Abstract
Genomic findings underscore the heterogeneity of head and neck squamous cell carcinoma (HNSCC)(1, 2). Identification of mutations that predict therapeutic response would be a major advance. We determined the mutationally altered, targetable mitogenic pathways in a large HNSCC cohort. Analysis of whole-exome sequencing data from 151 tumors revealed the PI3K pathway to be the most frequently mutated oncogenic pathway (30.5%). PI3K pathway-mutated HNSCC tumors harbored a significantly higher rate of mutations in known cancer genes. In a subset of HPV-positive tumors, PIK3CA or PIK3R1 was the only mutated cancer gene. Strikingly, all tumors with concurrent mutation of multiple PI3K pathway genes were advanced (stage IV), implicating concerted PI3K pathway aberrations in HNSCC progression. Patient-derived tumorgrafts with canonical and non-canonical PIK3CA mutations were sensitive to an m-TOR/PI3K inhibitor (BEZ-235) in contrast to PIK3CA wildtype tumorgrafts. These results suggest that PI3K pathway mutations may serve as predictive biomarkers for treatment selection.
Keywords: PI3K, mutation, BEZ-235, head and neck cancer
Introduction
Head and neck squamous cell carcinoma (HNSCC) is a frequently lethal cancer with few effective therapeutic options. Recent genomic findings in head and neck cancer revealed a wide spectrum of unexpected genetic aberrations(1, 2). This genomic heterogeneity of HNSCC tumors underscores an obstacle to the identification of effective molecular targeting agents likely to benefit the majority of HNSCC patients. To date, there is a translational gap between genomics and treatment selection for HNSCC patients. TP53 mutation is the single most common mutational event. Yet the loss of function of this tumor suppressor gene has remained challenging to exploit therapeutically. Mitogenic pathways are crucial for cancer development and progression. In other malignancies, mutations of growth pathway genes have been shown to result in pathway activation, enhanced tumor growth, and increased sensitivity to agents targeting the mutated pathway. However, the potential of genomics-based therapy selection has not been widely investigated in HNSCC.
We and others recently reported genomic mutational profiles of over 100 HNSCC tumors(1, 2). Here, we analyzed an additional 45 HNSCC tumors by whole exome sequencing using the Illumina platform. In an effort to identify mutationally altered, targetable mitogenic pathways in HNSCC, we combined all currently available mutational data (from whole-exome sequencing) of 151 HNSCC primary tumors and evaluated the mutational events of genes in three major mitogenic pathways that have been previously implicated in HNSCC pathophysiology, namely the MAPK(3), JAK/STAT(4), and the PI3K pathways(3). These key mitogenic pathways are targetable in human cancers with a variety of agents currently in various stages of clinical development.
Results
Nearly one third of HNSCC tumors harbor PI3K pathway mutations
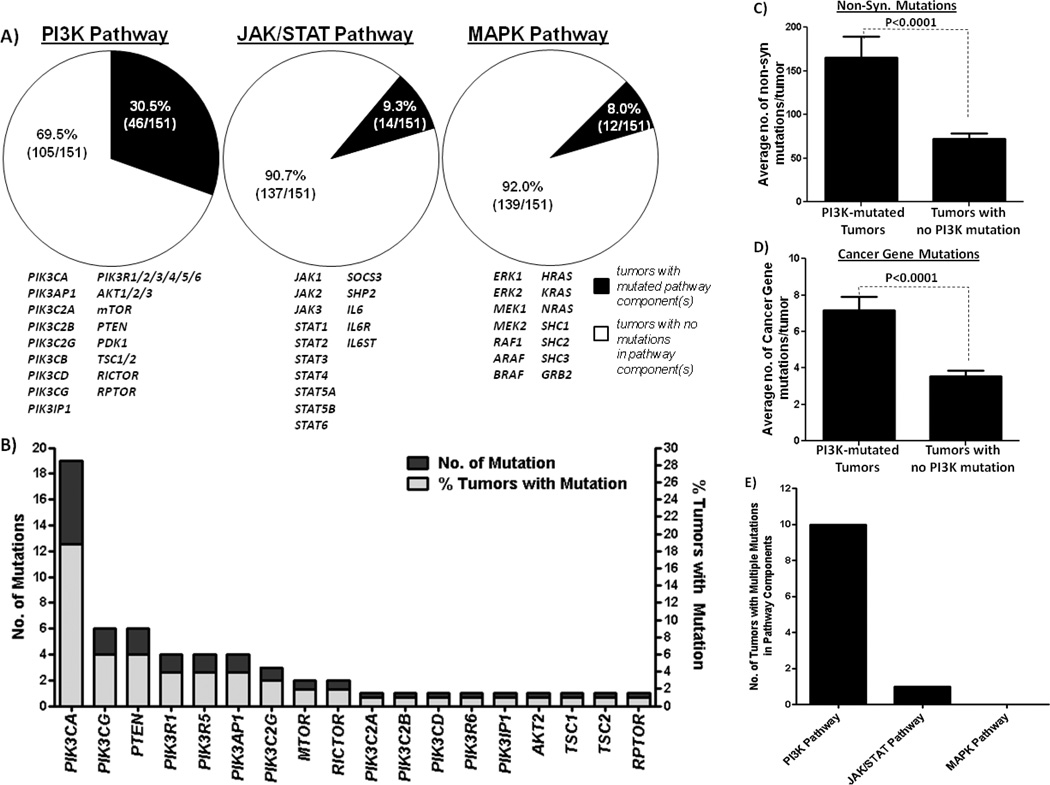
To date, whole-exome sequencing data of 106 HNSCC tumors are available. Here, we reported the whole-exome sequencing data of additional 45 HNSCC tumors collected at the University of Pittsburgh (Supplementary Table 1). Our results, similar to previous reports, showed a high degree of inter-tumor mutation heterogeneity, further confirming the complexity of HNSCC biology and the associated challenges in defining subsets of patients likely to respond to specific targeted therapies. With the aim of identifying mutationally altered, targetable mitogenic pathways in HNSCC, we assessed the mutational events of genes comprising three major mitogenic and targetable pathways in HNSCC; the JAK/STAT, MAPK and PI3K pathways. Pathway component genes were defined as follows: JAK/STAT pathway (JAK1, JAK2, JAK3, STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, STAT6, SOCS3, SHP2, IL6ST, IL6R and IL6), MAPK pathway (Erk1, Erk2, MEK1, MEK2, RAF1, ARAF, BRAF, HRAS, KRAS, NRAS, SHC1, SHC2, SHC3, and GRB2) and PI3K pathway (PIK3CA, PIK3AP1, PIK3C2A, PIK3C2B, PIK3C2G, PIK3CB, PIK3CD, PIK3CG, PIK3IP1, PIK3R1/2/3/4/5/6, AKT1/2/3, mTOR, PTEN, PDK1, TSC1/2, RICTOR and RPTOR). Strikingly, almost one third of all HNSCC tumors analyzed in our cohort (30.5%; 46/151 tumors) harbored PI3K-pathway mutations, while only 9.3% (14/151) and 8.0% (12/151) of tumors contained mutations in the JAK/STAT or the MAPK pathways, respectively (Figure 1A, Supplementary Tables 2A–E). In contrast to other smoking-related aerodigestive tract cancers where KRAS is frequently mutated(5), the HNSCC-MAPK mutational profile is characterized primarily by HRAS mutations, which accounted for 7 of the 12 pathway mutations identified. PIK3CA is the most commonly mutated gene in the HNSCC-PI3K mutational profile (Figure 1B). These results demonstrate that despite the genomic heterogeneity of HNSCC tumors, the PI3K pathway is the most frequently somatically mutated mitogenic pathway in HNSCC tumors, found in 30.5% of cases, providing a potential approach to treat a substantial subset of patients.
Figure 1.
Mutations in oncogenic signaling pathways in HNSCC. (A) Mutation rates of the major mitogenic pathways (the PI3K pathway, the MAPK pathway and the JAK/STAT pathway) in 151 HNSCC patient tumors determined by whole exome sequencing. Components of each pathway examined are displayed underneath each pie chart. (B) Bar graph detailing the number of mutations (dark bars) of each particular component of the PI3K pathway as well as the number of HNSCC tumors harboring these mutations (grey bars). (C) PI3K pathway-mutated HNSCC tumors have higher of non-synonymous mutations and (D) cancer gene mutations when compared to HNSCC tumors without any PI3K pathway mutations. Bar graph representing the average number of non-synonymous mutations per tumor (C) and the average number of cancer gene mutations (D) in 151 HNSCC tumors. Statistical significance was calculated by Fisher’s Exact test, P<0.0001 (N=151). (E) Graphical representation of the number of HNSCC tumors with mutation of multiple components of the PI3K, MAPK and the JAK/STAT pathways, respectively.
A detailed analysis of the PI3K pathway mutational events showed that 19 of the 151 tumors (12.6%) harbor a PIK3CA mutation (Figure 1B). This mutation rate is similar to that detected in prior reports of HNSCC tumors (7.4% and 10.8% rate(6, 7)). Further, we found PIK3CG and PTEN mutations in 4.0% (6/151) of HNSCC tumors, while PIK3R1 (also known as p85), PIK3R5 and PIK3AP1 were mutated in 2.7% tumors (4/151). Other components of the PI3K pathway were mutated in <2% cases (Figure 1B). Major downstream effectors of the PI3K pathway, including PDK1, AKT1 were not mutated, while AKT2 and mTOR were only mutated in 1.3% (2 mutations) of HNSCC tumors. Although PIK3CA gene amplification data is not available for the previously sequenced tumors, in the newly added cohort, PIK3CA was amplified in 24.4% (11/45) of the cases.
Previous reports noted loss of heterozygosity (LOH) of the tumor suppressor PTEN in HNSCC, by PCR-based microsatellite analysis primarily using D10S215 and/ D10S541 probes in relatively small HNSCC cohorts (e.g. 17 and 39 tumors, respectively)(8, 9). Although comprehensive analysis of PTEN copy number was not available in the published genomic HNSCC studies(1, 2), PTEN gene copy number change was analyzed using a highly sensitive Affymetrix Genome-Wide Human SNP Array 6.0 platform in our 45 newly sequenced HNSCC tumors. Our results showed that PTEN gene copy loss (≥1 copy loss) was only found in 8.16% of cases (4/45), indicating that PTEN loss is not likely to be the primary mediator of PI3K pathway alteration in HNSCC (unlike other cancers such as glioblastoma, where PTEN loss can be as high as 20–60%)(10, 11). However, all 4 tumors with PTEN gene copy loss expressed relatively low levels of PTEN protein when compared to HNSCC tumors without PTEN gene copy alteration (P<0.001, Supplementary Figure 1).
PI3K pathway-mutated HNSCC tumors demonstrate an increased rate of cancer gene mutations
To determine if HNSCC tumors harboring mutations in PI3K pathway genes contained a higher number of mutations in other cancer-associated genes, we compared the mutation rates of PI3K pathway-mutated tumors to non-PI3K pathway-mutated tumors. We found that tumors harboring PI3K pathway mutations have higher rates of mutation than non-PI3K-mutated HNSCC tumors. As shown in Figure 1C, PI3K pathway-mutated HNSCC tumors harbored 2.3 times more non-synonymous mutations (165.5±24.1 vs 72.1±6.6 mutations, P<0.0001) than tumors without PI3K mutations, indicating increased genomic instability in tumors harboring PI3K pathway mutations. Further, cancer gene filtering analysis showed that these PI3K pathway-mutated HNSCC tumors harbored twice as many cancer gene mutations than those without PI3K pathway mutations (Figure 1D, 7.2±0.8 vs 3.6±0.3, P<0.0001: defined by the Cancer Gene Census, COSMIC Database)(12). These results suggest that PI3K pathway mutations in HNSCC may facilitate the expansion or selection of tumor cells that are already genetically unstable and thus harbor more genomic aberrations including aberrations in known cancer genes. This contention is supported by further analysis demonstrating that DNA damage/repair genes (based on the DNA damage gene list in the cBio portal database(13), which includes ATM, ATR, CHEK1/2, BRCA1/2, FANCF, MLH1, MSH2, MDC1, PARP1 and RAD51) were found to be mutated at a significantly higher frequency in the PI3K-mutated tumors (average mutation rate of 37.0%; 17 mutations in 46 tumors) compared to tumors without PI3K pathway mutations (average mutation rate of 15.2%; 16 mutations in 105 tumors) (P=0.033).
The association between PI3K pathway mutations and genomic instability is observed in HNSCC derived from all anatomic sites in our cohort (e.g. oral cavity, pharynx and larynx, data not shown). Mutation rates in laryngeal tumors (186.3±27.1, n=32, data not shown) are significantly higher than the rates of mutation in tumors from the other anatomical locations (78.6±8.3, n=116, P=0.0005, data not shown). Additionally, the prevalence of PI3K pathway mutations is higher in laryngeal tumors (53.1%±9.0%, n=32, data not shown) compared with tumors from the other anatomical locations (25.0%±4.0%, n=116, P=0.0005, data not shown).
Co-mutation analysis showed that tumors with PI3K pathway mutation(s) are also associated with mutations of several known tumor suppressor genes including ARID1A, MLL and MLL3 (P<0.05; Supplementary Table 3), which contribute to chromatin remodeling and transcriptional regulation in cancers(14–17). Intriguingly, ARID1A has been shown to influence signaling through the PI3K pathway, suggesting that ARID1A may regulate the PI3K pathway and expand the number of tumors susceptible to targeting the PI3K pathway(18). Of note, PI3K pathway-mutated tumors are not associated with TP53 (P=1.0) or NOTCH1 mutations (P=0.34, data not shown).
Interestingly, we also identified 3 tumors where PIK3CA or PIK3R1 was the only known mutated cancer gene [HN_00361, HN_63027 and HN41PT with the respective PI3K-mutations of PIK3R1 (453_454insN), PIK3CA(E542K) and PIK3CA(H1047L)]. Strikingly, all three tumors were associated with infection by the human papillomavirus (HPV). Although the number of HPV-positive HNSCC tumors in this cohort is relatively small (15 cases, see Supplementary Table 4), 5 of these tumors harbored PI3K pathway mutations (33.3) suggesting that a subset of HPV-positive HNSCC tumors (20%; 3/15 cases) may be driven by PI3K-pathway mutation(s) alone, without an associated increase in underlying genomic instability.
Only advanced stage HNSCC tumors harbor multiple PI3K-pathway mutations
In HNSCC cancers containing PI3K pathway mutations, 21.7% (10/46) harbored mutations in more than one PI3K pathway member genes, indicating that genetic alterations at multiple levels in the PI3K pathway are relatively common in HNSCC (Table 1). In contrast, HNSCC tumors rarely, if ever, harbored multiple mutations in the MAPK pathway (0 tumors), or the JAK/STAT pathway (only one contained both JAK3 and STAT1 mutations; HN_63080) (Figure 1C). Strikingly, all of these HNSCC tumors (100%; 10/10 cases) with concurrent PI3K mutational events were advanced (Stage IV) (Table 1). None of these tumors was associated with HPV infection. These findings suggest that concerted PI3K pathway aberrations may contribute to HNSCC progression. This finding appears to be unique to HNSCC. Examination of recently published tumor datasets including breast, colon and lung SCC showed that only 1/25 breast tumors, 1/27 colon carcinoma, and 0/31 lung SCC tumors harbored multiple PI3K pathway mutations were stage IV (data not shown; cBio portal(13)). While all 10 tumors with multiple PI3K pathway mutations were advanced (Stage IV), there is no significant association between advanced disease and individual PI3K pathway mutations (data not shown). Additionally, mutation rates do not vary significantly between Stage IV and earlier Stage (I-III) disease (data not shown). In the absence of models assessing the specific contribution of each mutation to cell growth or survival, it is not possible to determine the precise effect of individual mutations in tumors that harbor more than one mutation of genes in the PI3K pathway.
Table 1. HNSCC Tumors with Multiple Mutations in a Single Mitogenic Pathway.
Describes all tumors in our cohort harboring mutations in more than one gene in a defined mitogenic pathway by mutation type and pathological stage.
| Pathway | Tumor | Annotated Genes |
Amino Acid Change |
TNM Stage/Overall Staging |
|---|---|---|---|---|
| PI3K | HN_00190 | PIK3C2G | p.V656L | T1N2bM0/ Stage 4 |
| Pathway | PTEN | p.D92E | ||
| HN_62421 | PIK3R1 | p.D560H | T4N0M0/ Stage 4 | |
| MTOR | p.L2260H | |||
| HN_62469 | PIK3CA | p.H1047R | T2N2bM0/ Stage 4 | |
| MTOR | p.R1161Q | |||
| HN_63039 | PIK3CA | p.H1047L | T4N2bM0/ Stage 4 | |
| PTEN | p.R335* | |||
| HN_22PT | PIK3CG | p.G491E | T4aN2bM0/ Stage 4 | |
| PIK3AP1 | p.G313R | |||
| HNPTS_1 | PTEN | p.R14S | T4N0M0/ Stage 4 | |
| PIK3IP1 | p.A144S | |||
| PIK3CA | p.E545K | |||
| HNPTS_29 | PIK3C2G | p.S1272L | T4N2bM0/ Stage 4 | |
| PIK3R5 | p.E322K | |||
| PIK3CA | p.E542K | |||
| HNPTS_38 | PIK3R5 | p.E60* | T4N2M0/ Stage 4 | |
| PIK3CA | p.H1047R | |||
| HNPTS_42 | TSC2 | p.S1514* | T4N2BM0/ Stage 4 | |
| PIK3CA | p.E545K | |||
| PIK3CG | p.A156V | |||
| HNPTS_45 | AKT2 | p.Y351C | T4N1M0/ Stage 4 | |
| PIK3CA | p.H1047R | |||
| JAK/STAT | HN_63080 | JAK3 | p.R948C | T4aN2bM0/ Stage 4 |
| Pathway | STAT1 | p.Q330K | ||
| MAPK | NONE | |||
| Pathway | ||||
PIK3CA canonical and novel mutations increase survival and pathway activation in HNSCC tumors
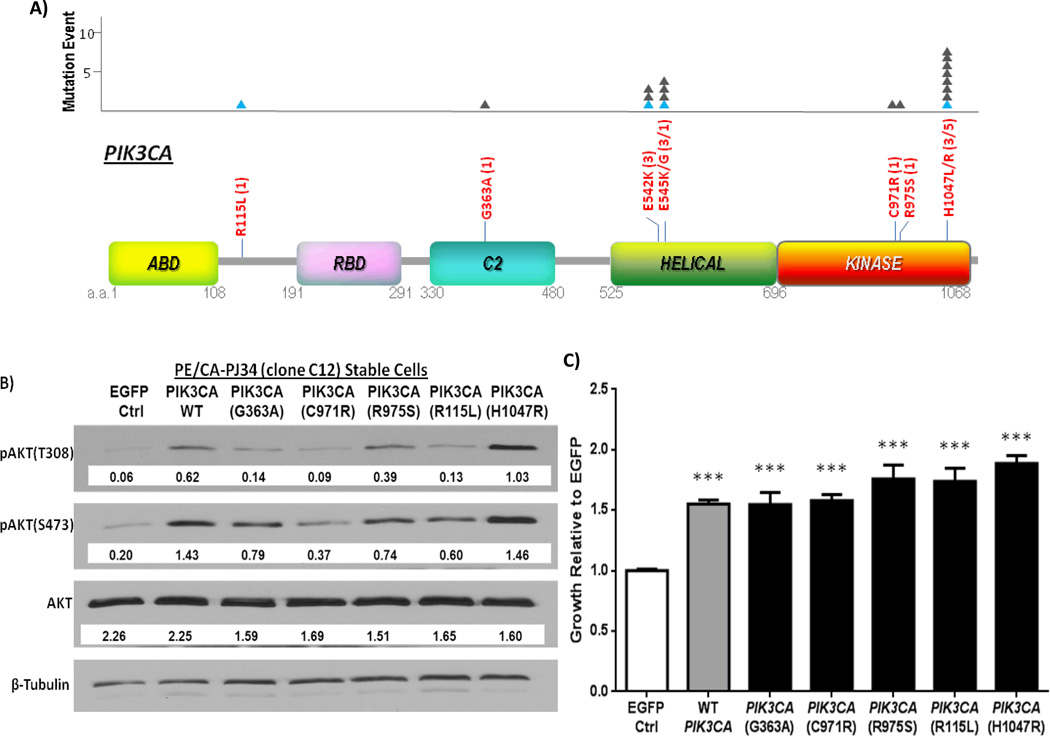
PIK3CA is a critical gene in the PI3K signaling pathway. In HNSCC tumors, the most common sites of PIK3CA mutations included H1047R/L (8 mutations total), E545K/G (4 mutations) and E542K (3 mutations) (Figure 2A). All of which represent previously reported canonical (“hotspot”) mutation sites. This HNSCC-PIK3CA mutation pattern (~90% of mutations found in the helical/kinase domains), is similar to that observed in cervical, breast and lung SCC cancers, but is distinct from other tumors such as endometrial cancer, lung adenocarcinoma, glioblastoma multiforme, and prostate carcinoma (Supplementary Table 5). In addition, we detected 4 previously unreported, novel PIK3CA mutations (R115L, G363A, C971R, R975S). To determine the functional consequences of these mutations we stably expressed, by retroviral infection, each of the novel mutations, and a hotspot mutation (H1047R), in a representative HNSCC cell line that is WT for all PI3K pathway components. Overexpression of WT PIK3CA (mimicking PIK3CA gene amplification), and expression of all the engineered PIK3CA mutants individually, resulted in enhanced growth compared to infection with EGFP control. Furthermore, the canonical hotspot mutation showed significantly enhanced growth compared to overexpression of WT PIK3CA (p = 0.0001). The novel mutations were found to confer moderate growth advantage compared to simulated WT amplification (R115L; p = 0.1174, G363A; p = 0.9637, C971R; p = 0.6503, R975S; p = 0.0958). Immunoblotting of cell lysates revealed that enhanced HNSCC growth conferred by introduction of the novel mutations was associated with increased PI3K pathway activation as reflected by elevated expression of phosphorylated AKT (Figures 2B & C). In the absence of complete functional characterization of these novel mutations, these findings should be considered supportive but not definitive evidence of oncogenic function.
Figure 2.
PIK3CA mutations in HNSCC tumors. A) Schematic of all PIK3CA mutations found in 151 HNSCC tumors by whole exome sequencing. The amino acid (a.a.) positions of each domain is shown in grey below each domain. The number of mutational events at each site is indicated by a filled triangle (▲) in the graph above. Blue triangles indicate mutations found in HPV/HNSCC tumors. Keys: ABD: p85 binding domain; RBD: Ras binding domain; C2 Superfamily; Helical: PIK domain; Kinase: Kinase domain of PIK3CA. (B) Effects of PIK3CA mutations on PI3K signaling in HNSCC cells. WT PIK3CA, hotspot mutant H1047R, and novel mutants: R115L, G363A, C971R, and R975S were stably expressed in an HNSCC cell line harboring no endogenous mutations in the PI3K pathway, PE/CA-PJ34 (clone C12) cells, by retroviral infection. Shown here is a representative Western blot with densitometry values normalized to beta-tubulin loading controls for each engineered cell line. Increased phosphorylation of AKT at the T308 and/or S473 residue was generally observed in HNSCC cells stably expressing WT or mutant PIK3CA constructs relative to the EGFP expressing HNSCC cells, indicating enhanced activation of the PI3K signaling pathway. (C) Effects of PIK3CA mutations on HNSCC cell growth. HNSCC cells stably expressing WT or mutant PIK3CA constructs demonstrated enhanced growth at 72 hours in media with 2% FBS by MTT assay compared to cells expressing EGFP vector control (p<0.0001***). PIK3CA(H1047R) expressing cells further demonstrated enhanced growth when compared to simulated WT PIK3CA amplification (p = 0.001). Data shown here represents growth studies from three sets of independent replicate cell lines (separate infections, n=18 for each group).
HNSCC patient tumorgrafts with PIK3CA mutations are exquisitely sensitive to BEZ-235
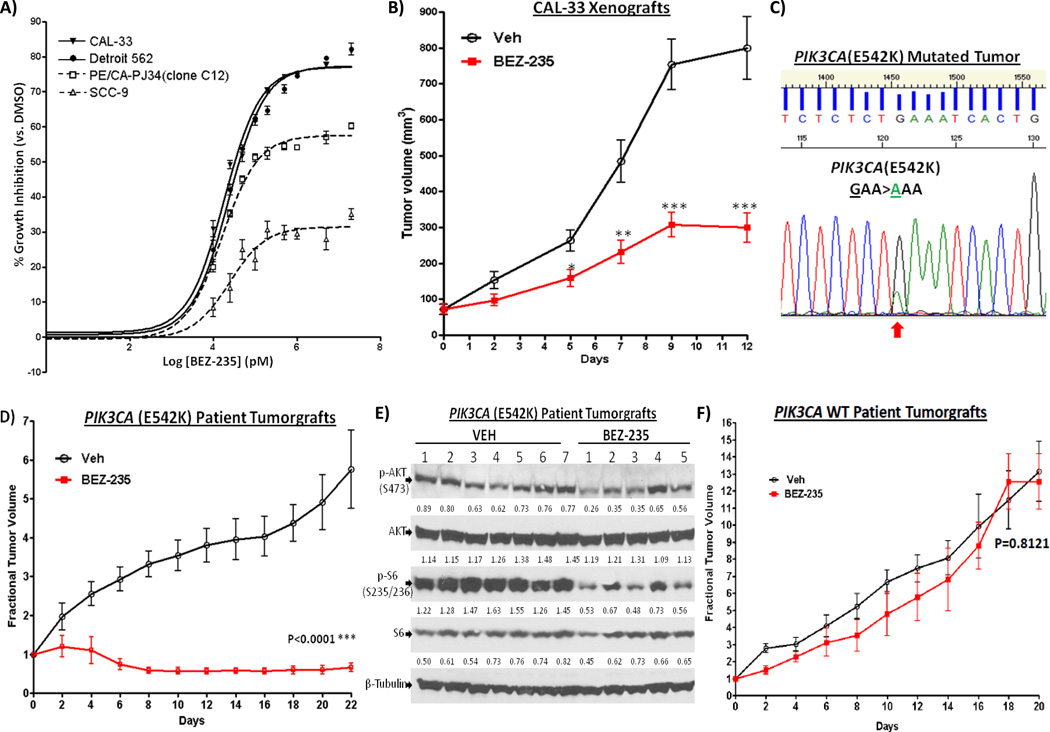
Reports in other cancers suggest that tumors with PI3K pathway activation may be more sensitive to PI3K pathway inhibitors (19). To determine the predictive value of PIK3CA mutational status in HNSCC, we examined the sensitivity of HNSCC cell lines that did and did not harbor intrinsic activating driver PIK3CA(H1047R) hotspot mutations to PI3K pathway inhibitors. As shown in Figure 3A, HNSCC cell lines containing endogenous PIK3CA(H1047R) mutations (CAL-33 and Detroit 562)(20) demonstrated increased sensitivity to PI3K pathway inhibition by the mTOR/PI3K inhibitor BEZ-235 compared to representative HNSCC cells with WT PIK3CA [SCC-9 and PE/CA-PJ34(clone C12)]. Next, mice bearing CAL-33 xenografts were found to be sensitive to BEZ-235 treatment in vivo when compared to vehicle control (Figure 3B). Due to the lack of HPV-HNSCC cell line models that contain PIK3CA mutations, we developed an HPV-positive PIK3CA-mutated HNSCC patient tumorgraft model (E542K) (Figure 3C) to determine the sensitivity of HPV-positive PIK3CA-mutated HNSCC tumors to PI3K pathway targeting. As shown in Figure 3D, BEZ-235 treatment (at 25mg/kg/day by oral gavage) significantly inhibited the growth of HPV-positive PIK3CA-mutated patient tumorgraft in vivo (P<0.0001). Inhibition of tumor growth was accompanied by decreased PI3K signaling as evident by down regulation of p-AKT(S473) (P=0.0124), and p-S6(S235/236) (P<0.0001) in the BEZ-235-treated tumors (Figure 3E). Another HNSCC patient-derived tumorgraft model (HPV-negative) harboring a PIK3CA mutation (E110K) was also found to be sensitive to BEZ-235 treatment (Supplementary Figure 2). In contrast, patient tumorgrafts with WT PIK3CA and low baseline p-AKT levels were not sensitive to the growth inhibitory effects of BEZ-235 (Figure 3F and Supplementary Figure 3). These results indicate that activating mutations of PI3K pathway have the potential to serve as biomarkers for treatment selection in HNSCC. Xenografts developed from a HNSCC cell line harboring a PIK3CA mutation (H1047R) were more sensitive to the combination of BEZ-235 plus cetuximab (the only FDA-approved molecular targeting agent in HNSCC) compared with cetuximab alone (Supplemental Figure 4), suggesting that targeting PI3K in the setting of PIK3CA mutant tumors can enhance treatment responses to cetuximab.
Figure 3.
PIK3CA mutation enhances sensitivity to PI3K pathway inhibition. (A) HNSCC cells containing endogenous PIK3CA mutation (H1047R) (CAL-33; Detroit 562) and cells containing WT PIK3CA [SCC-9, PE/CA-PJ34 (clone C12)] were treated with a PI3K/mTOR inhibitor, BEZ-235, followed by growth determinations at 48 hrs (n=4). Experiments were repeated 3 times with similar results. (B) BEZ-235 inhibited growth of CAL-33 xenografts [with endogenous PIK3CA(H1047R) mutation]. CAL-33 cells (0.5 ×106 cells) were inoculated into the flanks of nude mice. Treatment was started when the tumors became palpable 8 days after tumor cell inoculation. BEZ-235 (25mg/kg/day, n=9) or vehicle (n=10) was given by oral gavage. (C) Sanger sequencing results showing PIK3CA(E542K) mutation in the HPV-HNSCC patient tumor that were implanted the flanks of NOD SCIDγ mice. (D) PIK3CA-mutated tumorgrafts are sensitive to BEZ-235 treatment. HNSCC patient tumorgrafts were implanted into the flanks of NOD SCIDγ mice and treatment was started when tumors became palpable. BEZ-235 (25mg/kg) or vehicle control was given daily by oral gavage. Mice were given vehicle (n=7) or BEZ-235 (n=5) when tumors became palpable. Treatment with BEZ-235 significantly reduced the tumor size when compared to vehicle control (P<0.0001). (E) Western blots showing the effects of BEZ-235 (vs. vehicle control) on expression of PI3K signaling components in the PIK3CA-mutated tumorgrafts. Tumors were harvested for Western blotting at the end of the experiment on day 22. Densitometry values of band intensity are shown below each band (normalized to total beta-tubulin level). Phospho-AKT (S473) and phospho-S6 (S235/236) levels were significantly reduced upon BEZ-235 treatment when compared to the vehicle-treated tumors (P=0.0124 and P<0.0001, respectively). (F) HNSCC patient tumorgrafts from a WT PIK3CA tumor expressing low levels of pAKT were implanted into the flanks of NOD SCIDγ mice and treatment was started when tumors became palpable. BEZ-235 (25mg/kg) (n=6) or vehicle control (n=6) was given daily by oral gavage. Treatment with BEZ-235 failed to significantly reduce the tumor size when compared to vehicle control (P=0.300).
Discussion
The increase in targeted agents for cancer treatment results in an unprecedented opportunity for personalized cancer medicine. Selection of therapies based on mutation status of molecular targets has transformed clinical management and survival of several human malignancies. The EGFR monoclonal antibody cetuximab is the only targeted therapy that is FDA-approved to date for HNSCC treatment, yet there are no biomarkers that can be assessed in the primary tumor to predict clinical responses to this agent. The recent elucidation of HNSCC genomics offers an opportunity to identify genetic subgroups of HNSCC tumors to guide treatment decisions.
In this report, we employed a bioinformatic approach to identifying mutationally altered, targetable mitogenic pathways in HNSCC. Analyses of all currently available HNSCC whole-exome sequencing data (a total of 151 primary HNSCC tumors) revealed several key findings with important implications for HNSCC pathobiology and treatment. The PI3K pathway is the most frequently mutated oncogenic pathway in HNSCC, with the relative number of PI3K-mutated tumors compared to RAS/MAPK and JAK/STAT-mutated tumors being approximately 3-fold greater. Similar ratios of PI3K pathway mutations (relative to RAS/MAPK or JAK/STAT) are seen in squamous cell carcinoma of the lung and in cervical cancer; both of which share common risk factors with HNSCC, including tobacco and HPV infection, respectively. In contrast, the RAS/MAPK pathway is more frequently mutated than the PI3K pathway in colon and thyroid cancers, and both the PI3K and RAS/MAPK pathways are mutated at comparable rates in lung adenocarcinomas(13). The percentage of HNSCC tumors harboring multiple mutations in the PI3K pathway is similar to that observed in breast cancers (4.9%, 25/507 tumors) and glioblastomas (9.1%, 25/276 tumors) , higher than in thyroid cancer (0.3%, 1/323 tumors) and much lower than most other cancers, including uterine carcinoma (65.7%, 163/248 tumors), melanoma (24.9%, 63/253 tumors), and interestingly lung squamous cell carcinoma (17.4%, 31/178 tumors), which otherwise shares common risk factors and similar relative rates of pathway mutations with HNSCC.(13
Using novel patient-derived tumorgraft models with an oncogenic PIK3CA(E542K) mutation we demonstrated that these tumors are exquisitely sensitivity to a PI3K pathway inhibitor (BEZ-235). Similar results were demonstrated in another HNSCC patient-derived tumorgraft model with a PIK3CA (E110K) mutation, previously reported in breast cancer (21). In contrast, treatment of human-derived heterotopic tumorgrafts with wildtype PIK3CA and low basal expression levels of phospho-AKT, with a PI3K pathway inhibitor, was ineffective. These findings suggest that: 1) PI3K-pathway inhibitors can be effective for treating HNSCC tumors with PI3K mutations; and 2) mutation-guided treatment responses can be evaluated/monitored using patient-derived HNSCC tumorgraft models in vivo. In fact, early-phase clinical trial results showed that patients with solid tumors harboring a PIK3CA hotspot mutation (H1047R) were found to be responsive to PI3K pathway inhibitors(22). However, the effects of other PIK3CA mutations on mediating drug sensitivity in HNSCC preclinical models or clinical trials has not been previously reported. Findings from our study implicate that PIK3CA(E542K) mutation, as well as other non-hotspot mutations (such as E110K) may also identify an HNSCC subgroup potentially responsive to PI3K-pathway inhibitors. In particular, our results using HNSCC patient-derived tumorgrafts suggest that HNSCC tumors with activating PIK3CA mutations may be more sensitive to a dual PI3K/mTOR inhibitor (such as BEZ-235) compared to tumors with wildtype PIK3CA (Figures 3E and Supplementary Figure 3), as indicated by significant inhibition of p-S6 expression in the PIK3CA mutated, but not in the wildtype tumorgrafts. In fact, a recent report of five HNSCC cases found that mTOR-based targeted therapy may be more effective in HNSCC tumors harboring PIK3CA mutation and/or PTEN loss(23).
PI3K pathway-mutated HNSCC tumors were found to have a higher rate of non-synonymous mutations, including an increased number of defined cancer gene mutations, compared to tumors without PI3K pathway mutations. This observation implies that the PI3K pathway-mutated HNSCC tumors have “oncogenic” advantage even with genomic instability, and/or that PI3K-mutated HNSCC tumors intrinsically harbor a “mutator” phenotype rendering them more prone to mutation. The oncogenic advantage of PI3K pathway-mutated tumors can be partly explained by PIK3CA “driver” mutations’ growth-promoting activity(24) (Figure 2C), while the “mutator” phenotype of these tumors is supported by the our finding that PI3K pathway-mutated tumors are associated with ARID1A and MLL3 mutations, which are important tumor suppressor genes(15, 16, 25). It is possible that both the “oncogenic/growth” advantage and “mutator” phenotype associated with PI3K pathway-mutated HNSCC tumors are necessary for HNSCC progression; especially since PI3K pathway mutations in these tumors are not associated with TP53 mutation, a previously recognized tumor suppressor alteration that contributes to HNSCC carcinogenesis. Although the relationship of PI3K pathway mutations and TP53 mutation has not been carefully examined in most cancers, a recent study in bladder cancer showed that PIK3CA mutations were significantly more common in TP53 WT tumors(26). Hence, PI3K pathway mutations may mediate tumor progression in the absence of TP53 genetic alteration.
Our finding that all 10 HNSCC tumors with concurrent mutations of multiple PI3K-pathway genes were advanced stage cancers (Stage IV) suggests the potential involvement of concurrent alterations of multiple nodes of the PI3K pathway in HNSCC progression. This agrees with the recent report that in addition to PIK3CA mutation, other pathway components such as PIK3R1 and PIK3R2, when mutated, can also serve to drive cell growth/survival(27). Although the effects of multiple PI3K pathway mutations on cancer cell growth or progression has not been previously investigated, our results support the possibility that genetic alterations at multiple nodes in this oncogenic pathway, a common feature of many solid tumors, may identify a subgroup of cancer patients most likely to respond to PI3K pathway inhibitors. These cumulative findings identify the PI3K pathway as the most frequently mutated mitogenic pathway in HNSCC tumors. Prospective identification of patients whose tumors harbor these mutations is likely to identify a subgroup of individuals who may benefit from treatment with PI3K pathway inhibitors.
Materials and Methods
Additional methods are detailed as Supplementary Information.
Cell Cultures
The HNSCC cell lines, Detroit 562, SCC-9 cells were obtained from ATCC, Manassas, VA, while PE/CA-PJ34(clone C12) cells were obtained Sigma-Aldrich, St. Louis, MO. CAL-33 was a kind gift from Dr. Gerard Milano (University of Nice). All cell lines were genotypically verified. HNSCC cell lines were cultured in the respective culture medium containing 10% fetal calf serum, 1X penicillin/streptomycin solution (Invitrogen, Carlsbad, USA): CAL-33 and Detroit 562 in DMEM, SCC-9 in DMEM/F12 with 0.4 µg/ml hydrocortisone, and PE/CA-PJ34(clone C12) cells in IDMEM with 2 mM glutamine (Mediatech, Inc, Manassas, VA). All cell lines were maintained in a humidified cell incubator at 37°C, 5% CO2
Cancer Gene Census Comparison and Co-mutation Analysis
A mutation comparison program was written in Visual Basic for Microsoft Excel to compare the existence of HNSCC mutations vs. a reference list of mutations of interest (in this case, cancer genes). The program allows side-by-side comparison between multiple groups (2 or more) to find out common mutational events, as well as the number of common events in multiple groups. A cancer gene list was generated in each subgroup of tumors by comparing the Cancer Gene Census list (COSMIC Database) with non-synoynomous mutation gene list of each tumor subgroup (the PI3K-mutated tumors, tumors without PI3K-mutation, PIK3CA-mutated tumors, PIK3CA WT tumors) using this comparison program. This analysis allows us to find out the number of cancer genes mutated in each subgroup.
Mutation Validation by Sanger Sequencing
Sanger sequencing was performed on patient tumors that were grafted for tumorgraft studies. About 25–50mg of tumor tissues (pathologically confirmed HNSCC with >70% tumor cell contents) were used for extraction of DNA by QIAamp DNA Mini Kit (Qiagen, Inc, Valencia, CA). Sequencing primers for HNSCC-associated PIK3CA hotspot mutations were synthesized (Sigma-Aldrich, St. Louis, MO) and used for Sanger sequencing. The primer sequences for E542 site mutation are: 5'-cacgagatcctctctctaaaatcactgagcaggag-3' (forward) and 5'-ctcctgctcagtgattttagagagaggatctcgtg-3' (reverse). Sanger sequencing was performed at the Genomics and Proteomics Core Laboratories at the University of Pittsburgh.
HNSCC Tumorgraft Model and Drug Treatment
BEZ-235 was obtained as a kind gift from Novartis, USA. HPV-positive HNSCC patient tumorgrafts were derived under the auspices of an IRB-approved protocol, with PIK3CA WT or PIK3CA(E542K) mutation were implanted into the flanks of NOD SCIDγ mice and treatment was started when tumors became palpable. BEZ-235 (25mg/kg) or vehicle control was given daily by oral gavage. Tumor volumes were measured every two days.
Supplementary Material
Sentence Statement of Significance.
Treatment options for HNSCC are limited by an incomplete understanding of the targetable mutations that “drive” tumor growth. Here, we define a subgroup of HNSCC harboring activating mutations of genes in the PI3K pathway where targeting the pathway demonstrates antitumor efficacy. These results suggest that PI3K pathway mutation assessment may be used to guide HNSCC therapy.
Acknowledgements
Grant Support: The study is supported by P50 CA097190 and the American Cancer Society (to JRG), the Patricia L. Knebel Fund of the Pittsburgh Foundation (to VWYL), NCI K08 1K08CA163677 (to PSH), UPCI CCSG funding support P30CA047904ALL (to RM, NT); funding support for MLH (5T32DC000066-10 to JRG, Research Training in Otolaryngology, University of Pittsburgh).
Abbreviations
- HNSCC
Head and Neck squamous cell carcinoma
- PI3K
Phosphoinositide-3-kinase pathway
- PIK3CA
PI3-kinase p110 subunit alpha
- PIK3R1 or p85
PI3-kinase subunit p85-alpha
- PTEN
phosphatase and tensin-like protein
Footnotes
Disclosure of Potential Conflicts of Interest: JRG receives research support from Bristol-Myers Squibb, Novartis and Atellas (previously OSI Pharmaceuticals).
Author Contributions:
VWYL contributed to the original idea, experimental design and execution, and manuscript preparation
MLH, HL contributed to experimental design and execution, and manuscript preparation
YZ, MF, QZ, DX, YL contributed to experimental design and execution
MLH, BSV, KP, BG, SS, DD contributed to bioinformatics analysis
PKN designed computer programs for data-analysis
BD contributed to Sanger sequencing analysis
PH contributed to whole exome sequencing analysis
LW, SC, RS, TN, MR contributed to sample acquisition, preparation and collection of pathological data
JTJ, SK, UD, RLF contributed to HNSCC tumor collection
GB contributed to manuscript preparation
JRG contributed to experimental design and manuscript preparation
References
- 1.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. doi: 10.1126/science.1206923. Epub 2011/07/30. doi: 10.1126/science.1206923. PubMed PMID: 21798897; PubMed Central PMCID: PMC3162986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. Epub 2011/07/30. doi: 10.1126/science.1208130 science.1208130 [pii]. PubMed PMID: 21798893; PubMed Central PMCID: PMC3415217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bancroft CC, Chen Z, Yeh J, Sunwoo JB, Yeh NT, Jackson S, et al. Effects of pharmacologic antagonists of epidermal growth factor receptor, PI3K and MEK signal kinases on NF-kappaB and AP-1 activation and IL-8 and VEGF expression in human head and neck squamous cell carcinoma lines. Int J Cancer. 2002;99(4):538–548. doi: 10.1002/ijc.10398. PubMed PMID: 11992543. [DOI] [PubMed] [Google Scholar]
- 4.Grandis JR, Drenning SD, Chakraborty A, Zhou MY, Zeng Q, Pitt AS, et al. Requirement of Stat3 but not Stat1 activation for epidermal growth factor receptor-mediated cell growth in vitro. J Clin Invest. 1998;102(7):1385–1392. doi: 10.1172/JCI3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tam IY, Chung LP, Suen WS, Wang E, Wong MC, Ho KK, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12(5):1647–1653. doi: 10.1158/1078-0432.CCR-05-1981. Epub 2006/03/15. doi: 12/5/1647 [pii] 10.1158/1078-0432.CCR-05-1981. PubMed PMID: 16533793. [DOI] [PubMed] [Google Scholar]
- 6.Kozaki K, Imoto I, Pimkhaokham A, Hasegawa S, Tsuda H, Omura K, et al. PIK3CA mutation is an oncogenic aberration at advanced stages of oral squamous cell carcinoma. Cancer Sci. 2006;97(12):1351–1358. doi: 10.1111/j.1349-7006.2006.00343.x. Epub 2006/10/21. doi: CAS343 [pii] 10.1111/j.1349-7006.2006.00343.x PubMed PMID: 17052259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen Y, Goldenberg-Cohen N, Shalmon B, Shani T, Oren S, Amariglio N, et al. Mutational analysis of PTEN/PIK3CA/AKT pathway in oral squamous cell carcinoma. Oral Oncol. 2011;47(10):946–950. doi: 10.1016/j.oraloncology.2011.07.013. Epub 2011/08/10. doi: S1368-8375(11)00752-4 [pii] 10.1016/j.oraloncology.2011.07.013. PubMed PMID: 21824802. [DOI] [PubMed] [Google Scholar]
- 8.Okami K, Wu L, Riggins G, Cairns P, Goggins M, Evron E, et al. Analysis of PTEN/MMAC1 alterations in aerodigestive tract tumors. Cancer Res. 1998;58(3):509–511. Epub 1998/02/11. PubMed PMID: 9458098. [PubMed] [Google Scholar]
- 9.Shao X, Tandon R, Samara G, Kanki H, Yano H, Close LG, et al. Mutational analysis of the PTEN gene in head and neck squamous cell carcinoma. Int J Cancer. 1998;77(5):684–688. doi: 10.1002/(sici)1097-0215(19980831)77:5<684::aid-ijc4>3.0.co;2-r. Epub 1998/08/04. doi: 10.1002/(SICI)1097-0215(19980831)77:5<684::AID-IJC4>3.0.CO;2-R [pii]. PubMed PMID: 9688299. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275(5308):1943–1947. doi: 10.1126/science.275.5308.1943. Epub 1997/03/28. PubMed PMID: 9072974. [DOI] [PubMed] [Google Scholar]
- 11.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15(4):356–362. doi: 10.1038/ng0497-356. Epub 1997/04/01. doi: 10.1038/ng0497-356. PubMed PMID: 9090379. [DOI] [PubMed] [Google Scholar]
- 12.Rozenblatt-Rosen O, Deo RC, Padi M, Adelmant G, Calderwood MA, Rolland T, et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature. 2012;487(7408):491–495. doi: 10.1038/nature11288. Epub 2012/07/20. doi: 10.1038/nature11288 nature11288 [pii]. PubMed PMID: 22810586; PubMed Central PMCID: PMC3408847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. Epub 2012/05/17. doi: 10.1158/2159-8290.CD-12-0095 2/5/401 [pii]. PubMed PMID: 22588877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44(7):760–764. doi: 10.1038/ng.2291. Epub 2012/05/29. doi: 10.1038/ng.2291 ng.2291 [pii]. PubMed PMID: 22634756. [DOI] [PubMed] [Google Scholar]
- 15.Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330(6001):228–231. doi: 10.1126/science.1196333. Epub 2010/09/10. doi: 10.1126/science.1196333 science.1196333 [pii]. PubMed PMID: 20826764; PubMed Central PMCID: PMC3076894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43(12):1219–1223. doi: 10.1038/ng.982. Epub 2011/11/01. doi: 10.1038/ng.982 ng.982 [pii]. PubMed PMID: 22037554. [DOI] [PubMed] [Google Scholar]
- 17.Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44(5):570–574. doi: 10.1038/ng.2246. Epub 2012/04/10. doi: ng.2246 [pii] 10.1038/ng.2246. PubMed PMID: 22484628. [DOI] [PubMed] [Google Scholar]
- 18.Liang H, Cheung LW, Li J, Ju Z, Yu S, Stemke-Hale K, et al. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome Res. 2012;22(11):2120–2129. doi: 10.1101/gr.137596.112. Epub 2012/10/03. doi: 10.1101/gr.137596.112 gr.137596.112 [pii]. PubMed PMID: 23028188; PubMed Central PMCID: PMC3483541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groesser L, Herschberger E, Ruetten A, Ruivenkamp C, Lopriore E, Zutt M, et al. Postzygotic HRAS and KRAS mutations cause nevus sebaceous and Schimmelpenning syndrome. Nat Genet. 2012;44(7):783–787. doi: 10.1038/ng.2316. Epub 2012/06/12. doi: 10.1038/ng.2316 ng.2316 [pii]. PubMed PMID: 22683711. [DOI] [PubMed] [Google Scholar]
- 20.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. Epub 2012/03/31. doi: 10.1038/nature11003. PubMed PMID: 22460905; PubMed Central PMCID: PMC3320027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3(8):772–775. doi: 10.4161/cbt.3.8.994. Epub 2004/07/16. doi: 994 [pii]. PubMed PMID: 15254419. [DOI] [PubMed] [Google Scholar]
- 22.Janku F, Wheler JJ, Naing A, Falchook GS, Hong DS, Stepanek VM, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer Res. 2013;73(1):276–284. doi: 10.1158/0008-5472.CAN-12-1726. Epub 2012/10/16. doi: 10.1158/0008-5472.CAN-12-1726 0008-5472.CAN-12-1726 [pii]. PubMed PMID: 23066039; PubMed Central PMCID: PMC3537862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holsinger FC, Piha-Paul SA, Janku F, Hong DS, Atkins JT, Tsimberidou AM, et al. Biomarker-Directed Therapy of Squamous Carcinomas of the Head and Neck: Targeting PI3K/PTEN/mTOR Pathway. J Clin Oncol. 2013;31(9):e137–e140. doi: 10.1200/JCO.2012.43.2716. Epub 2013/01/30. doi: JCO.2012.43.2716 [pii] 10.1200/JCO.2012.43.2716. PubMed PMID: 23358976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murugan AK, Hong NT, Fukui Y, Munirajan AK, Tsuchida N. Oncogenic mutations of the PIK3CA gene in head and neck squamous cell carcinomas. Int J Oncol. 2008;32(1):101–111. Epub 2007/12/22. PubMed PMID: 18097548. [PubMed] [Google Scholar]
- 25.Watanabe Y, Castoro RJ, Kim HS, North B, Oikawa R, Hiraishi T, et al. Frequent alteration of MLL3 frameshift mutations in microsatellite deficient colorectal cancer. PLoS One. 2011;6(8):e23320. doi: 10.1371/journal.pone.0023320. Epub 2011/08/20. doi: 10.1371/journal.pone.0023320 PONE-D-11-03053 [pii]. PubMed PMID: 21853109; PubMed Central PMCID: PMC3154922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kompier LC, Lurkin I, van der Aa MN, van Rhijn BW, van der Kwast TH, Zwarthoff EC. FGFR3, HRAS, KRAS, NRAS and PIK3CA mutations in bladder cancer and their potential as biomarkers for surveillance and therapy. PLoS One. 2010;5(11):e13821. doi: 10.1371/journal.pone.0013821. Epub 2010/11/13. doi: 10.1371/journal.pone.0013821. PubMed PMID: 21072204; PubMed Central PMCID: PMC2972209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung LW, Hennessy BT, Li J, Yu S, Myers AP, Djordjevic B, et al. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer discovery. 2011;1(2):170–185. doi: 10.1158/2159-8290.CD-11-0039. Epub 2011/10/11. doi: 10.1158/2159-8290.CD-11-0039. PubMed PMID: 21984976; PubMed Central PMCID: PMC3187555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Q, Bhola NE, Lui VW, Siwak DR, Thomas SM, Gubish CT, et al. Antitumor mechanisms of combined gastrin-releasing peptide receptor and epidermal growth factor receptor targeting in head and neck cancer. Mol Cancer Ther. 2007;6(4):1414–1424. doi: 10.1158/1535-7163.MCT-06-0678. PubMed PMID: 17431120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.