Abstract
Critically short telomeres activate cellular senescence or apoptosis, as mediated by the tumor suppressor p53, but in the absence of this checkpoint response, telomere dysfunction engenders chromosomal aberrations and cancer. Here, analysis of p53-regulated genes activated in the setting of telomere dysfunction identified Zfp365 (ZNF365 in humans) as a direct p53 target that promotes genome stability. Germline polymorphisms in the ZNF365 locus are associated with increased cancer risk, including those associated with telomere dysfunction. On the mechanistic level, ZNF365 suppresses expression of a subset of common fragile sites (CFS) including telomeres. In the absence of ZNF365, defective telomeres engage in aberrant recombination of telomere ends, leading to increased telomere sister chromatid exchange (T-SCE) and formation of anaphase DNA bridges, including ultra-fine DNA bridges (UFB), and ultimately increased cytokinesis failure and aneuploidy. Thus, the p53-ZNF365 axis contributes to genomic stability in the setting of telomere dysfunction.
Keywords: ZNF365, telomeres, genomic instability, p53, aneuploidy
Introduction
Telomeres maintain chromosome structural integrity and thus promote genomic stability. Dysfunctional telomeres provoke inappropriate DNA repair followed by chromosome non-disjunction events and breakage-fusion-bridge cycles, and, therefore, are major instigators of the genomic instability in diverse human cancer types (reviewed in [1]). The multi-protein shelterin complex consisting of TRF1, TRF2, POT1, Rap1, TPP1 and TIN2 forms the telomere loop, or t-loop, that protects telomere ends by preventing their recognition as damaged DNA [2]. To date, studies have shown a number of proteins are recruited to replicate structurally constrained telomeres: TRF2 can recruit Topo2a and Apollo to resolve the positive superhelical strain and protect telomeres [3], and TRF1 plays an essential role in telomere replication [4]. In addition, the RecQ helicases WRN and BLM play essential roles in DNA replication, recombination, and repair, and in the maintenance of functional telomeres [5]. In particular, BLM is critical for the resolution of late-replicating structures as part of the BTR complex observed on UFBs [6]. Telomeres are often incompletely replicated until late anaphase, and intermediate structures are associated with Fanconi anemia proteins in response to replication inhibition [7].
The genetic interactions between telomere dysfunction and the molecular circuitry underlying the telomere checkpoint response continue to be active areas of investigation. Previous studies have demonstrated functional interactions between telomeres and DNA damage-sensing, transduction (ATM, ATR), and repair molecules (Ligase IV, DNA-PK, Ku70) at the organismal level, where combined deficiencies accelerated telomere attrition and dysfunction, culminating in genomic catastrophe and accelerated aging [8-11]. The acute loss of telomere function by expression of dominant-negative TRF2 (telomere repeat-capping protein) leads to the activation of Atm and p53 in human cell culture models, consistent with their roles in sensing telomere dysfunction and eliciting cellular checkpoint responses [12].
In the in vivo setting, genetic analysis of the interplay between telomere dysfunction and Atm or p53 has revealed opposing phenotypic consequences upon loss of Atm versus p53 in the telomerase knockout mouse. In late generation mTerc-/- Atm-/- mice and derivative cells, there is increased genomic instability and preservation of p53-mediated senescence and apoptosis, leading to severe tissue atrophy and degeneration, premature aging, and suppression of cancer compared with mTerc+/+ Atm-/- controls. In contrast, late generation mTerc-/- p53-/- mice and cells show restoration of cellular proliferation and survival, an increase in organ cellularity, and enhanced tumorigenesis with altered tumor spectrum, underscoring the critical role of p53 status in dictating genome integrity and cellular and organismal fates with regard to degeneration or cancer [10, 13, 14].
Many correlative studies in human cancer and numerous other studies have supported the view that persistent DNA damage signaling resulting from telomere dysfunction provides pressure to deactivate critical checkpoints and sets the stage for accumulation of chromosomal aberrations and aneuploidy [15]. A recent study demonstrated the molecular basis of tetraploidization by deprotected telomeres in the absence of Pot1 and p53, further implicating dysfunctional telomeres as a cause of genomic instability in human cancer [16].
Genomic instability is a prominent feature of hereditary and sporadic cancers in which we observe loss of DNA repair and checkpoint genes, increased replication stress associated with activated oncogenes, and erosion of telomeres (reviewed in [17]). Functional DNA repair and cellular checkpoint processes promote proper replication of the genome in part by resolving replication blockage. Hereditary defects in DNA repair pathway components such as WRN, BLM, and BRCA1/2 cause DNA breaks during replication, resulting in chromosomal rearrangement [18, 19]. In particular, inherently unstable CFSs tend to break or recombine following partial replication inhibition. Of relevance to our study, such genomic fragile sites include telomeres that are particularly sensitive to replication blockade [4, 6].
Despite the large body of well-described biological outcomes, the molecular mechanisms underlying the role of p53 in suppressing the genomic instability associated with telomere dysfunction are not well understood. In this current study, we attempt to dissect the circuitry of the p53-mediated checkpoint response by analyzing transcriptional changes associated with telomere dysfunction. Our study identified ZNF365 as a necessary target whose activation by p53 in the presence of critically short telomeres contributes to genomic stability. We provide evidence that loss of ZNF365 leads to increased expression of CFS and dysfunctional telomeres, aberrant sister telomere recombination, and increased aneuploidy. Furthermore, ZNF365 expression is downregulated in triple negative breast cancer (TNBC), in line with multiple genome-wide association studies defining ZNF365 as a major locus of breast cancer susceptibility in BRCA2-mutant patients [20, 21]. Together, these results support the view that ZNF365 is a novel player contributing to genomic stability.
Results
p53 reactivation in cells with telomere dysfunction causes robust gene expression changes resembling a cellular checkpoint response
To define the p53-mediated transcriptome associated with telomere dysfunction, we utilized a generation 4 telomerase-negative, Atm-negative (G4 mTerc-/- Atm-/-) skin fibroblast model that retains a robust p53-dependent telomere checkpoint. Consistent with previous work, G4 mTerc-/-Atm-/-p53-/- (triple knockout, TKO) skin fibroblasts show inactivation of cellular checkpoints that provide genomic stability. These cells exhibit frequent telomere signal-free ends and significantly shorter telomeres compared with p53 single KO cells [10] (Figure 1A-C).
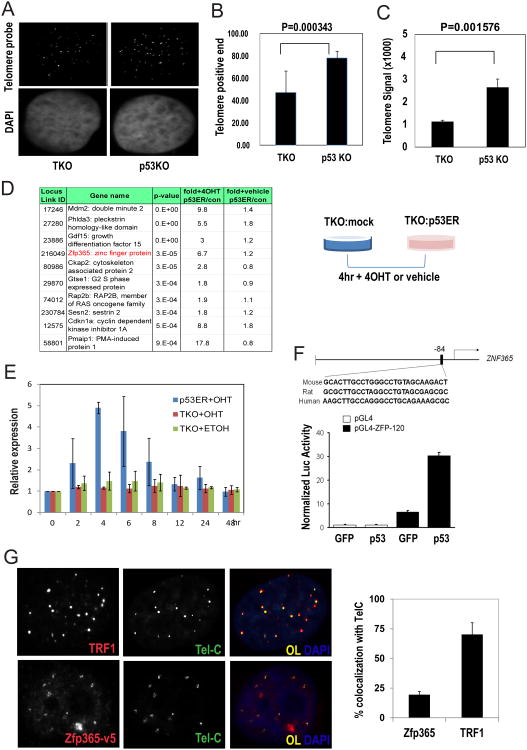
Figure 1. Significantly shortened telomeres in TKO in comparison with p53 KO fibroblasts and induction of Zfp365 expression by p53.
A. Representative photographs of TKO (left) and p53KO (right) nuclei stained for DNA (DAPI, lower) and telomere PNA-FISH (FITC, upper) B, C. Histograms that represent mean numbers of telomere positive ends (B) and mean integrals of PNA signals (signal area × signal intensity) per nucleus (C) (mean±SD, n=10) suggest the presence of shorter telomeres in the TKO model. D. TKO cells were transfected with p53ER and p53 was re-actived by 4-OHT. Top ranked genes induced by p53 are listed (FDR<0.05, p<0.0005 by two way ANOVA, n=5). E. Enforced p53 activation induced time-dependent Zfp365 expression in TKO cells. TKO cells were transduced with retroviral particles for p53ER and treated with 100 nM 4OHT for multiple time points. None transduced cells and ethanol treatment were used as controls. For indicated time points Zfp365 expression was analyzed by qRT-PCR. F. Upper, a putative p53 binding element conserved in multiple species locates at 84 bp upstream of Zfp365 transcription start site. Lower, luciferase activity assay using a 120 bp DNA fragment of the Zfp365 promoter region cloned into pGL4 vector. Either GFP or p53 encoding plasmids were co-transfected and analyzed for the luciferase activity after 72 h. G. Zfp365 partially localizes at telomeres after replication challenge with Aph. On the left, IF-FISH staining forZfp365-v5 (red), TRF1 (red), and telomeres (TelC-FITC) suggests greater co-localization of TRF1 with telomeres, as further illustrated by comparison of the % of telomeres positive for TRF1 or Zfp-v5 plotted on the right (mean±SD, n=30).
In the TKO cells, we introduced an inducible p53 allele encoding a p53-estrogen receptor fusion protein (p53ER) that becomes functional upon addition of 4-hydroxytamoxifen (4-OHT) [22]. We chose a time point of 4 hours post-4-OHT induction to catalog potential direct targets of p53 in the transcriptome, as known transcriptional targets of p21 and Mdm2 started to show robust induction (Figure S1A-B). Zfp365 (ZNF365 in human) ranked high among other genes (Mdm2, Phlda3, Gdf15, Ckap2, Gtse1, Sesn2, Cdkn1a, 2-way ANOVA, p<0.0005) that were previously linked to p53 biology (http://linkage.rockefeller.edu/p53) (Figure 1D, Supplemental Table 1).
Consistent with the microarray data, Zfp365 expression peaks at 4 hour upon p53 re-activation followed by restoration to normal basal levels (Figure 1E). A survey of the Zfp365 5kb promoter region revealed a putative p53 binding element at position -84bp relative to the transcriptional start site that is conserved across multiple species (Figure 1F). A 120 bp fragment, which encompasses the proximal promoter region containing this putative p53 binding element, showed an approximate 6-fold increase in luciferase activity with enforced p53 expression. Collectively, these results identify Zfp365 as a novel transcriptional target of p53 in the context of cells experiencing telomere dysfunction.
ZNF365 plays a role in telomere biology and is implicated in breast cancer risk
Next, we characterized Zfp365 in both telomere biology and in human cancers with dysfunctional telomeres. Zfp365 showed a partial co-localization with a fraction of telomeres by immunofluorescence coupled with telomere FISH. This is in contrast to TRF1 localization which is mostly telomeric. Its co-localization with telomeres becomes detectable only after aphidicolin (Aph) treatment-mediated replication challenges, suggesting it may transiently localize to the stalled replication sites including telomeric/subtelomeric regions (Figure 1G). We further examined its expression and localization pattern in multiple murine and human cell types. Among the human orthologs of Zfp365, ZNF365-A isoform, which bears 98% sequence homology, showed identical subcellular localization with Zfp365, while ZNF365-B or -C isoforms, which have different C-termini primarily localized to centrosomes (Figures S2A, B). Hereafter, we refer to both Zfp365 and ZNF365-A collectively as ZNF365 based on their indistinguishable features. Together, our results suggest that ZNF365 is regulated at the levels of expression and localization in the setting of telomere dysfunction, suggesting a potential role in telomere biology.
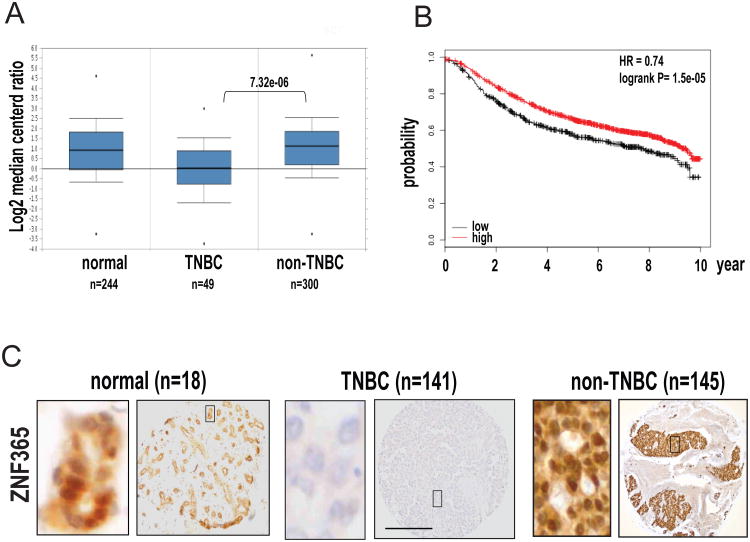
Critically short and dysfunctional telomeres are universally present in early lesions of human breast cancer [23]. Given recent reports alluding to ZNF365 as a major genetic locus for breast cancer risk in BRCA2 mutation carriers [20, 21], we surveyed its expression in triple negative breast cancer (TNBC) vs. non-TNBC in the Cancer Genome Atlas (TCGA) dataset. The former has been shown to exhibit ‘BRCAness’ (i.e., defective double-strand break (DSB) repair capacity) that is shared with BRCA mutation carriers [24]. Interestingly, expression analysis showed the lowest expression of ZNF365 in TNBC (n=49), while average expression levels remained high in non-TNBC (n=300), suggesting loss of ZNF365 expression may correlate with BRCAness (Figure 2A, t-test p=7.32e-06). Furthermore, the level of ZNF365 expression stratified TCGA cohort (n=2978) into two groups that exhibited a significant difference in 10-year, relapse-free survival by Kaplan-Meier analysis (Figure 2B; HR=0.74, log-rank p=1.5e-05).
Figure 2. ZNF365 expression varies in different subtypes of breast cancer.
A. Boxed plot of ZNF365 expression levels among human normal breast tissues (n=244), TNBC (n=49) and non-TNBC (n=300) in Oncomine TCGA data sets. B. Kaplan-Meier survival plot showed the prognostic effect of ZNF365 expression (206448_at) on the relapse free survival over 10 years. HR and Log-rank analysis were performed. C. Representative immunohistochemical staining of TMA with specific antibody against ZNF365. Higher magnification images of the boxed regions are shown on the left. Scale bar, 200 μm.
Next, we stained for ZNF365 in a set of tissue microarrays (TMA) comprising cohorts of normal breast tissue (n=18) and TNBC (n=141) as well as non-TNBC (luminal type A, n= 145). Consistently, ZNF365 exhibited nuclear expression in normal breast epithelium and non-TNBC, while its expression declined significantly in TNBC (Figure 2C, Table 1). Together, ZNF365 expression varies in different subtypes of breast cancer where distinct levels of telomere dysfunction are present.
Table 1.
Analysis of the expression level of ZNF365 by immunohistochemical staining in TMA with non-parametric Wilcoxon Rank Sum Test for H-score between TNBC and non-TNBC (LumA) shows difference (p < 0.001) at 0.05 significance level.
| Case | ||||
|---|---|---|---|---|
|
|
||||
| Variable | Non-TNBC Luma | TNBC | p-value | |
| H-Score | N | 145 | 141 | |
| Mean | 99.2 | 12.2 | ||
| StdDev | 62.5 | 24.4 | ||
| StdErr | 5.2 | 2.1 | ||
| Median | 100.0 | 2.0 | <0.001 | |
| Min | 0.0 | 0.0 | ||
| Max | 300.0 | 170.0 | ||
Loss of ZNF365 contributes to telomere dysfunction
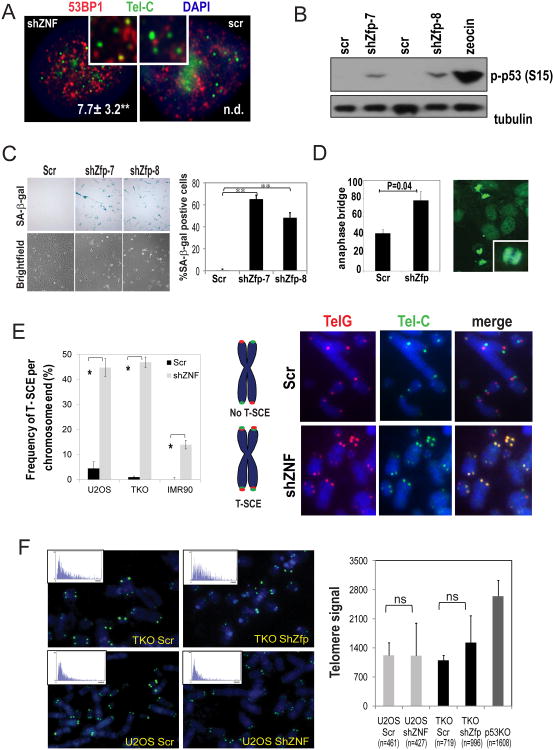
To further investigate ZNF365 function, we conducted loss of function studies with short hairpin-mediated knockdown in mouse and human cells (Figure S3A). Upon knockdown of ZNF365, numerous 53BP1 positive foci appeared with many localizing to telomeres, and cultures showed robust activation of p53 as well as early senescent phenotype (Figure 3A-C). In order to measure the degree of telomere dysfunction, we determined the anaphase bridge index (ABI) out of total late anaphase. ZNF365-depleted TKO cells exhibited a high ABI, indicating the presence of unseparated sister chromatids or DNA bridges (Figure 3D).
Figure 3. Depletion of ZNF365 causes damaged telomeres and cellular senescence.
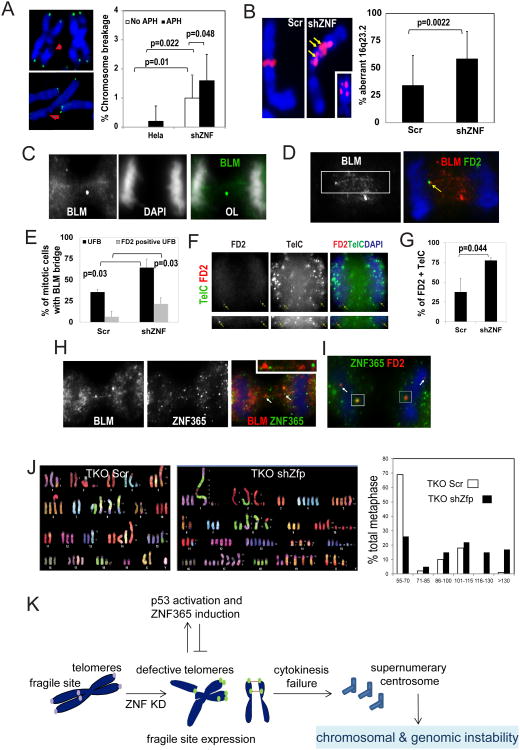
A. Depletion of ZNF365 increased 53BP1 positive telomeres. 53BP1 IF staining (red) combined with FISH for TelC (green) showed frequency of 53BP1-positive TelC telomeres out of total telomeres in Hela Scr (non-targeting scrambled sequence) and shZNF cells. The quantitative results are shown in the bottom right of the panel **, p=0.005. n.d., not determined. B. Activation of p53 by Zfp365 knockdown in p16Ink4a-null MEFs. Cultures were harvested 96 h post shRNA knock down and p-p53 (S15) immunoblotting was performed with Zeocin treated cell lysate as a positive control. C. Knockdown of Zfp365 in early passage MEFs (P=2) led to senescence (lower, bright field; upper, senescence associated beta-galactosidase (SA-β-gal) staining) phenotype. The percentage of the SA-β-gal-positive cells was plotted on the right side. **, p<0.05. D. A representative DAPI staining (green) image shows anaphase bridges from TKO shZfp culture. The anaphase bridge index (# of nuclei with anaphase bridges)/(total # of anaphase nuclei) is plotted on the left (mean±SD, 3 independent experiments). The inset shows the higher magnification of an anaphase bridge. E. Level of T-SCE in U2OS, TKO, and IMR90 Scr and ZNF365 knockdown cells. Left, frequency of T-SCE is plotted (n=5). Y axis represents mean±SD of total T-SCE events out of the total number of chromosomes ends counted. *, p<0.01. Center, diagram explains telomere patterns affected by T-SCE. Right, representative T-SCE images of metaphases hybridized with probes against the leading (TelG, red) and lagging (TelC, green) telomeres with DAPI staining from U2OS Scr and shZNF cells. F. Representative TEL-FISH (TelC, green) images in TKO Scr, TKO shZfp, U2OS Scr and U2OS shZNF cells. Insets show the distribution diagram of telomere length. Right, the average telomere length (mean±SD) of each cell type is plotted (number of telomeres analyzed. ns, not significant.
Disruption of a telomere structural component (i.e., TRF2, POT1a) activates DNA damage responses [12, 25, 26]. Consequently, aberrant repair of telomeres by the mechanism of non-homologous end joining mediates chromosomal instability induced by the break-fusion-break cycle [27, 28]. Thus, we posited that p53 activation and cellular senescence responses in ZNF365-depleted cells might be due to disrupted telomeres and ensuing recombination events on unprotected telomeres that lead to DNA-damage signaling. Our analysis of the metaphases showed increased p-p chromosomal and intra-chromatid fusions in shZfp TKO cells relative to Scr controls (Figure S3B).
Most strikingly, there was a significant increase in T-SCE and telomere defects in multiple cell types of U2OS, IMR90, and TKO upon ZNF365 knockdown with two independent shRNAs (Figure 3E, S4A-C). In early-passage IMR90 cells with sufficient telomere reserve, T-SCE occurred in fewer than 2-3 cell cycles after knocking down ZNF365 (Figure 3E, S4A). These results suggest that T-SCE is an outcome of an exacerbated dysfunctional state at telomeres and acute loss of ZNF365 is sufficient to cause such change in cells with longer telomeres (Supplemental Table 2). Furthermore, the average telomere length was not significantly affected in cells with T-SCE. Instead, we observed a high degree of variation in the distribution of telomere intensity (Figure 3F, note larger S.D. in ZNF365 KD cells). Moreover, no telomere signals co-localized with promyelocytic leukemia (PML) (Figure S4D), suggesting against upregulation of the alternative lengthening of telomere (ALT)-associated PML bodies (APB). Collectively, these results suggest that ZNF365 is necessary to suppress aberrant recombination on dysfunctional telomeres.
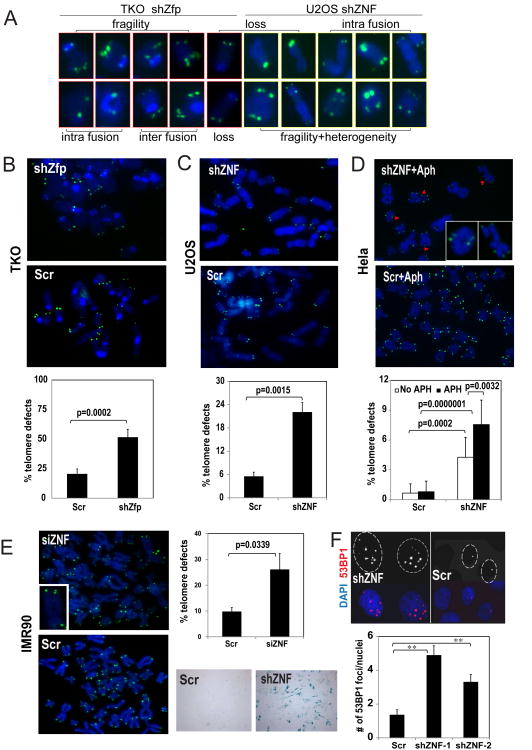
Loss of ZNF365 leads to defective telomere structures and FRA16D expression
Telomeres are topologically strained genomic regions where cellular DNA replication forks are subject to stalling. Interestingly, ZNF365 knockdown increased the number of telomeres that highly resemble “fragile telomeres,” as previously described for abnormally fused or amplified sister telomeres in TRF1-deficient cells (Figure 4A). In addition, telomere defects resulting from ZNF365 knockdown were classified into several structural classes: telomere heterogeneity, loss, fragility, and fusion (intra and inter chromosomal) (Figure 4A). ZNF365 depletion increased the incidence of telomere defect in TKO and U2OS cells compared with the control Scr cells (Figure 4B-C).
Figure 4. Loss of ZNF365 causes defective telomeres.
A. Representative images of defective telomeres (heterogeneity, loss, fragility, and intra and inter chromosomal fusions) observed in ZNF365 knockdown TKO and U2OS cells as in (B-C). B-C. Knockdown of ZNF365 increased defective telomeres in (B) TKO primary murine fibroblasts and (C) U2OS cells. Frequencies of defective telomeres are plotted in the lower panels. D. Pictures of fragile telomere structures shown in metaphase prepared from Hela Scr and shZNF cells treated with 0.3 μM Aph for 24 h. Frequency of telomere defects (pointed by red arrowheads, higher magnification images are shown in the inset) with or without Aph treatment are plotted in lower panels (n=10). E. ZNF365 siRNA treated early passage (P=5) IMR90 cells exhibit increased telomere defects. The inset shows a representative fragile telomere. Upper right, quantified telomere defects plot. Lower right, SA-β-gal staining in IMR90 Scr and shZNF cells showing increased senescence in the latter. F. 53BP1 IF staining (red, nuclei marked) shows increased DSB in two independent shZNF expressing U2OS cells compared with Scr control expressing ones. Number of 53BP1 positive foci per nuclei (mean±SD) is plotted (n=5). **, p<0.01.
The high incidence of these telomere defects in ZNF365-deficient cells prompted us to speculate that replication of telomeric regions was affected, as similarly observed in TRF1-deficient cells [4]. Therefore, we examined the effect of replication challenge on the frequency of defective telomeres. Consistent with previous studies, Hela cells treated with low-dose Aph showed fragile telomeres, albeit less prominently than ZNF365-depleted Hela cultures (Figure 4D). In ZNF365-deficient Hela cells, we mainly observed fragility of telomeres with infrequent fusion events. Overall the level of telomere defects was much less prominent than that in telomerase-negative cell types that we examined, suggesting telomerase may have resolved some of the telomere defects.
In order to determine whether fragility is due to shortened telomeres, knockdown of ZNF365 was performed in early-passage (P<5) IMR90 primary fibroblasts with sufficiently long telomeres [29]. In IMR90, acute knockdown of ZNF365 caused telomere defects, including heterogeneity and fragility, that were followed by early cellular senescence (Figure 4E). These changes were accompanied by large variations in telomere length (Scr, ranges from 440 to 7507 vs siZNF, 4 to 9867), suggesting telomere phenotypes might occur at the acutely shortened telomeres. These results demonstrate that ZNF365 is essential to prevent defective telomeres regardless of telomere length.
Our observation of increased 53BP1 foci in ZNF365-depleted cells, even in the absence of exogenous genotoxic stress, indicated the presence of widespread DNA damage (Figure 4F). These findings pointed to a potential role for ZNF365 in genomic stability for non-telomeric DNA. Notably, chromosomal breakage in ZNF365-depleted culture increased significantly upon Aph treatment, indicating conversion of stalled replication forks into DNA DSBs (Figure 5A). These increased DSBs point to the involvement of ZNF365 in the maintenance of non-telomeric and low dose Aph-sensitive fragile sites.
Figure 5. Loss of ZNF365 leads to common fragile site expression and aneuploidy.
A. Elevated chromosome breakage in shZNF Hela cells treated with 0.3 μM Aph for 24 h (n=25). The number of chromosome breakage per metaphase (mean±SD) is plotted on the right. Arrows point to representative chromosome breakages. B. Chromosomal aberrations in the FRA16D region. Left, FISH for 16q23.2 showing that the probe maps to the site of a chromosomal fragment originating from FRA16D in Hela shZNF cells treated with Aph. Arrows point to genomic regions with 16q23.2 amplification. Inset shows a representative FRA16D duplication observed in Hela shZNF cells. Right, frequency of aberrations (mean±SD) at 16q23.2 (n=25) in Hela Scr and shZNF cells. C. BLM-coated UFB structure frequently seen in Aph-treated anaphase U2OS cells. U2OS cells were treated with 0.3 μM Aph for 24 h and stained for BLM and counterstained with DAPI. OL, overlay of BLM and DAPI staining. D. FD2-associated BLM-coated bridge structure (pointed by yellow arrow) from Aph-treated ZNF365 depleted U2OS cells. U2OS shZNF cells were treated with 0.3 μM Aph for 24 h and stained for BLM (red) and FD2 (green) and counterstained with DAPI. E. Percentages of mitotic U2OS Scr or shZNF cells with BLM positive UFB alone or FD2 and BLM double-positive UFBare plotted. F. FD2 (red) and telomere (green) IF-FISH staining shows a fraction of telomeres are associated with FD2 foci in interphase nuclei (upper left panel) or FD2 sister foci in mitotic U2OS shZNF cells (lower left panel). Arrows point to double positive foci. G. Percentage of FD2 positive telomere sister foci in mitotic U2OS Scr and shZNF cells (n=17). H. ZNF365 and BLM IF staining shows a fraction of ZNF365 is localized to BLM positive UFBs in Aph-treated mitotic cells. Inset shows the arrow-pointed UFB with ZNF365 foci from an anaphase nucleus. I. ZNF365 partially co-localizes with FD2 sister foci in an anaphase nucleus. Insets show higher magnification of the arrow pointed double positive foci. DAPI-stained DNA structures are shown in blue. J. Spectral karyotyping analysis of metaphase spread from TKO Scr and shZfp cells. The latter displays significant level of aneuploidy. Right, percentages of total metaphases examined (Y-axis) vs. the chromosome number distribution (X-axis) are plotted. K. Schematic summary of proposed model for ZNF365 function. Defective telomeres induce ZNF365 expression by p53 activation. ZNF365 is necessary to suppress the expression of fragile sites including telomeres. In its absence there is increased formation of anaphase DNA bridges due to accumulation of unresolved replication intermediates, occluding cell division sites and inhibiting cytokinesis. Increased cytokinesis failure contributes to aneuploidy and genomic instability.
In order to examine the effect of ZNF365 deficiency on CFS expression, we surveyed the well-characterized genomic region of 16q23.2 (FRA16D). Consistent with our prediction, fragile site expression at 16q23.2 increased significantly in Aph-treated ZNF365-depleted Hela cells compared with control cells (Figure 5B). However, expression at other fragile sites FRA3B (3p14.2) and 12p12.1 was marginal, suggesting a site-specific role of ZNF365 (Figure S5A). Together, our results indicate that ZNF365 is a necessary element in the maintenance of a subset of genomic regions that are sensitive to replication stress, including telomeres.
Lack of ZNF365 increases mitotic UFB and aneuploidy
Next, we determined the outcome of ZNF365 deficiency in mitotic cells. Both Scr and shZNF U2OS cells treated with Aph increased UFBs, as determined by BLM staining positive structures (Figure 5C). Fanconi anemia proteins cooperate in DNA replication and recombination and are activated at stalled replication forks [30, 31]. In particular, persistent FANCD2 (FD2) foci localize to UFBs originating from CFS, including FRA16D [7]. Because knockdown of ZNF365 increased expression of a subset of CFS under replication challenge, we scored and compared the frequency of both FD2-associated and total UFBs. In ZNF365-depleted cells, an increased number of UFBs originated from unresolved DNA replication sites under Aph-induced replicative stress, as evidenced by associated FD2 foci on the structure (Figure 5D, E)[32]. This finding is similar to what has been observed in cells with RAD51 or Fanconi anemia gene depletion that are defective for recombination and repair [7].
To confirm that telomeres are indeed among the unresolved replication intermediates in ZNF365-depleted cells, IF-FISH of FD2 and TelC was performed. In interphase nuclei, FD2 co-localized with multiple TelC-positive foci, some of which formed sister foci in mitotic cells (Figure 5F). The number of FD2- and TelC-positive sister foci in mitotic cells increased, consistent with increased telomere defects in ZNF365-depleted cells (Figure 5G). Furthermore, co-staining with BLM or FD2 detected a fraction of ZNF365 on the UFBs and the mitotic FD2 sister foci (Figures 5H, I) as well as co-localized with TelC and FRA16D (data not shown), suggesting its potential role in resolution of replication intermediates at these sites. Together, our results demonstrate that ZNF365 deficiency suppresses timely resolution of replication intermediates, increasing FD2-associated UFBs.
Interestingly, numerous metaphases from ZNF365-silenced cells exhibited aneuploidy. In particular, cells with p53-deficient backgrounds became aneuploid upon ZNF365 depletion (Figure 5J). There was a concomitant increase in centrosome number and appearance of multinucleated cells in culture, suggesting frequent cytokinesis failure in ZNF365-depleted cells (Figure S5B-C). Similarly, the increased number of anaphase DNA bridges in Fanconi anemia mutant cells correlated with a higher rate of cytokinesis failure, resulting in binucleated cells and increased apoptosis observed in Fanconi anemia patients [33]. As mentioned earlier, the increased presence of anaphase DNA bridges including UFBs may contribute to chromosomal and genomic instability in ZNF365-depleted cells (Figure 3D). It is conceivable that DNA bridges are likely the outcome of failed resolution of stalled and incomplete replication of telomeres and CFS in the absence of ZNF365. Collectively, our results suggest that ZNF365 is necessary for the timely completion of replication in a subset of fragile sites and thus, suppresses cytokinesis failure and aneuploidy.
Discussion
In this study, we explored the p53 transcriptome in the context of telomere dysfunction and identified ZNF365 as a key effector of genome maintenance. Our characterization of ZNF365 provides novel insights into feedback regulation of genomic stability by a telomere dysfunction-induced checkpoint. Our findings gain added significance in light of the observation that the dysfunctional telomere is one of the hallmarks of degenerative and cancer-prone conditions [17].
In cells with telomere dysfunction, one of the most robustly up-regulated genes upon reconstitution of p53 activity was ZNF365.Our studies show that the biology of ZNF365 and p53 are intimately linked, as p53 regulates ZNF365 expression and loss of ZNF365 function activates p53 by exacerbating telomere defects, forging a feedback regulatory loop. This feedback regulatory pattern further corroborates p53's fail-safe action to preserve genomic stability as well as ZNF365's effector action (Figure 5K). Similarly, checkpoint activation in Pot1a-deficient cells promoted bypass of mitosis and endoreplication in p53-null cells, providing a common mechanism for the development of aneuploidy by dysfunctional telomeres [16].
A subset of telomerase-negative cancers utilize an ALT mechanism to maintain telomere length by adding telomeric repeats to chromosome ends [34, 35]. ALT features highly heterogeneous telomere lengths, rapid changes in telomere length and a greatly elevated level of recombination at telomeres [36-38]. Loss of ZNF365 leads to telomere phenotypes partially fulfilling these criteria in both ALT positive U2OS cells and ALT-negative MEFs and normal human fibroblasts. Furthermore, ZNF365 deficient cells did not exhibit upregulation of APBs which are a specialized type of PML bodies that contain telomeric fragments, telomere-binding proteins, and factors involved in recombination, and thus, provide a catalytic surface for telomere recombination [34, 39, 40]. However, it is formally possible that APB reveals only under conditions such as methionine starvation or ALT does not necessarily involve APBs as previously described [41, 42]. A lack of co-localization of telomeres and PML may suggest APBs are not the site for induced T-SCE in ZNF365 knockdown cells and loss of ZNF365 may upregulate ALT through alternative mechanisms. Furthermore, elevated T-SCE in ZNF365 knockdown cells could reflect defects in a repair of stalled replication forks or DNA breaks that maintains telomeres, resulting in recombination and rearrangement. Indeed, our characterization of ZNF365 suggests its potential role in DSB repair and replication fork recovery. We tested whether ZNF365 is involved in the recovery of stalled replication forks induced by hydroxyurea (HU) based on the possibility that increased DSB in knockdown cells is due to the conversion of arrested replication forks. Consistent with our hypothesis, ZNF365 depleted cells showed significantly decreased length of replication fork tract after HU-mediated replication arrest (Figure S6). Our results place ZNF365 at stalled forks where it promotes resolution and recovery of replication. It is noteworthy that defects in stalled fork recovery are not limited to telomeres suggesting a general role of ZNF365 in maintenance of genomic stability.
Unlike premature senescence after many rounds of cell division in WRN- or BLM-defective cells, knocking down ZNF365 causes an early growth arrest, a phenotype seen in cells with an acute loss of TRF1 or TRF2 function [4, 43]. In terms of telomere phenotype, ZNF365-depleted cells did not exhibit significant telomere end-to-end fusion unless there was pre-existing telomere attrition (e.g., TKO, Figure S3B), contrasting with that of TRF2-deleted cells [43]. Based on the similarity to the telomere phenotype of TRF1-deficient cells, defective telomeres seen in ZNF365-deficient cells arise most likely from replication problems. Indeed, telomeres pose a structural hindrance to the machinery of DNA replication and thus may constitute fragile sites where replication fork stalling may significantly increase the rate of recombination [4, 44], as evidenced by increased T-SCE in our study.
Our observation that ZNF365 maintains stability of a specific fragile site (i.e., FRA16D) aside from telomeres suggests that ZNF365 may have sequence-specific interactions with this CFS. A recent study demonstrated that TRF1 specifically stabilized 2q14, a CFS bearing an interstitial telomeric sequence TTAGGG, while it did not affect non-telomeric CFS, including FRA16D [45]. One plausible mechanism would be the presence of a ZNF365 DNA-binding motif within FRA16D and (sub)telomeric genomic regions. As a protein containing a single C2H2 zinc finger domain, ZNF365 homo-dimerizes (data not shown) and possibly tethers to specific DNA sequences. The exact mechanism of how ZNF365 participates in resolution of late-replicating CFS and telomeres warrants further investigation.
A previous study reported that misexpression of ZNF365 led to cytokinesis failure, suggesting that its deregulation may contribute to the development of genomic instability during tumorigenesis [46]. Of high relevance to our findings, recent genome-wide association studies have identified SNPs within ZNF365 that bear significant correlation with breast cancer susceptibility [47]. In particular, an independent study identified SNPs within the same intron as a significant modifier locus to BRCA2-mutant patient susceptibility to breast cancer [20]. It is possible that these SNPs have other relevant functional links apart from affecting ZNF365 expression or function. Nevertheless, our study provides a potential molecular mechanism by which alteration of ZNF365 instigates genomic instability. Whether ZNF365 functionally contributes to breast or other cancer pathogenesis warrants future investigation. A better understanding of this telomere-dysfunction-initiated cellular checkpoint response may illuminate novel therapeutic approaches for neoplastic and degenerative diseases linked to telomere biology.
Materials and Methods
Antibodies and Cell lines
We used antibodies against 53BP1 (Bethyl labs), TRF1 (gift from T. De Lange), FANCD2 (Novous), BLM (C-18; Santa Cruz), v5 (Invitrogen), ZNF365 (clone 3c23), lamin A/C and pericentrin (Abcam). U2OS, Saos-2, Hela, and IMR90 cells were obtained from the American Type Culture Collection (ATCC). Cells were tested for the absence of mycoplasma and authenticated by standard DNA microsatellite short tandem repeats (PowerPlex 1.2 System, Promega) and the resulting DNA fingerprints were matched to the reference published by ATCC.
Time-dependent transcriptome analysis after p53ER activation
Ear skin fibroblasts isolated from G4 mTerc-/- Atm-/- p53-/- TKO mice were used to study the effects of p53 reactivation. Cells were transduced with retroviral particles for pBabe-p53ERT2 (p53ER, provided by Dr. Gerard Evan at UCSF) and both uninfected cells and p53ER-expressing cells were treated with 100nM OHT for multiple time points to catalog the p53 transcriptome. We used the Affymetrix Mouse Genome 430 2.0 Array to analyze total RNA. To identify p53 targets, fold changes were calculated as a ratio of the mean expression values of the OHT-treated p53ER-infected or uninfected samples at 4 hours. For data analysis, the best-route combination of analysis methods was used. Briefly, background and PM/MM corrections were performed by Affymetrix Microarray Suite 5.0, (MAS5.0) and expression summary values for each probe set were calculated in Bioconductor (http://www.bioconductor.org/). In addition, hierarchical clustering was performed in dchip (http://biosun1.harvard.edu/complab/dchip/) for both microarray studies to assess similarities and differences in gene expression among samples. Two-way analysis of variance (ANOVA) was carried out on the expression profiles of all samples to assess the significance of interaction between p53-containing virus infection and OHT treatment, assigning each probe set a p-value. In order to exclude other factors potentially affecting p53 activity, we set the criteria for fold change by p53+4OHT/mock infected+4OHT of greater than 1.5; and this fold change over p53+vehicle/mock infected+vehicle is also greater than 1.5. In addition, fold change of p53+4OHT/mock infected+4OHT is at least 1.2 times over all other comparisons: p53-0 hr/mock infection-0 hr, p53+vehicle/p53-0 hr, mock infected+vehicle/mock infected-0 hr. Microarray data have been deposited to GEO with accession number GSE44599.
Assay for T-SCE
Cells were labeled with 10 μg/ml BrdU for 12 to 16 h and metaphase spreads were used for chromosome orientation fluorescence in situ hybridization (CO-FISH) assay to detect T-SCE. Slides were rehydrated in PBS and treated with 0.5 mg/ml RNase A for 10 min at 37°C followed by staining with 0.5 μg/ml Hoechst 33258 in 2× SSC for 15 min, and exposed to UV light. BrdU-substituted DNA strands were digested with 10 U/μl of Exonuclease III for 10 min. Slides were dehydrated in ethanol series and hybridized with TelG-Cy3 (1:5000, Panagene), for 1 h then further hybridized for 2 h with TelC-FITC (1:500, Bio-Synthesis, Inc.). After hybridization slides were dehydrated in ethanol series, air-dried and mounted.
Knockdown of ZNF365
The RNAi Consortium clone IDs for the shRNAs used in this study are as follows: shZNF-1, TRCN0000128973; shZNF-2, TRCN0000128835; shZfp-7, TRCN0000175966; shZfp-8, TRCN0000176181. To deplete ZNF365 acutely in IMR90 cells, cells were transfected with ZNF siRNA 100 nM (5′- CATACCAGATTTAGAAGCTTGTCAT-3′, Dharmacon) for two rounds, with a 48-h interval, using Lipofectamine 2000. Primer sequences for qRT-PCR analysis of ZNF365 expression are available upon request.
Tel-FISH
Cells were treated with 0.1 μg/ml colcemid and prepared for metaphase spread. Slides were treated with pepsin and RNase, washed and dehydrated through ethanol series and hybridized with a TelG-Cy3 or TelC-FITC PNA probe (1:5000 or 1:500, respectively) by denaturation at 80 °C for 3 min, then incubation for 1 h. Slides were washed with 70% formamide and 0.06XSSC, followed by PBS containing 0.02% (v/v) Tween-20 and mounted. The length of telomeres was quantified with TFL-TELO Software.
Fragile site-FISH
Cells were treated with Aph (0.3 μM) for 24 h. The telomere, 3p14, 12p12 and 16q23 probes were labeled by nick translation using digoxigenin-11-dUTP (Roche) and were detected by rhodamine-conjugated antibody. For fragile site analysis, the chromosomal bands involved in aberrations were matched to the location of published fragile sites. The fused, duplicated, broken aberrant signals were scored.
IF-FISH
Cells grown on coverslips were fixed in 2% paraformaldehyde, permeabilized in 0.2% Triton X-100, and incubated with primary followed by Alexa-conjugated secondary antibodies. The cover slips were fixed in 2% paraformaldehyde and dehydrated in ethanol series. Hybridizing solution containing TelC-FITC (1:500, Bio-Synthesis, Inc.) were dropped on the coverslip and denatured at 70-80ºC followed by hybridization for 2 h. Slides were washed with 70% formamide and 0.06XSSC, followed by PBS containing 0.02% (v/v) Tween-20 and mounted.
TMA analysis
In house constructed TMA sets for different subtypes of breast cancers were stained with monoclonal ZNF365 antibody. Slides were manually scored based on no signal (0), weak (1), moderate (2), strong (3) staining criteria and calculated as H-score as previously described [48]. Significance of difference between H-scores was determined by Non-parametric Wilcoxon Rank Sum Test.
Statistical Analysis
All data represent the average from at least three independent experiments, with at least 100 cells counted per experiment. The unpaired two-tail Student's t-test or one-way ANOVA test with Tukey HSD post-test was used to determine significance for all experiments unless noted otherwise. HR and Log-rank analysis were performed as in [49].
Supplementary Material
Significance.
The contribution of the p53-ZNF365-telomere axis in the suppression of genomic instability illuminates how alterations in this pathway may confer increased cancer risk for individuals harboring germline alterations in the ZNF365 locus.
Acknowledgments
The authors thank Drs. John Petrini, Dipanjan Chowdhury, and Pengbo Zhou for helpful comments, Drs. Gerard Evan, Titia De Lange, and Miguel Mosquera for p53-ERT2 plasmid, TRF1 antibody, and CFS probes, respectively. The authors also acknowledge Ms. T. McDonald for technical assistance and the Weill Cornell Clinical and Translational Science Center for editorial support (UL1 TR000457-06). This work and R.A.D. is supported by R01CA84628 and U01CA141508 grants from the NIH National Cancer Institute and the Belfer Foundation. R.A.D. is supported by an American Cancer Society Research Professorship. J-H. P is supported by the Weill Cornell Medical College, the Ellison Medical Foundation (AG-NS-0646-10) and the Sidney Kimmel foundation (SKF-092).
Financial support: R.A.D. is supported by NIH National Cancer Institute (R01CA84628, U01CA141508), and an American Cancer Society Research Professorship. J-H. P is supported by the Weill Cornell Medical College, the Ellison Medical Foundation (AG-NS-0646-10) and the Sidney Kimmel foundation (SKF-092).
Abbreviation
- CFS
common fragile sites
- TNBC
triple negative breast cancer
- ABI
anaphase bridge index
- DSBs
DNA double-strand breaks
- Aph
aphidicolin
- 4-OHT
4-hydroxytamoxifen
- PML
promyelocytic leukemia
- T-SCE
telomere sister chromatid exchange
- UFB
ultra-fine DNA bridges
- ALT
alternative lengthening of telomere
- APB
ALT-associated PML body
- HU
Hydroxyurea
- FD2
FANCD2
Footnotes
Author Contribution: Y.Z., D.L., E.I., F.F., H.Y., and H.Z. performed experiments. Y.Z. E.I., Y.X., A.P., S.S., Z. C., and J.P. analyzed data. Y.Z., R.D., and J.P. designed research and wrote the manuscript.
Conflict of Interest: None
References
- 1.De Lange T. Telomere-related genome instability in cancer. Cold Spring Harbor symposia on quantitative biology. 2005;70:197–204. doi: 10.1101/sqb.2005.70.032. [DOI] [PubMed] [Google Scholar]
- 2.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–34. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 3.Ye J, Lenain C, Bauwens S, Rizzo A, Saint-Leger A, Poulet A, et al. TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell. 142:230–42. doi: 10.1016/j.cell.2010.05.032. [DOI] [PubMed] [Google Scholar]
- 4.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Opresko PL, Mason PA, Podell ER, Lei M, Hickson ID, Cech TR, et al. POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. The Journal of biological chemistry. 2005;280:32069–80. doi: 10.1074/jbc.M505211200. [DOI] [PubMed] [Google Scholar]
- 6.Barefield C, Karlseder J. The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic acids research. 2012;40:7358–67. doi: 10.1093/nar/gks407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nature cell biology. 2009;11:753–60. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 8.Williams ES, Klingler R, Ponnaiya B, Hardt T, Schrock E, Lees-Miller SP, et al. Telomere dysfunction and DNA-PKcs deficiency: characterization and consequence. Cancer research. 2009;69:2100–7. doi: 10.1158/0008-5472.CAN-08-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nature cell biology. 2006;8:885–90. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- 10.Wong KK, Maser RS, Bachoo RM, Menon J, Carrasco DR, Gu Y, et al. Telomere dysfunction and Atm deficiency compromises organ homeostasis and accelerates ageing. Nature. 2003;421:643–8. doi: 10.1038/nature01385. [DOI] [PubMed] [Google Scholar]
- 11.Maser RS, Wong KK, Sahin E, Xia H, Naylor M, Hedberg HM, et al. DNA-dependent protein kinase catalytic subunit is not required for dysfunctional telomere fusion and checkpoint response in the telomerase-deficient mouse. Molecular and cellular biology. 2007;27:2253–65. doi: 10.1128/MCB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–5. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 13.Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ, et al. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–38. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 14.Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–5. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 15.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davoli T, Denchi EL, de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141:81–93. doi: 10.1016/j.cell.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nature reviews Molecular cell biology. 2010;11:220–8. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 18.Myung K, Datta A, Kolodner RD. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell. 2001;104:397–408. doi: 10.1016/s0092-8674(01)00227-6. [DOI] [PubMed] [Google Scholar]
- 19.Arlt MF, Casper AM, Glover TW. Common fragile sites. Cytogenetic and genome research. 2003;100:92–100. doi: 10.1159/000072843. [DOI] [PubMed] [Google Scholar]
- 20.Gaudet MM, Kirchhoff T, Green T, Vijai J, Korn JM, Guiducci C, et al. Common genetic variants and modification of penetrance of BRCA2-associated breast cancer. PLoS genetics. 2010;6:e1001183. doi: 10.1371/journal.pgen.1001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antoniou AC, Kuchenbaecker KB, Soucy P, Beesley J, Chen X, McGuffog L, et al. Common variants at 12p11, 12q24, 9p21, 9q31.2 and in ZNF365 are associated with breast cancer risk for BRCA1 and/or BRCA2 mutation carriers. Breast cancer research : BCR. 2012;14:R33. doi: 10.1186/bcr3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christophorou MA, Martin-Zanca D, Soucek L, Lawlor ER, Brown-Swigart L, Verschuren EW, et al. Temporal dissection of p53 function in vitro and in vivo. Nature genetics. 2005;37:718–26. doi: 10.1038/ng1572. [DOI] [PubMed] [Google Scholar]
- 23.Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, Rodriguez EG, et al. In situ analyses of genome instability in breast cancer. Nature genetics. 2004;36:984–8. doi: 10.1038/ng1409. [DOI] [PubMed] [Google Scholar]
- 24.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nature reviews Cancer. 2004;4:814–9. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 25.Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 26.Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 27.Espejel S, Franco S, Sgura A, Gae D, Bailey SM, Taccioli GE, et al. Functional interaction between DNA-PKcs and telomerase in telomere length maintenance. The EMBO journal. 2002;21:6275–87. doi: 10.1093/emboj/cdf593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan SW, Blackburn EH. Telomerase and ATM/Tel1p protect telomeres from nonhomologous end joining. Molecular cell. 2003;11:1379–87. doi: 10.1016/s1097-2765(03)00174-6. [DOI] [PubMed] [Google Scholar]
- 29.Henderson S, Allsopp R, Spector D, Wang SS, Harley C. In situ analysis of changes in telomere size during replicative aging and cell transformation. The Journal of cell biology. 1996;134:1–12. doi: 10.1083/jcb.134.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Molecular cell. 2001;7:249–62. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 31.D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nature reviews Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 32.Chan KL, Hickson ID. On the origins of ultra-fine anaphase bridges. Cell Cycle. 2009;8:3065–6. doi: 10.4161/cc.8.19.9513. [DOI] [PubMed] [Google Scholar]
- 33.Vinciguerra P, Godinho SA, Parmar K, Pellman D, D'Andrea AD. Cytokinesis failure occurs in Fanconi anemia pathway-deficient murine and human bone marrow hematopoietic cells. The Journal of clinical investigation. 2010;120:3834–42. doi: 10.1172/JCI43391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nature reviews Genetics. 2010;11:319–30. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 35.Hu J, Hwang SS, Liesa M, Gan B, Sahin E, Jaskelioff M, et al. Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell. 2012;148:651–63. doi: 10.1016/j.cell.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. The EMBO journal. 1995;14:4240–8. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perrem K, Colgin LM, Neumann AA, Yeager TR, Reddel RR. Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Molecular and cellular biology. 2001;21:3862–75. doi: 10.1128/MCB.21.12.3862-3875.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Londono-Vallejo JA, Der-Sarkissian H, Cazes L, Bacchetti S, Reddel RR. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer research. 2004;64:2324–7. doi: 10.1158/0008-5472.can-03-4035. [DOI] [PubMed] [Google Scholar]
- 39.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nature genetics. 2000;26:447–50. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 40.Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer research. 1999;59:4175–9. [PubMed] [Google Scholar]
- 41.Jiang WQ, Zhong ZH, Henson JD, Reddel RR. Identification of candidate alternative lengthening of telomeres genes by methionine restriction and RNA interference. Oncogene. 2007;26:4635–47. doi: 10.1038/sj.onc.1210260. [DOI] [PubMed] [Google Scholar]
- 42.Cerone MA, Autexier C, Londono-Vallejo JA, Bacchetti S. A human cell line that maintains telomeres in the absence of telomerase and of key markers of ALT. Oncogene. 2005;24:7893–901. doi: 10.1038/sj.onc.1208934. [DOI] [PubMed] [Google Scholar]
- 43.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nature cell biology. 2005;7:712–8. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 44.Glover TW, Stein CK. Induction of sister chromatid exchanges at common fragile sites. Am J Hum Genet. 1987;41:882–90. [PMC free article] [PubMed] [Google Scholar]
- 45.Bosco N, de Lange T. A TRF1-controlled common fragile site containing interstitial telomeric sequences. Chromosoma. 2012;121:465–74. doi: 10.1007/s00412-012-0377-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Q, Du X, Meinkoth J, Hirohashi Y, Zhang H, Liu Q, et al. Characterization of Su48, a centrosome protein essential for cell division. Proceedings of the National Academy of Sciences of the United States of America; 2006; pp. 6512–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindstrom S, Vachon CM, Li J, Varghese J, Thompson D, Warren R, et al. Common variants in ZNF365 are associated with both mammographic density and breast cancer risk. Nature genetics. 2011;43:185–7. doi: 10.1038/ng.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bacus S, Flowers JL, Press MF, Bacus JW, McCarty KS., Jr The evaluation of estrogen receptor in primary breast carcinoma by computer-assisted image analysis. American journal of clinical pathology. 1988;90:233–9. doi: 10.1093/ajcp/90.3.233. [DOI] [PubMed] [Google Scholar]
- 49.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast cancer research and treatment. 2010;123:725–31. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.