Abstract
Drug resistance is a major problem in the treatment of AIDS, due to the very high mutation rate of human immunodeficiency virus (HIV) and subsequent rapid development of resistance to new drugs. Identification of mutations associated with drug resistance is critical for both individualized treatment selection and new drug design. We have performed an automated mutation analysis of HIV Type 1 (HIV-1) protease and reverse transcriptase (RT) from approximately 40,000 AIDS patient plasma samples sequenced by Specialty Laboratories Inc. from 1999 to mid-2002. This data set provides a nearly complete mutagenesis of HIV protease and enables the calculation of statistically significant Ka/Ks values for each individual amino acid mutation in protease and RT. Positive selection (i.e., a Ka/Ks ratio of >1, indicating increased reproductive fitness) detected 19 of 23 known drug-resistant mutation positions in protease and 20 of 34 such positions in RT. We also discovered 163 new amino acid mutations in HIV protease and RT that are strong candidates for drug resistance or fitness. Our results match available independent data on protease mutations associated with specific drug treatments and mutations with positive reproductive fitness, with high statistical significance (the P values for the observed matches to occur by random chance are 10−5.2 and 10−16.6, respectively). Our mutation analysis provides a valuable resource for AIDS research and will be available to academic researchers upon publication at http://www.bioinformatics.ucla.edu/HIV. Our data indicate that positive selection mapping is an analysis that can yield powerful insights from high-throughput sequencing of rapidly mutating pathogens.
The emergence of drug-resistant mutants of human immunodeficiency virus type 1 (HIV-1) protease and reverse transcriptase (RT) genes is an ongoing problem in the fight against AIDS. HIV's mutation rate is high, approximately 4 × 10−5 mutations per base per replication cycle, or about one mutation every three generations, yielding at least 1014 mutations per day worldwide, based on available replication rates and AIDS population estimates (2, 12, 15). This can lead to the rapid development of resistance to new drug treatments. Researchers and clinicians have made enormous efforts to identify drug-resistant mutations in HIV protease and reverse transcriptase (RT), the molecular targets of the 18 antiretroviral drugs currently approved by the Food and Drug Administration. The discovery of a new drug-resistant mutation typically requires a combination of clinical data (e.g., AIDS patients displaying drug resistance), sequencing, and basic science (e.g., obtaining viral samples and performing phenotypic assays). Fast, automatic methods for identifying drug-resistant mutations could be of great value for researchers studying HIV and other pathogens.
Fortunately, the rapid evolution of HIV itself may provide a powerful tool for gaining understanding of its function in general and drug resistance in particular. HIV's high mutation rate is essentially performing a saturating mutagenesis experiment that in principle could reveal the detailed selection pressures for every possible mutation. The question is how to best read out this detailed information and make use of it.
In evolutionary biology, one important tool for characterizing selection pressure is the ratio of observed amino acid mutations over observed synonymous mutations (nucleotide mutations that do not change the amino acid translation), often referred to as Ka/Ks (amino acid mutations over synonymous mutations) or dn/ds (nonsynonymous mutations over synonymous mutations). Since amino acid mutations, but not synonymous mutations, experience selection pressure due to their effect on protein function, their ratio gives a straightforward measure of this selection pressure. Throughout this paper we will use the term Ka/Ks, which is normalized by the ratio expected under a random mutation model (i.e., in the absence of any selection pressure) (10). A Ka/Ks value of 1 indicates neutral selection, i.e., the observed ratio of mutations that cause amino acid changes versus those that do not exactly matches the ratio expected under a random mutation model. Thus, amino acid changes are neither being selected for nor against. A Ka/Ks value of <1 indicates negative selection pressure. That is, most amino acid changes are deleterious and are selected against, producing an imbalance in the observed mutations that favors synonymous mutations. Much less common is positive selection (Ka/Ks > 1), indicating that amino acid changes are favored, i.e., they increase the organism's fitness. This unusual condition may reflect a change in the function of a gene or a change in environmental conditions that forces the organism to adapt. For example, HIV mutations which confer resistance to new antiviral drugs might be expected to undergo positive selection in a patient population treated with these drugs.
In this paper we present a large-scale study of the value of positive selection for detecting drug-resistant mutations in HIV protease and RT. Ordinarily, Ka/Ks is measured as a single value for an entire gene (10). This can reveal very interesting positive selection events in the evolution of an organism, but unfortunately the overall Ka/Ks values for HIV protease and RT provide no extraordinary result (they are negative [18], as in most genes). A more interesting question is whether positive selection can be observed at the level of individual mutations rather than by pooling all data for the entire gene. However, this would require very large amounts of mutation data to obtain a statistically significant Ka/Ks result for each individual mutation.
To solve this problem, we have performed automated mutation analysis of approximately 40,000 HIV samples from AIDS patients sequenced by Specialty Laboratories from 1999 to mid-2002. This massive data set provides essentially complete mutagenesis in the regions sequenced (including protease codons 1 to 99 and RT codons 1 to 381). More importantly, it enables the calculation of accurate Ka/Ks values at each codon, and even for each individual amino acid within that codon, with high statistical significance. Using these data, we have found that positive selection detects most of the known drug-resistant mutations and discovered many new mutations that are strong candidates for drug resistance or other key functional changes in HIV. The positive selection map and complete mutation data for the 40,000 HIV samples can be of great value to the AIDS research community.
MATERIALS AND METHODS
Sequencing chromatogram preparation and analysis.
All sequencing was performed at Specialty Laboratories Inc. by using an HIV-1 GenotypR assay (22), from which individual patient identification information was removed prior to use in this study. The sequenced region included codons 1 to 99 of the protease gene and codons 1 to 381 of the RT gene (Fig. 1). Nucleotide sequences (six per patient), obtained from Specialty Laboratories' assay, were analyzed by using PHRED (3) to produce base calls and quality factors. In the cases of nucleotide mixtures, only the major (highest) peak was reported. The original clinical results of manual analysis were obtained at Specialty Laboratories by use of the Sequence Navigator software.
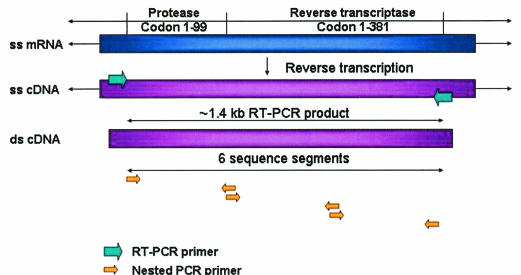
FIG. 1.
RT-PCR and sequencing of the HIV-1 protease and RT regions. HIV-1 RNA was isolated from AIDS patient plasma samples. Reverse transcription was performed to obtain the cDNA from single-stranded viral RNA. The HIV protease and RT region around 1.4 kb was amplified by PCR using two (forward and backward) unique primers. This was followed by a nested PCR, which split the target sequence into three shorter fragments with the use of six unique primers. These fragments were then cycle sequenced in forward and reverse directions.
Specialty Laboratories estimated that more than 99% of the samples that they sequenced are of subtype B. We compared all of the sequences in our data set against HIV-1 subtype reference sequences from the Los Alamos database by using the program BLAST and assigned each sample to the subtype with the highest identity. This analysis indicated that 99.28% of the samples are HIV-1 subtype B.
Single nucleotide polymorphism (snp) scoring and identification.
To identify real mutations and distinguish them reliably from possible sequencing errors, all six chromatogram reads for each sample were aligned against the subtype B reference sequence (GenBank accession no. GI9629357) and analyzed by the programs POA and snp_assess as previously described (5-7, 9). For each candidate mutation, snp_assess calculated the log odds ratio (LOD) of the probability that it is a true mutation versus the probability that it is a sequencing error. An LOD value of >3 implies that the likelihood of a sequencing error is less than 10−3 (Fig. 2).
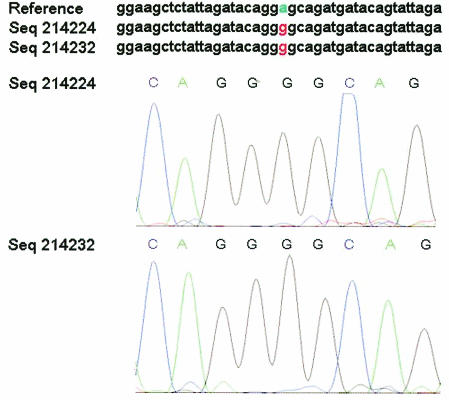
FIG. 2.
Chromatogram evidence for an HIV-1 protease mutation. The program snp_assess identified an A→G mutation with an LOD score of 11.6 (top). Chromatograms for the forward (Seq214224) and reverse (Seq214232) strand sequencing are shown in the lower panels. Seq214232 is shown in reverse complement for the purposes of comparison.
Manual verification of HIV mutations.
To validate the high-throughput mutation detection results, we randomly selected 4,043 samples and compared our results at 144 nucleotide positions with mutations previously reported for these samples by Specialty Laboratories (using manual examination of the base calls and chromatograms for clinical reporting of known drug-resistant mutations). Of 24,831 mutations detected by our procedure, 17,256 were independently verified by Specialty Laboratories' archived manual reports. Since the number of known clinically significant mutations and, hence, the number of positions manually scored by Specialty Laboratories varied during this period (from approximately 90 nucleotides in 1999 to all 144 in 2002), a 100% match was not expected. We randomly selected 40 of the 7,575 unverified mutations and manually examined the raw chromatogram data at Specialty Laboratories. All 40 were present and verified as real mutations, as judged by clear chromatogram peaks and agreement between sequencing of both strands. Thus, the false-positive rate in the mutation data reported in this paper is likely to be below 1%. Specialty Laboratories also reported 2,723 mutations at “mixture positions” (where the chromatogram indicated that a mix of more than one nucleotide was present). Of these, 1,236 mutations were also reported by our automatic procedure. Assuming that all 1,487 of the remaining mixture mutations are correct, this indicates a false-negative rate of under 6% [1,487/(24,831 + 1,487), where 24,831 is the number of mutations detected by our procedure].
Calculation of Ka/Ks for specific amino acid substitutions.
Our calculation is based on the definition of Ka/Ks developed by Li (10). The main differences of our approach are that (i) instead of calculating Ka/Ks for an individual gene or codon, we calculate an individual Ka/Ks value for each specific amino acid mutation; (ii) we follow the definition of Ka/Ks as normalized by a random mutation model (i.e., no selection pressure, described in detail below), unlike some treatments of dn/ds (25); (iii) HIV has a high transition/transversion ratio (20), which must be taken into account for an accurate Ka/Ks calculation. We first measured the transition and transversion frequencies ft and fv from the entire data set, according to the following formulas: ft = Nt/ntS and fv = Nv/nvS, where S is the total number of samples; Nt and Nv are the numbers of observed transition and transversion mutations, respectively; nt is the number of possible transitions in the region that was sequenced (simply equal to its length L in nucleotides); and nv is the number of possible transversions (equal to 2L). For this calculation, we used all of the nucleotides in the region that was sequenced. It is also possible to perform this calculation specifically on silent nucleotide positions (i.e., nucleotides where all possible mutations are synonymous); however, we have followed the more conservative approach of using all nucleotides, in keeping with previously published work (20). In this calculation (and all others below) we counted only single nucleotide substitutions; all other mutations were excluded.
The definition of Ka/Ks can be extended to a specific amino acid substitution (X→Y) at a codon by calculating the ratio of NY, the count of X→Y mutations observed at that codon, over Ns, the count of synonymous mutations observed at that codon. This NY/Ns ratio is then normalized by the ratio expected under a random mutation model (i.e., in the absence of any selection pressure), according to the following formula:
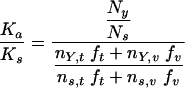 |
where nY,t is the number of possible transition mutations in the codon that would change X to Y, ns,t is the number of possible transition mutations in the codon that are synonymous, and nY,v and ns,v are the equivalent numbers for transversions. When the observed ratio (NY/NS) is greater than the expected ratio (in the denominator of the expression above), the selection pressure Ka/Ks is greater than 1 and we say that the mutation X→Y exhibits positive selection pressure.
We calculated an LOD confidence score for a mutation X→Y to be under positive selection pressure according to the following formula:
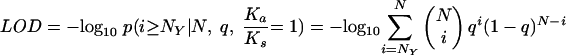 |
where N is the total number of mutations observed in the codon, and q is calculated as follows:
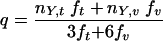 |
.
Drug resistance prediction P values.
Given n mutations with positive selection of a total of N mutations, we calculated the log probability of predicting at least m drug-resistant mutations by random chance (of the total number, M, of known drug-resistant mutations), according to the following hypergeometric distribution:
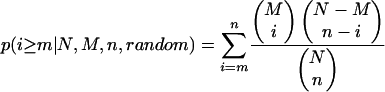 |
For example, of the 527 (N) amino acid changes in protease, we identified 69 (n) that displayed positive selection (Ka/Ks > 1) with LOD scores of 2 or greater (Table 1). Of a total of 52 (M) known drug-resistant mutations in protease, 25 (m) of these were identified among our 69 positively selected mutations. Thus, the P value for obtaining this result by random chance calculated from the above expression is 10−10.4.
TABLE 1.
Positive-selection pressure in protease
| Codon | Mutationa | Ka/Ks | LOD | Codon | Mutationa | Ka/Ks | LOD | |
|---|---|---|---|---|---|---|---|---|
| 7 | Anyb | 0.25 | ||||||
| Q7E | 5.82 | >300 | ||||||
| 10c | Any | 1.05 | 17.68 | |||||
| L101 | 16.97 | >300 | ||||||
| L10V | 2.59 | 10.79 | ||||||
| 12 | Any | 6.92 | 215.81 | |||||
| T12S | 48.69 | 11.30 | ||||||
| T12P | 34.36 | 11.63 | ||||||
| T12K | 12.01 | >300 | ||||||
| 13 | Any | 64.15 | 149.38 | |||||
| I13V | 221.34 | >300 | ||||||
| 15 | Any | 1.31 | 4.81 | |||||
| I15V | 4.46 | >300 | ||||||
| 19 | Any | 1.95 | 125.90 | |||||
| L19I | 17.37 | >300 | ||||||
| L19V | 3.73 | >300 | ||||||
| L19Q | 3.49 | >300 | ||||||
| 20 | Any | 0.50 | ||||||
| K20T | 2.52 | 11.13 | ||||||
| K20M | 2.11 | 11.26 | ||||||
| 24 | Any | 0.57 | ||||||
| L24I | 7.95 | 11.80 | ||||||
| 30 | Any | 1.33 | 9.75 | |||||
| D30N | 3.57 | >300 | ||||||
| 33 | Any | 3.56 | 218.37 | |||||
| L33V | 14.81 | 11.24 | ||||||
| L33F | 12.85 | >300 | ||||||
| L33I | 11.94 | >300 | ||||||
| 34 | Any | 0.18 | ||||||
| E34Q | 2.11 | 12.74 | ||||||
| 35 | Any | 10.23 | >300 | |||||
| E35D | 118.38 | >300 | ||||||
| 37 | Any | 107.57 | >300 | |||||
| S37N | 275.40 | >300 | ||||||
| 39 | Any | 2.12 | 26.08 | |||||
| P39Q | 11.74 | >300 | ||||||
| P39T | 4.26 | >300 | ||||||
| P39S | 3.18 | >300 | ||||||
| 41 | Any | 3.35 | >300 | |||||
| R41K | 8.59 | >300 | ||||||
| 43 | Any | 0.24 | ||||||
| K43T | 2.62 | >300 | ||||||
| 45 | Any | 1.49 | 5.55 | |||||
| K45R | 3.54 | 8.85 | ||||||
| K45Q | 3.27 | 139.96 | ||||||
| 47 | Any | 1.03 | 0.29 | |||||
| I47V | 3.52 | 142.44 | ||||||
| 48 | Any | 0.24 | ||||||
| G48V | 5.06 | 10.93 | ||||||
| 50 | Any | 0.44 | ||||||
| I50V | 1.12 | >300 | ||||||
| 53 | Any | 1.82 | 10.13 | |||||
| F53Y | 3.63 | 7.02 | ||||||
| F53L | 3.57 | 112.78 | ||||||
| 54 | Any | 2.12 | 91.45 | |||||
| I54L | 6.57 | 10.40 | ||||||
| I54M | 4.39 | 10.44 | ||||||
| I54V | 4.00 | >300 | ||||||
| 55 | Any | 0.78 | ||||||
| K55R | 1.98 | 116.02 | ||||||
| 57 | Any | 2.06 | 77.61 | |||||
| R57K | 5.30 | >300 | ||||||
| 58 | Any | 0.11 | ||||||
| Q58E | 2.47 | 10.96 | ||||||
| 59 | Any | 0.06 | ||||||
| Y59F | 1.25 | 5.81 | ||||||
| 60 | Any | 2.76 | 96.01 | |||||
| D60E | 32.13 | >300 | ||||||
| 61 | Any | 0.11 | ||||||
| Q61E | 1.92 | 10.77 | ||||||
| 62 | Any | 6.05 | 119.07 | |||||
| I62V | 20.78 | >300 | ||||||
| 63 | Any | 5.33 | >300 | |||||
| L63P | 12.68 | >300 | ||||||
| 64 | Any | 2.86 | 49.66 | |||||
| I64V | 8.13 | >300 | ||||||
| I64L | 5.17 | 10.30 | ||||||
| 65 | Any | 0.36 | ||||||
| E65D | 4.18 | 10.81 | ||||||
| 69 | Any | 0.95 | ||||||
| H69Q | 6.68 | 11.06 | ||||||
| 71 | Any | 4.79 | >300 | |||||
| A71V | 7.77 | >300 | ||||||
| A71T | 4.00 | 9.73 | ||||||
| 72 | Any | 0.90 | 0.01 | |||||
| I72V | 1.59 | >300 | ||||||
| I72L | 1.06 | 2.59 | ||||||
| 73 | Any | 2.13 | 49.67 | |||||
| G73S | 4.42 | >300 | ||||||
| G73C | 4.40 | >300 | ||||||
| G73A | 2.42 | 2.80 | ||||||
| 74 | Any | 5.84 | 143.17 | |||||
| T74S | 88.12 | >300 | ||||||
| T74P | 10.82 | >300 | ||||||
| 76 | Any | 0.19 | ||||||
| L76V | 2.79 | >300 | ||||||
| 77 | Any | 11.02 | >300 | |||||
| V77I | 27.08 | >300 | ||||||
| 82 | Any | 3.07 | 187.52 | |||||
| V82A | 6.29 | >300 | ||||||
| V82F | 2.99 | 2.73 | ||||||
| 84 | Any | 2.12 | 12.19 | |||||
| I84V | 7.33 | >300 | ||||||
| 85 | Any | 1.26 | 2.41 | |||||
| I85V | 3.22 | 149.56 | ||||||
| 88 | Any | 1.21 | 188.09 | |||||
| N88D | 1.21 | 188.09 | ||||||
| 89 | Any | 3.53 | 10.79 | |||||
| L89M | 3.53 | 10.79 | ||||||
| L89V | 1.89 | 10.91 | ||||||
| 90 | Any | 4.06 | >300 | |||||
| L90M | 248.57 | >300 | ||||||
| 92 | Any | 0.08 | ||||||
| K92K | 1.31 | 10.66 | ||||||
| 93 | Any | 13.27 | >300 | |||||
| I93L | 447.66 | >300 |
Known drug-resistant mutations are highlighted in bold.
Any, we calculated the Ka/Ks ratio for the total set of amino acid mutations at this codon.
Codon 10 has an LOD score of <2 when only single nucleotide substitutions are counted. But its LOD score is >2 when both single and multiple nucleotide substitutions are counted.
We used the following known drug-resistant mutation positions: in protease, positions 8, 10, 20, 24, 30, 32, 33, 36, 46, 47, 48, 50, 53, 54, 63, 71, 73, 77, 82, 84, 88, 90, and 93 (8, 19, 23, 26); in RT, positions 41, 44, 62, 65, 67, 69, 70, 74, 75, 77, 98, 100, 101, 103, 106, 108, 115, 116, 118, 151, 179, 181, 184, 188, 190, 210, 215, 219, 225, 227, 230, 234, 236, and 333 (17, 19).
RESULTS
Automated HIV mutation detection.
Raw chromatogram data from sequencing of 39,767 AIDS patient plasma samples were provided by Specialty Laboratories, obtained from HIV-1 GenotypR assays performed at Specialty Laboratories. Each base was sequenced at least twice (on complementary strands), and putative mutations were scored by using the program snp_assess (7), which takes into account local chromatogram quality, sequence context, the agreement between all the reads, and detailed sequencing error statistics as previously measured from 400 × 106 bp of chromatogram reads. Our data are from samples that are almost exclusively HIV-1 subtype B (see Materials and Methods).
We identified 1,923,620 candidate mutations in these samples, of which 1,830,097 had high LOD scores (LOD > 3; throughout this paper we will focus on mutations with LOD scores of >3). Manual verification indicates that the false-positive rate in the mutation data presented in this paper is less than 1% (see Materials and Methods). This sequencing covers the whole protease gene (297 bp) and the first 1,143 bp of the RT gene. The average number of mutations (compared to the subtype B reference) was 31.96 per kb overall, 29.57/kb in HIV protease, and 32.58/kb for RT. Approximately 349,000 mutations were detected in protease, and 1.48 million were detected in RT. This represented 1,148 distinct codon mutations in protease and 3,873 in RT. On average, each distinct mutation was observed in 364 independent samples, corresponding to an allele frequency of 0.92%. The overall ratio of transition to transversion was 8.75, indicating that the HIV-1 pol enzyme has a very strong bias towards transition substitutions. This result is consistent with a previous report (20).
We identified 232,299 amino acid mutations in protease and 586,192 mutations in RT. These subdivided into 528 distinct amino acid changes at 91 codon positions for protease and 1,964 distinct amino acid changes at 361 codon positions for RT. Thus, the average population frequency of each amino acid change was 1.1% in protease and 0.75% in RT. We detected 5.33 distinct amino acid changes per codon in protease and 5.15 per codon in RT.
Positive selection mapping of individual amino acid mutations.
To relate these polymorphism data to their potential impact on protein function, we mapped all mutations onto the HIV-1 subtype B protein sequences for the protease (amino acids 1 to 99) and RT (amino acids 1 to 381) proteins. Overall, the Ka/Ks value for this region is 0.2687, indicating that it is under negative selection. This is consistent with previous reported results (18).
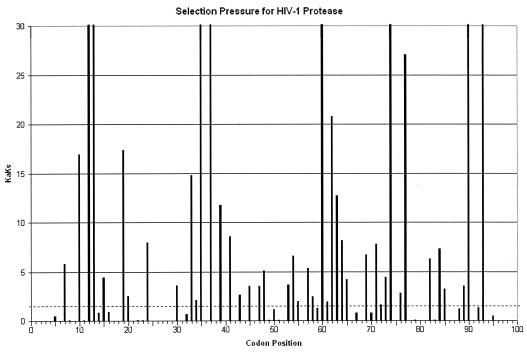
To seek drug-resistant mutations, we mapped positive selection pressure throughout the sequenced region by calculating a Ka/Ks value for each amino acid mutation. These results show dramatic differences in Ka/Ks at different positions in the proteins and strongly positive selection pressure at individual amino acids (Fig. 3 and 4). In marked contrast to the overall pattern of negative selection pressure in this region, we observed Ka/Ks values of >1 (i.e., positive selection) for 69 individual mutations in protease and 142 mutations in RT and Ka/Ks values of >10 for 20 mutations in protease and 47 mutations in RT. To assess the statistical significance of these results, we also calculated a P value for each mutation, giving the probability of the observed results under the assumption of neutral selection pressure (i.e., Ka/Ks = 1; see Materials and Methods for details of the P value calculation). By using a 1% statistical significance threshold, our positive-selection results are statistically significant (P values of <10−10 in most cases [Tables 1 and 2 ]).
FIG. 3.
Positive selection mapping of HIV-1 protease from 40,000 patient samples. The Ka/Ks value represents the greatest selection pressure among all the individual amino acid mutations at each codon. The dotted line indicates the Ka/Ks value of 1.
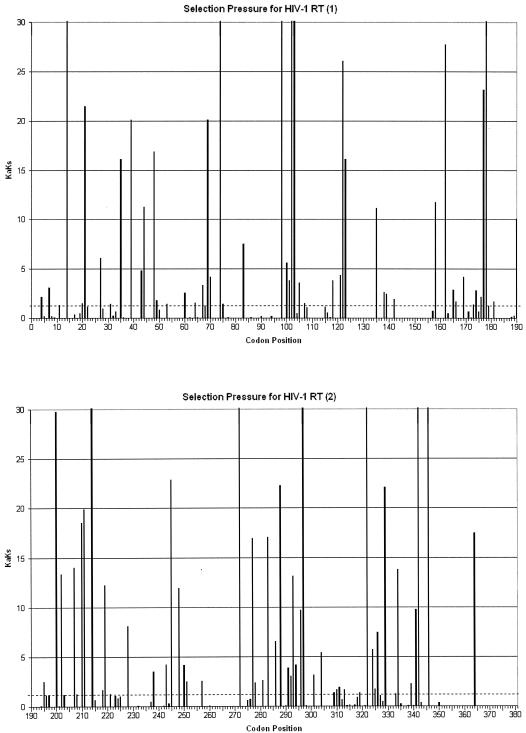
FIG. 4.
Positive selection mapping of HIV-1 RT from 40,000 patient samples. The Ka/Ks value represents the greatest selection pressure among all the individual amino acid mutations at each codon. The dotted line indicates the Ka/Ks value of 1.
TABLE 2.
Positive-selection pressure in RTa
| Codon | Mutationb | Ka/Ks | LOD | Codon | Mutationb | Ka/Ks | LOD | |
|---|---|---|---|---|---|---|---|---|
| 35 | Anyc | 8.35 | >300 | |||||
| V35I | 16.13 | >300 | ||||||
| V35L | 16.81 | >300 | ||||||
| 39 | Any | 9.14 | >300 | |||||
| T39A | 20.08 | >300 | ||||||
| T39K | 10.88 | 9.85 | ||||||
| T39S | 9.44 | 5.22 | ||||||
| 60d | Any | 1.05 | 2.02 | |||||
| V60I | 2.59 | >300 | ||||||
| 67 | Any | 1.34 | 32.92 | |||||
| D67N | 3.34 | >300 | ||||||
| 69 | Any | 1.23 | 6.78 | |||||
| T69N | 20.10 | >300 | ||||||
| T69S | 2.20 | >300 | ||||||
| 70 | Any | 1.58 | 53.74 | |||||
| K70R | 4.16 | >300 | ||||||
| 74 | Any | 2.97 | 281.60 | |||||
| L74V | 32.47 | >300 | ||||||
| L74I | 11.26 | >300 | ||||||
| 83 | Any | 2.91 | 288.93 | |||||
| R83K | 7.49 | >300 | ||||||
| 98 | Any | 2.68 | 159.41 | |||||
| A98S | 41.52 | >300 | ||||||
| A98G | 16.13 | 10.46 | ||||||
| 102 | Any | 26.13 | 254.90 | |||||
| K102Q | 394.42 | >300 | ||||||
| 103 | Any | 2.75 | >300 | |||||
| K103N | 30.46 | >300 | ||||||
| 118 | Any | 1.55 | 43.59 | |||||
| V118I | 3.81 | >300 | ||||||
| 122 | Any | 9.85 | >300 | |||||
| E122K | 26.07 | >300 | ||||||
| 123 | Any | 1.57 | 100.62 | |||||
| D123E | 16.10 | >300 | ||||||
| 135 | Any | 5.39 | 193.86 | |||||
| I135T | 11.15 | >300 | ||||||
| I135L | 6.71 | 9.95 | ||||||
| 162 | Any | 1.26 | 14.56 | |||||
| S162C | 27.73 | >300 | ||||||
| 165 | Any | 1.19 | 3.10 | |||||
| T165I | 2.87 | 261.12 | ||||||
| 177 | Any | 2.07 | 178.07 | |||||
| D177E | 23.12 | >300 | ||||||
| 178 | Any | 4.95 | 89.14 | |||||
| I178L | 30.36 | 10.26 | ||||||
| I178M | 9.60 | >300 | ||||||
| 200 | Any | 15.83 | >300 | |||||
| T200A | 29.77 | >300 | ||||||
| 202 | Any | 3.90 | 31.08 | |||||
| I202V | 13.38 | >300 | ||||||
| 210 | Any | 1.35 | 49.64 | |||||
| L210W | 18.55 | 9.80 | ||||||
| 211 | Any | 8.41 | >300 | |||||
| R211K | 19.93 | >300 | ||||||
| 214 | Any | 45.78 | >300 | |||||
| L214F | 112.47 | >300 | ||||||
| 219d | Any | 1.01 | 6.62 | |||||
| K219Q | 12.25 | 10.14 | ||||||
| K219N | 1.77 | 10.54 | ||||||
| 245 | Any | 1.87 | 152.83 | |||||
| V245E | 22.90 | 10.50 | ||||||
| V245M | 1.85 | >300 | ||||||
| 248 | Any | 1.09 | 2.07 | |||||
| E248D | 11.93 | >300 | ||||||
| 272 | Any | 47.06 | >300 | |||||
| P272A | 914.59 | >300 | ||||||
| 277 | Any | 6.60 | >300 | |||||
| R277K | 16.96 | >300 | ||||||
| 286 | Any | 2.79 | >300 | |||||
| T286A | 6.53 | >300 | ||||||
| T286P | 2.58 | 3.48 | ||||||
| 288 | Any | 1.27 | 13.49 | |||||
| A288S | 22.26 | >300 | ||||||
| 292 | Any | 1.23 | 8.57 | |||||
| V292I | 3.02 | >300 | ||||||
| 293 | Any | 3.80 | 195.08 | |||||
| I293V | 13.12 | >300 | ||||||
| 297 | Any | 7.19 | >300 | |||||
| E297A | 38.16 | 10.09 | ||||||
| E297K | 13.67 | >300 | ||||||
| E297Q | 7.95 | 10.23 | ||||||
| 322 | Any | 3.16 | 159.17 | |||||
| S322T | 53.16 | >300 | ||||||
| S322A | 13.33 | 10.45 | ||||||
| 326 | Any | 2.39 | 15.26 | |||||
| I326V | 7.45 | >300 | ||||||
| 329 | Any | 2.30 | 33.45 | |||||
| I329L | 22.07 | >300 | ||||||
| I329V | 2.90 | 10.34 | ||||||
| 334 | Any | 1.32 | 23.67 | |||||
| Q334L | 13.77 | 10.46 | ||||||
| Q334E | 10.49 | 10.46 | ||||||
| Q334H | 2.43 | 10.51 |
Due to the great number of amino acid mutations (142) under positive-selection pressure, only those (55) at codons with a Ka/Ks value of >1 for the total set of mutations at that codon are listed here.
Known drug-resistant mutations are highlighted in bold.
Any, we calculated the Ka/Ks ratio for the total set of amino acid mutations at this codon.
Codon has an LOD score of <2 when only single nucleotide substitutions are counted. But its LOD score is >2 when both single and multiple nucleotide substitutions are counted.
These positive selection results were also highly specific. We observed very different Ka/Ks values for different mutations at a given individual position (Tables 1 and 2). For example, at Ile 93 in protease, the mutation I93L had a Ka/Ks value of 447.66 (P < 10−300), but other mutations at this position did not show statistically significant positive selection pressure (Ka/Ks values of 4.53, 0.05, 0.04, 0.01 for I93M, I93F, I93V, and I93T, respectively). Indeed, these results demonstrate the importance of detecting positive selection pressure at the level of individual mutations rather than for an entire codon as has been previously described (4, 21, 25). We compared our results with Ka/Ks values calculated for each individual codon (by pooling the observation counts for all nonsynonymous mutations at that codon; see Materials and Methods for details). In some cases, a positive selection detected for an individual mutation could also be detected at the codon level (e.g., the T12S mutation had a Ka/Ks value of 49, whereas a mutation of T12 to any amino acid had a codon Ka/Ks value of 6.9). However, in many other cases the codon Ka/Ks calculation failed to detect strong positive selection that was easily detectable at the level of individual mutations (e.g., the G48V mutation had a Ka/Ks value of 5.1, but the mutation of G48 to any amino acid had a codon Ka/Ks value of 0.24). For protease, of 47 positions for which we detected positive selection (Ka/Ks values of >1 for individual mutations), 19 were not detected by the codon-based calculation (40%). This is perhaps not surprising. Different amino acids at the same position are expected to experience different selection pressures. Calculating an overall Ka/Ks for an entire codon can obscure positive selection of a single amino acid at that codon. If other amino acid replacements at that position are negatively selected, the overall Ka/Ks for the codon might indicate negative selection.
Positive selection of drug-resistant mutations.
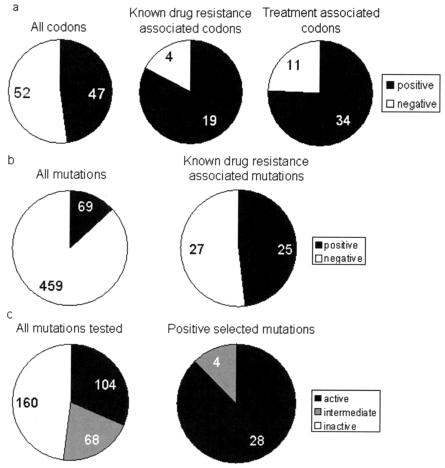
Positive selection mapping identified the majority of drug-resistant mutation positions identified in the published literature for HIV protease (Fig. 5a). We identified 47 positions in protease that showed positive selection of individual mutations. Notably, 19 of these 47 positions are known to be associated with drug resistance. Thus, positive selection mapping identified most (83%) of the 23 known drug-resistant mutation positions in protease (Fig. 5a). This is a statistically significant match. The P value for obtaining this result by random chance is 10−3.3. Moreover, the known drug-resistant mutations at these positions matched the amino acid changes that we observed to be positively selected (Table 1). It should be noted that at two of the four known drug-resistant mutations that we missed (Met 36 and Met 46), no synonymous mutations are possible (all mutations change the amino acid), and therefore we could not even calculate a Ka/Ks ratio there.
FIG. 5.
Positive selection identifies drug resistance and positive fitness mutations. (a) Identification of codons with positive selection, either from the set of all positions in HIV protease (All codons), positions reported in the literature as sites of drug-resistant mutations (Known drug resistance associated codons), or positions reported as sites of mutations specifically associated with adaptation to drug treatment (Treatment associated codons). (b) Identification of specific amino acid mutations with positive selection, either from the set of all HIV protease mutations found in our data set (All mutations), or mutations reported in the literature as causing drug resistance (Known drug resistance associated mutations). (c) Phenotypic fitness, as measured by a protease activity assay by Loeb et al., for a random sample of HIV protease mutants (All mutations tested), or the subset of those mutations found to have positive selection in our study (Positive selected mutations). active, protease mutants with normal or greater-than-wild-type proteolytic activity; intermediate, partial cleavage was observed in the assay; inactive, no proteolytic cleavage was observed.
Positive selection mapping yielded similar results in RT. One hundred ten positions stood out from the background of negative selection, with strongly positive Ka/Ks values and high LOD scores. Twenty of these positions correspond to known drug-resistant mutations (there are 34 known drug resistance-associated positions in RT). The P value for obtaining this result by random chance is 10−3.9. The strongest Ka/Ks values were at positions 272 (914.59), 102 (394.42), and 214 (112.47). These are novel results. The functional significance of positive selection at these positions is unknown, although two of the positions are immediately adjacent to known drug-resistant mutations (K101Q, K103N, and T215F).
The statistical significance of our results becomes even stronger when evaluated at the level of individual mutations. Of the 527 amino acid changes in protease, we identified 69 that displayed positive selection (Ka/Ks > 1) with LOD scores greater than 2 (Table 1). Twenty-five of these corresponded to known drug-resistant mutations, of a total of 52 known to exist in protease. This is a statistically significant match: the P value for obtaining this result by random chance is 10−10.4. Of the 2,255 amino acid changes that we identified in RT, 142 demonstrated positive selection (LOD score > 2) (Table 2). Of these, 23 matched known drug-resistant mutations. The P value for obtaining this result (of the 55 known drug-resistant mutations in RT, by random chance) is 10−13.9.
Comparison with independent drug treatment studies for HIV protease.
One basic weakness of our data set is the lack of drug treatment histories for the individual patients. Not only do we lack information about what specific treatment a patient received, but also many of the samples may come from patients who have not been treated with any HIV drugs. We therefore compared our results with a carefully controlled independent study that identified mutations associated with specific drug treatments (24). Rather than calculating Ka/Ks, this study kept a detailed drug treatment history for each patient and measured the change in the frequency of each mutation among patients treated with a given set of drugs from that of patients not treated with those drugs. By comparing 1,004 HIV isolates from untreated patients with 1,240 HIV isolates from patients treated with one or more protease inhibitors, Wu et al. identified 45 positions in HIV protease where mutations were specifically associated with drug treatment.
Our Ka/Ks data match the results of Wu et al. closely. Of the 47 positions in protease identified by our positive selection mapping, 34 matched those found by Wu et al. (Fig. 5a). This is a statistically significant result (P < 10−5.2). It is striking that Ka/Ks mapping of a random sample of HIV sequences, with no drug treatment information whatsoever, finds the majority (76%) of drug-resistant mutations identified by a careful study of specific drug treatments (24).
Comparison with independent assays of phenotypic fitness for HIV protease.
Positive selection mapping should yield important information not only about drug resistance but also about mutations that improve viral fitness in other ways. To test this hypothesis, we also compared our results to the exhaustive site-directed mutagenesis results of Loeb et al., who constructed and assayed the biochemical activity of approximately 50% of all point mutants of HIV-1 subtype B protease (11). These data demonstrate that our positive selection mapping detects not only drug resistance but also key determinants of fitness (Fig. 5c). While the set of all mutations tested by Loeb et al. was strongly biased towards negative activity (no detectable protease activity), with a smaller number of positive (normal) activity and intermediate activity, the mutations detected by our positive selection metric were almost entirely of normal or increased activity. This is a significant result, with a P value of 10−16.6.
DISCUSSION
Large-scale clinical mutation database for HIV-1 protease and RT.
We have produced a large-scale analysis of polymorphisms in the HIV-1 protease and RT regions, based on sequencing of clinical samples from the United States, representative of HIV-1 subtype B. Our present data set includes 39,767 individual AIDS patient plasma samples and 1,830,097 detected HIV mutations. This database provides a much larger data set for understanding recent HIV evolution and functional pressure than has previously been available (for two recent examples, see references 16 and 24) and can be useful for many different applications. Our mutation analysis will be available upon publication to academic researchers at http://www.bioinformatics.ucla.edu/HIV.
A major difficulty in anti-HIV drug development is the rapid selection of mutations in the viral genome that confer drug resistance by means of resultant changes within the protein target. Current clinical and academic research has been focused on some known codon positions that are associated with drug resistance. It is very important to detect and understand the pattern of these mutations. Due to the polymorphic nature of the HIV virus genome and the complexity of the drug resistance mechanisms, other codon positions may also play a role in the development of drug resistance. Our calculation of selection pressure for all the codons on the HIV-1 protease and RT regions can help to identify important codon positions that might affect drug resistance. Despite the fact that the Specialty Laboratories data set included no drug treatment information for the patients, our Ka/Ks calculations successfully identified 76% of mutations found to be associated with drug resistance through clinical studies.
Our approach differs from previous work in several ways. First, a number of studies have examined the problem of calculating selection pressure for individual sites in a protein (4, 14, 21, 25). However, our approach calculates Ka/Ks for each observed amino acid mutation, instead of combining all observed mutations for a site into a single Ka/Ks value. Our data for protease indicate that pooling multiple mutations for a site can obscure a large fraction (40%) of the positively selected sites that can be detected at the level of individual amino acid mutations. Second, we have not made use of any drug treatment information; our method does not require it, and the Specialty Laboratories data set did not include it. By contrast, Wu et al. identified drug-resistant mutations without considering Ka/Ks by comparing the frequency of each mutation in two groups: patients that received a specific drug treatment regime and a control population of untreated patients (24). Third, because our data reflect a single subtype (B), we have not considered phylogeny relationships or ancestral genotype in our analysis. For populations that contain important phylogenetic structure, it would be better to measure Ka/Ks in a way that takes this structure into account, as has been previously described (4). Finally, the base-calling software that we used (PHRED) does not report minor peaks when two or more nucleotide bases are present as a mixture at a given position in the chromatogram. Such minor peaks can be of great interest and deserve further analysis.
Impact of mutation on protein function.
There are 23 known HIV-1 protease inhibitor (PI) drug-resistant mutations that have been mapped onto the terminal domain (positions 8, 88, 90, and 93), the core domain (positions 10, 20, 24, 30, 32, 63, 71, 73, 77, 82, and 84), and the flap domain (positions 33, 36, 46, 47, 48, 50, 53, and 54), respectively (8, 18, 21, 24). Nineteen of them were detected by positive selection mapping in our data. The high proportion of known drug-resistant positions detected by this approach (76 to 83%) suggests that it could provide a relatively useful and reliable new tool for detecting important new drug-resistant mutations.
Indeed, our positive selection analysis does detect 28 novel positions in protease that may be important functional determinants but the significance of which is currently unknown. Some of these positions (e.g., 35, 37, 62, 64, 72, 74, and 85) are adjacent to codon positions known to be associated with drug resistance (e.g., 36, 63, 71, 73, and 85). But most of the newly identified positions are not near the active site. Instead, they are primarily located in the core domain (e.g., positions 12, 13, 15, 19, 60, 62, and 64) and flap domain (e.g., positions 35, 37, 39, 41, 45, and 57). They may affect either enzyme catalysis or dimer stability or reshape the active site through long-range structural perturbations (19). It is possible that some of the positively selected mutations may act as accessory mutations that improve viral fitness rather than directly interfering with drug binding (1).
Our comparison with the protease activity data of Loeb et al. (11) demonstrated that our approach also provides a useful window on important fitness determinants in the evolving viral population. Given that Ka/Ks measures reproductive fitness fairly directly, this is not unexpected. Most of the positively selected mutations observed were conservative amino acid changes (Table 1). Thus, while natural selection evidently favors amino acid changes at these positions, such changes appear to be constrained to preserve structure and function. New experiments will be required to assess whether any of these mutations acts as a primary cause of drug resistance or contributes to drug resistance via a secondary effect.
The situation for RT is even more complicated. Most of the known drug-resistant mutations are in the 5′ polymerase coding region, particularly in the “fingers” (codons 1 to 85 and 118 to 155) and “palm” (codons 86 to 117 and 156 to 237) subdomains (19). We have identified codon positions with positive selection pressure not only in the fingers and palm subdomains but also in the “thumb” subdomain (codons 238 to 318), which has seldom been mentioned in research on drug-resistant mutations before. In addition to affecting drug resistance and virus fitness, some of the newly identified positions might be epitopes for cell-mediated immunity (13). The importance of all these codon positions needs to be experimentally examined.
The amino acid-specific Ka/Ks data show distinct patterns of positive selection. For example, at protease codon 12 several amino acid changes were positively selected (T12K, T12P, and T12S), resulting in an overall Ka/Ks value of 6.92 for the codon. By contrast, at position 48 a single amino acid change (G48V) was positively selected, while the other possible amino acid changes were negatively selected, resulting in an overall Ka/Ks for the codon of 0.24 (negative selection). Such specificity may reveal important functional constraints in the evolution of the enzyme. G48V is strongly selected for (Ka/Ks value of 5.06) and has been shown to cause drug resistance (19). It is striking that other amino acid replacements at this position are not also favored, implying a significant functional constraint.
Acknowledgments
We thank A. Bakker, B. Boyadzhyan, I. Chen, M. Patnaik, C. Ramirez Kitchen, T. Schutzbank, R. Woodhall, and N. Wylie for valuable discussion and comments on this work.
This work was supported by U.C. Life Science Informatics grant 01-10090 and by funding from Specialty Laboratories, Inc. to C. Lee and L. Chen. C. Lee was supported by NIH grant 1P20MH065166-01.
REFERENCES
- 1.Bally, F., R. Martinez, S. Peters, P. Sudre, and A. Telenti. 2000. Polymorphism of HIV type 1 gag p7/p1 and p1/p6 cleavage sites: clinical significance and implications for resistance to protease inhibitors. AIDS Res. Hum. Retrovir. 16:1209-1213. [DOI] [PubMed] [Google Scholar]
- 2.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 3.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 4.Fitch, W. M., R. M. Bush, C. A. Bender, and N. J. Cox. 1997. Long term trends in the evolution of H(3) HA1 human influenza type A. Proc. Natl. Acad. Sci. USA 94:7712-7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu, G., B. Modrek, H. M. F. R. Stensland, J. Saarela, P. Pajukanta, V. Kustanovich, L. Peltonen, S. F. Nelson, and C. Lee. 2002. Efficient discovery of single-nucleotide polymorphisms in coding regions of human genes. Pharmacogenomics J. 2:236-242. [DOI] [PubMed] [Google Scholar]
- 6.Irizarry, K., G. Hu, M. L. Wong, J. Licinio, and C. Lee. 2001. Single nucleotide polymorphism identification in candidate gene systems of obesity. Pharmacogenomics J. 1:193-203. [DOI] [PubMed] [Google Scholar]
- 7.Irizarry, K., V. Kustanovich, C. Li, N. Brown, S. Nelson, W. Wong, and C. Lee. 2000. Genome-wide analysis of single-nucleotide polymorphisms in human expressed sequences. Nat. Genet. 26:233-236. [DOI] [PubMed] [Google Scholar]
- 8.Kempf, D. J., J. D. Isaacson, M. S. King, S. C. Brun, Y. Xu, K. Real, B. M. Bernstein, A. J. Japour, E. Sun, and R. A. Rode. 2001. Identification of genotypic changes in human immunodeficiency virus protease that correlate with reduced susceptibility to the protease inhibitor lopinavir among viral isolates from protease inhibitor-experienced patients. J. Virol. 75:7462-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, C., C. Grasso, and M. Sharlow. 2002. Multiple sequence alignment using partial order graphs. Bioinformatics 18:452-464. [DOI] [PubMed] [Google Scholar]
- 10.Li, W. H. 1993. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J. Mol. Evol. 36:96-99. [DOI] [PubMed] [Google Scholar]
- 11.Loeb, D. D., R. Swanstrom, L. Everitt, M. Manchester, S. E. Stamper, and C. A. I. Hutchinson. 1989. Complete mutagenesis of the HIV-1 protease. Nature 340:397-400. [DOI] [PubMed] [Google Scholar]
- 12.Mansky, L. M., and H. M. Temin. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439-1443. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen, R., and Z. Yang. 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148:929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582-1586. [DOI] [PubMed] [Google Scholar]
- 16.Rhee, S. Y., M. J. Gonzales, R. Kantor, B. J. Betts, J. Ravela, and R. W. Shafer. 2003. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 31:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schinazi, R. F., B. A. Larder, and J. W. Mellors. 1997. Mutations in retroviral genes associated with drug resistance. Int. Antivir. News 5:129-142. [Google Scholar]
- 18.Seibert, S. A., C. Y. Howell, M. K. Hughes, and A. L. Hughes. 1995. Natural selection on the gag, pol, and env genes of human immunodeficiency virus 1 (HIV-1). Mol. Biol. Evol. 12:803-813. [DOI] [PubMed] [Google Scholar]
- 19.Shafer, R. W., K. Dupnik, M. A. Winters, and S. H. Eshleman. 2001. A guide to HIV-1 reverse transcriptase and protease sequencing for drug resistance studies. In C. Kuiken, F. McCutchan, B. Foley, J. W. Mellors, B. Hahn, J. Mullins, P. Marx, and S. Wolinsky (ed.), HIV sequence compendium. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.M. [PMC free article] [PubMed]
- 20.Sharp, P. M., E. Bailes, F. Gao, B. E. Beer, V. M. Hirsch, and B. H. Hahn. 2000. Origins and evolution of AIDS viruses: estimating the time-scale. Biochem. Soc. Trans. 28:275-282. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki, Y., and T. Gojobori. 1999. A method for detecting positive selection at single amino acid sites. Mol. Biol. Evol. 16:1315-1328. [DOI] [PubMed] [Google Scholar]
- 22.Wilson, J. W., P. Bean, T. Robins, F. Graziano, and D. H. Persing. 2000. Comparative evaluation of three human immunodeficiency virus genotyping systems: the HIV-GenotypR method, the HIV PRT GeneChip assay, and the HIV-1 RT line probe assay. J. Clin. Microbiol. 38:3022-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winters, M. A., and T. C. Merigan. 2001. Variants other than aspartic acid at codon 69 of the human immunodeficiency virus type 1 reverse transcriptase gene affect susceptibility to nucleoside analogs. Antimicrob. Agents Chemother. 45:2276-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu, T. D., C. A. Schiffer, M. J. Gonzales, J. Taylor, R. Kantor, S. Chou, D. Israelski, A. R. Zolopa, W. J. Fessel, and R. W. Shafer. 2003. Mutation patterns and structural correlates in human immunodeficiency virus type 1 protease following different protease inhibitor treatments. J. Virol. 77:4836-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang, Z., R. Nielsen, N. Goldman, and A. M. Pedersen. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yusa, K., M. F. Kavlick, P. Kosalaraksa, and H. Mitsuya. 1997. HIV-1 acquires resistance to two classes of antiviral drugs through homologous recombination. Antivir. Res. 36:179-189. [DOI] [PubMed] [Google Scholar]