Abstract
The mammalian gut ecosystem has significant influence on host physiology1–4, but the mechanisms that sustain this complex environment in the face of different stresses remain obscure. Perturbations to this ecosystem, such as through antibiotic treatment or diet, are currently interpreted at the level of bacterial phylogeny5–7. Less is known about the contributions of the abundant population of phage to this ecological network. Here, we explore the phageome as a potential genetic reservoir for bacterial adaptation by sequencing murine fecal phage populations following antibiotic perturbation. We show that antibiotic treatment leads to the enrichment of phage-encoded genes that confer resistance via disparate mechanisms to the administered drug as well as genes that confer resistance to antibiotics unrelated to the administered drug, and we demonstrate experimentally that phage from treated mice afford aerobically cultured naïve microbiota increased resistance. Systems-wide analyses uncover post-treatment phage-encoded processes related to host colonization and growth adaptation, indicating that the phageome broadly enriches for functionally beneficial genes under stress-related conditions. We also show that antibiotic treatment expands the interactions between phage and bacterial species, leading to a more highly connected phage-bacterial network for gene exchange. Our work implicates the phageome in the emergence of multidrug resistance and indicates that the adaptive capacity of the phageome may represent a community-based mechanism for protecting the gut microflora, preserving its functional robustness during antibiotic stress.
Antibiotic treatment, an important and often necessary therapeutic intervention, can negatively impact the mammalian gut environment, potentially giving rise to immune2 and metabolic deficiencies8. Studies on the disruption of intestinal homeostasis have focused on the resulting alterations in microbial composition6,7. However, investigation of the gut ecosystem has evinced a myriad of resident phage9, and it remains unclear how perturbation of the gut environment affects these symbionts. Phage can contribute genes that are advantageous to their microbial hosts10,11, in turn promoting their own survival and propagation12. This gene flow suggests that phage may play an important role in the adaptation of the microbiome to stressful environments. In this work, we used a comparative metagenomic approach to explore the effects of antibiotic perturbation on functions encoded in the phageome as well as to examine how antibiotic treatment alters the phage-bacterial ecological network.
We treated groups of young adult mice (n = 5) orally with physiologically relevant concentrations of ciprofloxacin (a quinolone that inhibits DNA synthesis) or ampicillin (a β-lactam that inhibits cell-wall synthesis), each with a respective control. We obtained collective fecal samples from each group after eight weeks of treatment and purified phage as previously described9,13. DNA was extracted from phage and whole-genome amplified before performing shotgun 454 GS FLX+ pyrosequencing. We obtained a total of 440,792 quality reads, with a median read length of 477 nt (210 Mb total; Fig. S1). Evaluation of contamination by quantification of bacterial 16S ribosomal RNA genes indicated that contaminating bacterial sequences represented less than 0.1% of our data, which was subsequently accounted for in all statistical analyses (see Supplementary Discussion and Fig. S2).
Phage DNA sequences were compared to the non-redundant NCBI protein and environmental protein databases (BLASTX; E-value < 10−5). Approximately 70% of reads were not assigned to previously sequenced genes (Fig. S3), suggesting that the mouse phageome, like many other viral communities14,15, harbors uncharacterized genetic material. We used the most significant BLAST alignment of a sequence, when available, to determine its phylogenetic origin. Most of the identifiable phage in our mouse phageomes (Fig. S4a) were from the Caudovirales order, comprising the tailed phage families Siphoviridae, Podoviridae, and Myoviridae, many of which are known to have a temperate life cycle. Since phage genomes incorporate bacterial genes, we also identified bacterial taxa; we found that 97% of phage-encoded bacterial genes were attributed to the four phyla known to dominate the gut (the Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria; Fig. S4b), consistent with the known hosts of the phage we detected.
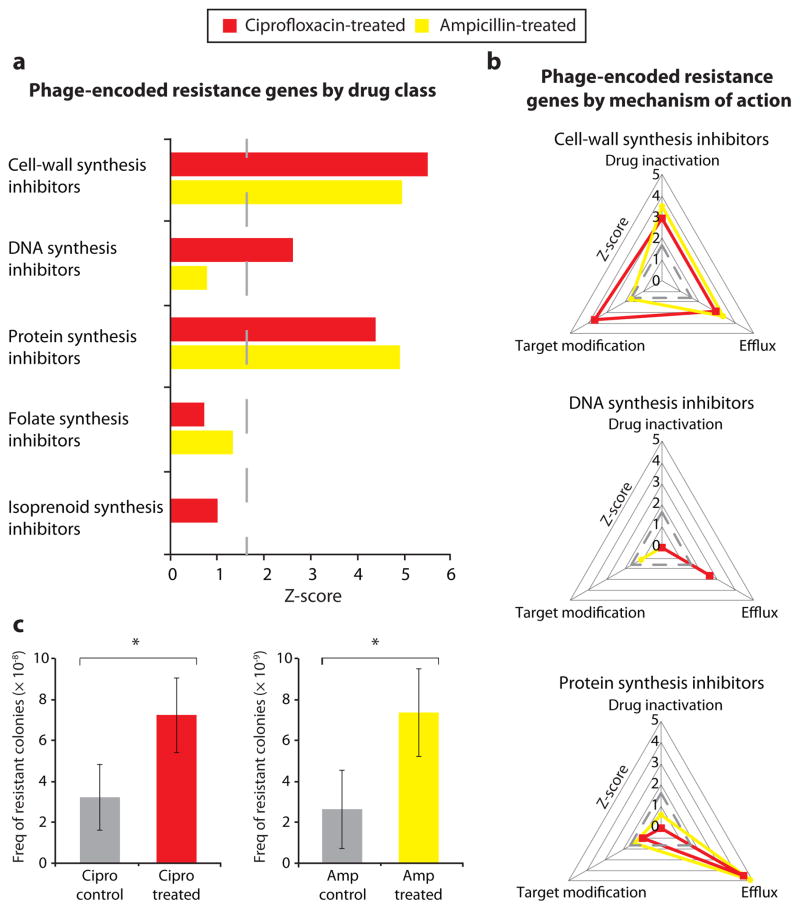
We hypothesized that antibiotic treatment leads to increases in phage-encoded genes for drug resistance. To investigate this, we compared DNA sequences in the phageome to an assembled database of antibiotic resistance proteins (BLASTX; E-value < 10−3, see Methods). We found that reads annotated as antibiotic resistance genes were highly enriched in phage metagenomes from mice treated with ciprofloxacin or ampicillin compared to those from control mice (Z = 7.3 and Z = 7.0, respectively; Fig. S5; read annotations enumerated in Table S1). We catalogued the resistance reservoir by annotating phage-encoded genes based on the drug class to which they confer resistance (Fig. 1a). Our analysis revealed that resistance to the administered drug class was enriched in phage metagenomes from antibiotic-treated mice, such that resistance to DNA synthesis inhibitors was enriched in ciprofloxacin treatment (Z = 2.6), and resistance to cell-wall synthesis inhibitors was enriched in ampicillin treatment (Z = 5.0). Additionally, upon drug treatment, new resistance genes were found in the phageome. For example, phage from ciprofloxacin-treated mice carried genes encoding numerous quinolone efflux pumps (e.g., norM, mexD, mexF), and phage from ampicillin-treated mice carried genes encoding sensor and response regulators of cell-wall synthesis inhibitors (e.g., vanRS).
Figure 1. Antibiotic resistance is enriched in phage metagenomes following drug perturbation in mice.
a, b, Z-scores are shown for sequencing reads annotated as antibiotic resistance genes in phage from ciprofloxacin-treated mice (red) and phage from ampicillin-treated mice (yellow) in comparison to respective control mice. Dashed lines correspond to a Z-score of 1.65 (P = 0.05). Phage-encoded resistance genes were classified according to the drug class to which they confer resistance (a) and by their mechanism of resistance (b). c, Frequency of colonies resistant to ciprofloxacin (1 μg/ml) upon infection of microbiota with phage from ciprofloxacin-treated mice or phage from control mice (left), and frequency of colonies resistant to ampicillin (4 μg/ml) upon infection of microbiota with phage from ampicillin-treated mice or phage from control mice (right). P-values from Mann-Whitney U-test; n > 12. Mean ± s.e.m. *P < 0.05.
Of note, resistance to other, orthogonal drug classes was also overrepresented in the antibiotic-perturbed phageomes. Both treatments led to significant enrichment of resistance to antibiotics that target protein synthesis, and ciprofloxacin treatment also led to significant enrichment of resistance to cell-wall synthesis inhibitors (Fig. 1a). This cross-resistance was mediated by drug-specific inactivators (e.g., chloramphenicol acetyltransferases), as well as multidrug resistance exporters (e.g., mdtK; Table S1). Together, these findings implicate the phage metagenome as a potential source of multidrug resistance during antibiotic treatment of the host.
We aimed to understand the specific mechanisms represented in phage-encoded genes conferring resistance to the most significantly enriched classes of antibiotics: inhibitors of cell-wall synthesis, DNA synthesis, and protein synthesis. We categorized resistance genes according to primary resistance mechanisms, which include modification or protection of the drug target (target modification), enzymatic inactivation of the drug (drug inactivation), and transport of the drug out of the cell (efflux)16. Using this framework to classify phage-encoded resistance genes, our analysis revealed that antibiotic treatment led to disparate resistance mechanism profiles for each drug class (Fig. 1b). Analysis of resistance to cell-wall synthesis inhibitors showed that all types of resistance mechanisms were significantly enriched under both ciprofloxacin treatment and ampicillin treatment. In contrast, resistance to DNA synthesis and protein synthesis inhibitors occurred predominantly by efflux. Resistance to protein synthesis inhibitors occurred by target modification and drug inactivation mechanisms at low levels, and, in accord with its rarity, resistance to DNA synthesis inhibitors via drug inactivation was not detected. These data likely reflect resistance mechanisms that are both environmentally available in the gut ecosystem and impose sustainable in vivo fitness costs. As continued treatment with an antibiotic invariably leads to its own resistance, mechanisms that enable cross-resistance encoded by the phageome may be an important consideration when selecting subsequent therapeutics.
We next sought to demonstrate that phage from antibiotic-treated mice confer increased drug resistance to the host-associated bacterial community. We assessed the frequency of resistant isolates from aerobically cultured naïve microbiota that were infected ex vivo with phage from antibiotic-treated or control (untreated) mice. Our results show that this fraction of microbiota infected with phage from mice administered ciprofloxacin or ampicillin yielded two to three times more colonies resistant to the respective drug than the aerobically cultured fraction of microbiota infected with phage from control mice (Fig. 1c). These data indicate that phage from antibiotic-treated mice can contribute relevant functional advantages to their microbial hosts.
We next took a systems-level approach and classified other phage-encoded genes into functional pathways described by the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. We depicted enriched functional changes in phage metagenomes following antibiotic treatment as a network diagram (Fig. 2; abundances shown in Fig. S6). Among the most significantly enriched pathways were functional properties related to the mode of action of the administered drug (Table S2). Phageomes from ampicillin-treated mice were enriched for the amino sugar and nucleotide metabolism pathway (part of the broader carbohydrate metabolism process; Z = 5.6) indicating overrepresentation of genes related to synthesis of cell wall constituents, and increases in these components have been shown to be requisite for drug resistance in clinical isolates17. Additionally, we found that phageomes from ciprofloxacin-treated mice were enriched for replication- and repair-related pathways, including base excision repair (Z = 6.1), nucleotide excision repair (Z = 7.4), and homologous recombination (Z = 11.2). Included in these pathways are members of the GO system for the repair of DNA oxidative lesions, which has been demonstrated to reduce cytotoxicity due to a range of antibiotic classes18. Also represented are members of the DNA damage-inducible SOS system, known to provide protection against antibiotic-mediated cell death and induce the development of resistance-conferring mutations19. Furthermore, hyper-recombination has been shown to promote multidrug resistance phenotypes20. These results show that under drug treatment, the phageome encodes diverse mechanisms for modulating antibiotic susceptibility.
Figure 2. Broad bacterial functions are enriched in phage metagenomes following drug perturbation in mice.
Network depicts KEGG pathways significantly enriched with antibiotic treatment compared to controls. Treatments are represented by large nodes; enriched pathways are represented by small nodes, grouped by their higher-level processes, and colored by the treatment condition (red – ciprofloxacin; yellow – ampicillin; orange – common to both treatments). In total, we identified 24 out of 188 pathways enriched with ciprofloxacin treatment (Z ≥ 3.46, Bonferroni corrected) and 18 out of 178 pathways enriched with ampicillin treatment (Z ≥ 3.43, Bonferroni corrected).
We also observed that phage metagenomes from antibiotic-treated mice were enriched for microbial functions that contribute to host metabolism (Fig. 2 and Table S2). Phageomes from ciprofloxacin-treated mice were uniquely enriched for pathways relevant to the metabolism of cofactors and vitamins, including thiamine, an essential nutrient provided by the microbiome. Microbiota ferment polysaccharides indigestible by the host alone; metabolism of these sugars enables bacterial survival in and colonization of the gut environment21 and, as a beneficial consequence, provides energy to the host4. We found that polysaccharide degradation genes, specifically related to metabolism of starch, cellulose, lactose, and fructans (plant-derived fructose polymers), were enriched with antibiotic treatment (Fig. 3a). Genes coding for carbohydrate active enzymes (CAZymes), which enable bacteria to ferment a variety of dietary and host-sourced glycans, were also enriched under antibiotic treatment and represented by a range of glycoside hydrolase and glycosyltransferase families (Fig. S7 and Table S3). Because many gut microbes express only a specific array of carbohydrate-degrading enzymes22, bacteria which acquire these genes from the phage reservoir may gain additional foraging capacity and, consequently, a selective growth advantage. These results suggest that the phageome may be an adaptive repository for functions important for the host-commensal relationship that may otherwise be depleted by antibiotic perturbation. (See Supplementary Information for additional discussion.)
Figure 3. Investigation of bacterial functions encoded in phage.
a, Bacterial enzymes from sugar metabolism to glycolysis (left) with corresponding Z-scores in phage from drug-treated mice in comparison to control mice (right). Dashed line corresponds to a Z-score of 1.65 (P = 0.05). b, Class-level taxonomic distribution of all sequences of bacterial origin identified in phage sequencing data (far left) and sequences annotated with enriched functions following drug perturbation. “Other” constitutes taxa that contributed less than 1% to all distributions.
We next aimed to elucidate the phylogenetic basis of these phage-encoded bacterial functions. In Fig. 3b, we illustrate the taxonomic composition of all sequences of bacterial origin and sequences annotated with enriched functions following drug perturbation. Examining bacterial phylotypes that contribute antibiotic resistance genes to the phage metagenome, we found a comparatively high representation of the Clostridia class and a low representation of the Bacilli class. Notably, a large fraction of CAZyme-annotated sequences originated from the Bacteroidetes class, which comprises members found to have diverse capabilities for carbohydrate metabolism22. Investigation of thiamine metabolism reveals that the Bacilli and Verrucomicrobia classes constitute a large proportion of these annotations. Since the phageome reflects emergent properties of its environment15, phylogenetic analyses of phage-encoded elements may more broadly enable the identification of bacteria actively contributing to specific functions in the gut environment.
Our results show that the phageome harbors a diversity of potentially beneficial functional elements in the face of antibiotic perturbation. However, the extent to which this genetic reservoir is accessible to members of the microbiota remains unclear23. To investigate this, we sought to elucidate the phage-bacterial ecological network and how it changes under antibiotic treatment. We approximated the network of phage-microbe interactions with relationships identified through the reconstruction and analysis of individual viral genomes. De novo assembly was accomplished using stringent parameters (see Methods). Reconstructed viral genomes are composed of a mosaic of bacterial genes, and we utilized the phylogenetic origins of these sequences to determine putative phage-bacterial associations (Fig. 4). Our resulting network recapitulated known interactions, including the lysogenic relationships of foodborne pathogens, such as bacteriophage phi3626 infection of C. perfringens24 and Siphoviridae Listeria phage A500 infection of L. monocytogenes25. Importantly, antibiotic treatment leads to widespread restructuring of the phage-bacterial ecological network (Fig. 4). These data show that new links between phage and bacteria are formed with drug treatment, giving rise to significantly greater network connectivity (Fig. S8). This increased connectivity is reflective of phage broadly, as more bacterial species are associated with a given phage (Fig. S9). These results suggest that antibiotic treatment increases the frequency of phage integration and stimulates broad host range, which promotes a functional reservoir that is both genetically diverse and highly accessible by gut bacteria.
Figure 4. Phage-bacterial ecological network.
Dash marks represent associations between virotypes and bacterial species identified from phylogenetic analysis of reconstructed phage genomes. Phage-bacterial associations only in control metagenomes (black), only in drug-treated metagenomes (red), and commonly identified in control and drug-treated metagenomes (gray). Data are the union of associations identified in 50 assemblies of randomly sampled reads from each treatment.
While the phageome is a highly connected network for gene exchange, the functional consequence of acquiring genetic material from this reservoir depends on the molecular context of the host bacterium. Acquisition of a single gene may enhance an existing function by gene dosage or enable a novel phenotype. As some proteins rely on additional machinery, subsequent horizontal gene transfer events may be required to produce a phenotype. Moreover, redundantly acquired genes may gain new functionality through paralogous evolution.
Here we demonstrate that antibiotic treatment enriches the phage metagenome for stress-specific and niche-specific functions, while mediating changes in the topology of the phage-bacterial ecological network to potentiate accessibility of these genetic elements. Functional resilience of the microbiome following environmental perturbation has been empirically documented and has engendered interest in the restorative forces that return the commensal flora to its pre-perturbed state6,7,26. Our results implicate phage encapsulation of adaptive signatures as a community-based mechanism for functional robustness in the gut environment during stress. Of note, antibiotic treatment can also prime the gut environment for pathogen invasion2, and our findings pose potential implications for the emergence of drug resistance and evasion strategies in pathogenic populations. Cohabitation of phage and bacteria in the gut ecosystem is likely governed by complex and dynamic interactions, particularly during stress perturbation. Additional work is needed to discern the selective pressures imposed on each member of this community and the resulting mechanisms that influence the encoding and progressive enrichment of functionally beneficial genes in the phageome. Phage-mediated gene flow may be an important phenotypic buffer for bacterial communities, and further investigation of the adaptive reservoir of the phageome and the dynamic nature of the phage-bacterial ecological network may prove critical to understanding the influence of the gut ecosystem on host physiology.
Methods
Mouse study
All experiments involving animals were pre-reviewed and approved by the Boston Children’s Hospital Institutional Animal Care and Use Committee (IACUC). Groups of separately housed 6-week-old female FVB/NJ mice (n = 5; Jackson Laboratory) were treated with ampicillin (142.5 mg/L) or ciprofloxacin (62.5 mg/L) in their drinking water. The corresponding dosage was 28.5 mg/kg/day ampicillin and 12.5 mg/kg/day ciprofloxacin, based on the average mouse weight of 20 g and an approximate intake of 4 ml per day. Control mouse groups were supplied with standard drinking water (ampicillin) or alkaline water (ciprofloxacin). Treatments were refreshed twice per week. Mice were housed in sterile conditions and received autoclaved chow during the course of this study. We harvested fresh collective fecal samples from each group to obtain ample material (3–4 g) for purification. Samples were stored at −80 °C prior to use.
Viral purification and preparation of genomic DNA
Viral purification from fecal samples of mice after eight weeks was performed as previously described9,13. An aliquot of the viral preparation (1.5 g ml−1 layer from ultracentrifugation) was stained with SYBR gold and visualized with epifluorescence microscopy to verify the absence of bacterial contamination. Viral particles were concentrated and desalted using an Ultra-4 Centrifugal Filter Unit (Ultracel-30K MWCO; Millipore) to a volume of ~200 μl. Concentrated viral samples were treated with DNase (0.2 mg/ml) and samples were passed through a 0.22 μM filter to ensure no procedural contamination was introduced. Genomic DNA was extracted using the QiaAMP DNA mini kit (Qiagen) using the protocol for viral DNA detailed in the manual. Genomic DNA was amplified using the Illustra Genomiphi v2 kit (GE) according to manufacturer’s instructions. For each sample, we pooled amplified DNA from three separate reactions to minimize bias.
Next-generation sequencing
Viral DNA was submitted to the Tufts Genomic Core for library preparation and shotgun sequencing. Equimolar concentrations of multiplexed (Rapid Library MID) samples were pooled on a single plate and pyrosequenced using the 454 GS FLX+ platform. Resulting sequences were filtered by removing duplicates using the tool available at http://microbiomes.msu.edu/replicates/28 with the following parameters: sequence identity cutoff = 97%; length difference requirement = 0; number of beginning base pairs to check = 20.
Antibiotic resistance annotations
To facilitate annotation of antibiotic resistance genes, we assembled a database consisting of the Antibiotic Resistance Genes Database27 and UniProt proteins annotated with the GO term “antibiotic breakdown” (GO: 0017001) to achieve a total of 12,687 protein sequences. The functional annotations of the proteins in this database are supported by either experimental validation or high-quality computational prediction. Viral DNA sequences were compared to this database using BLASTX, and sequences with an E-value < 10−3 were deemed significant. This cutoff was selected to maintain a consistent stringency, by accounting for database size, with functional annotation to the NCBI databases.
Functional annotations
Phage DNA sequences were compared to the non-redundant NCBI protein (nr) and environmental protein (env_nr) databases (BLASTX; E-value < 10−5). Sequences were annotated with KEGG29 (v61.0) using custom Perl scripts interfaced with the KEGG API. The most significant BLAST hit that resulted in a KEGG annotation was used, and we included all annotations in the event that a hit had multiple annotations. KEGG ortholog annotations were compared to glycoside hydrolases and glycosyltransferases found on www.cazy.org30 to identify CAZyme-encoding genes.
Statistical testing
To compare the functional annotations of two metagenomic datasets, A and B, we generated a distribution for each A and B reflecting the number of annotations from 10,000 trials of random sampling with replacement, sampled at the number of reads in the comparison dataset. To account for the effects of contaminating bacterial DNA in our comparative metagenomic analyses, we assumed that contamination would be uniformly distributed and therefore we randomly discarded bacterial reads in each sampling trial according to the amount of contamination detected by quantitative PCR. We compared the number of annotations identified in a given sample to the comparison distribution and determined a Z-score, calculated as (x−μ)/σ, where, for example, x is the raw number of annotations in sample A, μ is the mean number of annotations in the distribution for sample B, and σ is the standard deviation of B’s distribution. In essence, this results in two Z-scores, one comparing A to B and one comparing B to A, and consequently the minimum |Z| was used. According to the Central Limit Theorem, random sampling with replacement results in a normal distribution, so Z-scores > 1.65 (P < 0.05) were considered enriched. In KEGG pathway analysis, Bonferroni was used to correct for multiple hypotheses, where the P-value cutoff 0.05/n (n was the number of total third-level pathways identified in our phage metagenomes) was converted to a Z-score.
Phage infection of microbiota
Microbiota were isolated from fecal samples of naïve mice as previously described31, except PBS + 0.1% cysteine supernatants were plated on four separate LB agar plates. Colonies were grown aerobically for 24 hours before plates were scraped with 2 ml LB, amassed, and cultured at 37 °C and 300 RPM for two hours. This mixture was stored as 150 μl aliquots in 15% glycerol at −80 °C. For each experiment, an entire aliquot was used as the inoculum to minimize growth biases. Phage were isolated as described above. Due to limited sample availability, we harvested phage from mice that had been treated for five weeks for ciprofloxacin experiments and phage from mice that had been treated for three weeks for ampicillin experiments along with phage from control mice, respectively. Phage preparations from drug-treated mice and control mice were diluted to the same volume, split equivalently, and incubated with 0.25 ml microbiota (cultured to exponential phase in 0.2% maltose) with 5 mM CaCl2 and 10 mM MgSO4. Phage-microbiota mixtures were allowed to adsorb for one hour at 37 °C (no shaking). Phage-infected microbiota were then pelleted, resuspended in fresh LB, and plated on LB agar plates with ciprofloxacin (1 μg/ml) or LB agar plates with ampicillin (4 μg/ml). A 10 μl aliquot was serially diluted and plated onto no-drug LB agar plates. Frequency was calculated as: # of colonies on drug plate divided by # of colonies on no-drug plate. Additionally, the basal frequency of resistant isolates from microbiota was measured as above in the absence of phage, and this frequency was confirmed to be lower than that from microbiota infected with phage from either treated or untreated mice.
Quantification of 16S rRNA
Quantitative PCR was used to measure levels of the 16S rRNA gene in our viral preparations. We used the universal primers 8F and 338R9, and qPCR was performed using the SYBR Green I Master kit and the LightCycler 480 (Roche) according to manufacturer’s instructions.
Contig assembly and identification of phage-bacterial associations
Contigs were assembled using the Roche 454 GS De Novo Assembler with default parameters, except for a minimum overlap of 100 bp and a minimum identity of 100% to minimize erroneous alignments. Phage-bacterial associations were determined by computing the combination of phage phylogenetic annotations and bacterial phylogenetic annotations on a given contig, and non-redundant phage-bacterial associations were amassed for all contigs. To evenly compare the number of phage-bacterial associations across samples, we performed 50 assemblies using 60,000 randomly selected sequences with replacement from each sample. The data presented in Fig. 4 are representative of these 50 assemblies such that an association was illustrated if it was present in at least one assembly analysis. We computed the mean number of phage-bacterial associations for a given sample (Fig. S8) from the number of non-redundant associations found in each assembly trial. Significance was determined by comparing sample values using the Mann-Whitney U-test. We computed the mean bacteria-to-phage ratio for a given sample (Fig. S9) from the number of bacterial species associated with a given phage for all phage using the union of associations from 50 assemblies shown in Fig. 4. Significance was determined by comparing sample values using the Mann-Whitney U-test.
Supplementary Material
Acknowledgments
We thank K. Bodi and J. Schiemer at the Tufts Genomic Core for their sequencing assistance and A. Green and K. Pardee for ultracentrifugation help. This work was supported by the Howard Hughes Medical Institute and the NIH Director’s Pioneer Award Program.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions
All authors designed the study. C.S.S. oversaw the mouse work. S.R.M. and H.H.L. performed and analyzed the experiments with conceptual insight provided by J.J.C. S.R.M., H.H.L., and J.J.C. prepared the manuscript.
Virome data sets reported here are accessible in the NCBI Short Read Archive under accession number SRA073587.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandl K, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smillie CS, et al. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480:241–244. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 5.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108 (Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antunes LC, et al. Effect of antibiotic treatment on the intestinal metabolome. Antimicrob Agents Chemother. 2011;55:1494–1503. doi: 10.1128/AAC.01664-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliver KM, Degnan PH, Hunter MS, Moran NA. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science. 2009;325:992–994. doi: 10.1126/science.1174463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Novick RP. Phage-mediated intergeneric transfer of toxin genes. Science. 2009;323:139–141. doi: 10.1126/science.1164783. [DOI] [PubMed] [Google Scholar]
- 12.Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. Photosynthesis genes in marine viruses yield proteins during host infection. Nature. 2005;438:86–89. doi: 10.1038/nature04111. [DOI] [PubMed] [Google Scholar]
- 13.Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F. Laboratory procedures to generate viral metagenomes. Nat Protoc. 2009;4:470–483. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- 14.Breitbart M, et al. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinsdale EA, et al. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- 16.Walsh C. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000;406:775–781. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- 17.Hanaki H, et al. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J Antimicrob Chemother. 1998;42:199–209. doi: 10.1093/jac/42.2.199. [DOI] [PubMed] [Google Scholar]
- 18.Foti JJ, Devadoss B, Winkler JA, Collins JJ, Walker GC. Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics. Science. 2012;336:315–319. doi: 10.1126/science.1219192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol. 2010;8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanage WP, Fraser C, Tang J, Connor TR, Corander J. Hyper-recombination, diversity, and antibiotic resistance in pneumococcus. Science. 2009;324:1454–1457. doi: 10.1126/science.1171908. [DOI] [PubMed] [Google Scholar]
- 21.Chang DE, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci U S A. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu J, Gordon JI. Honor thy symbionts. Proc Natl Acad Sci U S A. 2003;100:10452–10459. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stern A, Mick E, Tirosh I, Sagy O, Sorek R. CRISPR targeting reveals a reservoir of common phages associated with the human gut microbiome. Genome Res. 2012 doi: 10.1101/gr.138297.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmer M, Scherer S, Loessner MJ. Genomic analysis of Clostridium perfringens bacteriophage phi3626, which integrates into guaA and possibly affects sporulation. J Bacteriol. 2002;184:4359–4368. doi: 10.1128/JB.184.16.4359-4368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorscht J, et al. Comparative genome analysis of Listeria bacteriophages reveals extensive mosaicism, programmed translational frameshifting, and a novel prophage insertion site. J Bacteriol. 2009;191:7206–7215. doi: 10.1128/JB.01041-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu B, Pop M. ARDB--Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009;37:D443–447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. Isme J. 2009;3:1314–1317. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- 29.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodman AL, et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.