Abstract
Hepatitis C virus (HCV) persists in the majority of those infected despite host immune responses. Evidence has accrued that selectively fixed mutations in the envelope genes (E1 and E2) are associated with viral persistence, particularly those that occur within the first hypervariable region of E2 (HVR1). However, the individual amino acid residues under selection have not been identified, nor have their selection pressures been measured, despite the importance of this information for understanding disease pathogenesis and for vaccine design. We performed a high-resolution analysis of published gene sequence data from individuals undergoing acute HCV infection, employing two phylogenetic methods to determine site-specific selection pressures. Strikingly, we found a statistically significant association between the number of sites selected and disease outcome, with the fewest selected sites in fulminant HCV cases and the greatest number of selected sites in rapid progressors, reflecting the duration and intensity of the arms race between host and virus. Moreover, sites outside the HVR1 appear to play a major role in viral evolution and pathogenesis, although there was no association between viral persistence and specific mutations in E1 and E2. Our analysis therefore allows fine dissection of immune selection pressures, which may be more diverse than previously thought. Such analyses could play a similarly informative role in studies of other persistent virus infections, such as human immunodeficiency virus.
Hepatitis C virus (HCV) is a major health problem worldwide, infecting an estimated 170 million people (34). Although a minority spontaneously clear the virus, HCV persists in approximately 80% of those infected, causing liver inflammation. A significant proportion of these individuals go on to develop liver cirrhosis and hepatocellular carcinoma; the annual death toll due to HCV in the United States is projected to exceed that of AIDS in the next few years (5). Conversely, acute HCV illness is often asymptomatic and patients rarely present in this stage. Hence, there is only a limited understanding of early disease, despite the general view that the initial immune response may be critical in determining whether a persistent infection is established (29).
Several studies have documented strong and broad cellular immune responses, mediated by CD4+ and CD8+ T cells, in acute-phase patients who go on to resolve the disease (6, 14, 15, 29). In persistent (chronic) infection, however, such responses are generally weak. It is therefore possible that differences in the initial interaction between HCV and the cellular immune response determine outcome. The role of antibodies at this time is also unclear. While they may offer some protection, their production does not prevent progression to persistent disease and antibodies are found in the sera of chronically infected patients (3). The role of cellular versus humoral responses in determining both initial control and long-term progression of the disease, and whether the virus evolves to escape these responses, has yet to be clearly established.
HCV is a member of the Flaviviridae, a family of positive-sense RNA viruses that evolve rapidly due to high rates of mutation and replication in the absence of a proofreading polymerase (12). It exists as six genotypes (each containing multiple subtypes), which differ in geographical distribution (26). Its 9.6-kb genome encodes a single polyprotein, which is cleaved into 6 nonstructural and 3 structural proteins: the core, plus two envelope glycoproteins, E1 and E2. These form a heterodimer, which is likely to mediate attachment to hepatocytes (and potentially other cells), and is targeted by antibodies. The N terminus of E2 contains the hypervariable region 1 (HVR1), which varies markedly between isolates and is thought to contain an immunodominant linear B-cell epitope recognized by neutralizing antibodies (8, 10, 23, 24). It has been proposed that escape mutations in HVR1 evade the limited cross-reactivity of the antibody responses and play a key role in the establishment of persistent infection (2, 20). This is supported by the finding that hypogammaglobulinemic patients, who lack a functional humoral immune system and the associated antibody selection pressure, show far lower rates of nucleotide substitution in HVR1 than do control patients (2).
More evidence for the action of natural selection on HVR1 is that there are often proportionally more nonsynonymous (dN) than synonymous (dS) substitutions per site. Several studies have examined the dN/dS ratio from patients with different disease outcomes and found that patients with persistent disease tend to have a higher dN/dS ratios in HVR1 than patients who resolved infection, suggesting the action of continual immune-driven positive selection (9, 17, 19). Ray et al. (19) sequenced clonal variants from the first PCR-positive sample of patients whose eventual outcome (clearance versus persistence) was known. The E1 and N-terminal half of E2 (326 amino acids) were amplified in 10 persisters and 5 resolvers. Using a sliding window analysis, persistence was associated with a higher dN/dS ratio in HVR1, while clearers seemed to show a high dN/dS ratio in the E1 region that spanned amino acids 290 to 340 (all sites are given as amino acid positions from the start of the translated region, taking the beginning of HVR1 as amino acid 384). Similarly, Farci et al. (9) analyzed E1E2 sequences, 186 amino acids in length, from 2 patients with fulminant hepatitis, 3 patients who spontaneously resolved the infection, 3 slow progressors, and 3 rapid progressors. In all cases, the sequences were longitudinally sampled for several months of acute disease. Their results suggested that progressive disease was accompanied by a higher dN/dS ratio, especially in HVR1, which correlated temporally with antibody seroconversion, such that the humoral immune system was proposed as the driving force for viral adaptation.
Although informative, the analyses of selection pressures in HCV undertaken to date do not allow identification of specific amino acid sites that are under positive selection; hence, the role of individual sites in immune escape cannot be explicitly determined. Further, the interpretation of a single dN/dS value for a whole gene region is confounded by the action of negative selection on many sites. To address these issues for the first time in HCV, we employed a phylogenetic analysis of site-specific selection pressures. This allowed us to determine the particular sites involved in immune escape and assess how the intrahost evolution of HCV relates to disease outcome.
MATERIALS AND METHODS
Sequence data.
Sequences from the Ray et al. (19) and Farci et al. (9) data sets, covering the E1-E2 region, were downloaded from GenBank; after removal of identical sequences, 63 sequences of 978 bp and 380 sequences of 558 bp remained, respectively. These sequences were aligned manually by using the program SE-AL (http://evolve.zoo.ox.ac.uk).
Phylogenetic and selection analysis.
Maximum-likelihood (ML) phylogenetic trees were estimated by using the PAUP* package (28a). In all cases, the Hasegawa-Kishino-Yano (HKY85) model of nucleotide substitution was used, with the ratio of transitions to transversions, as well as a gamma distribution of rate variation among sites (with 4 rate categories), estimated from the data. All parameter values are available from the authors upon request. Potential positively selected sites were identified by using two phylogenetic methods; an ML approach, utilizing the CODEML program from the PAML package (35), and a parsimony approach undertaken by using the MacClade program (16a). The cross-sectional (i.e., single time point) nature of the Ray et al. data made it suitable for the ML analysis alone, employed separately on the progressors and resolvers. There were insufficient sequences per patient to allow further resolution. In contrast, the longitudinal (i.e., multi-time point) Farci et al. data was analyzed by both approaches, as the greater number of sequences per patient allowed each to be considered separately.
The ML method implemented in the CODEML program fits various models of codon evolution to sequence data connected by a phylogenetic tree and considers selection pressures at individual codon sites. The models of codon evolution differ in their distribution of dN/dS values among codons. For simplicity, we employed two models: M7, which assumes a beta distribution with 10 categories of dN/dS across codons, but where the dN/dS ratio is constrained to be <1 in each category, so that the model only specifies neutral evolution, and M8, which differs from M7 only in that an extra class of codons is added to account for positive selection (i.e., dN/dS >1) (35). As these two models are nested, their likelihoods can be compared by using a likelihood ratio test, with significant evidence for positive selection provided if M8 rejects M7 and contains a class of codons with a dN/dS ratio of >1. An empirical Bayesian approach, also available in CODEML, was then employed to identify individual codons subject to positive selection, with a posterior probability of >95% taken to indicate positive selection unless otherwise stated.
In the case of the Farci et al. (9) data, where longitudinally sampled sequences from individual patients are available, we also employed a parsimony approach to analyze selection pressures. This approach is favored over the dN/dS ratio analysis in this case because it considers the change in allele frequencies through time, which can only be observed in longitudinal data, rather than assuming that all amino acid changes are fixed in the population. To perform this analysis, an ML phylogeny was estimated (as described above) and all amino acid changes were mapped on this tree by using the parsimony algorithm implemented in the MacClade program. Sites had to fulfill one of two criteria to be deemed candidates for positive selection: (i) there were synapomorphic changes, occurring on the internal branches of the tree, which indicates that they have been transmitted through the population, possibly because they represent a fitness increase, or (ii) the site showed mutation to the same amino acid on more than one terminal branch, which is likely to indicate a convergent selective change that has arisen independently in multiple lineages.
RESULTS
Analysis of the Ray et al. data set.
Ray et al. (19) found that progressors had an elevated dN/dS ratio in HVR1, whereas resolvers displayed a higher dN/dS ratio in E1. Our ML analysis revealed that both progressors and resolvers showed evidence for positive selection. Specifically, the M8 model was significantly favored over M7 (P < 0.001) in all cases, and several positively selected sites were identified in each subset. Moreover, we found that the majority (8 of 9) of residues identified as under selection in both progressors and resolvers were located within HVR1, whereas only one site was identified as selected outside this region. Therefore, rather than the whole HVR1 being under positive selection, a few sites have a dN/dS ratio that is significantly greater than 1 while the majority are conserved. This supports the widely held view that although amino acid replacements are accepted at some sites, others are subject to relatively strong selective constraints (27).
Crucially, our analysis also indicated that selection pressures may differ according to disease outcome. Specifically, resolvers exhibited fewer selected sites in E1-E2, with only two selected sites at a 95% posterior probability level, while progressors have seven such sites (Fig. 1a). It is also possible that different residues are selected depending on disease outcome, as the two putative selected sites in resolvers (HVR1 residues 11 and 21) were not identified at the 95% level in those that developed persistent disease. Finally, Ray et al. found that resolvers had a high dN/dS ratio in a region of E1 which was even stronger than that in HVR1 from progressors. In our analysis, this selection in E1 was in fact restricted to two sites. Both selected sites were of marginal significance (positions 301 [P = 0.917] and 334 [P = 0.8696]).
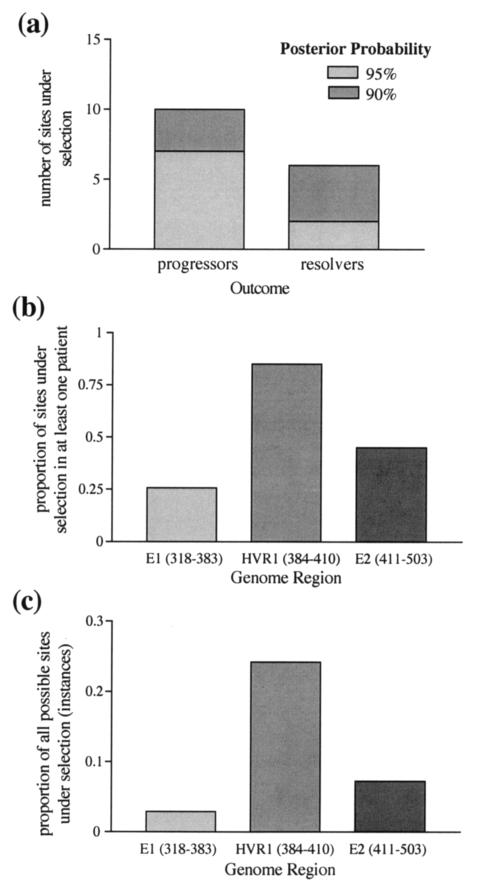
FIG. 1.
Summary of the estimated number of selected sites in each data set. (a) Ray et al. (19) data set. The figure shows the number of sites with a >90 or >95% posterior probability of being selected, estimated by using CODEML. More selected sites were found in sequences from patients with progressing HCV infection. (b and c) Farci et al. (9) data set. (b) Proportion of codons under selection in at least one patient. (c) Proportion of instances (number of patients × number of codons) under selection. There is greater evidence for positive selection in HVR1 than in the surrounding regions. Selected sites were identified by using both MacClade and CODEML.
Analysis of the Farci et al. data set.
In the Farci et al. study (9), an important difference was seen between the selection pressures in HVR1 and those in the surrounding region, and between patients who resolved disease as opposed to those who developed progressive hepatitis, suggesting stronger host immune selection pressure in the HVR1 of persisters. Our ML analysis identified selected sites in 7 of the 11 patients (P4, P5, P7, P9, P10, P11, and P12), whereas MacClade detected selection in every patient, although the number of sites identified differed markedly between patients, ranging from 1 to 33 residues. Consequently, CODEML did not identify many of the putative selected sites detected by MacClade. In sum, 167 selected sites were detected by either method from all 11 patients studied; of these, MacClade identified 165 sites and CODEML identified 45 sites.
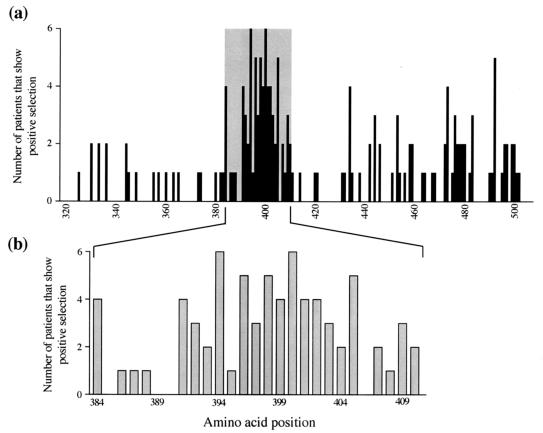
With these results in hand, we used the Farci et al. data to address three crucial questions regarding adaptive evolution in HCV. (i) Is HVR1 under stronger positive selective pressure than the rest of E1 and E2? (ii) Which sites are selected? (iii) Is there any correlation between disease outcome and the strength of selection? With respect to the first question, our analysis confirms that HVR1 indeed exhibits a greater proportion of selected sites (Fig. 1b). Similarly, HVR1 also contains more instances of selection (Fig. 1c). This refers to the fact that multiple patients often showed evidence of selection at the same site and is, in effect, the sum of all selected sites in all patients. Some sites are also more subject to positive selection than others. To assess site-specific selection pressures, we determined the frequency with which each site is selected across all 11 patients. This analysis revealed that few patients display selected sites in E1, whereas HVR1 shows many highly selected sites in many patients, and there is some spatial clustering of selection in E2 (Fig. 2a). A more detailed examination of HVR1 reveals a mix of positively and negatively selected sites (Fig. 2b). Specifically, sites in the central region of HVR1 are more often subject to adaptive evolution while sites 385T, 389G, 390G, and 406G are not selected in any patient and are almost invariant, suggesting that they are of great functional importance.
FIG. 2.
Site-by-site analysis of positive selection in the Farci et al. (9) data set. Selected sites were identified by using both MacClade and CODEML. (a) Number of patients that showed positive selection at each codon position. The shaded area represents HVR1. E1 is to the left of this region, and E2 is to the right. (b) A close-up of the results for HVR1, revealing heterogeneous selection among sites within the HVR.
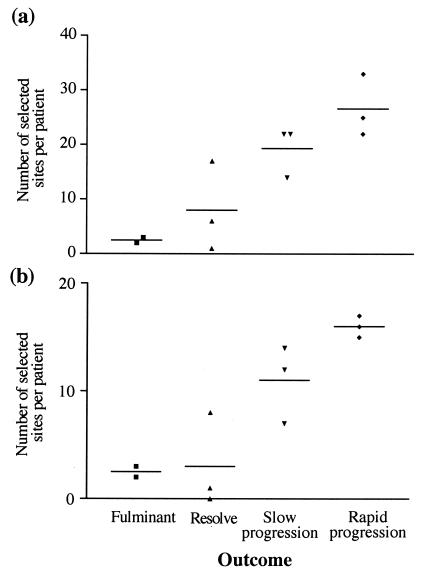
By far the most striking observation from our analysis of the Farci et al. data set was the strong correlation between the number of selected sites per patient and the outcome of disease. When the whole E1-E2 region is analyzed, the four different disease outcomes have statistically significant differences in the number of selected sites per patient (P = 0.0087, one way analysis of variance), according to the following order: fulminant patients < resolvers < slow progressors < rapid progressors (Fig. 3a). However, analysis of HVR1 alone gives no correlation with outcome (P = 0.1143). Although this lack of correlation between disease endpoint and HVR1 evolution could reflect the short length of sequence, it is striking that greatest significance is seen if HVR1 is excluded from the analysis (P = 0.0035) (Fig. 3b). This implies that it is the region outside HVR1 that is most closely correlated with outcome and that inclusion of HVR1 may even dilute the analysis. Finally, there was no correlation between disease outcome and specific selected sites, as no sites showed an association with any one outcome.
FIG. 3.
Relationship between disease progression and the number of putative selected sites identified in each patient for the Farci et al. (9) data set. Sites were identified by using both MacClade and CODEML. (a) Number of selected sites per patient in the whole region sequenced. (b) Number of selected sites per patient in E1 and E2, excluding HVR1.
DISCUSSION
The HCV envelope glycoproteins E1 and E2 are exposed to antibody attack before entry into the host cell. Although the viral population within individual patients exhibits considerable diversity in these proteins, this in itself is not indicative of adaptation to the immune system; not only could a site have hitchhiked to fixation, giving a false signal of positive selection but it may also simply reflect a region under weak selective constraint. Analysis of sequences on a site-by-site basis allows selection pressures to be examined at a greater resolution, avoiding the problems faced by bulk analyses, namely that individual targets of selection cannot be identified and that nearby sites with few selective constraints may eclipse adaptive evolution at single codons. Although the absence of well-established in vitro neutralization assays for HCV make it difficult to prove immune escape, this can be clarified by correlation of selected sites with regions that interact with the host, either through the immune response or via cell surface receptors.
Distribution of selected sites in E1-E2.
In both data sets, we found that positive selection was concentrated within HVR1, suggesting that it is subject to a high level of immune pressure. Several studies have documented the existence of an immunodominant B-cell epitope within HVR1, between amino acids 398 to 410 (10, 24), so antibody-mediated selection pressure could be the main selective force. The C terminus of HVR1 is also recognized by CD4+ T cells, which may provide help for antibody production (25). Hence, the positive selection we demonstrate may facilitate viral escape from antibody responses, although HCV can readily establish a chronic infection in the face of continual production of neutralizing antibodies (3). HVR1 may therefore act as a decoy, generating a strong but ineffective antibody response that provides the selective force for viral adaptation but is unable to achieve clearance (19). A second hypervariable region, HVR2, is thought to be located upstream, within amino acids 474 to 482, in viruses of genotype 1b. However, amino acids in this region showed no evidence for positive selection.
The remainder of E2 showed widespread selection, with numerous sites identified by multiple patients in the Farci et al. data set. Selected sites are less frequent in E1. This may be explained partly by the poor ability of E1 to act as an immunogen (11), so that it may be subject to less immune pressure. An analogous distribution of selection pressures has been documented in the human immunodeficiency virus (HIV) type 1 env gene. A study which used site-specific analytical methods on sequences from HIV type 1-infected patients with different outcomes also found selected sites throughout the gene, in addition to detecting a cluster of positively selected sites within the epitope-rich V3 region (21).
Selection pressures in E1-E2.
There may be multiple selection pressures acting on E1-E2. For example, selection may arise from as yet uncharacterized epitopes, and it seems likely that conformational antibody targets exist outside HVR1 (31). Detailed knowledge of the structure of these proteins would reveal whether the selected sites occur in regions that are more exposed. However, the heterodimerization of the E1 and E2 means that it has so far proved impossible to express these proteins in vitro for structural investigation.
Specific virus-cell receptor interactions may also result in strong selection. Although interaction with the tetraspanin CD81 receptor does not seem to mediate viral entry (20), binding of E2 to CD81 may have an important immunomodulatory role. E2-CD81 ligation on T cells reduces the threshold for their activation, resulting in enhanced proliferation and cytokine production, potentially contributing to liver damage (31). E2-mediated CD81 cross-linking can also result in the inhibition of interferon, which inhibits viral replication and skews the immune response to a Th1 profile (30). The human scavenger receptor class B type 1 has also been identified as a putative receptor for HCV entry into host cells (22). This protein belongs to the CD36 superfamily and is highly expressed on hepatocytes, potentially accounting for the observed liver tropism. E2 also binds the C-type lectins DC-SIGN and DC-SIGNR; although not expressed on hepatocytes, these are likely to be attachment factors for dendritic cells that may increase infection efficiency (18). E2 binding sites for these receptors may therefore be under selection pressure to optimize binding affinity and so facilitate immunomodulation and cell entry.
It is also possible that selection pressures in E1-E2 are related to glycosylation. The HCV envelope proteins are highly glycosylated, with 5 potential N-linked glycosylation sites in E1 and 11 potential sites in E2. Glycosylation can limit the antibody response to E1 due to the masking of potential epitopes by steric hindrance (11). Selection for escape from neutralizing antibodies could therefore lead to mutation of N-linked glycosylation sites, resulting in the repeated rearrangement of the surface sugar moieties (33). This mechanism, dubbed the glycan shield, would provide evolving protection against the binding of neutralizing antibodies and has recently been postulated to be important in HIV escape, after mutation of N-linked glycosylation sites was found to be more common than variation at epitopes (32). This would allow immune escape but avoid excessive variation due to selection pressure at the epitopes or receptor binding regions themselves, which may affect viral fitness. However, we found no evidence of selection for either acquisition or loss of N-linked glycosylation sites, indicating that in these two data sets at least, there is no evolution of the glycan shield.
Finally, despite the strong selection observed in HVR1, some sites appear to be structurally conserved. The restricted pattern of amino acid replacement at some sites corresponds to those documented as largely invariant in a review of almost 300 sequences (27), although two of these generally conserved sites were selected by patients from the Farci et al. (9) study (positions 20 and 26 of HVR1). These constraints suggest a structural role for HVR1 mediated by the ability to maintain E2 receptor recognition (22). Variation within this framework may not only facilitate immune escape but may also have an immunomodulatory role.
Viral evolution and disease outcome.
Both data sets reveal a strong correlation between the outcome of disease and the number of selected sites. This association is particularly apparent in the Farci et al. data, where patients have been differentiated into four disease endpoints. However, only the Ray et al. data pointed toward the selection of different sites according to outcome, although this may be a function of the smaller sample size.
Analysis of the whole region sequenced by Farci et al. (9) indicates that the four categories of disease outcome show significantly different numbers of selected sites, with fulminant patients possessing the least number of sites, clearers showing an intermediate number of sites, and slow and rapid progressors displaying a high frequency of selected residues. While analysis of HVR1 alone does not yield a significant result, exclusion of this region reveals that the number of selected sites in E1 and E2 outside HVR1 is still significantly associated with disease outcome. This implies that although selection is less condensed outside HVR1, it is the remainder of the envelope proteins that may provide insight as to reasons behind the different outcomes.
Why is there a correlation between viral adaptation and disease outcome? It has been suggested that fulminant disease occurs when a strong cytotoxic T lymphocyte (CTL) immune response encounters a large number of infected hepatocytes, causing massive liver damage in the process of removing the virus. This has been quantitatively assessed in the murine model of lymphocytic choriomeningitis virus (7). It is possible that rapid viral replication kinetics are initially established due to a weak innate immune response early in infection, in turn followed by a CTL response which is responsible for the immunopathology; a balanced immune response may thus be a crucial feature of a successful outcome (13). It is unlikely that a large viral load per se directly causes pathology, as has been previously suggested (28): the determinants of immunopathology are a complex interaction between the dynamics and distribution of the host immune response and local viral replication (1). The scarcity of selected sites in these patients therefore corresponds with a very short period of selection: any virus-host arms race would be of short duration due to a rapidly emerging adaptive immune response that removes the virus too quickly for significant selection to occur. In contrast, resolvers show a number of selected sites, compatible with some degree of selection before the host immune system wins, eliminating the virus.
It is also notable that progressors have more selected sites than either resolvers or fulminant patients and that those who progress rapidly to disease have more selected sites than those who progress more slowly. This suggests that slow progressors stimulate a weaker host selective response than fast progressors and so are under less pressure to fix mutants of increased fitness, whereas rapid progressors experience strong and lasting selection pressure, resulting in an intense arms race requiring multiple amino acid changes. The host selective pressure may be immunological, mediated by both cellular and humoral components, or may be generated independently, for example due to the potential increase in viral fitness by altering binding affinity or immunomodulatory effects.
If adaptive evolution outside HVR1 is driven by immune selection pressure, there are two hypotheses for how the variation seen relates to disease outcome. If selection were largely antibody mediated, the humoral response would dictate whether a patient progresses to persistent disease. The spectrum of disease outcomes could then be due to interpatient variability in either the antibody quality or duration of selection. From our study, it seems more likely to be the latter, as there was no evidence that resolvers recognize different epitopes from those where virus persists: the outcomes were not separated on the basis of which sites were selected, indicating that patients are targeting similar viral constituents. Thus, a highly effective antibody response may enable some patients to resolve. In turn, this generates fewer selected sites than seen in persisters because the arms race acts over a shorter period. Alternatively, outcome may depend predominantly on cellular mechanisms; this seems very plausible given the apparent importance of the early CD4 helper and CTL response (15). Antibodies exert some selection pressure on envelope proteins but may be unable to control infection without T-cell assistance. Failure of the cellular immune system to remove the virus due to initial inadequacy or viral escape may itself result in persistence; if the battle is already lost, further antibody responses, although generating selective pressure, may be ineffective in achieving clearance. This may explain why we continue to see humoral responses during chronic infection. Resolution, on the other hand, could be achieved if the early T-cell response is broad and multispecific, while also providing help for the antibody response (13). Similar results have been observed in the murine lymphocytic choriomeningitis virus model (4). In future experiments it will be important to match sequence changes with both the cellular and humoral responses.
Lastly, it is possible that the relationship between the frequency of selection across the envelope proteins and disease outcome is in part due to differences in sampling, as more sequences have been taken from patients who progressed. However, the correlation between the number of selected sites identified and the number of sequences analyzed may itself be artifactual, as samples containing variant viruses would have been available for a longer period in patients with progressive hepatitis and, hence, more sequences were obtainable. This can only be clarified by analysis of sequences in an experiment with a similar number of patients of each outcome, over multiple time points, and with an equal number of sequences collected over the same duration of infection.
Detecting viral adaptation.
The use of two methods for analyzing selection pressures in the Farci et al. (9) data set not only makes our study more robust but it also allows a methodological comparison between the two approaches. The ML approach (CODEML program) is valuable in that it gives a quantitative measure of the strength of selection at each site in the form of a dN/dS ratio. However, it is not ideal for intrapatient data from single patients, as in the Ray et al. (19) data, as these contain sequence polymorphisms rather than just those changes that have reached fixation, so the dN component will also contain transient deleterious mutations. Similarly, it will fail to detect selective sweeps between time points in longitudinally sampled data, as in the Farci et al. (9) data, if this selection involves only small numbers of nonsynonymous changes that are swamped by later synonymous substitutions. The method is therefore overly conservative, and the selected sites identified form a subset of those found by the parsimony analysis (MacClade program), which takes into account the dynamics of allele fixation.
Patient 8 of the Farci et al. (9) data provides an important illustration of the inadequacies of CODEML in the analysis of intrapatient data. This method overlooks 13 codon positions that have an amino acid change in the fourth and final time points only, which may indicate the fixation of advantageous mutations, and which are identified as selected by the MacClade analysis. It is also noteworthy that there is a single amino acid insertion immediately before HVR1 in the first three time points only. This indicates that there was a selective sweep between the third and fourth time points which resulted in the removal of viral genotypes with the insertion and the establishment of a very different viral population. It seems highly unlikely that this population could have evolved from the viral clones sequenced from previous time points due to the many unique substitutions. Therefore, the patient either experienced two separate transmission events or the second strain was at a very low frequency initially, rendering it undetectable until other variants were cleared by the immune system, allowing it to proliferate. Either scenario would indicate the existence of little cross-reactivity between strains.
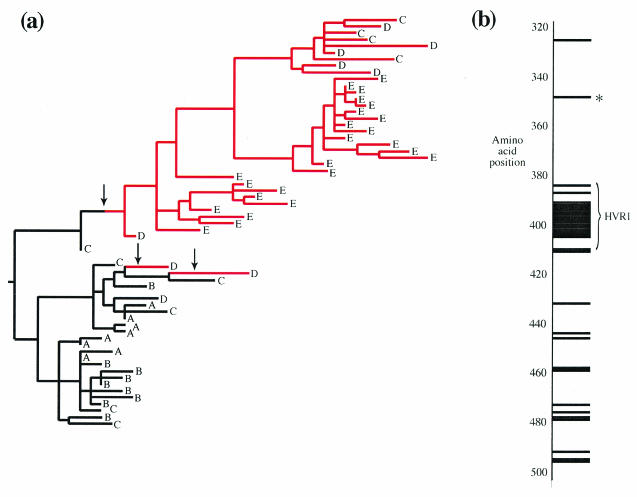
Conversely, the MacClade analysis can be used to detect both multiple amino acid changes across patients and intrapatient selective sweeps. However, it is a qualitative approach and requires arbitrary definition of what constitutes a selected site. As an example, we present the MacClade results from patient 10, a rapid progressor. The reconstruction of a single amino acid change (I-348-V) onto the ML phylogenetic tree (Fig. 4a) illustrates how a mutation may arise and rapidly spread to fixation. The mutation appears at least twice between time points C and D, although all time point E sequences are descended from only one of these occurrences, suggesting that this ancestor had a superior genetic background. Figure 4b shows the distribution of selected sites across the region sequenced. Again, CODEML detects only a subset of the sites identified by MacClade (18 of 33).
FIG. 4.
Viral adaptation in patient 10 of the Farci et al. (9) data set. Samples from this patient, who rapidly progressed to chronic hepatitis, were obtained at 5 time points during acute infection, indicated by the letters A to E, which correspond to weeks 3, 8, 13, 16, and 21 after infection, respectively. (a) Parsimony reconstruction of amino acid changes at codon 348, superimposed upon the ML phylogeny. Arrows indicate amino acid changes. Three mutations from isoleucine (black) to valine (red) occur between time points C and D, of which one spreads to fixation at time point E. One other equally parsimonious reconstruction is possible (data not shown). (b) Location of the 33 selected sites in patient 10. MacClade identified all 33 sites, whereas CODEML detected only 18 of these. Codon 348 is marked by an asterisk.
In sum, our study shows the importance of high-resolution selection analyses in providing detailed information about the evolutionary forces acting on the HCV genome. Such analyses could be readily extended to other sites thought to be under selection from the immune system, providing strong evidence of viral adaptation and escape if viewed in concert with immunological data on the status of both humoral and cellular responses. Since immunology and escapology are often taught by viruses (16, 36), such reverse mapping of epitopes could provide a powerful tool for identification of relevant immune responses and aid vaccine design.
Acknowledgments
This work was sponsored by the Wellcome Trust.
We thank Sean Wise for contributions to the initial stages of this work and David Thomas and Stuart Ray for assistance in data preparation.
REFERENCES
- 1.Bocharov, G., P. Klenerman, and S. Ehl. 2003. Modelling the dynamics of LCMV infection in mice. II. Compartmental structure and immunopathology. J. Theor. Biol. 221:349-378. [DOI] [PubMed] [Google Scholar]
- 2.Booth, J. C., U. Kumar, D. Webster, J. Monjardino, and H. C. Thomas. 1998. Comparison of the rate of sequence variation in the hypervariable region of E2/NS1 region of hepatitis C virus in normal and hypogammaglobulinemic patients. Hepatology 27:223-227. [DOI] [PubMed] [Google Scholar]
- 3.Cerino, A., M. Bissolati, A. Cividini, A. Nicosia, M. Esumi, N. Hayashi, K. Mizuno, R. Slobbe, P. Oudshoorn, E. Silini, M. Asti, and M. U. Mondelli. 1997. Antibody responses to the hepatitis C virus E2 protein: relationship to viraemia and prevalence in anti-HCV seronegative subjects. J. Med. Virol. 51:1-5. [PubMed] [Google Scholar]
- 4.Ciurea, A., P. Klenerman, L. Hunziker, E. Horvath, B. Odermatt, A. F. Ochsenbein, H. Hengartner, and R. M. Zinkernagel. 1999. Persistence of lymphocytic choriomeningitis virus at very low levels in immune mice. Proc. Natl. Acad. Sci. USA 96:11964-11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen, J. 1999. The scientific challenge of hepatitis C. Science 285:26-30. [DOI] [PubMed] [Google Scholar]
- 6.Day, C. L., G. M. Lauer, G. K. Robbins, B. McGovern, A. G. Wurcel, R. T. Gandhi, R. T. Chung, and B. D. Walker. 2002. Broad specificity of virus-specific CD4+ T-helper-cell responses in resolved hepatitis C virus infection. J. Virol. 76:12584-12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehl, S., P. Klenerman, R. M. Zinkernagel, and G. Bocharov. 1998. The impact of variation in the number of CD8(+) T-cell precursors on the outcome of virus infection. Cell. Immunol. 189:67-73. [DOI] [PubMed] [Google Scholar]
- 8.Farci, P., H. J. Alter, D. C. Wong, R. H. Miller, S. Govindarajan, R. Engle, M. Shapiro, and R. H. Purcell. 1994. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc. Natl. Acad. Sci. USA 91:7792-7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farci, P., A. Shimoda, A. Coiana, G. Diaz, G. Peddis, J. C. Melpolder, A. Strazzera, D. Y. Chien, S. J. Munoz, A. Balestrieri, R. H. Purcell, and H. J. Alter. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339-344. [DOI] [PubMed] [Google Scholar]
- 10.Farci, P., A. Shimoda, D. Wong, T. Cabezon, D. De Gioannis, A. Strazzera, Y. Shimizu, M. Shapiro, H. J. Alter, and R. H. Purcell. 1996. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc. Natl. Acad. Sci. USA 93:15394-15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fournillier, A., C. Wychowski, D. Boucreux, T. F. Baumert, J. C. Meunier, D. Jacobs, S. Muguet, E. Depla, and G. Inchauspe. 2001. Induction of hepatitis C virus E1 envelope protein-specific immune response can be enhanced by mutation of N-glycosylation sites. J. Virol. 75:12088-12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenkins, G. M., A. Rambaut, O. G. Pybus, and E. C. Holmes. 2002. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. E vol. 54:156-165. [DOI] [PubMed] [Google Scholar]
- 13.Klenerman, P., F. Lechner, M. Kantzanou, A. Ciurea, H. Hengartner, and R. Zinkernagel. 2000. Viral escape and the failure of cellular immune responses. Science 289:2003. [DOI] [PubMed] [Google Scholar]
- 14.Lechner, F., N. H. Gruener, S. Urbani, J. Uggeri, T. Santantonio, A. R. Kammer, A. Cerny, R. Phillips, C. Ferrari, G. R. Pape, and P. Klenerman. 2000. CD8+ T lymphocyte responses are induced during acute hepatitis C virus infection but are not sustained. Eur. J. Immunol. 30:2479-2487. [DOI] [PubMed] [Google Scholar]
- 15.Lechner, F., D. K. Wong, P. R. Dunbar, R. Chapman, R. T. Chung, P. Dohrenwend, G. Robbins, R. Phillips, P. Klenerman, and B. D. Walker. 2000. Analysis of successful immune responses in persons infected with hepatitis C virus. J. Exp Med. 191:1499-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucas, M., U. Karrer, A. Lucas, and P. Klenerman. 2001. Viral escape mechanisms-escapology taught by viruses. Int. J. Exp. Pathol. 82:269-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Maddison, D. R., and W. P. Maddison. 2000. MacClade. Analysis of phylogeny and character evolution, version 4. Sinauer Associates, Sunderland, Mass.
- 17.Manzin, A., L. Solforosi, E. Petrelli, G. Macarri, G. Tosone, M. Piazza, and M. Clementi. 1998. Evolution of hypervariable region 1 of hepatitis C virus in primary infection. J. Virol. 72:6271-6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pohlmann, S., J. Zhang, F. Baribaud, Z. Chen, G. J. Leslie, G. Lin, A. Granelli-Piperno, R. W. Doms, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J. Virol. 77:4070-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ray, S. C., Y. M. Wang, O. Laeyendecker, J. R. Ticehurst, S. A. Villano, and D. L. Thomas. 1999. Acute hepatitis C virus structural gene sequences as predictors of persistent viremia: hypervariable region 1 as a decoy. J. Virol. 73:2938-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roccasecca, R., H. Ansuini, A. Vitelli, A. Meola, E. Scarselli, S. Acali, M. Pezzanera, B. B. Ercole, J. McKeating, A. Yagnik, A. Lahm, A. Tramontano, R. Cortese, and A. Nicosia. 2003. Binding of the hepatitis C virus E2 glycoprotein to CD81 is strain specific and is modulated by a complex interplay between hypervariable regions 1 and 2. J. Virol. 77:1856-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross, H. A., and A. G. Rodrigo. 2002. Immune-mediated positive selection drives human immunodeficiency virus type 1 molecular variation and predicts disease duration. J. Virol. 76:11715-11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimizu, Y. K., M. Hijikata, A. Iwamoto, H. J. Alter, R. H. Purcell, and H. Yoshikura. 1994. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J. Virol. 68:1494-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimizu, Y. K., H. Igarashi, T. Kiyohara, T. Cabezon, P. Farci, R. H. Purcell, and H. Yoshikura. 1996. A hyperimmune serum against a synthetic peptide corresponding to the hypervariable region 1 of hepatitis C virus can prevent viral infection in cell cultures. Virology 223:409-412. [DOI] [PubMed] [Google Scholar]
- 25.Shirai, M., T. Arichi, M. Chen, T. Masaki, M. Nishioka, K. Ikeda, H. Takahashi, N. Enomoto, T. Saito, M. E. Major, T. Nakazawa, T. Akatsuka, S. M. Feinstone, and J. A. Berzofsky. 1999. T cell recognition of hypervariable region-1 from hepatitis C virus envelope protein with multiple class II MHC molecules in mice and humans: preferential help for induction of antibodies to the hypervariable region. J. Immunol. 162:568-576. [PubMed] [Google Scholar]
- 26.Simmonds, P., E. C. Holmes, T. A. Cha, S. W. Chan, F. McOmish, B. Irvine, E. Beall, P. L. Yap, J. Kolberg, and M. S. Urdea. 1993. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J. Gen. Virol. 74(Pt 11):2391-2399. [DOI] [PubMed] [Google Scholar]
- 27.Smith, D. B. 1999. Evolution of the hypervariable region of hepatitis C virus. J. Viral Hepat. 6(Suppl. 1):41-46. [DOI] [PubMed] [Google Scholar]
- 28.Stumpf, M. P., and O. G. Pybus. 2002. Genetic diversity and models of viral evolution for the hepatitis C virus. FEMS Microbiol. Lett. 214:143-152. [DOI] [PubMed] [Google Scholar]
- 28a.Swofford, D. L. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods), 4th ed. Sinauer Associates, Sunderland, Mass.
- 29.Thimme, R., D. Oldach, K. M. Chang, C. Steiger, S. C. Ray, and F. V. Chisari. 2001. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J. Exp. Med. 194:1395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tseng, C. T., and G. R. Klimpel. 2002. Binding of the hepatitis C virus envelope protein E2 to CD81 inhibits natural killer cell functions. J. Exp. Med. 195:43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wack, A., E. Soldaini, C. Tseng, S. Nuti, G. Klimpel, and S. Abrignani. 2001. Binding of the hepatitis C virus envelope protein E2 to CD81 provides a co-stimulatory signal for human T cells. Eur. J. Immunol. 31:166-175. [DOI] [PubMed] [Google Scholar]
- 32.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 33.Weiner, A. J., D. Y. Chien, Q. Choo, S. R. Coates, G. Kuo, and M. Houghton. 2000. Humoral responses to hepatitis C. Academic Press, New York, N.Y.
- 34.World Health Organization. 1997. Hepatitis C: global prevalence. Wkly. Epidemiol. Rec. 72:341-344. [PubMed] [Google Scholar]
- 35.Yang, Z., R. Nielsen, N. Goldman, and A. M. Pedersen. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zinkernagel, R. M. 1996. Immunology taught by viruses. Science 271:173-178. [DOI] [PubMed] [Google Scholar]