Summary
Migrating neurons are bipolar, with a leading process and a trailing process [1]. The proximal region of the leading process displays a concentration of F-actin that contributes to the advance of the soma and the centrosome [2–7]. Here we show that kinesin-6, a microtubule-based motor protein best known for its role in cytokinesis, also concentrates in this region. Depletion of kinesin-6 results in multipolar neurons that are either stationary or continuously change their direction of movement. In such neurons, F-actin no longer concentrates in a single process. During cytokinesis, kinesin-6 forms a complex with a Rho family GTPase activating protein called MgcRacGAP to signal to the actin cytoskeleton so that cortical movements are concentrated in the cleavage furrow [8–13]. During neuronal migration, MgcRacGap also concentrates in the proximal region of the leading process, and inhibition of its activity results in a phenotype similar to kinesin-6 depletion. We conclude that neuronal migration utilizes a cytoskeletal pathway analogous to cytokinesis, with kinesin-6 signaling through MgcRacGap to the actin cytoskeleton to constrain process number and restrict protrusive activity to a single leading process, thus resulting in a bipolar neuron able to move in a directed fashion.
Results and discussion
Kinesin-6 depletion results in altered migration and morphology of cortical neurons in vivo
Three EGFP-containing plasmids, kinesin-6-shRNA-coding, corresponding scrambled control or empty vector, were individually electroporated into cortical ventricular zones of E14 mice (see Experimental Procedures for details and of this and other procedures in Supplemental section). (efficiency of knockdown shown in figure 1, E). Similar numbers of fluorescent cells appeared in all groups, but with slightly fewer in the kinesin-6 shRNA group, consistent with cytokinesis failure leading to cell death of neural progenitors [14, 15]. No differences in radial glial cells were observed among the groups (data not shown), indicating that observed effects on neuronal migration were not due to alterations in the scaffold along which migration occurs.
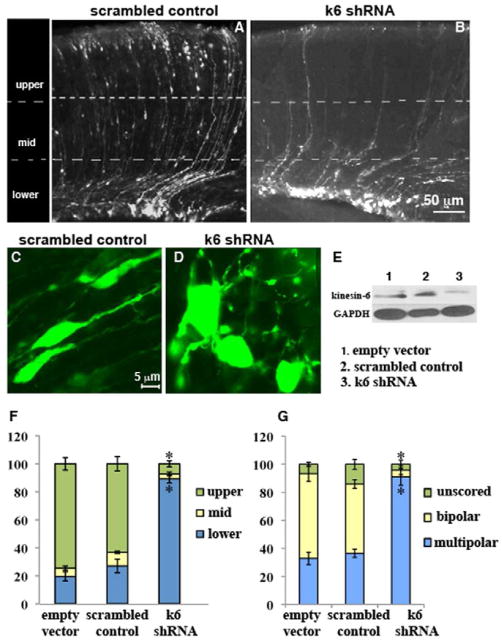
Figure 1.
Altered distribution and morphology of cortical neurons after in utero electroporation of kinesin-6 shRNA. Coronal sections from brains that had been electroporated with scrambled control shRNA (A) or kinesin-6 shRNA (B) at E14 and harvested at P1. Quantification of cell distribution in the 3 bins (F). For the empty vector group, n = 4 brains, 1152 cells from 6 sections were counted. For the scrambled control group, n = 3 brains, 739 cells from 5 sections were counted. For the kinesin-6 shRNA group, n = 4 brains, 524 cells from 10 sections were counted. (C, D) Morphology of cells in subventricular zone region in coronal sections generated after in utero electroporation of scrambled control shRNA, and kinesin-6 shRNA respectively. (G) Quantification of cell morphology. Error bars represent SEM. (E) Western blot for kinesin-6 and glyceraldehyde-3-phosphate (GAPDH) (loading control) in NIH/3T3 cells transfected with control empty vector, scrambled control shRNA or kinesin-6 shRNA after 72 hr showing approximately 70% depletion of kinesin-6 protein. Migration of cortical neurons was also analyzed in vitro (see supplemental figure 1).
Each section was divided into 3 bins: upper, middle and lower. As expected [16, 17], most cells reached the upper bin in the case of the empty vector and scrambled control (figure 1, A, F) groups (74.5 ± 4.5% and 63.2 ± 5.1 % respectively; no significant difference). In the kinesin-6 shRNA group, most cells were stalled in the lower bin (figure 1, B, F); only 7.3 ± 2.2 % cells were found in the upper bin (significant difference with the empty vector and scrambled control in both the upper bin [p<0.0001, p<0.0001 respectively] and the lower bin [(p<0.0001, p<0.0001 respectively, student’s t-test]).
As they enter the cortical intermediate zone region from the subventricular zone region, neurons normally transform from a multipolar to a bipolar morphology suitable for directed migration [18, 19]. In the kinesin-6 shRNA group, most cells in this region were multipolar (figure 1, D, G) (91.1 ± 6%) unlike the empty vector and the scrambled control groups (figure 1, C, G) (32.8 ± 4.4% and 36.6 ± 3% were multipolar respectively) (no significant difference between the empty vector and scrambled control groups; kinesin-6 shRNA group was significantly different from each of the other 2 groups [p, 0.008 and p, 0.007 respectively, student’s t-test]). Many cells were bipolar in the empty vector and the scrambled control groups (60.4 ± 5.5% and 49.3 ± 3.1% respectively), but very few were bipolar in this region in the kinesin-6 shRNA group (4.2 ± 3%).
Kinesin-6 depletion alters cortical neuron migration along fiber lattice in vitro
In a culture system of cortical neurons migrating along glial fibers in vitro [20], neurons transfected with kinesin-6 shRNA assumed a multipolar morphology and were more-or-less immobile (supplemental figure 1, C, D), whereas cells transfected with scrambled control migrated rapidly (supplemental figure 1, A, B). Mean migration speed of control neurons was 40.4 μm/hr (± 4.8, n = 14), while that of kinesin-6 shRNA neurons was 15.6 μm/hr (± 3.7, n = 11) (significant difference [p = 0.0007, student’s t-test])
Kinesin-6 depletion affects migration of cerebellar granule neurons in vitro
For mechanistic studies, cerebellar granule neurons were transfected with siRNA (control or kinesin-6) and pEGFP-C1 and then plated on glass. Efficiency of knockdown is shown in figure 2, A. Time-lapse imaging indicated that while control neurons moved in a unidirectional fashion (figure 2, B), kinesin-6-depleted neurons frequently changed direction of migration (figure 2, C) (also see Video 1 in supplemental). While control neurons exhibited an average net displacement of 26.3 μm/hr ± 2.0 (n = 17), kinesin-6-depleted neurons exhibited little average net displacement (6.7 μm/hr ± 1.0, n = 14, p<0.0001 student’s t-test) (figure 2, D). Thus kinesin-6 depletion did not render these neurons stationary as with the cortical neurons, but perturbed directed movement, as reflected by a decrease in the persistence of movement (figure 2, E, F).
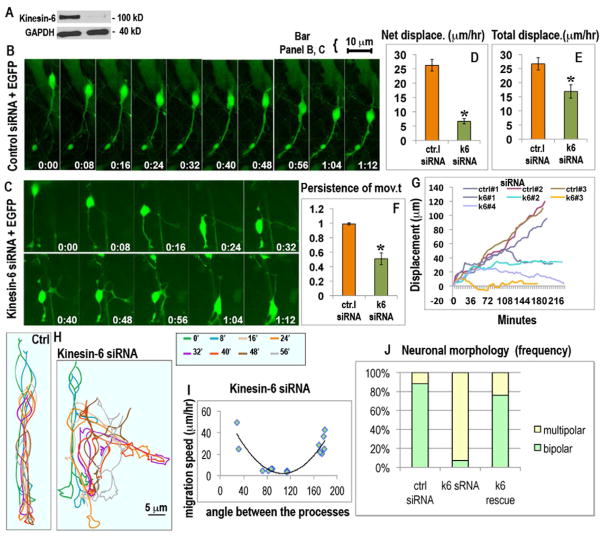
Figure 2.
Effect of siRNA-mediated depletion of kinesin-6 on migration and morphology of cerebellar granule neurons in vitro. (A) Western blot for kinesin-6 and GAPDH in dissociated cerebellar cells transfected with control siRNA (left lane) or kinesin-6 siRNA (right lane) for 72 hr, showing more than 90% depletion of kinesin-6 protein. (B) Migration of cerebellar neurons co-transfected with control siRNA, EGFP and (C) kinesin-6 siRNA, EGFP as represented by images captured 4 min apart; see also video 1. (D, E, F) Average net displacement per hr, average total displacement per hr and average persistence of movement for control and kinesin-6-depleted neurons. Error bars represent SEM. (G) Tracings of movements of somas of 3 control neurons (upper 3 lines) and 4 kinesin-6-depleted neurons (lower four lines). (H) Tracings of cell shape of a migrating neuron transfected with control siRNA (on the left) or kinesin-6 siRNA (on the right). Different time frames were traced in different colors and this color code is explained in the box on the side. (I) Correlation between the angle between the processes and migration speeds for individual cells for the kinesin-6 siRNA group. (J) Quantification of cell morphology in control, kinesin-6 siRNA and “kinesin-6 siRNA plus kinesin-6-GFP” (rescue) groups. See supplemental figure 2 for the effect of kinesin-6 depletion on centrosome movements and the kinesin-6 overexpression phenotype.
While most control neurons exhibited a bipolar morphology with a prominent leading process and a thin trailing process, kinesin-6-depleted neurons were usually multipolar (figure 2, H, J). Most often these cells had 3 processes, 2 of which were morphologically similar, and a third which was thinner and more similar to the trailing process of control neurons (see figure 3, D, for morphological analyses on fixed cells). Morphological analyses on live cells indicated that the proportion of multipolar cells was significantly higher at 92.9% (n = 14) in the kinesin-6 siRNA group compared to the control group, in which the proportion was 7.1% (n = 17) (figure 2, J, p<0.0001, chi square test). This phenotype was rescued by expression of siRNA-resistant kinesin-6-GFP (figure 2, J, n = 25). When the angle between the 2 similar processes (plotted against the speed of migration on a per cell basis; see figure 2, I) was closer to 180 degrees (or 0 degrees), migration speeds were higher. When the angle was 60 to 120 degrees, the speeds were lower, suggesting that altered morphology is at least partially responsible for altered migration. Overexpression of kinesin-6 caused slight bundling of microtubules and reduced the length of the leading process by 36.3% (supplemental figure 2, F, F′; n = 8 in each group, p<0.0001).
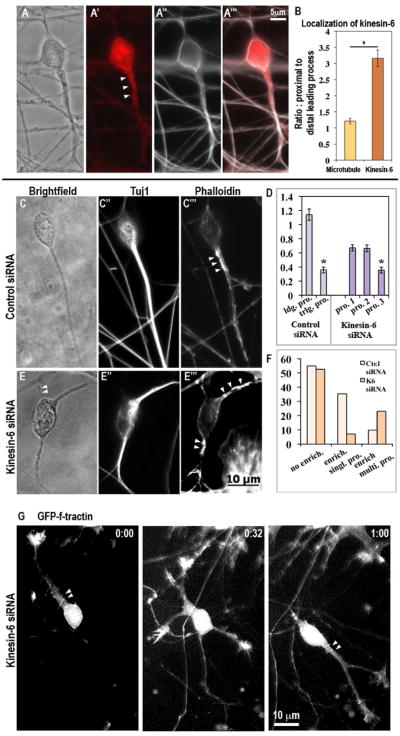
Figure 3.
Kinesin-6 immunostaining in cultured cerebellar granule neurons and effect on actin distribution. (A-A‴, B) Kinesin-6 and alpha-tubulin (to reveal microtubules) co-immunostaining in cerebellar granule neuron. Note that alpha-tubulin (microtubule) staining does not show enrichment in the proximal leading process whereas kinesin-6 staining is enriched. Immunostaining for Tuj1 (neuron-specific beta-III tubulin) and simultaneous staining with phalloidin to detect F-actin on neurons transfected with control siRNA (C-C‴) or kinesin-6 siRNA (E-E‴) after 72 hr. In the control siRNA group, a subset of cells showed strong F-actin enrichment in specific subcellular regions. In this subset, this enrichment was typically restricted to the proximal region of a single leading process, whereas in the kinesin-6 siRNA group, F-actin enrichment was often observed simultaneously in multiple processes. Arrowheads indicate F-actin enriched regions (D) Quantification of cellular morphology in terms of process thickness for cells transfected with control or kinesin-6 siRNA. In the control group, neurons possessed a single leading process, which can be defined by greater thickness of this process, whereas in the kinesin-6 group, neurons typically possessed 3 processes, 2 of which were of similar thickness. Effect of kinesin-6 depletion on microtubule dynamics was also analyzed (see supplemental figure 3). (F) Quantification of the proportion of neurons showing F-actin enrichment in the total population, and pattern of actin enrichment within this subset, in cultures that were transfected with control siRNA or kinesin-6 siRNA for 72 hr. (G) Live imaging of F-actin using GFP-F-tractin probe in kinesin-6 depleted cells.
Movement of the centrosome, observed with RFP (red fluorescent protein)-tagged pericentrin, consistently preceded that of the caudal tip in the control siRNA group (supplemental figure 2, A, A′). By contrast, in the kinesin-6-depleted group, the caudal tip and the centrosome sometimes overlapped (supplemental figure 2, B, B′). In the control siRNA group, for 88.9% of the cells, the centrosome was consistently situated in the anterior soma, but that was true for only 10% of the cells in the kinesin-6 siRNA group (n = 27 for control siRNA group, n = 20 for kinesin-6 siRNA group, p< 0.0001, Fisher’s exact test). (See supplemental figure 2, C).
Kinesin-6 is enriched in the proximal region of the leading process
Immunostain signal for kinesin-6 was more intense in the proximal region of the leading process compared to its distal region (figure 3, A–A‴). The regions rich in kinesin-6 were typically less concentrated with microtubules than other regions of the leading process, indicating a true enrichment of kinesin-6 relative to microtubule mass (figure 3, B). This was further confirmed by comparison to volume marker, EGFP (not shown). In most of these neurons, the centrosome was situated within this enriched region (92.9%) (see supplemental figure 2, D–D″, and E-E″ for micrographs). Correlation between kinesin-6 enrichment and centrosome localization was significant (p, 0.001, Pearson’s correlation coefficient, between the “centrosome only” group and “both” group; and p, 0.005 between the “enrichment only” and “both” group, total n = 16). Interestingly, there was a strong correlation between kinesin-6 and F-actin enrichments (p, 0.0008, Pearson’s correlation coefficient, between the “actin only” and “both” groups and also between the “kinesin-6 only” and “both” groups), which might relate to the fact that kinesin-6 has an actin-binding domain [21].
F-actin is mis-localized in kinesin-6-depleted granule neurons
Phalloidin-staining showed F-actin enriched in the proximal leading process (figure 3, C–C‴) of control neurons. Because kinesin-6-depleted neurons were generally multipolar, no one process was clearly identifiable as the leader. While in control cells, the leading process was consistently and significantly thicker than the trailing process (mean thickness for leading process, 1.2 μm ± 0.01 and mean thickness for trailing process, 0.4 μm ± 0.01, n = 36. See figure 3, D), kinesin-6-depleted cells typically had 3 processes of which 2 were of comparable thickness while the third process was usually thinner (mean thickness for processes, 0.7 μm ± 0.01, 0.7 μm ± 0.01, and 0.4 μm ± 0.01, n = 34). In phalloidin-stained control cells, 54.9% cells did not show any F-actin enrichment, 35.3% cells showed enrichment in the leading process and 9.8% cells showed enrichment in multiple processes (n = 51). This is consistant with previous studies indicating that proximal F-actin enrichment only occurs when the cell is making a forward step, and hence would not be expected to be in every cell at all times [3]. In kinesin-6-depleted cultures, 52.6% cells did not show any F-actin enrichment, 7.0% cells showed enrichment in a single process whereas, 23% cells showed enrichment in multiple processes (n = 57. See figure 3, C–C‴, E-E‴ and also F). The difference between the control siRNA and kinesin-6 siRNA group, with regard to percentage of cells showing F-actin enrichment in a single process and that in multiple processes, was found to be significant by using 2×2 contingency table test (p<0.0001). Rate of F-actin turnover in the proximal leading process is higher than within other cellular compartments [3]. No detectable differences in the dynamics of the actin enrichments in control and kinesin-6-depleted neurons were observed when F-actin dynamics were probed using time-lapse microscopy on GFP-F-tractin transfected neurons (figure 3, G, see Supplemental Experimental Procedures).
Changes in microtubule organization in kinesin-6-depleted migratory neurons
Neurons were co-transfected with kinesin-6 or control siRNA, RFP-Pct (to label the centrosome), and EGFP-EB3 (to label growing microtubule plus ends, which appear as fluorescent comets). Comets filled the leading process and curved around the nucleus in the soma (still images in supplemental figure 3, A, B). Comet speed was indistinguishable in controls (0.21 μm/sec ± 0.02) and kinesin-6-depleted cells (0.23 μm/sec ± 0.01). In the control leading process and soma, most comets emerged and projected away from the centrosome (93.8 ± 1% and 93.7 ± 3.3% respectively). In kinesin-6-depleted cells, the proportion of comets directed away from the centrosome in the soma region was similar to controls (95.3 ± 1.7%, p, 0.7), but there was a significant decrease in comets directed away from the centrosome in the leading process (70.2 ± 1.5%, p, 0.02) (supplemental figure 3, C) (n = 9 cells in each group). Notably, no significant difference was observed in the orientation of comets directed away from the centrosome, in the process that was not the leader at that point of time, in comparison with the leading process in controls (92.5 ± 2.5%, p, 0.8, student’s t-test) (n = 9 cells in each group). Thus, unlike the actin enrichment, which could appear simultaneously in 2 processes, the microtubule defect resulting from kinesin-6 depletion was specific to the process serving as the leader at any moment in time. We suspect that this reflects a role for kinesin-6 in either maintaining microtubule organization in the face of major alterations in the actin cytoskeleton, and/or in directing which process will take possession of the actin enrichment thus becoming the leader.
MgcRacGAP in migratory neurons
A Rho family GTPase-activating protein called MgcRacGAP combines during cytokinesis with kinesin-6 to form a complex called centralspindlin that binds to microtubules in the spindle midzone and regulates F-actin localization via a downstream signaling cascade [9–13]. If this same situation obtains in migratory neurons, the two proteins should track together. Conducting time-lapse imaging on neurons transfected with a DsRed-tagged MgcRacGAP construct, we found the tagged protein to localize in the leading process and soma in a distribution that co-localized with kinesin-6 in immunostain analyses. As the neurons migrated, we observed the tagged MgcRacGAP moving through these cellular compartments, with co-transfected EGFP marking the outline of the cell. For multiple cells (n = 14 cells, n = 27 instances of forward stepping of soma), quantitative analyses of the tagged MgcRacGAP revealed it moving to concentrate in the proximal region of the process serving as the leader, in similar fashion to the F-actin enrichment (see supplemental figure 4 and its legend for details). In kinesin-6-depleted neurons that displayed normal bipolar morphology, MgcRacGAP distribution resembled that of controls, whereas kinesin-6-depleted multipolar cells showed MgcRacGAP in more than one process simultaneously (supplemental figure 4, C).
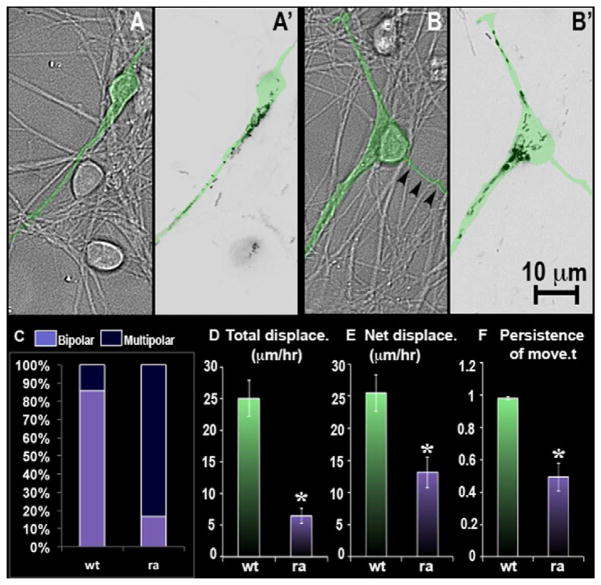
A GAP-inactive version of the construct called MgcRacGAP-RA, is known to act as a dominant-negative [22]. Comparison of control neurons (i.e. not kinesin-6 depleted) transfected with wild-type MgcRacGAP or MgcRacGAP-RA indicated that while the wild-type protein was localized in the leading process, the mutant protein was distributed in 2 different processes (figure 4, A, A′, B, B′). Time-lapse imaging indicated that while neurons transfected with wild-type MgcRacGAP exhibited net displacement of 25 μm/hr (± 2.9, n = 14 cells), and total displacement of 25.5 μm/hr (± 2.8), the MgcRacGAP-RA group exhibited net displacement and total displacement of 6.5 μm/hr (±1.2, n = 12 cells) and 13.2 μm/hr (± 2.4) respectively. Persistence of movement was also different for these two groups (figure 4, D, E, F). A significantly higher proportion of cells transfected with MgcRacGAP-RA exhibited multipolar morphology (83.3%) compared to those transfected with wild-type MgcRacGAP (14.3%) (figure 4, C). These effects on migration closely resembled those resulting from kinesin-6 depletion, consistent with a cytokinesis-like pathway functioning in migrating neurons. This view is buoyed by previous findings that RhoA (the target of MgcRacGAP signaling) also accumulates in the leading process [23].
Figure 4.
Expression of DsRed-tagged wild-type or mutant (dominant/negative) MgcRacGAP in cultured cerebellar granule neurons. (A, A′) DIC and inverted epifluorescence still images respectively of neurons transfected with wild-type MgcRacGAP construct. (B, B′) DIC and inverted epifluorescence still images respectively of neurons transfected with mutant (ra) MgcRacGAP construct. Arrowheads in B indicate the thinner of the 3 processes, with the MgcRacGAP entering the other 2. Transfected neuron is digitally colored green in top four panels. (C) Analyses of morphology of migrating neurons transfected with the wild-type or mutant construct. (D, E) Quantification for average total displacement and average net displacement per hr for wild-type and mutant groups. (F) Quantification of persistence of movement for wild-type and mutant groups. See also supplemental figure 4.
We previously reported that kinesin-6 helps establish the mixed orientation of dendritic microtubules [24, 25], presumably via a sliding microtubule mechanism consistent with the properties of kinesin-6 to transport minus-ends of microtubules toward plus-ends of other microtubules [26]. Our present findings, consistent with the strong expression of kinesin-6 in migratory neurons [27], indicate that kinesin-6 plays another, earlier role in neuronal development. In migratory neurons, kinesin-6 is not essential to concentrate F-actin in the proximal region of processes, but it is essential to constrain the F-actin concentration to a single process. Such constraint appears to be critical for enabling the neuron to designate a single process as the leader, and hence to enable the centrosome to follow that process in an orderly fashion. The role of kinesin-6 in neuronal migration borrows a mechanistic theme from cytokinesis, one of the most fundamental cytoskeletal pathways in nature, with the proximal region of the leading process being analogous to the cleavage furrow.
Supplementary Material
Highlights.
Depletion of kinesin-6 perturbs directed neuronal migration
Kinesin-6 localizes in and directs actin filaments to the proximal leading process
Depletion of kinesin-6 results in a multi-polar neuron with no leading process
Mechanisms that regulate cytokinesis are re-purposed for neuronal migration
Acknowledgments
This work was funded by grants from the National Institutes of Health (R01 NS28785) and the National Science Foundation (0841245) to P.W.B., as well as from the Department of Biotechnology, Government of India to S.T., and from the Priority Academic Program Development of Jiangsu Higher Education Institutions and National Natural Science Foundation Grant (31171007) to M.L. We thank K. Kadam of the Tata Institute for performing the in utero electroporations. For helpful discussions, we thank I. Fischer, X. Yuan, K. Toyo-oka, and W. Yu, all of Drexel University. A.F. is the recipient of the Doris Willig, M.D. Award from Drexel University College of Medicine, Institute for Women’s Health and Leadership. S.T. is the recipient of the Shanti Swarup Bhatnagar prize. The authors declare no competing financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Edmondson JC, Hatten ME. Glial-guided granule neuron migration in vitro: a high-resolution time-lapse video microscopic study. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1987;7:1928–1934. doi: 10.1523/JNEUROSCI.07-06-01928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME. Par6alpha signaling controls glial-guided neuronal migration. Nature neuroscience. 2004;7:1195–1203. doi: 10.1038/nn1332. [DOI] [PubMed] [Google Scholar]
- 3.Solecki DJ, Trivedi N, Govek EE, Kerekes RA, Gleason SS, Hatten ME. Myosin II motors and F-actin dynamics drive the coordinated movement of the centrosome and soma during CNS glial-guided neuronal migration. Neuron. 2009;63:63–80. doi: 10.1016/j.neuron.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nature neuroscience. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- 5.Bellion A, Baudoin JP, Alvarez C, Bornens M, Metin C. Nucleokinesis in tangentially migrating neurons comprises two alternating phases: forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2005;25:5691–5699. doi: 10.1523/JNEUROSCI.1030-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma X, Kawamoto S, Hara Y, Adelstein RS. A point mutation in the motor domain of nonmuscle myosin II-B impairs migration of distinct groups of neurons. Molecular biology of the cell. 2004;15:2568–2579. doi: 10.1091/mbc.E03-11-0836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaar BT, McConnell SK. Cytoskeletal coordination during neuronal migration. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13652–13657. doi: 10.1073/pnas.0506008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glotzer M. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nature reviews Molecular cell biology. 2009;10:9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mishima M, Kaitna S, Glotzer M. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Developmental cell. 2002;2:41–54. doi: 10.1016/s1534-5807(01)00110-1. [DOI] [PubMed] [Google Scholar]
- 10.Pavicic-Kaltenbrunner V, Mishima M, Glotzer M. Cooperative assembly of CYK-4/MgcRacGAP and ZEN-4/MKLP1 to form the centralspindlin complex. Molecular biology of the cell. 2007;18:4992–5003. doi: 10.1091/mbc.E07-05-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 12.Bement WM, Benink HA, von Dassow G. A microtubule-dependent zone of active RhoA during cleavage plane specification. The Journal of cell biology. 2005;170:91–101. doi: 10.1083/jcb.200501131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Somers WG, Saint R. A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Developmental cell. 2003;4:29–39. doi: 10.1016/s1534-5807(02)00402-1. [DOI] [PubMed] [Google Scholar]
- 14.Sarkisian MR, Frenkel M, Li W, Oborski JA, LoTurco JJ. Altered interneuron development in the cerebral cortex of the flathead mutant. Cereb Cortex. 2001;11:734–743. doi: 10.1093/cercor/11.8.734. [DOI] [PubMed] [Google Scholar]
- 15.Di Cunto F, Imarisio S, Hirsch E, Broccoli V, Bulfone A, Migheli A, Atzori C, Turco E, Triolo R, Dotto GP, et al. Defective neurogenesis in citron kinase knockout mice by altered cytokinesis and massive apoptosis. Neuron. 2000;28:115–127. doi: 10.1016/s0896-6273(00)00090-8. [DOI] [PubMed] [Google Scholar]
- 16.Tsai JW, Chen Y, Kriegstein AR, Vallee RB. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. The Journal of cell biology. 2005;170:935–945. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falnikar A, Tole S, Baas PW. Kinesin-5, a mitotic microtubule-associated motor protein, modulates neuronal migration. Molecular biology of the cell. 2011;22:1561–1574. doi: 10.1091/mbc.E10-11-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nature neuroscience. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 19.Tabata H, Nakajima K. Multipolar migration: the third mode of radial neuronal migration in the developing cerebral cortex. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2003;23:9996–10001. doi: 10.1523/JNEUROSCI.23-31-09996.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nichols AJ, Carney LH, Olson EC. Comparison of slow and fast neocortical neuron migration using a new in vitro model. BMC neuroscience. 2008;9:50. doi: 10.1186/1471-2202-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuriyama R, Gustus C, Terada Y, Uetake Y, Matuliene J. CHO1, a mammalian kinesin-like protein, interacts with F-actin and is involved in the terminal phase of cytokinesis. The Journal of cell biology. 2002;156:783–790. doi: 10.1083/jcb.200109090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirose K, Kawashima T, Iwamoto I, Nosaka T, Kitamura T. MgcRacGAP is involved in cytokinesis through associating with mitotic spindle and midbody. The Journal of biological chemistry. 2001;276:5821–5828. doi: 10.1074/jbc.M007252200. [DOI] [PubMed] [Google Scholar]
- 23.Guan CB, Xu HT, Jin M, Yuan XB, Poo MM. Long-range Ca2+ signaling from growth cone to soma mediates reversal of neuronal migration induced by slit-2. Cell. 2007;129:385–395. doi: 10.1016/j.cell.2007.01.051. [DOI] [PubMed] [Google Scholar]
- 24.Lin S, Liu M, Mozgova OI, Yu W, Baas PW. Mitotic motors coregulate microtubule patterns in axons and dendrites. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32:14033–14049. doi: 10.1523/JNEUROSCI.3070-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharp DJ, Yu W, Ferhat L, Kuriyama R, Rueger DC, Baas PW. Identification of a microtubule-associated motor protein essential for dendritic differentiation. J Cell Biol. 1997;138:833–843. doi: 10.1083/jcb.138.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nislow C, Lombillo VA, Kuriyama R, McIntosh JR. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature. 1992;359:543–547. doi: 10.1038/359543a0. [DOI] [PubMed] [Google Scholar]
- 27.Ferhat L, Kuriyama R, Lyons GE, Micales B, Baas PW. Expression of the mitotic motor protein CHO1/MKLP1 in postmitotic neurons. The European journal of neuroscience. 1998;10:1383–1393. doi: 10.1046/j.1460-9568.1998.00159.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.