, suggesting that this part of the protein is expressed on the capsid surface (1). Our data confirm and extend this hypothesis. Based on the previous estimated by Stewart et al. that the main mass of pIX is located ca. 65 Å below the tops of the hexon capsomers (21, 45), incorporation of spacers up to 75 Å would suffice to lift the ligands and expose them at the surface of the capsid. In line with this hypothesis is our observation that the increasing spacer-length is directly correlated with increasing ligand accessibility. We observed an improved antibody labeling of the MYC peptide-containing viruses with longer spacers. Since there was no significant effect of the spacer length on the efficiency of pIX incorporation into the capsid, these data suggests that the spacers make the MYC epitope more accessible for antigen-specific immunoglobulins.
Using an integrin-interacting RGD4c motif fused to the spacers, we evaluated the relative infection efficiencies of spacer-bearing pIX virions on mouse endothelioma cells (44). Wild-type hAd5 is largely unable to infect these cells, which have little if any CAR on their cell surface. However, incorporation of the RGD motif within pIX, particularly when a spacer was also incorporated, significantly increased transduction efficiency of CAR-negative cells.
Wild-type pIX is confined to the nucleus. Here we demonstrate that the pIX fusion proteins have a subcellular localization that is indistinguishable from wt.pIX in 911 cells. Hence, neither the presence of the RGD4c motif nor the presence of the various spacers prevents nuclear localization and accumulation in pIX bodies.
Modification of pIX can be used for vector targeting in two ways, the pIX gene can be modified in the vector backbone; alternatively, pIX variants can be synthesized in pIX-expressing helper cell lines (10). Although it has been suggested that pIX synthesis is toxic for mammalian cells (30), careful selection procedures yield cell lines that can efficiently complement pIX-deficient viruses.
Thus far the retargeting via pIX modification is not very efficient (comparing the number of transduced Eoma cells with that of the HeLa cells). Additional modifications may improve the efficacy and specificity of retargeting. For example, the ligand used in our experiments is the RGD4c motif, which is a small peptide with relatively low affinity for integrin receptors. Substitution of this ligand with other, higher-affinity ligands, such as single-chain antibodies, affibodies, or domain antibodies (4), may increase the infection efficiency. In addition, the natural viral entry mechanisms should be reduced or ablated to increase retargeting selectivity. In the present study we used wt.hAd5CMV-GFP virus, which still retains its natural CAR and αvβ integrins binding sites in the fiber knob and penton base. Removal of these sites may be essential to achieve more specific retargeting. Furthermore, the presence of the long hAd5 fibers may hinder pIX-mediated targeting as these long and protruding fibers may limit access of the cellular receptors to the heterologous ligands. The use of short fibers from other serotypes (34, 35, 51) or, alternatively, artificially shortened hAd5 fibers may therefore improve the efficiency of cell targeting via pIX-linked ligands.
Considering the increased accessibility achieved with the spacers between pIX and the fused ligand, we envisage that the alpha-helical spacers lift the ligands toward the hexon top and as a result expose them on the capsid surface. Here they are easily reached by cell surface receptors. Our data demonstrate the feasibility of using pIX as an anchor to link peptide ligands to retarget Ad vectors.
Acknowledgments
We thank Lee G. Fradkin and Henk K. Koerten for critically reading the manuscript and Bob J. Scholte and Bertie de Leeuw for scientific discussions. In addition, we gratefully acknowledge the useful suggestions by the anonymous reviewers.
REFERENCES
- 1.Akalu, A., H. Liebermann, U. Bauer, H. Granzow, and W. Seidel. 1999. The subgenus-specific C-terminal region of protein IX is located on the surface of the adenovirus capsid. J. Virol. 73:6182-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnberg, N., K. Edlund, A. H. Kidd, and G. Wadell. 2000. Adenovirus type 37 uses sialic acid as a cellular receptor. J. Virol. 74:42-48. [PMC free article] [PubMed] [Google Scholar]
- 3.Assa-Munt, N., X. Jia, P. Laakkonen, and E. Ruoslahti. 2001. Solution structures and integrin binding activities of an RGD peptide with two isomers. Biochemistry 40:2373-2378. [DOI] [PubMed] [Google Scholar]
- 4.Barnett, B. G., C. J. Crews, and J. T. Douglas. 2002. Targeted adenoviral vectors. Biochim. Biophys. Acta 1575:1-14. [DOI] [PubMed] [Google Scholar]
- 5.Belousova, N., V. Krendelchtchikova, D. T. Curiel, and V. Krasnykh. 2002. Modulation of adenovirus vector tropism via incorporation of polypeptide ligands into the fiber protein. J. Virol. 76:8621-8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 7.Bernards, R., P. I. Schrier, J. L. Bos, and A. J. van der Eb. 1983. Role of adenovirus types 5 and 12 early region 1b tumor antigens in oncogenic transformation. Virology 127:45-53. [DOI] [PubMed] [Google Scholar]
- 8.Boudin, M. L., J. C. D'Halluin, C. Cousin, and P. Boulanger. 1980. Human adenovirus type 2 protein IIIa. II. Maturation and encapsidation. Virology 101:144-156. [DOI] [PubMed] [Google Scholar]
- 9.Boulanger, P., P. Lemay, G. E. Blair, and W. C. Russell. 1979. Characterization of adenovirus protein IX. J. Gen. Virol. 44:783-800. [DOI] [PubMed] [Google Scholar]
- 10.Caravokyri, C., and K. N. Leppard. 1995. Constitutive episomal expression of polypeptide IX (pIX) in a 293-based cell line complements the deficiency of pIX mutant adenovirus type 5. J. Virol. 69:6627-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castel, S., R. Pagan, F. Mitjans, J. Piulats, S. Goodman, A. Jonczyk, F. Huber, S. Vilaro, and M. Reina. 2001. RGD peptides and monoclonal antibodies, antagonists of αv-integrin, enter the cells by independent endocytic pathways. Lab. Investig. 81:1615-1626. [DOI] [PubMed] [Google Scholar]
- 12.Chatterjee, P. K., M. E. Vayda, and S. J. Flint. 1985. Interactions among the three adenovirus core proteins. J. Virol. 55:379-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen, C. J., J. T. Shieh, R. J. Pickles, T. Okegawa, J. T. Hsieh, and J. M. Bergelson. 2001. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc. Natl. Acad. Sci. USA 98:15191-15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colby, W. W., and T. Shenk. 1981. Adenovirus type 5 virions can be assembled in vivo in the absence of detectable polypeptide IX. J. Virol. 39:977-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Jong, J. C., A. G. Wermenbol, M. W. Verweij-Uijterwaal, K. W. Slaterus, P. Wertheim-Van Dillen, G. J. Van Doornum, S. H. Khoo, and J. C. Hierholzer. 1999. Adenoviruses from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotypes Ad50 and Ad51 of species B1 and D, respectively. J. Clin. Microbiol. 37:3940-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dmitriev, I. P., E. A. Kashentseva, and D. T. Curiel. 2002. Engineering of adenovirus vectors containing heterologous peptide sequences in the C terminus of capsid protein IX. J. Virol. 76:6893-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everitt, E., B. Sundquist, U. Pettersson, and L. Philipson. 1973. Structural proteins of adenoviruses. X. Isolation and topography of low molecular weight antigens from the virion of adenovirus type 2. Virology 52:130-147. [DOI] [PubMed] [Google Scholar]
- 18.Fallaux, F. J., A. Bout, D. V. Van, I., D. J. van den Wollenberg, K. M. Hehir, J. Keegan, C. Auger, S. J. Cramer, H. van Ormondt, A. J. van der Eb, D. Valerio, and R. C. Hoeben. 1998. New helper cells and matched early region 1-deleted adenovirus vectors prevent generation of replication-competent adenoviruses. Hum. Gene Ther. 9:1909-1917. [DOI] [PubMed] [Google Scholar]
- 19.Fallaux, F. J., O. Kranenburg, S. J. Cramer, A. Houweling, H. van Ormondt, R. C. Hoeben, and A. J. van der Eb. 1996. Characterization of 911: a new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors. Hum. Gene Ther. 7:215-222. [DOI] [PubMed] [Google Scholar]
- 20.Fortunati, E., E. Ehlert, N. D. van Loo, C. Wyman, J. A. Eble, F. Grosveld, and B. J. Scholte. 2000. A multi-domain protein for beta1 integrin-targeted DNA delivery. Gene Ther. 7:1505-1515. [DOI] [PubMed] [Google Scholar]
- 21.Furcinitti, P. S., J. van Oostrum, and R. M. Burnett. 1989. Adenovirus polypeptide IX revealed as capsid cement by difference images from electron microscopy and crystallography. EMBO J. 8:3563-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh-Choudhury, G., Y. Haj-Ahmad, and F. L. Graham. 1987. Protein IX, a minor component of the human adenovirus capsid, is essential for the packaging of full-length genomes. EMBO J. 6:1733-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong, Y., H. X. Zhou, M. Guo, and N. R. Kallenbach. 1995. Structural analysis of the N and C termini in a peptide with consensus sequence. Protein Sci. 4:1446-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 25.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henning, P., M. K. Magnusson, E. Gunneriusson, S. S. Hong, P. Boulanger, P. A. Nygren, and L. Lindholm. 2002. Genetic modification of adenovirus 5 tropism by a novel class of ligands based on a three-helix bundle scaffold derived from staphylococcal protein A. Hum. Gene Ther. 13:1427-1439. [DOI] [PubMed] [Google Scholar]
- 27.Hong, S. S., B. Gay, L. Karayan, M. C. Dabauvalle, and P. Boulanger. 1999. Cellular uptake and nuclear delivery of recombinant adenovirus penton base. Virology 262:163-177. [DOI] [PubMed] [Google Scholar]
- 28.Krasnykh, V., N. Belousova, N. Korokhov, G. Mikheeva, and D. T. Curiel. 2001. Genetic targeting of an adenovirus vector via replacement of the fiber protein with the phage T4 fibritin. J. Virol. 75:4176-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krasnykh, V. N., G. V. Mikheeva, J. T. Douglas, and D. T. Curiel. 1996. Generation of recombinant adenovirus vectors with modified fibers for altering viral tropism. J. Virol. 70:6839-6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krougliak, V., and F. L. Graham. 1995. Development of cell lines capable of complementing E1, E4, and protein IX defective adenovirus type 5 mutants. Hum. Gene Ther. 6:1575-1586. [DOI] [PubMed] [Google Scholar]
- 31.Magnusson, M. K., S. S. Hong, P. Henning, P. Boulanger, and L. Lindholm. 2002. Genetic retargeting of adenovirus vectors: functionality of targeting ligands and their influence on virus viability. J. Gene Med. 4:356-370. [DOI] [PubMed] [Google Scholar]
- 32.Miceli, R. M., M. E. DeGraaf, and H. D. Fischer. 1994. Two-stage selection of sequences from a random phage display library delineates both core residues and permitted structural range within an epitope. J. Immunol. Methods 167:279-287. [DOI] [PubMed] [Google Scholar]
- 33.Mittereder, N., K. L. March, and B. C. Trapnell. 1996. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J. Virol. 70:7498-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyazawa, N., P. L. Leopold, N. R. Hackett, B. Ferris, S. Worgall, E. Falck-Pedersen, and R. G. Crystal. 1999. Fiber swap between adenovirus subgroups B and C alters intracellular trafficking of adenovirus gene transfer vectors. J. Virol. 73:6056-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizuguchi, H., and T. Hayakawa. 2002. Adenovirus vectors containing chimeric type 5 and type 35 fiber proteins exhibit altered and expanded tropism and increase the size limit of foreign genes. Gene 285:69-77. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura, T., K. Sato, and H. Hamada. 2003. Reduction of natural adenovirus tropism to the liver by both ablation of fiber-coxsackievirus and adenovirus receptor interaction and use of replaceable short fiber. J. Virol. 77:2512-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nemerow, G. R. 2000. Cell receptors involved in adenovirus entry. Virology 274:1-4. [DOI] [PubMed] [Google Scholar]
- 38.Obeso, J., J. Weber, and R. Auerbach. 1990. A hemangioendothelioma-derived cell line: its use as a model for the study of endothelial cell biology. Lab. Investig. 63:259-269. [PubMed] [Google Scholar]
- 39.Pasqualini, R., E. Koivunen, and E. Ruoslahti. 1997. Alpha v integrins as receptors for tumor targeting by circulating ligands. Nat. Biotechnol. 15:542-546. [DOI] [PubMed] [Google Scholar]
- 40.Roelvink, P. W., A. Lizonova, J. G. Lee, Y. Li, J. M. Bergelson, R. W. Finberg, D. E. Brough, I. Kovesdi, and T. J. Wickham. 1998. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 72:7909-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosa-Calatrava, M., L. Grave, F. Puvion-Dutilleul, B. Chatton, and C. Kedinger. 2001. Functional analysis of adenovirus protein IX identifies domains involved in capsid stability, transcriptional activity, and nuclear reorganization. J. Virol. 75:7131-7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russell, W. C. 2000. Update on adenovirus and its vectors. J. Gen. Virol. 81:2573-2604. [DOI] [PubMed] [Google Scholar]
- 43.Schoggins, J. W., J. G. Gall, and E. Falck-Pedersen. 2003. Subgroup B and F fiber chimeras eliminate normal adenovirus type 5 vector transduction in vitro and in vivo. J. Virol. 77:1039-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schraa, A. J., R. J. Kok, A. D. Berendsen, H. E. Moorlag, E. J. Bos, D. K. Meijer, L. F. de Leij, and G. Molema. 2002. Endothelial cells internalize and degrade RGD-modified proteins developed for tumor vasculature targeting. J. Control Release 83:241-251. [DOI] [PubMed] [Google Scholar]
- 45.Stewart, P. L., R. M. Burnett, M. Cyrklaff, and S. D. Fuller. 1991. Image reconstruction reveals the complex molecular organization of adenovirus. Cell 67:145-154. [DOI] [PubMed] [Google Scholar]
- 46.Stewart, P. L., S. D. Fuller, and R. M. Burnett. 1993. Difference imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 12:2589-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valentine, R. C., and H. G. Pereira. 1965. Antigens and structure of the adenovirus. J. Mol. Biol. 13:13-20. [DOI] [PubMed] [Google Scholar]
- 48.van Oostrum, J., and R. M. Burnett. 1985. Molecular composition of the adenovirus type 2 virion. J. Virol. 56:439-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Von Seggern, D. J., S. Huang, S. K. Fleck, S. C. Stevenson, and G. R. Nemerow. 2000. Adenovirus vector pseudotyping in fiber-expressing cell lines: improved transduction of Epstein-Barr virus-transformed B cells. J. Virol. 74:354-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wickham, T. J. 2000. Targeting adenovirus. Gene Ther. 7:110-114. [DOI] [PubMed] [Google Scholar]
- 51.Wickham, T. J., E. Tzeng, L. L. Shears, P. W. Roelvink, Y. Li, G. M. Lee, D. E. Brough, A. Lizonova, and I. Kovesdi. 1997. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J. Virol. 71:8221-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zitzmann, S., V. Ehemann, and M. Schwab. 2002. Arginine-glycine-aspartic acid (RGD)-peptide binds to both tumor and tumor-endothelial cells in vivo. Cancer Res. 62:5139-5143. [PubMed] [Google Scholar]