Abstract
Foot-and-mouth disease virus (FMDV) induces a very rapid inhibition of host cell protein synthesis within infected cells. This is accompanied by the cleavage of the eukaryotic translation initiation factor 4GI (eIF4GI). The cleavage of the related protein eIF4GII has now been analyzed. Within FMDV-infected cells, cleavage of eIF4GI and eIF4GII occurs with similar kinetics. Cleavage of eIF4GII is induced in cells and in cell extracts by the FMDV leader protease (Lpro) alone, generating cleavage products similar to those induced by enterovirus and rhinovirus 2A protease (2Apro). By the use of a fusion protein containing residues 445 to 744 of human eIF4GII, it was demonstrated that the FMDV Lpro specifically cleaves this protein between residues G700 and S701, immediately adjacent to the site (V699/G700) cleaved by rhinovirus 2Apro in vitro. The G700/S701 cleavage site does not correspond, by amino acid sequence alignment, to that cleaved in eIF4GI by the FMDV Lpro in vitro. Knowledge of the cleavage sites and the three-dimensional structures of the FMDV Lpro and rhinovirus 2Apro enabled mutant forms of the eIF4GII sequence to be generated that are differentially resistant to either one of these proteases. These results confirmed the specificity of each protease and showed that the mutant forms of the fusion protein substrate retained their correct sensitivity to other proteases.
Foot-and-mouth disease virus (FMDV) is a member of the family Picornaviridae. Like other picornaviruses, FMDV has a positive-sense RNA genome that is infectious. The genome, about 8.3 kb, encodes a large polyprotein that is processed, mainly by virus-encoded proteases, to generate each of the mature virus proteins. Two unrelated trans-acting proteases encoded within the FMDV genome have been identified, the leader protease (Lpro) and the 3C protease (3Cpro). The Lpro is related to the papain-like cysteine proteases (15, 32, 34), whereas the 3Cpro is related to the chymotrypsin-like serine proteases (1). The Lpro, located at the N terminus of the polyprotein, is produced in two forms (termed Lab and Lb) from the viral RNA. The two proteins differ in their N termini since the initiation of protein synthesis on the viral RNA occurs at two distinct sites, separated by 84 nucleotides (2, 35). Both forms of the Lpro are able to cleave the L/P1 junction in trans (24) and probably also in cis (see reference 10). The Lpro also induces the cleavage of the eukaryotic translation initiation factor 4G (eIF4G), a component of the cap-binding complex eIF4F (7, 24). The eIF4F complex consists of three different polypeptides: eIF4E (which binds to the cap structure at the 5′ terminus of all cytoplasmic cellular mRNAs), eIF4A (an RNA helicase), and eIF4G. The last component acts as a scaffold protein that also interacts with eIF3 (associated with the small ribosomal subunit), the poly(A) binding protein, and Mnk-1 (an eIF4E kinase) (see review in reference 9).
Usually, cleavage of eIF4G roughly correlates with the loss of cellular cap-dependent protein synthesis within cells infected with poliovirus (PV) or FMDV and many other picornaviruses (see review in reference 3). However, under certain circumstances, when virus replication is inhibited, the complete loss of intact eIF4G could be observed while cellular protein synthesis was maintained, albeit at a much-reduced level (5, 30). This apparent paradox was resolved when it was discovered that a second species of eIF4G, now termed eIF4GII, is also present within cells (12).
The major form of eIF4G is now referred to as eIF4GI; this protein is about 41% identical to eIF4GII, and the two proteins appear functionally equivalent (12). Within cells infected by PV or human rhinovirus 14 (HRV14) there is a differential rate of cleavage of eIF4GI and eIF4GII (13, 39). The cleavage of eIF4GI occurs rapidly, while proteolysis of eIF4GII is observed only later. Both forms of eIF4G must be cleaved to achieve complete loss of cap-dependent protein synthesis.
In contrast to the observations made within PV- or HRV14-infected cells, the cleavage of eIF4GI and eIF4GII occurs simultaneously within cells infected by HRV2 (36) or the closely related HRV16 (14). The protein responsible for inducing cleavage of both forms of eIF4G within cells infected with PV or HRV is the 2A protease (2Apro) (14, 19, 22). The difference in kinetics of eIF4GI and eIF4GII cleavage within cells infected with particular enteroviruses or rhinoviruses presumably reflects specific differences in the substrate binding efficiencies of the distinct 2Apro. The enterovirus and rhinovirus 2Apro is also related to the chymotrypsin-like family of proteases and hence are unrelated to the FMDV Lpro (31, 34, 37).
The cleavage of eIF4GI that is induced by the enterovirus 2Apro or by the FMDV Lpro occurs very early in infection and can be achieved in the absence of viral RNA replication (4, 5, 30). Recently, it has been shown that the 3Cpro of FMDV can also induce cleavage of eIF4GI within baby hamster kidney (BHK) cells (4). Subsequently it has been demonstrated that sequential cleavage of eIF4GI by the Lpro and then by 3Cpro occurs within BHK cells infected with wild-type (wt) FMDV (R. Strong and G. J. Belsham, unpublished results). The cleavage of eIF4GI by FMDV 3C occurs on the C-terminal side of the cleavage site induced by the Lpro (4). However, to date, there has been no identification of the precise cleavage sites in eIF4GI or eIF4GII within FMDV-infected cells.
The site within rabbit eIF4GI that is cleaved by incubation with recombinant FMDV Lpro in vitro has been identified as between G674 and R675 (18). (Note that throughout this paper the residues in eIF4GI have been numbered according to the system of Byrd et al. [6] for the largest form of eIF4GI, termed eIF4GIa, which is expressed from pSP4GI.) This site is just seven residues apart from the cleavage site (between R681 and G682) that is generated by 2Apro from enteroviruses and HRV2 in vitro (20, 41). This latter site has been authenticated by analysis of the C-terminal cleavage product of eIF4GI generated within PV-infected cells (41; A. Gradi and N. Sonenberg, unpublished results). Recent studies have identified the site in eIF4GII that is cleaved by the 2Apro in vitro (14). The scissile bond is between V699 and G700. It is noteworthy that this site does not correspond, by amino acid sequence alignment, to the position on eIF4GI that is cleaved by the 2Apro.
It has recently been demonstrated that eIF4GII can be cleaved by the FMDV Lpro in vitro (23, 27). However, neither the Lpro cleavage site nor the in vivo kinetics of eIF4GII cleavage have been reported. Here, we investigate these issues and discuss the results in relation to the known specificity and three-dimensional structure of the Lbpro (15).
MATERIALS AND METHODS
BHK cells were infected with FMDV (O1Kaufbeuren B64) as described previously (4). When appropriate, cells were labeled with [35S]methionine and [35S]cysteine in methionine- and cysteine-free Dulbecco modified Eagle medium for 15 min. Cell extracts were prepared and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography or for unlabeled extracts by immunoblotting with antibodies specific for the N- or C-terminal regions of eIF4GI or eIF4GII (see references 11 and 12) with appropriate peroxidase-labeled antispecies antibodies and chemiluminescence reagents.
Transient-expression assays within cells infected with the recombinant vaccinia virus vTF7-3, which expresses the T7 RNA polymerase (8), were performed as described previously (33). Briefly, human 293 cells were infected with vTF7-3 and transfected, with the use of Lipofectin (8 μg; Life Technologies), with plasmid DNA (2.5 μg) encoding the PV 2Apro (pAΔ802) or the FMDV Lbpro (pLb) as used previously (33). After 20 h cell extracts were prepared and analyzed by SDS-PAGE and immunoblotting as described above.
The expression and purification of recombinant eIF4E, FMDV Lbpro, and the 2Apro from Coxsackie virus B4 (CVB4) and HRV2 have been described previously (18, 21, 29).
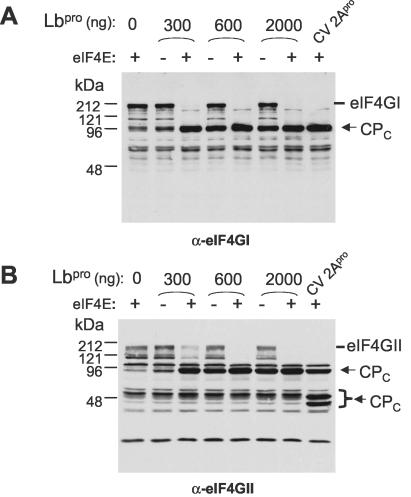
Cytoplasmic extracts from uninfected 293 cells were prepared in the absence of protease inhibitors as described previously (12, 13). Cell extracts (100 μg) were incubated for 60 min at 30°C in buffer A (20 mM HEPES-KOH [pH 7.6], 150 mM KCl, 1 mM dithiothreitol, 1 mM EDTA) in a total volume of 35 μl in the presence or absence of purified recombinant FMDV Lbpro. Before addition of the protease, the samples were preincubated for 5 min at 30°C with buffer alone or with recombinant eIF4E (1 μg). Following incubation, the reaction mixtures were divided into two samples and analyzed by SDS-PAGE and immunoblotting with polyclonal antibodies to the C-terminal cleavage products (CPC) of eIF4GI and eIF4GII as described above.
The construction and expression of the wt, G700E, and Δ674-702 mutant glutathione S-transferase (GST)-eIF4GII-FLAG fusion proteins have been described previously (14). Additional mutant forms of the expression plasmid were prepared using overlap PCR mutagenesis. The S701P mutant was made in several stages. One reaction mixture was prepared using the wt GST-eIF4GII-FLAG plasmid as template with the 4GIIfor primer (dGTTTGGTGGTGGCGACCATC) and the 4GIIrevSP primer (dGATCTTCGTGGCCCAACATTCAAC) with Pfu DNA polymerase. A second reaction mixture contained the same template with the primers 4GIIforSP (dGTTGAATGTTGGGCCACGAAGATC) and 4GIIrev (dCTCCGGGAGCTGCATGTGTC). The two products were gel purified and mixed, and a third PCR was performed using just the 4GIIfor and 4GIIrev primers. Similarly the G700K mutant was made using the same strategy with the 4GIIfor and 4GIIrev primers in conjunction with the mutant oligonucleotides 4GIIforGK (dGTTGAATGTTAAGTCACGAAGATC) and 4GIIrevGK (dGATCTTCGTGACTTAACATTCAAC). In each case the final products were digested with EcoRI and NotI, and the fragments of ca. 900 nucleotides were used to replace the wt sequences within the similarly digested plasmid encoding the GST-eIF4GII-FLAG protein (14). The presence of the required mutations was confirmed by DNA sequencing, and the mutant fusion proteins were isolated as described above. For analytical assays, purified recombinant GST-eIF4GII-FLAG protein (2.5 μg) or the mutant derivatives were incubated in buffer A with purified FMDV Lbpro or HRV2 2Apro (the amount of proteases used is indicated in the figure legends) at 30°C for 60 min in the presence or absence of recombinant eIF4E (1 μg) as indicated. Samples were analyzed by SDS-PAGE (15% acrylamide) and immunoblotting with anti-FLAG M2 antibodies (Sigma or Upstate Biotechnology Inc.).
For the identification of the cleavage site, recombinant GST-eIF4GII-FLAG protein (35 μg) was incubated with purified FMDV Lbpro (4 μg) at 30°C for 60 min in the presence of recombinant eIF4E (3 μg). The C-terminal FLAG-tagged cleavage fragment was captured using anti-FLAG M2 antibody resin (Sigma) as described previously (25) and purified using SDS-PAGE (15.5% acrylamide). After electrophoresis, the proteins were transferred in cyclohexylaminopropanesulfonic acid buffer onto a membrane (ProBlott; 0.2-μm pore size) and stained with 0.1% Coomassie blue R-250 in 50% methanol. The band of interest was isolated and subjected to automated Edman degradation. The N-terminal sequence analysis was performed by the Sheldon Biotechnology Center, McGill University.
RESULTS
Cleavage of eIF4GII within FMDV-infected cells.
The inhibition of host cell protein synthesis occurs very rapidly within FMDV-infected BHK cells (see reference 4). This is accompanied by the cleavage of eIF4GI. To examine the status of eIF4GII within FMDV-infected cells, a time course experiment was performed (Fig. 1). As observed previously (4), within 2 to 3 h postinfection, a dramatic decrease in host cell protein synthesis was detected by pulse-labeling of infected cells with [35S]methionine-cysteine (Fig. 1A). Consistent with our previous results, cleavage of eIF4GI occurred very rapidly (Fig. 1B). In the same extracts we also monitored the status of eIF4GII. The loss of the intact eIF4GII protein was also readily detected early in infection (Fig. 1C). The rabbit anti-C-terminal antibody detected two distinct cleavage products (with apparent sizes of about 85 and 33 kDa) that were derived from eIF4GII in extracts of cells harvested at 2 h postinfection. An additional product of 45 kDa accumulated by 3 h postinfection and probably represents a breakdown product of the 85-kDa species. The kinetics of primary cleavage for eIF4GI and eIF4GII were very similar. In both cases, cleavage had started at 1 h postinfection and no intact protein was detectable by 3 h postinfection. The recognition of eIF4GII from BHK cells by the antibody was not as efficient as is observed with human cell extracts; presumably this reflects a difference in the sequences of the proteins from the two species.
FIG. 1.
Cleavage of eIF4GI and eIF4GII occurs simultaneously within FMDV-infected cells. BHK cells were infected with FMDV. (A) At the indicated times (hours postinfection) cells were incubated with 35S-labeled amino acids for a further 15 min. Cell extracts were prepared and analyzed by SDS-PAGE (10% polyacrylamide) and autoradiography. (B and C) Cells infected in parallel were harvested at the indicated times, and cell extracts were analyzed by SDS-PAGE (6% polyacrylamide) and immunoblotting with rabbit polyclonal antibodies specific for eIF4GI (B) and eIF4GII (C) as described in Materials and Methods. Detection was achieved using peroxidase-labeled anti-rabbit antibodies and chemiluminescence reagents. The mobilities of molecular mass standards (kilodaltons) are indicated.
Cleavage of eIF4GII by the FMDV Lpro within cells.
We wished to determine whether the cleavage of eIF4GII was induced within cells by the expression of FMDV Lbpro alone. A transient-expression assay was performed within human 293 cells (infected with the recombinant vaccinia virus vTF7-3 [8], which expresses the T7 RNA polymerase) by using plasmids that expressed either the PV 2Apro or the FMDV Lbpro (Fig. 2). The status of both eIF4GI and eIF4GII was examined in cell extracts by immunoblotting with antisera to the N-terminal cleavage products. As observed previously (4), eIF4GI cleavage was very efficiently induced by the PV 2Apro and by the FMDV Lbpro (Fig. 2A). Furthermore, it was apparent that both proteases also induced efficient cleavage of eIF4GII (Fig. 2B). The major cleavage products of eIF4GII generated by the expression of the two different proteases and detected by the anti-N-terminal antibody were indistinguishable, suggesting that the cleavage sites generated by these two proteases are near each other. However, it should be noted that an additional smaller product (marked by an asterisk in Fig. 2B) was also induced by the FMDV Lbpro, presumably reflecting a second cleavage event within the N-terminal portion of the protein.
FIG. 2.
The FMDV Lbpro induces cleavage of eIF4GI and eIF4GII within cells. Human 293 cells were infected with the recombinant vaccinia virus vTF7-3 and transfected with the indicated plasmids that express FMDV Lbpro (pLb) or the PV 2Apro (pAΔ802) with Lipofectin. After 20 h, cell extracts were prepared and analyzed by SDS-PAGE and immunoblotting with antisera specific for the N termini of eIF4GI (A) and eIF4GII (B). The cleavage products and the full-length proteins are indicated. An additional cleavage product detected in extracts containing the FMDV Lbpro is highlighted by an asterisk.
In vitro cleavage of eIF4GI and eIF4GII by the FMDV Lpro.
The addition of eIF4E greatly stimulates the cleavage of eIF4GI and eIF4GII by HRV 2Apro in vitro (12, 16). Similarly, the cleavage of eIF4GI by FMDV Lbpro is stimulated by the presence of eIF4E (26). We wished to determine whether the cleavage of eIF4GII by the FMDV Lbpro in vitro was also modified by the addition of eIF4E. Cell extracts were prepared from uninfected human 293 cells and incubated with purified recombinant FMDV Lbpro in the presence or absence of recombinant cap-binding protein eIF4E. A single C-terminal cleavage product (CPC) of about 95 kDa was observed from both eIF4GI and eIF4GII in the presence of the FMDV Lbpro. The products observed were indistinguishable from those generated by incubation with recombinant 2Apro from CVB4 (Fig. 3) as used previously (14). The presence of added eIF4E greatly enhanced the cleavage of eIF4GI by the Lbpro in vitro, consistent with previous results (26). Furthermore, addition of eIF4E also strongly stimulated the cleavage of eIF4GII by the Lbpro in these assays (Fig. 3B).
FIG. 3.
Cleavage of eIF4GI and eIF4GII by the FMDV Lbpro in cell extracts is enhanced by the presence of added eIF4E. Extracts of uninfected human 293 cells (100 μg of protein) were incubated with the indicated amounts of recombinant FMDV Lbpro or CVB4 2Apro (7 μg) in the presence (+) or absence (−) of recombinant eIF4E (1 μg) as indicated. Samples were analyzed by SDS-PAGE (10% polyacrylamide) and immunoblotting with antibodies specific for the C terminus of eIF4GI (A) or eIF4GII (B). Proteins were detected using chemiluminescence reagents as in Fig. 1.
In vitro cleavage of recombinant eIF4GII fusion proteins by the FMDV Lbpro.
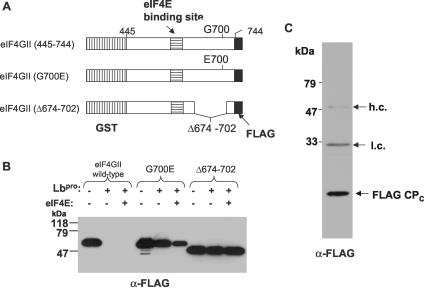
A recombinant fusion protein, GST-eIF4GII-FLAG (Fig. 4A), which contains an internal region (residues 445 to 744) of human eIF4GII (including the eIF4E binding site) linked to both GST and FLAG tags, was expressed in Escherichia coli and purified to homogeneity on glutathione-Sepharose as described previously (14). The protein was incubated in vitro with recombinant FMDV Lbpro in the presence or absence of added recombinant eIF4E. The GST-eIF4GII-FLAG fusion protein was completely cleaved by Lbpro under both conditions (Fig. 4B). We also analyzed the sensitivities to FMDV Lbpro of two similar fusion proteins (Fig. 4A) that contain a modified region of eIF4GII (Fig. 4B). These two mutant fusion proteins were initially prepared as part of an analysis of the in vitro cleavage of eIF4GII by the HRV2 2Apro (14). One mutant protein has a single point mutation, G700E, which confers resistance to cleavage by 2Apro, since the cleavage site is between residues 699 and 700. The second mutant protein has a deletion of 27 residues (between 674 and 702); this protein was also fully resistant to the 2Apro since the deletion removes the cleavage site (14). Surprisingly, both mutant proteins were also highly resistant to the action of the FMDV Lbpro in both the presence and the absence of eIF4E (Fig. 4B). A small yield of the C-terminal FLAG-tagged cleavage product was observed from the G700E mutant following incubation with a high level of the Lbpro (data not shown); however, most of the fusion protein remained intact. These results suggested that the cleavage sites for 2Apro and Lbpro on eIF4GII are near each other and are consistent with the generation, by these two different proteases, of indistinguishable eIF4GII cleavage products within cells (see above).
FIG. 4.
In vitro cleavage of a fragment of eIF4GII by FMDV Lbpro permits identification of the cleavage site. (A) Structure of the GST-eIF4GII-FLAG (abbreviated GST-4GII-FLAG) fusion proteins as described previously (14). (B) Recombinant GST-4GII-FLAG protein (2.5 μg) or mutant forms with a G700E substitution or deletion of residues 674 to 702 were incubated in the presence (+) or absence (−) of recombinant FMDV Lbpro (1 μg) alone or with recombinant eIF4E (1 μg) as indicated. Samples were analyzed by SDS-PAGE (15% polyacrylamide) and immunoblotting with anti-FLAG M2 antibodies. Detection was achieved using chemiluminescence reagents. (C) The C-terminal cleavage product of the wt GST-4GII-FLAG protein, generated by the FMDV Lbpro, was isolated using an anti-FLAG M2 affinity resin. An aliquot of the material was analyzed by SDS-PAGE (15.5% polyacrylamide) and immunoblotting with anti-FLAG M2 antibodies as for panel B. The presence of antibody heavy chains (h.c.) and light chains (l.c.) as well as the FLAG-tagged C-terminal cleavage product (CPC) is indicated.
To investigate this further, we set out to identify precisely the cleavage site in eIF4GII generated by the FMDV Lbpro in vitro. The epitope-tagged C-terminal cleavage product was captured using anti-FLAG affinity resin from a preparative-scale digestion of GST-eIF4GII-FLAG protein with the FMDV Lbpro. Western blot analysis, with anti-FLAG antibodies, identified the FLAG-tagged product in an aliquot of the sample (Fig. 4C). The residual sample was transferred to a polyvinylidene difluoride membrane, and the stained fragment was isolated and subjected to N-terminal amino acid sequence analysis. Five cycles of Edman degradation analysis identified the sequence SRRSQ, which exactly corresponds to residues 701 to 705 of eIF4GII (Fig. 5). This identified the FMDV Lbpro cleavage site on human eIF4GII as between residues G700 and S701, just 1 amino acid C terminal of the 2Apro cleavage site (14). This result is entirely consistent with the resistance of the G700E point mutant and the Δ674-702 deletion mutant to the action of the FMDV Lbpro (Fig. 4B).
FIG. 5.
Comparison of in vitro cleavage sites in eIF4GI and eIF4GII by FMDV Lbpro and enterovirus 2Apro. An alignment of a central region of human eIF4GI and eIF4GII including the eIF4E binding sites together with the regions of the proteins cleaved by FMDV Lbpro and enterovirus-rhinovirus 2Apro is shown. The positions of the cleavage sites determined from in vitro analyses are indicated within the boxes. The five amino acid residues, determined by N-terminal sequence analysis, of the C-terminal cleavage product obtained from the GST-eIF4GII-FLAG fusion protein following incubation with recombinant FMDV Lbpro are underlined with a dashed bar.
Mutant eIF4GII proteins with differential susceptibilities to 2Apro and Lbpro.
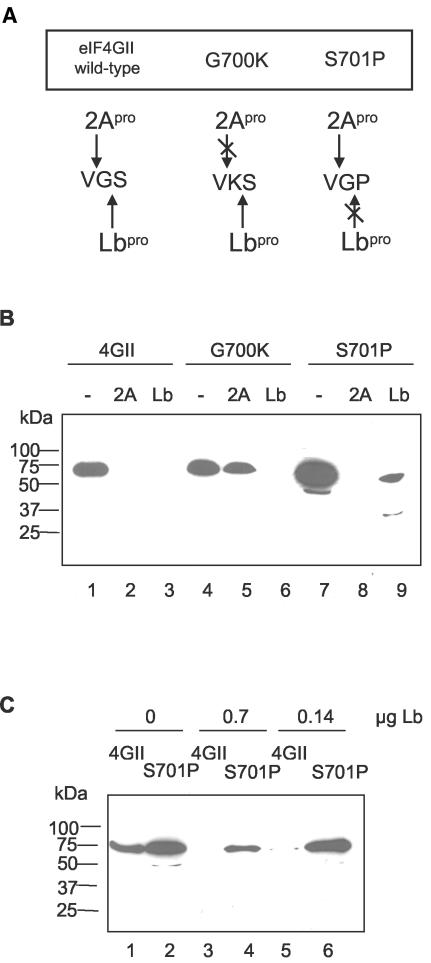
It is possible that introduction of a single mutation into the fusion protein substrate could significantly alter its structure and render it sufficiently different from the native molecule that the normal cleavage site might no longer be exposed and thus could give an incorrect “apparent” resistance. One way to test this would be to determine whether the resistant mutant substrate retained its specific susceptibility to a second protease. The results described above indicated that the cleavage sites of HRV2 2Apro and FMDV Lbpro on eIF4GII overlap. We wished to use our knowledge of the cleavage specificities of these two viral proteinases to introduce specific mutations aimed at generating mutant proteins which were refractory to one of the enzymes, while still permitting cleavage by the second. The completely different substrate requirements of HRV2 2Apro and FMDV Lbpro strengthened our belief that such mutant proteins could be designed (Fig. 6A). For the HRV2 2Apro, much evidence shows that the presence of glycine at P1′ (the first residue on the C-terminal side of the cleavage site) is an absolute requirement (38). Indeed, no cleavage site with an amino acid other than glycine at this position has been identified. Therefore, we decided to replace glycine at residue 700 with lysine (G700K) to prevent HRV2 2Apro cleavage. Lysine was chosen because residue 700 represents the P1 residue (N-terminal side of the cleavage site) for Lbpro on eIF4GII. Thus, the Lbpro cleavage site in this mutant becomes VK*S (Fig. 6A). This sequence is very reminiscent of the site (LK*G) in the viral polyprotein (see below) at which Lbpro cleaves. We were therefore confident that the eIF4GII mutant protein G700K would still be cleaved by FMDV Lbpro.
FIG. 6.
Specific mutations in eIF4GII have differential effects on 2Apro and Lbpro cleavage. (A) Predicted cleavage activity of 2Apro and Lbpro on eIF4GII mutant proteins. (B) The indicated fusion proteins were incubated for 1 h at 30°C in buffer A alone (lanes 1, 4, and 7), with purified HRV2 2Apro (1.2 μg; lanes 2, 5, and 8), or with purified FMDV Lbpro (1.43 μg; lanes 3, 6, and 9). Protein samples were analyzed by SDS-PAGE and immunoblotting with a monoclonal antibody against the C-terminal FLAG tag (Upstate Biotechnology Inc.). (C) The wt and S701P mutant GST-eIF4GII-FLAG fusion proteins were incubated with the indicated concentrations of purified FMDV Lbpro for 1 h at 30°C. Protein samples were analyzed by SDS-PAGE and immunoblotting as described for panel B.
To create an eIF4GII mutant protein that is refractory to cleavage by Lbpro but not by 2Apro, we replaced serine at residue 701 with proline (S701P; Fig. 6A). Proteinases rarely accept a proline residue at the P1′ position; furthermore, the architecture of the Lbpro substrate binding site makes it unlikely that this substrate would be efficiently cleaved. Specifically, the tight turn introduced by proline into the polypeptide chain would be expected to clash unfavorably with residues in proximity to the active site. For cleavage by 2Apro, the S701P mutation would change the P2′ residue in the eIF4GII substrate to a proline. At the cleavage site on the viral polyprotein, a proline residue is observed in this position. This led us to believe that the S701P mutant protein would still be a substrate for HRV2 2Apro (Fig. 6A).
The indicated mutations were therefore introduced into the GST-eIF4GII-FLAG plasmid, and the fusion proteins were expressed and purified by affinity chromatography on glutathione-Sepharose. The wt and mutant fusion proteins were then incubated alone or with one of the viral proteinases. The products were analyzed by SDS-PAGE and immunoblotting with anti-FLAG antibodies (Fig. 6B). As expected, the wt protein was fully digested by both proteinases (lanes 2 and 3). The G700K mutant was indeed resistant to 2Apro of HRV2 as predicted (lane 5), whereas, in contrast, the S701P mutant was still efficiently cleaved by this protease. As anticipated, the G700K protein was still cleaved, like the wt construct, by the Lbpro. However, unexpectedly the S701P mutant protein was not completely resistant to cleavage by Lbpro; a small amount of a novel cleavage product of about 30 kDa was detected (lane 9). The production of this 30-kDa protein appeared to represent an aberrant cleavage. Hence, we attempted to determine whether the S701P protein and the wt protein would exhibit markedly different sensitivities to the FMDV Lbpro at lower concentrations of the enzyme. Thus, the concentration of Lpro was reduced twofold (Fig. 6C, lanes 3 and 4) or fourfold (Fig. 6C, lanes 5 and 6) compared to that used in Fig. 6B. Under these conditions, the S701P fusion protein remained intact whereas the wt fusion protein was almost completely processed (Fig. 6C, lanes 5 and 6). In addition, the 30-kDa aberrant cleavage product was not detected. We conclude that the G700/P701 site is resistant to Lbpro as predicted. Thus, the two mutant forms of the GST-eIF4GII-FLAG fusion protein exhibit sensitivity to one protease while being resistant to the other. These results support the view that the structure of the fusion proteins is similar in each case and it is a reflection of the particular specificity of each protease that the mutated sites confer resistance.
DISCUSSION
The results presented here demonstrate that cleavage of both eIF4GI and eIF4GII occurs rapidly, simultaneously, and completely within FMDV-infected cells. These results are reminiscent of those obtained with HRV2 (36) and HRV16 (14) but contrast with the differential rates of cleavage of these proteins observed within cells infected by PV or HRV14 (13, 39). Transient-expression assays demonstrated that the FMDV Lbpro alone is able to induce cleavage of eIF4GII by itself within cells (Fig. 2); these results mirror earlier studies with eIF4GI (4, 24).
The direct cleavage of eIF4GI by the FMDV Lbpro in vitro was demonstrated by Kirchweger et al. (18), who also identified the cleavage site as between residues G674 and R675. The cleavage of purified eIF4GI is rather inefficient and requires high levels of Lbpro. In contrast, cleavage of eIF4GI within FMDV-infected cells occurs early in infection before detectable virus protein expression and can be achieved even in the absence of RNA replication (see reference 4). These data indicate that a low level of protease is required to initiate cleavage of eIF4GI within cells. It has been suggested that the 2Apro of enteroviruses and rhinoviruses and the Lpro of FMDV may induce cleavage of eIF4G indirectly within cells by activating a cellular protease (see review in reference 3). It is certainly the case that cellular proteases can cleave eIF4G (41). However, it has also been demonstrated that the addition of eIF4E to eIF4G greatly enhances the efficiency with which eIF4G can be cleaved by 2Apro and FMDV Lbpro in vitro (12, 14, 16, 26). These results suggested that the conformation of eIF4G may be altered by this (and possibly other) interaction(s), resulting in a complex that may be more susceptible to cleavage than the isolated protein is. Indeed, an earlier observation had indicated that eIF3 (which also interacts with eIF4G) also enhanced the susceptibility of eIF4G to 2Apro (40). The results presented here (Fig. 3) demonstrated that eIF4E greatly enhanced the cleavage of eIF4GI and eIF4GII within cell lysates by recombinant FMDV Lbpro; these results confirm and extend those of Ohlmann et al. (26), who analyzed only eIF4GI cleavage. Interestingly, when a fragment of eIF4GII (residues 445 to 744) was used as part of a GST fusion protein, it was found that this fragment was susceptible to cleavage by FMDV Lbpro in the presence or absence of eIF4E (Fig. 4B).
The cleavage of this GST-eIF4GII-FLAG fusion protein by Lbpro in vitro was used to identify the cleavage site within eIF4GII as between G700 and S701. This site is just 1 amino acid C terminal of the site cleaved by the HRV 2Apro in vitro (14). The identification of this site was entirely consistent with the resistance of the G700E mutant form of the GST-eIF4GII-FLAG protein to cleavage by Lbpro. Similarly, the deletion mutant lacking residues between 674 and 702 from eIF4GII was also completely resistant to cleavage by Lbpro. There have been no reports of successful expression of full-length recombinant eIF4GII in cells, but a His-tagged truncated form of this protein, lacking the first 143 residues, has been produced with the baculovirus expression system (see reference17a). Treatment of this protein (kindly supplied by S. Morley, University of Sussex, United Kingdom) with the recombinant FMDV Lbpro generated cleavage products (N. Foeger and T. Skern, unpublished results), but attempts to determine N-terminal sequence information from these products were unsuccessful. Thus, we have been unable to confirm the location of the cleavage site within the full-length protein. However, there are additional strands of evidence that support the view that the cleavage site identified in our work is authentic. Firstly, we demonstrated that the cleavage products of eIF4GII generated in cells and in cell extracts by the PV (and CVB4) 2Apro and the FMDV Lbpro comigrate (Fig. 2 and 3); this clearly suggests that the cleavage sites are near each other. Secondly, in the GST-eIF4GII-FLAG fusion protein (Fig. 4 and 6), we are using about 300 amino acids from eIF4GII, and yet individual modification of a single amino acid on either side of the identified scissile bond (G700/S701) was enough to confer resistance to cleavage by Lbpro.
On the basis of the known properties of 2Apro and Lbpro, it was possible to design mutant forms of the eIF4GII sequence which were differentially resistant to either one of these proteases. Thus, although it is apparent that multiple sites within hamster eIF4GII may be cleaved by proteases within FMDV-infected cells (Fig. 1), there is high specificity in the cleavage of the GST-eIF4GII-FLAG fusion protein by the Lbpro in vitro. It was interesting, however, that the S701P mutant protein (which was no longer cleaved by Lbpro at the usual position) was cleaved at a novel site with the use of a high concentration of the protease. An additional cleavage event in the N-terminal region of eIF4GII by the Lbpro was detected in human cells that expressed this protease (Fig. 2B). We have also observed that FMDV 3Cpro can cleave eIF4GII in vitro and within cells (A. Gradi and G. J. Belsham, unpublished results), and hence, this protease could contribute to the additional fragments of eIF4GII observed within FMDV-infected cells.
Comparison of the Lbpro cleavage site in eIF4GII (PLLNVG700 SRRSQP) with those in eIF4GI (SFANLG674 RTTLST [18]) and the viral polyprotein (strain O1K; VQRKLK201 GAGQSS [28]) reveals a number of differences. Most strikingly, the eIF4GII cleavage site does not possess a basic residue at either the P1 or P1′ position that flanks the scissile bond (although it can accept one, as in the G700K mutant). Until now, the presence of such a residue had been regarded as a major determinant of Lbpro specificity (10, 15). It seems possible, however, that the arginine (R) residue at P2′ may provide the positive charge required for Lbpro substrate binding. In addition, the presence of a serine (S) residue has not previously been observed at P1′ in an Lbpro cleavage site. On the P (N-terminal) side of the cleavage site, the presence of asparagine (N) at P3, valine (V) at P2, and glycine (G) at P1 fits well into the pattern of residues observed at these positions in the other cleavage sites.
It is noteworthy that the site cleaved in eIF4GII by the Lbpro does not correspond, by amino acid sequence alignment, to the position cleaved by this protease within eIF4GI (Fig. 5). The same is true for the sites in eIF4GI and eIF4GII that are cleaved by HRV 2Apro (14). The resistance of the eIF4GII mutant proteins G700E and S701P to Lbpro cleavage shows that the region of eIF4GII that corresponds to the cleavage site on eIF4GI (Fig. 5) is not recognized by Lbpro, even when the correct site is blocked. Why should this be so? Despite the availability of the crystal structure and other studies of Lbpro specificity, this question cannot be answered fully at present. Given that lysine (K) or asparagine (N) is found in the other cleavage sites at the P3 position, it seems possible that the presence of aspartate (D683) in the eIF4GII sequence might be detrimental to cleavage. In addition, although leucine (L) and phenylalanine (F) are often regarded as having similar properties, it is possible that the Lbpro can distinguish between the presence of these two residues at the P2 position. However, as papain accepts both residues well at P2 (17) and has a P2 binding pocket almost identical to that of Lbpro (15), it remains to be investigated whether Lbpro can discriminate so finely at this position.
Acknowledgments
R.S. gratefully acknowledges a studentship from the Biotechnology and Biological Sciences Research Council. This work was also supported in part by a grant (P-16189) from the Austrian Science Foundation (to T.S.) and by a grant from the Canadian Institute of Health Research (to N.S.).
We thank Elena Vassilieva for purified HRV2 2Apro, Simon Morley for His-tagged eIF4GII, Colin Lister for excellent technical assistance, and Marcos Di Falco (Sheldon Biotechnology Center, McGill University) for performing the amino acid analysis and for helpful discussions.
REFERENCES
- 1.Allaire, M., M. M. Chernaia, B. A. Malcolm, and M. N. G. James. 1994. Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like serine proteinases. Nature 369:72-76. [DOI] [PubMed] [Google Scholar]
- 2.Belsham, G. J. 1992. Dual initiation sites of protein synthesis on foot-and-mouth disease virus RNA are selected following internal entry and scanning of ribosomes in vivo. EMBO J. 11:1105-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belsham, G. J., and R. J. Jackson. 2000. Translation initiation on picornavirus RNA. Cold Spring Harbor Monogr. Ser. 39:869-900. [Google Scholar]
- 4.Belsham, G. J., G. M. McInerney, and N. Ross-Smith. 2000. Foot-and-mouth disease virus 3C protease induces cleavage of translation initiation factors eIF4A and eIF4G within infected cells. J. Virol. 74:272-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonneau, A. M., and N. Sonenberg. 1987. Proteolysis of the p220 component of the cap-binding protein complex is not sufficient for complete inhibition of host cell protein synthesis after poliovirus infection. J. Virol. 61:986-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrd, M. P., M. Zamora, and R. E. Lloyd. 2002. Generation of multiple isoforms of eukaryotic translation initiation factor 4GI by use of alternate translation initiation codons. Mol. Cell. Biol. 22:4499-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devaney, M. A., V. N. Vakharia, R. E. Lloyd, E. Ehrenfeld, and M. J. Grubman. 1988. Leader protein of foot-and-mouth-disease virus is required for cleavage of the p220 component of the cap-binding protein complex. J. Virol. 62:4407-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 10.Glaser, W., R. Cencic, and T. Skern. 2001. Foot-and-mouth disease virus leader proteinase: involvement of C-terminal residues in self-processing and cleavage of eIF4GI. J. Biol. Chem. 276:35473-35481. [DOI] [PubMed] [Google Scholar]
- 11.Goldstaub, D., A. Gradi, Z. Bercovitch, Z. Grosmann, Y. Nophar, S. Luria, N. Sonenberg, and C. Kahana. 2000. Poliovirus 2A protease induces apoptotic cell death. Mol. Cell. Biol. 20:1271-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gradi, A., H. Imataka, Y. V. Svitkin, E. Rom, B. Raught, S. Morino, and N. Sonenberg. 1998. A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18:334-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gradi, A., Y. V. Svitkin, H. Imataka, and N. Sonenberg. 1998. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. USA 95:11089-11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gradi, A., Y. V. Svitkin, W. Sommergruber, H. Imataka, S. Morino, T. Skern, and N. Sonenberg. 2003. Human rhinovirus 2A proteinase cleavage sites in eukaryotic initiation factor (eIF) 4GI and eIF4GII are different. J. Virol. 77:5026-5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guarné, A., J. Tormo, R. Kirchweger, D. Pfistermueller, I. Fita, and T. Skern. 1998. Structure of the foot-and-mouth disease virus leader protease: a papain-like fold adapted for self-processing and eIF4G recognition. EMBO J. 17:7469-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haghighat, A., Y. Svitkin, I. Novoa, E. Kuechler, T. Skern, and N. Sonenberg. 1996. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J. Virol. 70:8444-8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris, J. L., B. J. Backes, F. Leonetti, S. Mahrus, J. A. Ellman, and C. S. Craik. 2000. Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. Proc. Natl. Acad. Sci. USA 97:7754-7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Imataka, H., A. Gradi, and N. Sonenberg. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 17:7480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirchweger, R., E. Ziegler, B. J. Lamphear, D. Waters, H. D. Liebig, W. Sommergruber, F. Sobrino, C. Hohenadl, D. Blaas, R. E. Rhoads, and T. Skern. 1994. Foot-and-mouth disease virus leader proteinase: purification of the Lb form and determination of its cleavage site on eIF-4γ. J. Virol. 68:5677-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kräusslich, H.-G., M. J. Nicklin, H. Toyoda, D. Etchison, and E. Wimmer. 1987. Poliovirus proteinase 2A induces cleavage of eucaryotic initiation factor 4F polypeptide p220. J. Virol. 61:2711-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamphear, B. J., R. Q. Yan, F. Yang, D. Waters, H. D. Liebig, H. Klump, E. Kuechler, T. Skern, and R. E. Rhoads. 1993. Mapping the cleavage site in protein synthesis initiation factor eIF-4γ of the 2A proteases from human coxsackievirus and rhinovirus. J. Biol. Chem. 268:19200-19203. [PubMed] [Google Scholar]
- 21.Liebig, H.-D., E. Ziegler, R. Yan, K. Hartmuth, H. Klump, H. Kowalski, D. Blaas, S. W. L. Frasel, B. Lamphear, R. Rhoads, E. Kuechler, and T. Skern. 1993. Purification of two picornaviral 2A proteinases: interaction with eIF-4G and influence on translation. Biochemistry 32:7581-7588. [DOI] [PubMed] [Google Scholar]
- 22.Lloyd, R. E., M. J. Grubman, and E. Ehrenfeld. 1988. Relationship of p220 cleavage during picornavirus infection to 2A proteinase sequencing. J. Virol. 62:4216-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López de Quinto, S. L., E. Lafuente, and E. Martinez-Salas. 2001. IRES interaction with translation initiation factors: functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA 7:1213-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medina, M., E. Domingo, J. K. Brangwyn, and G. J. Belsham. 1993. The two species of the foot-and-mouth disease virus leader protein, expressed individually, exhibit the same activities. Virology 194:355-359. [DOI] [PubMed] [Google Scholar]
- 25.Morino, S., H. Imataka, Y. V. Svitkin, T. V. Pestova, and N. Sonenberg. 2000. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol. 20:468-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohlmann, T., V. M. Pain, W. Wood, M. Rau, and S. J. Morley. 1997. The proteolytic cleavage of eukaryotic initiation factor (eIF) 4G is prevented by eIF4E binding protein (PHAS-I; 4E-BP1) in the reticulocyte lysate. EMBO J. 16:844-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohlmann, T., D. Prevot, D. Decimo, F. Roux, J. Garin, S. J. Morley, and J. L. Darlix. 2002. In vitro cleavage of eIF4GI but not eIF4GII by HIV-1 protease and its effects on translation in the rabbit reticulocyte lysate system. J. Mol. Biol. 318:9-20. [DOI] [PubMed] [Google Scholar]
- 28.Palmenberg, A. 1989. Sequence alignments of picornaviral capsid proteins, p. 211-241. In B. L. Semler and E. Ehrenfeld (ed.), Molecular aspects of picornavirus infection and detection. American Society for Microbiology, Washington, D.C.
- 29.Pause, A., G. J. Belsham, A.-C. Gingras, O. Donze, T.-A. Lin, J. C. Lawrence, Jr., and N. Sonenberg. 1994. Insulin dependent stimulation of protein synthesis by phosphorylation of a novel regulator of 5′-cap function. Nature (London) 371:762-767. [DOI] [PubMed] [Google Scholar]
- 30.Perez, L., and L. Carrasco. 1992. Lack of direct correlation between p220 cleavage and the shut-off of host translation after poliovirus infection. Virology 189:178-186. [DOI] [PubMed] [Google Scholar]
- 31.Petersen, J. F., M. M. Cherney, H. D. Liebig, T. Skern, E. Kuechler, and M. N. James. 1999. The structure of the 2A proteinase from a common cold virus: a proteinase responsible for the shut-off of host-cell protein synthesis. EMBO J. 18:5463-5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts, P., and G. J. Belsham. 1995. Identification of critical amino acids within the foot-and-mouth disease virus leader protein, a cysteine protease. Virology 213:140-146. [DOI] [PubMed] [Google Scholar]
- 33.Roberts, L. O., R. A. Seamons, and G. J. Belsham. 1998. Recognition of picornavirus internal ribosome entry sites within cells; influence of cellular and viral proteins. RNA 4:520-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryan, M. D., and M. Flint. 1997. Virus-encoded proteinases of the picornavirus super-group. J. Gen. Virol. 78:699-723. [DOI] [PubMed] [Google Scholar]
- 35.Sangar, D. V., S. E. Newton, D. J. Rowlands, and B. E. Clarke. 1987. All foot and mouth disease serotypes initiate protein synthesis at two separate AUGs. Nucleic Acids Res. 15:3305-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seipelt, J., H. D. Liebig, W. Sommergruber, C. Gerner, and E. Kuechler. 2000. 2A proteinase of human rhinovirus cleaves cytokeratin 8 in infected HeLa cells. J. Biol. Chem. 275:20084-20089. [DOI] [PubMed] [Google Scholar]
- 37.Skern, T., B. Hampölz, A. Guarné, I. Fita, E. Bergmann, J. Petersen, and M. N. G. James. 2002. Structure and function of picornavirus proteinases, p. 199-212. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 38.Sommergruber, W., H. Ahorn, A. Zophel, I. Maurer-Fogy, F. Fessl, G. Schnorrenberg, H. D. Liebig, D. Blaas, E. Kuechler, and T. Skern. 1992. Cleavage specificity on synthetic peptide substrates of human rhinovirus 2 proteinase 2A. J. Biol. Chem. 267:22639-22644. [PubMed] [Google Scholar]
- 39.Svitkin, Y. V., A. Gradi, H. Imataka, S. Morino, and N. Sonenberg. 1999. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J. Virol. 73:3467-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wyckoff, E. E., R. E. Lloyd, and E. Ehrenfeld. 1992. Relationship of eukaryotic initiation factor 3 to poliovirus-induced p220 cleavage activity. J. Virol. 66:2943-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zamora, M., W. E. Marissen, and R. E. Lloyd. 2002. Multiple eIF4GI-specific protease activities present in uninfected and poliovirus-infected cells. J. Virol. 76:165-177. [DOI] [PMC free article] [PubMed] [Google Scholar]