Abstract
Hepatitis C virus (HCV) RNA replication is dependent on the enzymatic activities of the viral RNA-dependent RNA polymerase NS5B, which is a membrane-anchored protein. Recombinant NS5B lacking the C-terminal transmembrane domain (21 amino acids) is enzymatically active. To address the role of this domain in HCV replication in vivo, we introduced a series of mutations into the NS5B of an HCV subgenomic replicon and examined the replication capabilities of the resultant mutants by a colony formation assay. Replicons lacking the transmembrane domain did not yield any colonies. Furthermore, when Huh-7 cells harboring the HCV subgenomic replicon were treated with a synthetic peptide consisting of the NS5B transmembrane domain fused to the antennapedia peptide, the membrane association of NS5B was completely disrupted. Correspondingly, the HCV RNA titer was reduced by approximately 50%. A scrambled peptide used as a control did not have any effects. These findings suggest that the membrane association of NS5B facilitates HCV RNA synthesis. However, a related transmembrane domain derived from bovine viral diarrhea virus could not replace the HCV NS5B transmembrane segment. This finding suggests that the C-terminal 21 amino acids not only have a membrane-anchoring function but also may perform additional functions for RNA synthesis in vivo.
Hepatitis C virus (HCV) is a significant cause of morbidity and mortality, infecting over 170 million people worldwide (1, 8). Despite recent progress, the current method of treatment for HCV infection remains inadequate for the majority of patients (24, 29, 39). HCV is an enveloped, positive-sense, single-stranded RNA virus (25). Its 9.6-kb genome encodes a single polyprotein, which is proteolytically processed by a combination of host- and virus-encoded proteinases into 10 structural and nonstructural (NS) proteins (16, 26).
The establishment of the HCV subgenomic replicons and the subsequent studies of them revealed that most of the HCV NS proteins are involved in HCV RNA replication (2, 15, 28, 35). NS3 is a helicase and a serine protease whose function is dependent on NS4A. Chimpanzee studies showed that the enzymatic activities of NS3-4A serine protease and NS3 helicase are essential for the productive replication of HCV (21). Thus, these enzymatic activities may be key components in the replication of HCV subgenomic replicons. NS4B is an integral membrane protein which induces intracellular membrane alterations (9, 18). NS4B may play an important role in the formation of the HCV RNA replication complex (14a). NS5A is also involved in HCV RNA replication, as adaptive mutations in NS5A are associated with increased replication efficiency of HCV RNA (2, 22, 27). Its membrane association through its N-terminal transmembrane domain is essential for RNA replication of the subgenomic replicon (11). NS5B is an RNA-dependent RNA polymerase, a key enzyme involved in viral replication (21). NS5B contains a hydrophobic domain (21 amino acids [amino acids 571 to 591]) at its C terminus, which is a transmembrane segment (20, 34). All of these NS proteins, together with host proteins, are believed to form a membrane-associated RNA replication complex (14a).
Replication of most positive-stranded RNA viruses occurs in large complexes associated with intracellular membranes (4). Previous studies have revealed that HCV NS3 (30), NS4A (30), NS4B (18), NS5A (3, 11, 36, 37), and NS5B (19, 20, 34) proteins are associated with the endoplasmic reticulum and/or the Golgi apparatus when they are expressed alone, in the context of the entire polyprotein, or in the context of subgenomic replicon. NS4B, NS5A, and NS5B have all been found to be integral membrane proteins; the membrane anchorage domains of NS5A and NS5B have also been defined (11, 20, 34). We previously demonstrated that the HCV replication complex was cofractionated with a detergent-resistant, cavolin-2-rich membrane (35). All of the evidence so far is consistent with the formation of a membrane-associated HCV RNA replication complex that contains most of the HCV NS proteins.
HCV NS5B is a tail-anchored transmembrane protein (20, 34). It is likely that the membrane association of NS5B plays an important role in HCV replication. However, the recombinant NS5B lacking the C-terminal transmembrane domain is an active RNA-dependent RNA polymerase in vitro (13, 31, 32). Thus, the role of the transmembrane domain of HCV NS5B in HCV replication is so far not clear.
The C-terminal transmembrane domain of NS5B is essential for HCV replicon replication.
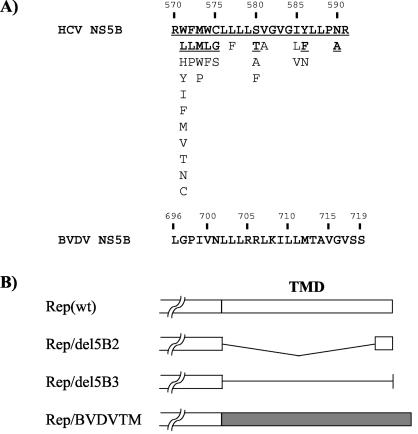
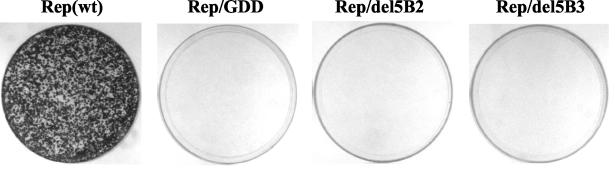
To address the role of the NS5B transmembrane domain in HCV RNA replication in vivo, we introduced a series of mutations into the NS5B of an HCV subgenomic replicon (Fig. 1). To generate a wild-type subgenomic replicon, Rep(wt), we constructed plasmid pUC-Rep/S1179I as follows. The cDNA based on HCV 1b replicon I377/NS3-3′ (GenBank accession no. AJ242652) 28) was assembled from chemically synthesized DNA oligonucleotides according to a published procedure and placed behind a T7 RNA polymerase promoter (2). A previously described adaptive mutation in NS5A, S1179I, was incorporated into this sequence (2). As a negative control, a mutation in the catalytic motif of NS5B (GDD→AAA) was generated by using the Quickchange mutagenesis kit according to the manufacturer's recommendations (Stratagene, La Jolla, Calif.). Rep/del5B2 was constructed by introducing an in-frame deletion of 19 amino acids (amino acids 571 to 589) into the NS5B transmembrane domain. All of the mutations were confirmed by DNA sequencing. The in vitro transcribed RNA was transfected into Huh-7 cells, and G418-resistant colonies were stained at 3 weeks posttransfection according to a procedure previously reported (17). As shown in Fig. 2, Rep(wt) gave rise to a large number of colonies, whereas Rep/GDD and Rep/del5B2 did not yield any colonies. This result suggests that the HCV NS5B transmembrane domain is indispensable for HCV RNA replication in the subgenomic replicon system.
FIG. 1.
(A) Amino acid sequences of the C-terminal transmembrane domain (amino acids 571 to 591) of HCV and BVDV NS5Bs (23, 34). Underlined boldface letters correspond to residues observed in more than 10% of the 307 HCV isolates (34). The rest occurred infrequently. Sequences are listed in decreasing order of observed frequency from top to bottom. Numbers above amino acid sequences represent amino acid positions in HCV and BVDV NS5Bs. (B) Schematic diagrams of the NS5B mutants in various replicons used in this study. Open and gray boxes represent HCV and BVDV NS5B transmembrane domains (TMD), respectively. Angled and straight lines represent the deletion and a noncoding RNA, respectively.
FIG. 2.
Deletion of the transmembrane domain of HCV NS5B abolishes replication of subgenomic replicons in Huh-7 cells. In vitro-transcribed replicons were transfected into Huh-7 cells, and G418-resistant colonies were stained with 0.2% crystal violet 3 weeks after transfection. For Rep/GDD, the GDD catalytic motif of HCV NS5B was changed to AAA.
It has been suggested that there are six evolutionarily conserved stem-loop structures in the NS5B-encoding region (38). One of them, SL9118, is located in the C-terminal transmembrane domain of NS5B. This structure is also located in the replicon from nucleotides 7717 to 7747 (nucleotide numbering is according to the published replicon sequence [GenBank accession no. AJ242652]), which is partially deleted in Rep/del5B2. To rule out the possibility that this deletion altered the cis-acting element of HCV RNA, we constructed Rep/del5B3 by introducing two consecutive stop codons at amino acid 571 (nucleotides 7707, 7709, and 7710 were mutated to adenosine to generate stop codons); the resulting construct expressed a truncated NS5B protein lacking the C-terminal 21 amino acids without altering the RNA structure (Fig. 1). Rep/del5B3 also did not yield any colony (Fig. 2). This result suggests that the C-terminal transmembrane domain, rather than the coding RNA element, is important for RNA replication.
The synthetic peptide consisting of the C-terminal transmembrane domain of NS5B fused to the antennapedia peptide partially disrupted the membrane association of NS5B and HCV replication.
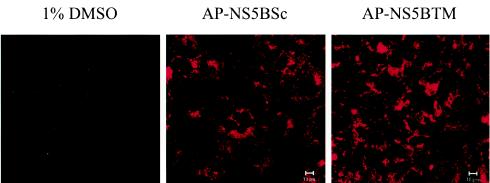
To examine whether the membrane-anchoring property or the primary sequence of the 21 amino acids is important for HCV RNA replication, we employed a cell-permeable peptide to attempt to disrupt the membrane association of NS5B without altering the RNA or protein sequence of the replicon (14). We used a peptide (AP-NS5BTM) corresponding to the C-terminal transmembrane domain (amino acids 571 to 590) of HCV NS5B fused to a portion of the homeodomain of Antennapedia (AP) (16 amino acids), a Drosophila transcription factor, which has been reported to facilitate the uptake of peptides into cultured mammalian cells through a nonendocytic, nondegradative pathway with 100% efficiency (5-7). The control peptide (AP-NS5BSc) was of the same length and amino acid composition but was scrambled in sequences of the NS5B transmembrane domain. We first determined the localization of the peptides in Huh-7/replicon cells by using the peptides labeled with rhodamine at their N termini. Huh-7/replicon cells were treated with the specific peptides at 2.5 μM. Two days after treatment, slides were washed in phosphate-buffered saline and fixed in 4% formaldehyde for 20 min at room temperature, followed by mounting in ProLong Antifade (Molecular Probes, Eugene, Oreg.). Both AP-NS5BTM and AP-NS5BSc were localized mainly in the cytoplasm, although there was slight staining in the nucleus (Fig. 3). We did not notice any difference between the two peptides with regard to intracellular localization.
FIG. 3.
Intracellular localization of the cell-permeable peptides. The various synthetic peptides were labeled with rhodamine at the N terminus. Huh-7/replicon cells were plated onto chamber slides and treated with the indicated peptides at 2.5 μM for 48 h. Cells were then washed with phosphate-buffered saline and fixed in 4% paraformaldehyde. Peptides were visualized by fluorescence microscopy.
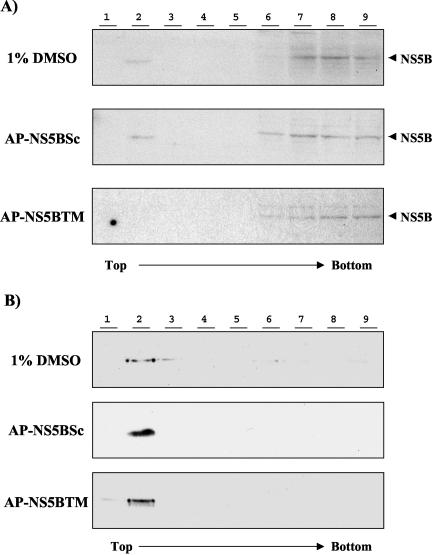
To examine the possible effect of these synthetic peptides on the membrane association of HCV NS5B, we treated Huh-7/replicon cells with synthetic peptides at 25 μM. Forty-eight hours later, membrane flotation analysis of cell lysates was performed as previously described (33). Under the conditions involved in this analysis, the membrane-containing materials float to the top of the sucrose gradient, while the cytosolic fractions remain at the bottom. The presence of NS5B in each fraction was determined by immunoblotting by using a monoclonal anti-NS5B antibody. The result showed that NS5B was in both the membrane fraction (fraction 2) and the cytosolic fractions (fractions 6, 7, 8, and 9) in the cells treated with either 1% dimethyl sulfoxide (DMSO) or the AP-NS5BSc peptide (Fig. 4A, top and middle panels). In contrast, when cells were treated with the AP-NS5BTM peptide, almost no NS5B was found in the membrane fractions (Fig. 4A, bottom panel). This result was confirmed in another replicon cell line, 1bneo/delS, in which NS5B was predominantly associated with the membrane (35; data not shown). In contrast, calnexin, which is an endoplasmic reticulum transmembrane protein (10), was not affected by the peptide treatment (Fig. 4B), indicating the specificity of these peptides. This result suggests that the synthetic peptide specifically and selectively inhibited the membrane anchorage of NS5B protein in the Huh-7/replicon cells. It is possible that AP-NS5BTM blocked the host cell membrane receptor for NS5B or that AP-NS5BTM perturbed the lipid membrane itself so as to alter efficient binding by NS5B. Finally, AP-NS5BTM might directly bind to NS5B and thereby prevent the latter from membrane binding.
FIG. 4.
The C-terminal transmembrane peptide affects the membrane association of HCV NS5B in Huh-7/replicon cells. Cells were treated with the indicated peptide at 25 μM or with DMSO alone for 48 h. Postnuclear lysates were analyzed by a membrane flotation assay. Fractions were collected from the top to the bottom of the gradient, and each fraction was analyzed for HCV NS5B (A) or calnexin (B) by immunoblotting. Experiments were repeated three times, and the results of representative experiments are shown.
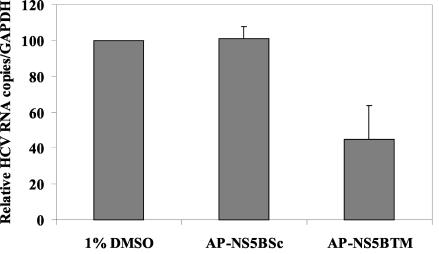
It has previously been shown that the membrane fraction of Huh-7/replicon cells contained RdRp activity, while cytosolic fractions did not (12; H. Aizaki, K. J. Lee, V. M.-H. Sung, H. Ishiko, and M. M. C. Lai, submitted for publication). Therefore, we examined whether changes in the membrane association of NS5B would affect HCV RNA replication. Huh-7/replicon cells were treated with the appropriate peptides at 25 μM for 48 h, and total cellular RNA was isolated with TRI-reagent (Molecular Research Center, Inc., Cincinnati, Ohio). HCV-specific RNA levels were determined by real-time reverse transcription (RT)-PCR amplifications with primers specific for the HCV 5′ untranslated region (Aizaki et al., submitted): 5′-GAGTGTCGTGCAGCCTCCAG-3′ (sense, 10 μM), 5′-CACTCGCAAGCACCCTATCA-3′ (antisense, 10 μM), and 5′-FAM-CCCGCAAGACTGCTAGCCGAGTAGTGTTGG-TAMRA-3′ (probe, 10 μM; Biosearch Technologies, Inc., Novato, Calif.). RT reaction mixtures were incubated for 50 min at 60°C, followed by inactivation of the reverse transcriptase coupled with activation of Taq polymerase for 5 min at 95°C. Forty cycles of PCR were performed with cycling conditions of 15 s at 94°C, 10 s at 55°C, and 1 min at 69°C. The real-time PCR signals were normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Applied Biosystems, Foster City, Calif.). As shown in Fig. 5, AP-NS5BTM peptide treatment reduced the amount of HCV RNA by more than 50% compared to that in the cells treated with AP-NS5BSc or 1% DMSO alone. This result suggests that the membrane association of HCV NS5B is important for HCV RNA synthesis. Thus, the membrane association of NS5B facilitates HCV RNA synthesis. It is not clear why the peptide did not completely inhibit RNA accumulation. It is possible that the NS5B not associated with the membrane still retained some polymerase activity in vivo. It is also possible that AP-NS5BTM peptide inhibits membrane association of newly synthesized NS5B while not disrupting already-associated NS5B. NS5B was found predominantly in the membrane fraction when the 1bneo/delS replicon was used in a previous analysis (14a, 17, 35; Aizaki et al., submitted). In contrast, the NS5B from the replicon used in this study, which is based on the HCV ConI strain (28), was found in both the membrane and the cytosolic fractions. Furthermore, the amount of NS5B in the cytosolic fractions was larger than that in the membrane fractions. It is possible that the difference in the replicon sequences may alter the property of NS5B.
FIG. 5.
The C-terminal transmembrane peptide inhibits HCV RNA replication in Huh-7/replicon cells. Cells were treated with the peptides as in the experiments shown in Fig. 4. Total cellular RNA was analyzed for HCV and GAPDH RNA by real-time Taqman PCR. Relative HCV RNA copies per GAPDH RNA are presented. The RNA level in 1% DMSO-treated cells is represented as 100%. Experiments were repeated four times, and the average values for the experiments are shown.
A related transmembrane domain derived from BVDV could not replace the HCV NS5B transmembrane domain.
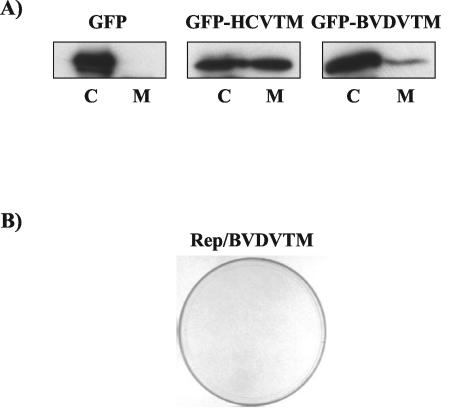
We next examined whether the transmembrane domain of NS5B could be replaced with the comparable transmembrane domains of other viruses, such as bovine viral diarrhea virus (BVDV). A hydrophobicity profile comparison between BVDV and HCV revealed a highly hydrophobic domain of 24 amino acids at the C terminus of BVDV NS5B (23) (Fig. 1A). We first studied whether the hydrophobic domain of BVDV NS5B could confer the membrane-binding property. The hydrophobic domain of BVDV NS5B and the corresponding transmembrane domain of HCV NS5B were fused to the C terminus of green fluorescence protein (GFP) (Clontech, Palo Alto, Calif.). Huh-7 cells were transfected with plasmids expressing these GFP fusion proteins or GFP alone. The membrane flotation assay was performed. Fractions 1 to 4 and 5 to 9 from the top of the gradient were pooled separately, and immunoblotting was performed with an anti-GFP antibody. GFP was localized exclusively in the cytosolic fraction (Fig. 6A, left panel), whereas GFP fusion proteins with either the HCV or the BVDV transmembrane domain were detected in both cytosolic and membrane fractions (Fig. 6A, middle and right panels), although the transmembrane domain of BVDV NS5B appeared to be weaker than that of HCV NS5B in membrane-anchoring activity. This result suggests that the transmembrane domains from both HCV and BVDV have a membrane-anchoring property. To address whether these two transmembrane domains have similar functions, we replaced the transmembrane domain of HCV NS5B with the corresponding domain of BVDV NS5B. The results showed that the chimeric NS5B (Rep/BVDVTM) did not support HCV replication (Fig. 6B); thus, the transmembrane domain from a related virus cannot replace that of HCV NS5B, suggesting that the transmembrane domain of HCV NS5B not only functions as a membrane anchor but may have other functions as well. However, we cannot completely rule out the possibility that the chimeric RNA may have altered RNA structures. We have recently found that the full-length and the C-terminal (21-amino-acid) truncated HCV NS5Bs differed in several enzymatic properties in vitro, such as template binding, nucleotide substrate selection, modulation of terminal transferase activity, and the optimum enzymatic conditions, indicating that the C-terminal 21 amino acids may affect the catalytic center of NS5B (N. Vo and M. M. C. Lai, unpublished data). Thus, it is conceivable that the C-terminal 21-amino-acid region interacts with the remaining parts of NS5B in a sequence-specific manner. It is also possible that this domain interacts with cellular proteins specifically. Our unpublished results showed that a cellular vesicle membrane transport protein, hVAP-33, binds to the N terminus of NS5A as well as the C terminus of NS5B. NS5A is a type II membrane protein, with its N terminus being the transmembrane domain (14a). In contrast, NS5B is a type I membrane protein. Thus, the NS5A- and NS5B-interacting domains of hVAP-33 are likely to be close to the membrane. We have shown that small interfering RNA of hVAP-33 could inhibit membrane association of HCV NS proteins and correspondingly inhibit HCV RNA replication (Gaoet al., submitted). It has also previously been reported that various single amino acid substitutions within the C-terminal transmembrane domain or extensions of the C-terminal transmembrane domain did not affect membrane association but dramatically impaired replication competence (20). Together, these findings indicate that the C-terminal transmembrane domain, in addition to being a membrane anchor, has other specific functions. It is also possible that Rep/BVDVTM replicated weakly but could not form any colonies under our selective condition (an active dose of 0.5 mg of G418/ml).
FIG. 6.
A related transmembrane domain from BVDV NS5B cannot replace the transmembrane domain of HCV NS5B. (A) The transmembrane domains of HCV and BVDV NS5Bs were fused to the C terminus of GFP and transfected into Huh-7 cells. A membrane flotation assay was performed 48 h after transfection. The membrane fractions (M; fractions 1 to 4) and the cytosolic fractions (C; fractions 5 to 9) were separately pooled and analyzed by immunoblotting with an anti-GFP rabbit polyclonal antibody. (B) Replication of the chimeric HCV-BVDV NS5B (Fig. 1B, Rep/BVDVTM). A colony formation assay was performed as described in the legend to Fig. 2.
In summary, our findings here showed that the C-terminal 21-amino-acid domain of NS5B likely serves dual functions in viral RNA replication in vivo, both as a membrane anchor and in participation in RNA synthesis in a sequence-specific manner. This requirement was not seen in the in vitro polymerase assay of NS5B.
REFERENCES
- 1.Alter, M. J. 1994. Transmission of hepatitis C virus—route, dose, and titer. N. Engl. J. Med. 330:784-786. [DOI] [PubMed] [Google Scholar]
- 2.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 3.Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. [DOI] [PubMed] [Google Scholar]
- 4.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derossi, D., S. Calvet, A. Trembleau, A. Brunissen, G. Chassaing, and A. Prochiantz. 1996. Cell internalization of the third helix of the Antennapedia homeodomain is receptor-independent. J. Biol. Chem. 271:18188-18193. [DOI] [PubMed] [Google Scholar]
- 6.Derossi, D., G. Chassaing, and A. Prochiantz. 1998. Trojan peptides: the penetratin system for intracellular delivery. Trends. Cell Biol. 8:84-87. [PubMed] [Google Scholar]
- 7.Derossi, D., A. H. Joliot, G. Chassaing, and A. Prochiantz. 1994. The third helix of the Antennapedia homeodomain translocates through biological membranes. J. Biol. Chem. 269:10444-10450. [PubMed] [Google Scholar]
- 8.Di Bisceglie, A. M., H. L. Bonkovsky, S. Chopra, S. Flamm, R. K. Reddy, N. Grace, P. Killenberg, C. Hunt, C. Tamburro, A. S. Tavill, R. Ferguson, E. Krawitt, B. Banner, and B. R. Bacon. 2000. Iron reduction as an adjuvant to interferon therapy in patients with chronic hepatitis C who have previously not responded to interferon: a multicenter, prospective, randomized, controlled trial. Hepatology 32:135-138. [DOI] [PubMed] [Google Scholar]
- 9.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elagoz, A., M. Callejo, J. Armstrong, and L. A. Rokeach. 1999. Although calnexin is essential in S. pombe, its highly conserved central domain is dispensable for viability. J. Cell Sci. 112:4449-4460. [DOI] [PubMed] [Google Scholar]
- 11.Elazar, M., K. H. Cheong, P. Liu, H. B. Greenberg, C. M. Rice, and J. S. Glenn. 2003. Amphipathic helix-dependent localization of NS5A mediates hepatitis C virus RNA replication. J. Virol. 77:6055-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Hage, N., and G. Luo. 2003. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J. Gen. Virol. 84:2761-2769. [DOI] [PubMed] [Google Scholar]
- 13.Ferrari, E., J. Wright-Minogue, J. W. S. Fang, B. M. Baroudy, J. Y. N. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallouzi, I. E., and J. A. Steitz. 2001. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 294:1895-1901. [DOI] [PubMed] [Google Scholar]
- 14a.Gao, L., H. Aizaki, J.-W. He, M. M. C. Lai. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78:3480−3488. [DOI] [PMC free article] [PubMed]
- 15.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67:2832-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hugle, T., F. Fehrmann, E. Bieck, M. Kohara, H. G. Krausslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70-81. [DOI] [PubMed] [Google Scholar]
- 19.Hwang, S. B., K. J. Park, Y. S. Kim, Y. C. Sung, and M. M. Lai. 1997. Hepatitis C virus NS5B protein is a membrane-associated phosphoprotein with a predominantly perinuclear localization. Virology 227:439-446. [DOI] [PubMed] [Google Scholar]
- 20.Ivashkina, N., B. Wolk, V. Lohmann, R. Bartenschlager, H. E. Blum, F. Penin, and D. Moradpour. 2002. The hepatitis C virus RNA-dependent RNA polymerase membrane insertion sequence is a transmembrane segment. J. Virol. 76:13088-13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai, V. C., C. C. Kao, E. Ferrari, J. Park, A. S. Uss, J. Wright-Minogue, Z. Hong, and J. Y. Lau. 1999. Mutational analysis of bovine viral diarrhea virus RNA-dependent RNA polymerase. J. Virol. 73:10129-10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang, T. J., B. Rehermann, L. B. Seeff, and J. H. Hoofnagle. 2000. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann. Intern. Med. 132:296-305. [DOI] [PubMed] [Google Scholar]
- 25.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 26.Lohmann, V., J. O. Koch, and R. Bartenschlager. 1996. Processing pathways of the hepatitis C virus proteins. J. Hepatol. 24:11-19. [PubMed] [Google Scholar]
- 27.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 29.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, J. K. Albrecht, et al. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 30.Mottola, G., G. Cardinali, A. Ceccacci, C. Trozzi, L. Bartholomew, M. R. Torrisi, E. Pedrazzini, S. Bonatti, and G. Migliaccio. 2002. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology 293:31-43. [DOI] [PubMed] [Google Scholar]
- 31.Oh, J. W., G. T. Sheu, and M. M. Lai. 2000. Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J. Biol. Chem. 275:17710-17717. [DOI] [PubMed] [Google Scholar]
- 32.Ranjith-Kumar, C. T., L. Gutshall, M. J. Kim, R. T. Sarisky, and C. C. Kao. 2002. Requirements for de novo initiation of RNA synthesis by recombinant flaviviral RNA-dependent RNA polymerases. J. Virol. 76:12526-12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanderson, C. M., H. H. Wu, and D. P. Nayak. 1994. Sendai virus M protein binds independently to either the F or the HN glycoprotein in vivo. J. Virol. 68:69-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt-Mende, J., E. Bieck, T. Hugle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276:44052-44063. [DOI] [PubMed] [Google Scholar]
- 35.Shi, S. T., K. J. Lee, H. Aizaki, S. B. Hwang, and M. M. Lai. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 77:4160-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi, S. T., S. J. Polyak, H. Tu, D. R. Taylor, D. R. Gretch, and M. M. Lai. 2002. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology 292:198-210. [DOI] [PubMed] [Google Scholar]
- 37.Tu, H., L. Gao, S. T. Shi, D. R. Taylor, T. Yang, A. K. Mircheff, Y. Wen, A. E. Gorbalenya, S. B. Hwang, and M. M. Lai. 1999. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology 263:30-41. [DOI] [PubMed] [Google Scholar]
- 38.Tuplin, A., J. Wood, D. J. Evans, A. H. Patel, and P. Simmonds. 2002. Thermodynamic and phylogenetic prediction of RNA secondary structure in the coding region of hepatitis C virus. RNA 8:824-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeuzem, S., S. V. Feinman, J. Rasenack, E. J. Heathcote, M. Y. Lai, E. Gane, J. O'Grady, J. Reichen, M. Diago, A. Lin, J. Hoffman, and M. J. Brunda. 2000. Peginterferon alfa-2a in patients with chronic hepatitis C. N. Engl. J. Med. 343:1666-1672. [DOI] [PubMed] [Google Scholar]