Abstract
Rhinoviruses are the most common infectious agents of humans. They are the principal etiologic agents of afebrile viral upper-respiratory-tract infections (the common cold). Human rhinoviruses (HRVs) comprise a genus within the family Picornaviridae. There are >100 serotypically distinct members of this genus. In order to better understand their phylogenetic relationship, the nucleotide sequence for the major surface protein of the virus capsid, VP1, was determined for all known HRV serotypes and one untyped isolate (HRV-Hanks). Phylogenetic analysis of deduced amino acid sequence data support previous studies subdividing the genus into two species containing all but one HRV serotype (HRV-87). Seventy-five HRV serotypes and HRV-Hanks belong to species HRV-A, and twenty-five HRV serotypes belong to species HRV-B. Located within VP1 is a hydrophobic pocket into which small-molecule antiviral compounds such as pleconaril bind and inhibit functions associated with the virus capsid. Analyses of the amino acids that constitute this pocket indicate that the sequence correlates strongly with virus susceptibility to pleconaril inhibition. Further, amino acid changes observed in reduced susceptibility variant viruses recovered from patients enrolled in clinical trials with pleconaril were distinct from those that confer natural phenotypic resistance to the drug. These observations suggest that it is possible to differentiate rhinoviruses naturally resistant to capsid function inhibitors from those that emerge from susceptible virus populations as a result of antiviral drug selection pressure based on sequence analysis of the drug-binding pocket.
Human rhinoviruses are the most common viral infectious agents in humans (9). In the United States, rhinoviruses account for more than one billion cases annually of viral respiratory tract infections (VRTI; i.e., the common cold) (34). The National Center for Health Statistics estimated that in 1996, 62 million cases of the common cold required medical attention or resulted in restricted activity in the United States (33). During this period, colds caused 45 million days of restricted activities and 22 million days lost from school. Roughly one in six colds results in a doctor's office visit, and up to 50% of these visits result in an antibiotic prescription (15, 29, 35, 47). In 1998, more antibiotic prescriptions were written for presumed VRTIs than for bacterial infections, at a cost of approximately $726 million (14). Such inappropriate antibiotic prescribing is a major contributing factor to the development of drug-resistant bacterial pathogens (4, 28, 35, 48). The total economic impact of non-influenza-related VRTI in terms of both direct (health care resource use) and indirect (productivity loss) costs approaches $40 billion annually (11).
HRVs comprise a genus within the family Picornaviridae. Picornaviruses are small, nonenveloped viruses containing a single strand of message sense RNA. The RNA is protected by a protein coat or capsid composed of 60 copies of each of four structural proteins, designated VP1 to VP4. The virus capsid provides several critical functions, including protection of the viral RNA from degradation during transport between cells, cell tropism through receptor specificity, and controlled release of the viral RNA from within the capsid to the cell cytoplasm during the uncoating process. The three-dimensional crystal structures of several human picornaviruses have been solved at atomic resolution, revealing details of capsid architecture and the arrangement of these proteins in the capsid (3, 21, 26, 38, 42, 51). VP1 is the largest and most surface-exposed protein of the virus capsid. The core structure of VP1, VP2, and VP3 is an eight-stranded antiparallel β-barrel. Within the β-barrel of VP1 is a surface-accessible hydrophobic pocket to which several classes of low-molecular-weight compounds, including pleconaril, have been reported to bind (25, 45). Upon binding, these compounds affect functions associated with the virus capsid, including virus attachment to cell receptors and uncoating of viral RNA (10, 13, 39, 44, 52). Pleconaril has been shown in phase 3 human clinical testing to be effective for treating the common cold of a picornavirus etiology (19).
There are 101 distinct HRV serotypes recognized currently, although several share significant antigenic cross-reactivity (6-8, 18, 24, 25). These viruses have been grouped according to several parameters, including receptor specificity (16, 20, 46, 49), susceptibility to antiviral compounds (1), and nucleotide sequence relatedness in the 5′-untranslated (31) and VP4/VP2 capsid protein-coding (5, 22, 23, 43, 50) regions of the genome.
The present study was conducted to generate complete molecular sequence information in the region of the genome encoding VP1 for all HRV serotypes in order to explore the basis for future HRV molecular serotyping efforts analogous to those reported for enteroviruses (36, 37). Further, the determination of the amino acids that comprise the VP1 drug-binding pocket of all HRV serotypes was sought in order to better understand the relationship between primary amino acid sequence and virus susceptibility to capsid function inhibitors such as pleconaril. To this end, the VP1 sequence of all 101 HRV serotypes and HRV-Hanks is reported here. The data are discussed in the context of phylogenetic relationship among members of the HRV genus, as well as the role of pocket amino acid residues in virus susceptibility to pleconaril.
MATERIALS AND METHODS
Viruses and cells.
All numbered prototype HRV serotypes were purchased from the American Type Culture Collection, Rockville, Md. HRV-Hanks was a gift from Frederick Hayden, University of Virginia at Charlottesville. HRVs were grown either on HeLa Ohio-I (HeLa-I) cells, which were a gift from Hayden, or on HeLa Ohio-Wis (HeLa-Wis) cells, which were a gift from Roland Rueckert, University of Wisconsin at Madison. HeLa-I cells were grown in Eagle minimal essential medium (EMEM; Sigma) supplemented with 5% fetal bovine serum (US Bio-Technologies, Inc., Parkerford, Pa.), 5% Fetalclone (HyClone, Logan, Utah), glutamine (Gibco), and antibiotics (vancomycin, amphotericin B, and gentamicin, Sigma; hereafter referred to as complete EMEM). HeLa-Wis cells were grown in EMEM supplemented with 10% fetal bovine serum and antibiotics (Pen/Strep, Sigma). Viruses were grown in cells at 33°C until a complete cytopathic effect (CPE) was observed. The stocks were frozen and thawed, clarified by low-speed centrifugation, and stored frozen as 1-ml aliquots at −80°C.
Drug susceptibility testing.
Antiviral activity in cell culture was measured in an assay evaluating protection from virus-induced CPE. HeLa-I or HeLa-Wis cells were seeded into 96-well tissue culture cluster plates at 4 × 104 cells/well, followed by overnight incubation at 37°C. The next day, the cells were infected with a dilution of an HRV serotype that had been shown to produce ≥85% CPE after 3 days of incubation. Virus was allowed to absorb to cells for 1 h at 33°C, followed by the addition of serial 0.5-log10 dilutions of pleconaril (in a final dimethyl sulfoxide concentration of 0.5%) in quadruplicate. After 3 days of incubation at 33°C, the plates were fixed by the addition of 5% glutaraldehyde and stained with crystal violet (0.5% solution in water). After the cells were rinsed and dried, residual cell staining was measured spectrophotometrically at an optical density of 570 nm on a Molecular Devices Vmax tunable plate reader (Sunnyvale, Calif.). The serial dilution data were subjected to analysis by a four-parameter curve-fitting program, from which a 50% effective concentration (EC50) value was derived. The EC50 value is defined as the compound concentration that affords 50% protection of a cell monolayer from virus-induced CPE.
Amplification and sequencing of capsid-coding region of HRV prototype strains. (i) RNA extraction.
Viral RNA was extracted from 140 or 280 μl of the virus stock by using QIAamp Viral RNA Mini Kit (Qiagen, Inc., Valencia, Calif.) according to the manufacturer's instructions. RNA was eluted from the spin column in a total volume of 60 μl of RNase-free water containing 0.04% sodium azide and stored frozen at −80°C.
(ii) RT-PCR amplification.
The region of the viral RNA encoding the structural proteins of all HRV serotypes was amplified by reverse transcription-PCR (RT-PCR). The 5′ primer utilized for PCR was 5′-CTA CTT TGG GTG TCC G-3′, a highly conserved sequence within the 5′-noncoding region of the viral genome (nucleotides 536 to 551 of HRV-16). There is no similarly conserved sequence immediately downstream of VP1. Consequently, two degenerate antisense oligonucleotides were prepared based on amino acid sequence scanning within the 2Apro-coding region (immediately downstream of VP1) of both published HRV sequences and those that had been generated in our laboratory. The downstream oligonucleotides utilized were 5′-TGT KCG RTA WAT GAT TAR ATC-3′ (the reverse complement of nucleotides 3287 to 3307 in HRV-16) and 5′-YCC WCC ACA RTC WCC WGG TTC-3′ (the reverse complement of nucleotides 3488 to 3508 in HRV-16), where K = G or T, R = A or G, W = A or T, and Y = C or T. The former oligonucleotide amplified HRV-A viruses exclusively, whereas the latter was effective for viruses belonging to both species. A third antisense oligonucleotide represented a conserved region in the nonstructural protein 2B (5′-TTI AGA AAY CTC CAI GGI GAI CC-3′; reverse complement of nucleotides 3857 to 3879 of HRV14; where I = inosine). This oligonucleotide was most effective for amplification of HRV-B viruses, although it was also useful for a small number of HRV-A viruses. In all instances, at least one of these three oligonucleotides proved suitable for use in priming of the RT reaction and subsequent PCR. (A document detailing the primers utilized to sequence each HRV prototype strain is available from D.C.P. upon request by E-mail.)
A two-step RT-PCR process was utilized to amplify the capsid-coding region of HRV prototype strains. RT was conducted with the Invitrogen Thermoscript RT system (Invitrogen, Carlsbad, Calif.) with 1 μg of RNA in a final reaction volume of 20 μl. RNA and primer were first heated to 65°C for 5 min, followed by addition of the RT reaction components. The reaction was incubated at 50°C for 1 h and terminated by incubation at 85°C for 5 min. The RNA was then degraded by incubation at 37°C for 20 min with 2 U of RNase H (Invitrogen).
One microliter of the RT reaction product was subjected to PCR amplification by using the Roche Expand High Fidelity PCR system (Roche Applied Science, Indianapolis, Ind.) according to the manufacturer's protocol. The PCR cycling conditions were as follows: 1 cycle of 94°C for 2 min; 10 cycles of 94°C for 30 s, 42°C for 90 s, and 68°C for 4 min; 20 cycles of 94°C for 30 s, 42°C for 90 s, and 68°C for 4 min plus 10 s/cycle; and a final cycle of 72°C for 10 min, followed by holding at 4°C.
Because degenerate primers were utilized in both the RT assay and PCR, multiple bands were often observed when the amplification products were subjected to agarose gel electrophoresis. Consequently, PCR products of the expected molecular mass were gel extracted by using the QiaQuick gel extraction kit (Qiagen) and eluted in 30 μl of diethyl pyrocarbonate-treated water. Purified products were stored at −20°C.
(iii) Sequencing and analysis of RT-PCR amplicons.
PCR products were sequenced with fluorescent-labeled nucleotides on an ABI 377 sequencer at Biotech Core Incorporated (Mountain View, Calif.). Sequencing primers included those used in the RT-PCR, as well as those derived from genome walking. Each nucleotide assignment was confirmed by sequence analysis of both strands of cDNA. The GenBank accession numbers for these sequences are AY355180 to AY355281.
Sequences were analyzed by using programs in the Lasergene suite (version 5.03) of sequence analysis software (DNASTAR, Inc., Madison, Wis.). Text data files were imported into EditSeq for conversion to sequence data and for predicted amino acid translation. Contigs were built by using SeqMan. Multiple viral cDNA and predicted amino acid translation sequences were aligned by using MegAlign. Phylogenic analysis of both the nucleotide and amino acid sequences were conducted by using the Phylip (Phylogeny Inference) package (version 3.6a3). The programs DNADIST (for nucleotide) and PROTDIST (for amino acid) were used to generate the distance matrix by using the maximum-likelihood model of nucleotide and amino acid substitution. Dendrograms were drawn by using NEIGHBOR, the neighbor-joining option in PHYLIP. Bootstrap analysis was conducted by using SEQBOOT with 100 replicates, and the consensus tree was chosen from the 100 generated by CONSENSE. The trees were visualized in TREE VIEW (version 1.6.6). To estimate branch lengths from the consensus tree, a distance matrix was generated by using PROTDIST from the original alignment of the VP1 region. Branch lengths for the user-defined consensus tree were estimated from the calculated distance matrix in FITCH.
(iv) Computational modeling.
Tyr152 to Phe and Val191 to Leu modeling experiments were conducted with rotamer exploration to characterize the available conformational space of the mutated side chains and the resultant volume changes. Furthermore, all residues within 5 Å of any mutated amino acid of either the empty or the pleconaril-occupied pocket of wild-type and variant viruses were allowed to relax to probe possible shape changes; however, no meaningful changes were observed.Alternatively, pleconaril and methyl ether (as a very small dummy ligand) were docked into wild-type and variant pockets with both ligand and receptor flexible (all amino acids within 5 Å of any ligand atom); again, no geometry deviations could be observed. Computations were carried out with the MMFF94s force field in MOE (version 2003.02, Chemical Computing Group, Inc., Montreal, Quebec, Canada).
RESULTS
Amplification and sequencing of VP1.
Prior to the present study, the nucleotide sequence encoding VP1 of about 20 HRV serotypes was available. The strategy we used to complete the sequencing of all HRV serotypes involved the amplification of the entire capsid protein-coding region of each prototype virus, followed by sequencing of both the VP4/VP2 junction as described by Savolainen et al. (43) and the VP1 region. This allowed for comparison of the respective virus collections by examination of VP4/VP2 sequence data and for assessment of virus phylogenetic relationships based on sequence data in the two distinct regions of the HRV genome.
Phylogenetic grouping of HRV serotypes based on VP1 deduced amino acid sequence.
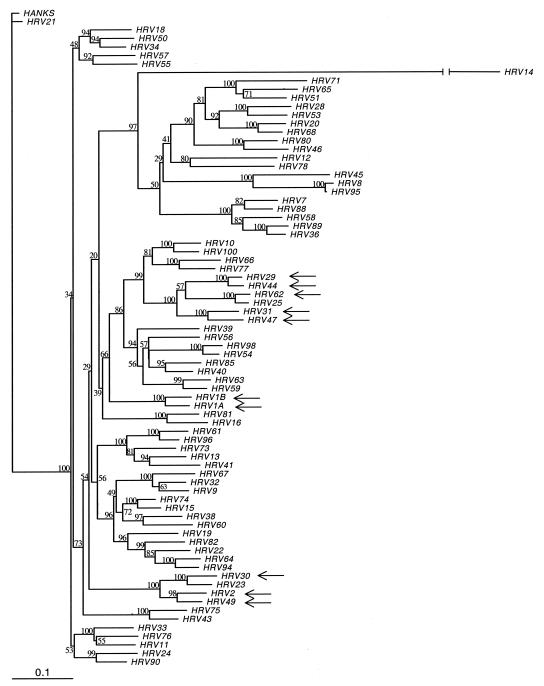
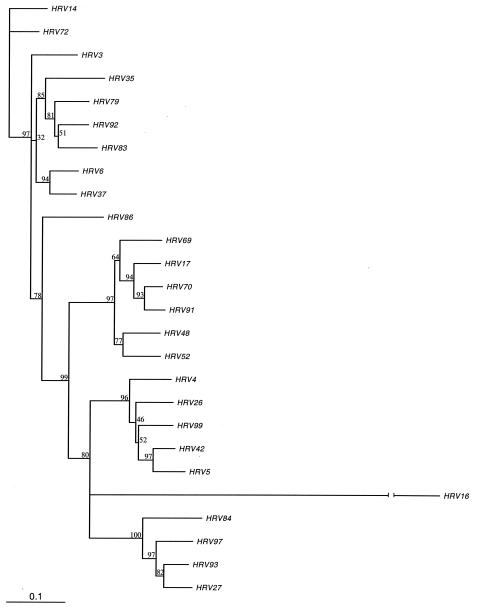
Complete VP1 nucleotide sequence data were generated for all 101 HRV serotypes and HRV-Hanks. The data generated here confirm the segregation of all but one of the 101 numbered prototype HRV strains into two genetic groups or species as described previously (5, 22, 31, 43) Based on VP1 sequence, 75 of the 101 HRV serotypes cluster within the previously designated HRV species A (HRV-A), which includes 65 HRVs that utilize ICAM-1 to attach to cells (the major receptor binding group) (16, 46, 49) and all 10 HRVs that utilize the low-density lipoprotein receptor family (the minor receptor binding group; Fig. 1) (20). HRV-Hanks also belongs to HRV-A. Twenty-five viruses cluster within HRV species B (HRV-B; Fig. 2). One virus, HRV-87, clusters with human enterovirus 68 as reported previously (5, 23, 43).
FIG. 1.
Phylogenetic tree of HRV-A species members based on VP1 deduced amino acid sequence. The neighbor-joining dendrogram is based on a maximum-likelihood distance matrix calculated in PHYLIP (see Materials and Methods). The distance matrix was generated in PROTDIST, and the dendrogram was generated by using NEIGHBOR. The amino acids of the VP1 region were aligned in MegAlign by using the CLUSTAL W algorithm. Branch numbers represent bootstrap values for 100 trials, and arrows identify minor receptor-binding group viruses. To estimate branch lengths from the consensus tree, a distance matrix was generated by using PROTDIST from the original alignment of the VP1 region. Branch lengths for the user-defined consensus tree were estimated from the calculated distance matrix in FITCH. A single HRV-B species member (HRV-14) is included to provide perspective on the interspecies relationship within the genus. The branch length for HRV-14, which is 0.98, has been truncated for clarity.
FIG. 2.
Phylogenetic tree of HRV-B species members based on VP1 deduced amino acid sequence. The neighbor-joining dendrogram is based on a maximum-likelihood distance matrix calculated in PHYLIP (see Materials and Methods). The distance matrix was generated in PROTDIST, and the dendrogram was generated by using NEIGHBOR. The amino acids of the VP1 region were aligned in MegAlign by using the CLUSTAL W algorithm. Branch numbers represent bootstrap values for 100 trials. To estimate branch lengths from the consensus tree, a distance matrix was generated by using PROTDIST from the original alignment of the VP1 region. Branch lengths for the user-defined consensus tree were estimated from the calculated distance matrix in FITCH. A single HRV-A species member (HRV-16) is included to provide perspective on the interspecies relationship within the genus. The branch length for HRV-16, which is 1.07, has been truncated for clarity.
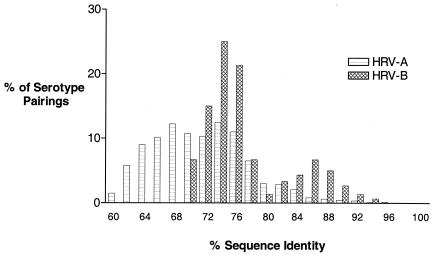
Within HRV-A, the level of VP1 deduced amino acid identity ranged from a low of 57.7% (HRV serotypes 29 and 45) to a high of 98.6% (HRV-8 and HRV-95). Nineteen serotype pairs had amino acid sequence identity of 90% or more (Table 1). Deduced amino acid sequence identity in HRV-B ranged from a low of 68.3% (several serotype pairs) to a high of 93.6% (HRV-70 and HRV-91). Seven serotype pairs were identical by 90% or more (Table 1). Overall, the level of interserotypic amino acid identity across HRV-A prototype strains was significantly lower than that of HRV-B (Fig. 3). Median identities in the respective groups were 70.2% versus 74.3% (P < 0.0001 as determined by Mann-Whitney nonparametric test).
TABLE 1.
HRV serotype pairs with a VP1 deduced amino acid identity of ≥90%a
| HRV serotype pairs | % aa identity | HRV serotype pairs | % aa identity | |
|---|---|---|---|---|
| HRV-A species | ||||
| 8 and 95 | 98.6 | |||
| 21 and Hanks | 97.2 | |||
| 25 and 62 | 95.4 | |||
| 29 and 44 | 95.0 | |||
| 54 and 98 | 94.4 | |||
| 36 and 89 | 94.1 | |||
| 61 and 96 | 92.7 | |||
| 64 and 94 | 92.7 | |||
| 59 and 63 | 92.3 | |||
| 15 and 74 | 92.0 | |||
| 10 and 100 | 92.0 | |||
| 1A and 1B | 92.0 | |||
| 23 and 30 | 91.9 | |||
| 2 and 49 | 91.9 | |||
| 20 and 68 | 91.9 | |||
| 9 and 32 | 91.0 | |||
| 34 and 50 | 90.9 | |||
| 7 and 88 | 90.3 | |||
| 40 and 85 | 90.2 | |||
| HRV-B species | ||||
| 70 and 91 | 93.6 | |||
| 17 and 70 | 92.2 | |||
| 5 and 42 | 91.7 | |||
| 27 and 93 | 91.6 | |||
| 6 and 37 | 90.3 | |||
| 17 and 91 | 90.2 | |||
| 14 and 72 | 90.0 |
The percent deduced amino acid (aa) sequence identity values were calculated by using the CLUSTAL W alignment algorithm and the sequence distance option from MegAlign.
FIG. 3.
Interserotypic amino acid identity in HRV-A and HRV-B species members across VP1. The percent deduced amino acid sequence identity values were calculated by using the CLUSTAL W alignment algorithm and sequence distance option from MegAlign.
Phylogenetic relationships based on comparison of VP4/VP2 junction and VP1.
In addition to sequencing of VP1, we also sequenced the VP4/VP2 junction of each virus and compared phylogenetic trees generated from the deduced amino acid sequence of these two regions. Overall, the trees were in excellent agreement with one another, with only minor shifts observed (data not shown). In order to confirm the accuracy of virus stock serotype designations, the VP4/VP2 DNA sequence results were compared to those published by Savolainen et al. (43). The results from the two laboratories were in excellent agreement with the single exception of HRV-31, for which isolates from the two laboratories differed by >20% at the nucleotide level. The identity of the HRV-31 strain used here was confirmed by neutralization assay with monospecific antiserum from the American Type Culture Collection (data not shown). The results indicate that the HRV-31 sequence data reported by Savolainen et al. are incorrect and that this is in fact HRV-32. Based on our sequence data for the VP4/VP2 junction, HRV-31 clusters with HRV-47 with a nucleotide identity score of 91.7%, and HRV-32 clusters with HRV-9 with a nucleotide identity score of 85.5%.
Cross-neutralization testing.
The high level of VP1 amino acid identity between HRV-8 and HRV-95 (98.6%) prompted us to reconfirm the reported serotypic uniqueness of these two viruses. In addition, antiserum to HRV-21 was evaluated for its ability to neutralize the untyped HRV-Hanks (97.2% VP1 amino acid identity). Monospecific antiserum raised against HRV-8 fully neutralized both HRV-8 and HRV-95 at a similar dilution of 1:1,280 to 1:2,560 (data not shown). Similarly, HRV-21 antiserum efficiently neutralized both HRV-21 and HRV-Hanks. We therefore conclude that HRV-8 and HRV-95 are the same serotype and that HRV-Hanks (which was previously untyped) is in fact HRV-21.
Amino acid residues in drug-binding pocket.
Antiviral compounds, such as pleconaril, that inhibit capsid functions integrate into a highly conserved hydrophobic pocket comprised almost exclusively of amino acid residues located within VP1. Prototype HRV strains exhibit a broad range of susceptibility to pleconaril inhibition in cell culture (Tables 2 and 3). The level of interserotypic amino acid conservation within this pocket was assessed in order to gain insight into why such a broad range of susceptibility existed to pleconaril. In addition, the identity of the amino acids in the pocket could serve as predictors of isolates naturally resistant to pleconaril, thus differentiating these viruses from those that might emerge in response to drug treatment. Such information could serve as the basis for a treatment-emergent resistance-monitoring plan should pleconaril be used broadly in a population.
TABLE 2.
Alignment of amino acids in drug-binding pocket of HRV-A virusesa
| Serotype | EC50 (μM)b | Amino acid at position:
|
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 98 | 99 | 100 | 101 | 118 | 120 | 122 | 124 | 142 | 143 | 144 | 166 | 167 | 168 | 179 | 181 | 184 | 190 | 192 | 208 | 212 | 214 | 217 | 238 | 260 | ||
| Majority sequence | I | N | L | Q | F | S | I | L | Y | M | Y | A | S | V | F | L | L | Y | M | T | N | M | L | H | H | |
| Serotype | ||||||||||||||||||||||||||
| 39 | 0.01 | - | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | V | - | - | - | - | N |
| 47 | 0.01 | - | - | - | - | - | - | V | M | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | G |
| 55 | 0.02 | V | - | I | - | - | - | V | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | - |
| 71 | 0.02 | - | T | - | K | - | - | - | I | - | - | - | - | - | - | - | I | I | - | - | S | - | - | - | - | Y |
| Hanks | 0.02 | - | - | I | K | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | A | - | - | - | - | - |
| 68 | 0.03 | - | T | I | K | - | - | - | I | - | - | - | - | - | - | - | I | M | - | - | S | - | - | - | - | R |
| 74 | 0.03 | - | - | - | K | - | - | V | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 100 | 0.03 | - | T | - | - | - | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - |
| 2 | 0.04 | - | - | I | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 21 | 0.04 | - | - | I | K | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | A | - | - | - | - | - |
| 22 | 0.04 | - | - | - | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | - | - | - | - | - |
| 58 | 0.04 | - | S | - | - | - | - | - | I | F | - | - | - | - | - | - | I | M | - | - | S | - | - | I | - | - |
| 7 | 0.05 | - | S | - | - | - | - | - | I | F | - | - | - | - | I | - | I | M | - | - | S | - | - | I | - | - |
| 51 | 0.05 | - | T | - | K | - | - | - | I | - | - | - | - | - | - | - | I | V | - | - | S | - | - | - | - | Y |
| 85 | 0.05 | - | T | I | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | R |
| 89 | 0.05 | V | S | - | - | - | - | - | I | F | - | - | - | - | - | - | I | M | - | - | S | - | - | I | - | - |
| 20 | 0.06 | - | T | I | K | - | - | - | I | - | - | - | - | - | - | - | I | M | - | - | S | - | - | - | - | K |
| 40 | 0.07 | - | T | I | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 78 | 0.08 | - | - | - | - | - | - | - | - | F | - | F | - | - | - | - | I | S | - | - | - | - | - | - | - | Y |
| 34 | 0.09 | - | - | - | - | - | - | V | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | - |
| 96 | 0.09 | - | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 25 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | D |
| 38 | 0.10 | - | - | - | K | - | - | V | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 88 | 0.10 | - | S | - | - | - | - | - | I | F | - | - | - | - | - | - | I | M | - | - | S | - | - | I | - | - |
| 36 | 0.11 | - | S | - | - | - | - | - | I | F | - | - | - | - | - | - | I | M | - | - | S | - | - | I | - | - |
| 44 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | G |
| 56 | 0.11 | - | T | - | - | - | - | V | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 66 | 0.11 | - | T | - | - | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | N |
| 75 | 0.11 | - | - | - | - | - | - | V | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | R |
| 10 | 0.13 | - | T | - | - | - | - | V | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 31 | 0.13 | - | - | - | - | - | - | V | I | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | G |
| 11 | 0.15 | - | - | - | - | - | - | - | - | - | - | - | - | - | I | - | - | M | - | - | N | - | - | - | - | - |
| 33 | 0.16 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | N | - | - | - | - | - |
| 46 | 0.16 | - | T | - | L | - | - | - | I | - | - | - | - | - | - | - | I | M | - | - | - | - | - | I | - | Y |
| 62 | 0.16 | - | - | - | - | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | D |
| 63 | 0.16 | - | T | - | - | - | - | V | - | - | - | - | - | - | I | - | - | - | - | - | V | - | - | - | - | Y |
| 1B | 0.16 | - | T | - | - | - | - | V | - | - | - | - | M | - | I | - | - | - | - | - | S | - | - | I | - | R |
| 49 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 57 | 0.17 | - | - | - | - | - | - | V | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | - |
| 80 | 0.17 | - | T | - | M | - | - | - | I | - | - | - | - | - | - | - | I | M | - | - | - | - | - | - | - | Y |
| 90 | 0.19 | - | - | - | - | - | - | - | I | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | - |
| 19 | 0.20 | - | - | - | K | - | - | V | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | - |
| 28 | 0.20 | - | T | - | K | - | - | - | I | - | - | - | - | - | - | - | I | M | - | - | A | - | - | - | - | Y |
| 24 | 0.21 | - | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | - | S | - | - | - | - | - |
| 61 | 0.21 | - | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 73 | 0.21 | - | S | - | - | - | - | V | I | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 9 | 0.22 | - | - | I | K | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | - |
| 82 | 0.22 | - | S | I | K | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | |
| 30 | 0.23 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| 43 | 0.23 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| 76 | 0.23 | - | - | - | - | - | - | V | - | - | - | - | - | - | I | - | - | - | - | - | N | - | - | - | - | - |
| 18 | 0.24 | - | - | - | - | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | - |
| 67 | 0.25 | - | - | I | K | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | - |
| 29 | 0.27 | V | - | - | - | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | - |
| 53 | 0.28 | - | T | - | K | - | - | - | I | - | - | - | - | - | - | - | I | M | - | - | A | - | - | - | - | - |
| 81 | 0.30 | - | - | - | - | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | - |
| 23 | 0.32 | - | - | - | - | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | G |
| 64 | 0.32 | - | T | - | K | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | A | - | - | - | - | Y |
| 32 | 0.33 | - | - | I | K | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | - |
| 65 | 0.33 | - | T | - | K | - | - | - | I | - | - | - | - | - | - | - | I | V | - | - | S | - | - | - | - | - |
| 50 | 0.35 | - | - | - | - | - | - | V | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | N |
| 12 | 0.39 | - | - | - | K | - | - | - | I | - | - | - | - | - | - | - | I | I | - | - | - | - | - | - | - | - |
| 1A | 0.43 | - | T | - | - | - | - | - | - | - | - | - | M | - | I | - | - | - | - | - | S | - | - | I | - | Y |
| 54 | 0.46 | - | T | - | - | - | - | - | - | F | - | - | P | - | I | - | - | - | - | - | - | - | - | - | - | - |
| 60 | 0.48 | - | - | - | K | - | - | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 77 | 0.55 | - | T | - | - | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | - |
| 16 | 0.57 | - | - | - | - | - | - | - | M | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | R |
| 13 | 0.58 | - | S | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 15 | 0.59 | - | - | - | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D |
| 59 | 0.78 | - | T | - | - | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | V | - | - | - | - | - |
| 95 | 0.81 | - | - | I | - | - | - | - | I | - | - | - | - | - | - | - | I | S | - | - | I | - | - | I | L | - |
| 94 | 1.10 | - | T | - | K | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | - | - | - | - | - | - |
| 41 | 1.75 | - | S | - | - | - | - | - | I | - | - | F | - | - | - | - | - | - | - | - | - | - | - | - | - | Y |
| 98 | 1.80 | - | T | - | - | - | - | V | - | F | - | - | P | - | I | - | - | - | - | - | - | - | - | - | - | Y |
| 8 | 3.42 | - | - | I | - | - | - | - | I | - | - | - | - | - | - | - | I | S | - | - | I | - | - | I | L | - |
| 45 | 4.30 | - | S | - | - | - | - | - | I | - | - | F | - | - | - | - | I | S | - | - | V | - | - | I | L | - |
Amino acid residues in the drug-binding pocket are defined as those for which any atom is within 4 Å of a panel of compounds bound in HRV-1A or HRV-14 (27). Amino acid numbering is based on VP1 sequence of HRV-16 (38). The majority sequence is defined as the amino acid residue that appears most frequently in the group of viruses analyzed. Amino acids that are conserved across all HRV-A serotypes are in boldface, while those that vary by only one residue are underscored. -, Same amino acid as in the majority sequence.
EC50 is the concentration of pleconaril that protected 50% of a cell monolayer from virus-induced cytopathology. Values represent means of two or more independent tests.
TABLE 3.
Alignment of amino acids in drug-binding pocket of HRV-B virusesa
| Serotype | EC50 (μM)b | Amino acid at position:
|
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 104 | 105 | 106 | 107 | 124 | 126 | 128 | 130 | 150 | 151 | 152 | 174 | 175 | 176 | 186 | 188 | 191 | 197 | 199 | 215 | 219 | 221 | 224 | 245 | 267 | ||
| Majority sequence | I | N | L | S | F | S | Y | I | A | M | Y | P | S | V | F | V | V | Y | C | I | N | M | M | H | G | |
| Serotype | ||||||||||||||||||||||||||
| 83 | 0.02 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 92 | 0.02 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 35 | 0.04 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 79 | 0.04 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 6 | 0.06 | - | - | F | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | I | - | - |
| 37 | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 3 | 0.09 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 86 | 0.13 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | I | - | - |
| 14 | 0.16 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 17 | 0.23 | - | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | I | - | - |
| 70 | 0.30 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | I | - | - |
| 91 | 0.73 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | I | - | - | - | - | - | I | - | - |
| 72 | 0.94 | - | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 26 | 1.60 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | L | - | - | - | - | - | - | - | - |
| 48 | 1.80 | - | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | - | - | - | - | - | I | - | - |
| 52 | 2.49 | V | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | I | - | - |
| 27 | 5.90 | - | - | - | - | - | - | - | - | - | - | F | - | - | - | - | - | L | - | N | - | - | - | I | - | - |
| 69 | 6.68 | - | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | - | - | - | - | - | I | - | - |
| 4 | >12.5 | - | S | - | - | - | - | - | - | - | - | F | - | - | - | - | - | L | - | - | - | - | - | I | - | - |
| 5 | >12.5 | - | - | - | - | - | - | - | - | - | - | F | - | - | - | - | - | L | - | - | - | - | - | - | - | - |
| 42 | >12.5 | - | - | - | - | - | - | - | - | - | - | F | - | - | - | - | - | L | - | - | - | - | - | - | - | - |
| 84 | >12.5 | - | - | - | - | - | - | - | - | - | - | F | - | - | - | - | - | L | - | N | - | - | - | - | - | - |
| 93 | >12.5 | - | - | - | - | - | - | - | - | - | - | F | - | - | I | - | - | L | - | N | - | - | - | I | - | - |
| 97 | >12.5 | - | - | - | - | - | - | - | - | - | - | F | - | - | - | - | - | L | - | N | - | - | - | - | - | - |
| 99 | >12.5 | - | - | - | - | - | - | - | - | - | - | F | - | - | - | - | - | L | - | - | - | - | - | I | - | - |
Amino acid residues in the drug-binding pocket are defined as those for which any atom is within 4 Å of compounds bound in HRV-14 (27). Amino acid numbering is based on VP1 sequence of HRV-14 (27). The majority sequence is defined as the amino acid residue that appears most frequently in the group of viruses analyzed. Amino acids that are conserved across all HRV-B serotypes are in boldface, while those that vary by only one residue are underscored. -, Same amino acid as in the majority sequence.
See Table 2, footnote b.
Alignment of amino acids in the drug-binding pockets of HRV-A and HRV-B species members are shown in Tables 2 and 3, respectively. The amino acids chosen for this analysis are those with any atom within 4 Å of a collection of related antiviral compounds as determined by X-ray crystallography in HRV-1A and HRV-14 (27). Amino acid numbering is based on the sequence of HRV-16 (HRV-A) and HRV-14 (HRV-B). Viruses are listed by order of decreasing susceptibility to pleconaril.
Although the pleconaril susceptibility range of virus serotypes of the HRV-A genetic group was broad (EC50 = 0.01 to 4.3 μM), all species members were inhibited in cell culture at nontoxic drug concentrations. Of the 25 amino acid residues in the drug-binding pocket, 8 are conserved across all HRV-A viruses (Phe118, Ser120, Met143, Ser167, Tyr190, Met192, Asn212, and Met214; Table 2 and Fig. 4). All but one of these amino acid positions (Met192) is also conserved in HRV-B viruses (the equivalent positions in HRV-B viruses are Phe124, Ser126, Met151, Ser175, Tyr197, Asn219, and Met221, respectively). Ten additional positions in the pocket of HRV-A viruses are occupied by one of only two amino acid residues, including VP1 amino acid position 98 (Ile or Val in all viruses), 100 (Leu or Ile), 142 (Tyr or Phe), 144 (Tyr or Phe), 168 (Val or Ile), 179 (Phe or Ile), 181 (Leu or Ile), 217 (Leu or Ile), and 238 (His or Leu). Overall, the drug-binding pocket amino acid sequence of the majority of HRV-A viruses is highly conserved, with a median number of amino acid differences between serotype pairs of 5.0.
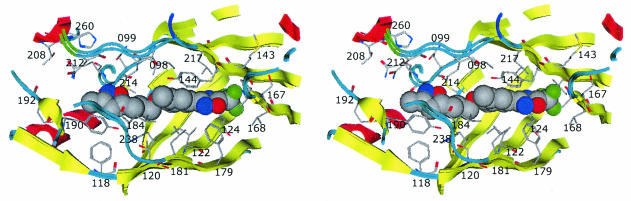
FIG. 4.
Stereo pair ribbon diagram of drug-binding pocket with bound pleconaril. The crystal structures of HRV-14 and HRV-16 are superimposed. Amino acid residues of HRV-16 are shown. The image was generated in MOE.
HRV-B species members similarly span a broad pleconaril susceptibility range, with EC50 values of 0.02 to >12.5 μM (Table 3). However, the amino acid sequence diversity in the VP1 drug-binding pocket among the HRV-B viruses appeared less than that observed with the HRV-A viruses. Of the 25 amino acids in HRV-B viruses, 17 were identical across all prototype strains. Of the eight positions that varied, only one position (i.e., position 191) was occupied by more than two different amino acid residues in prototype HRV strains. The median number of pocket amino acid differences between HRV-B species members was only 3.0. All seven prototype viruses that were not susceptible to inhibition by pleconaril in cell culture (EC50 > 12.5 μM) differed from the predominant sequence at amino acid positions 152 (Phe in naturally resistant serotypes and Tyr in susceptible serotypes) and 191 (Leu in naturally resistant serotypes and Val in susceptible serotypes). An eighth prototype, HRV-27, which was marginally susceptible to pleconaril (EC50 = 5.9 μM), shared this same Phe152/Leu191 pattern. These results suggest that presence of Phe152/Leu191 is “diagnostic” of naturally occurring resistance or weak susceptibility to pleconaril in prototype HRV-B viruses.
Relationship between drug susceptibility and pocket amino acid sequence.
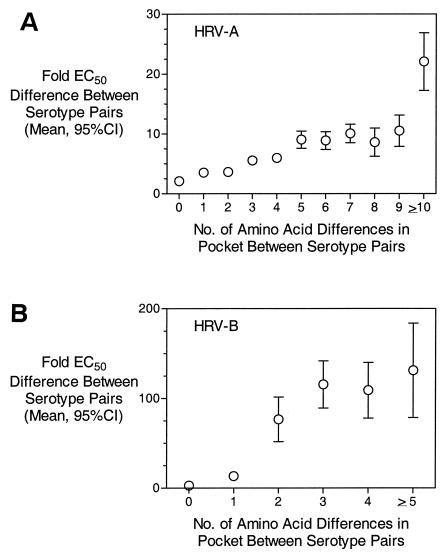
Pleconaril interacts almost exclusively with amino acids in the drug-binding pocket of VP1 (Fig. 4). This pocket is buried beneath the surface of the virus and is accessed via a narrow pore that extends to the base of the virus canyon (45). Amino acid residues that line this pore, or those immediately outside of the pocket, could have a role in determining drug susceptibility to compounds such as pleconaril. To gain a better understanding of the relationship between pocket amino acid residues and drug susceptibility, a pairwise comparison was made for all HRV-A viruses at all 25 amino acid positions in the drug-binding pocket. The number of nonconserved amino acids was then determined and plotted against the mean (with a 95% confidence intervals [CI]) fold difference in EC50 value to pleconaril between the two isolates. As shown in Fig. 5A, when the drug-binding pockets of two HRV-A viruses were identical, the mean difference in susceptibility (EC50 values) to pleconaril was 2.1-fold (95% CI = 1.7- to 2.5-fold). This indicates that when two viruses have identical drug-binding pocket sequences, they share a nearly identical susceptibility to pleconaril. As the number of differences between viruses in the drug-binding pocket increases, the predictive power for drug susceptibility decreases. For example, with only one amino acid difference, the mean fold difference in EC50 value was 3.5 (95% CI = 3.0 to 4.0). When viruses differed by 5 or ≥10 amino acids, the mean fold differences in EC50 value were 9.0 (95% CI = 7.6 to 10.5) and 22.1 (95% CI = 17.3 to 26.9), respectively. Even more dramatic results were observed with HRV-B species viruses (Fig. 5B). Viruses with identical drug-binding pocket sequences had a mean fold difference in EC50 value of 2.8 (95% CI = 2.3 to 3.4). With only a single amino acid change in the drug-binding pocket, the mean fold-difference in EC50 value jumped to 13.2 (95% CI = 9.6 to 16.9). These results indicate that primary amino acid sequence in the drug-binding pocket is the major driver for and predictor of drug susceptibility.
FIG. 5.
Relationship of amino acid conservation in drug-binding pocket and virus susceptibility to pleconaril inhibition. The mean fold difference (with 95% CI) in pleconaril EC50 values between serotype pairs is plotted as a function of the number of amino acid differences in the drug-binding pocket.
DISCUSSION
Phylogenetic relationship of HRV serotypes based on VP1 sequence.
The present study reports the sequence for the entire VP1 capsid protein-coding region of all prototypic HRV strains. As reported first by Andries et al., using patterns of susceptibilities to antiviral compounds and later by several laboratories using sequencing of other regions of the genome, 101 members of the HRV genus can be subdivided into two distinct genetic groupings or species based on nucleotide and/or amino acid sequence relatedness in VP1 (1). In the present study, 75 of the 101 serotypes (plus HRV-Hanks) cluster in species A (HRV-A), 25 cluster into species B (HRV-B), and a single serotype, HRV-87, best fits with enterovirus species D (5, 36, 37, 43). Interserotypic amino acid identity scores in VP1 were as low as 57.7 and 68.3% within genetic groups HRV-A and HRV-B, respectively. In contrast, intergroup amino acid identity scores in VP1 ranged from 35.1 to 43.6%.
Phylogenetic grouping of HRV serotypes based on the VP1 deduced amino acid sequence yielded results comparable to those reported for sequencing across the VP4/VP2 junction (our results and reference 43). Sequence data generated across the VP4/VP2 junction in the present study were in excellent agreement with those reported previously with the single exception of serotype HRV-31 (43). Although Savolainen et al. reported this virus to be highly homologous to HRV-32, our data suggest that these two viruses differ significantly from one another both in the VP4/VP2 junction and in VP1 (43). The serotypic identity of the HRV-31 strain studied here was confirmed with monospecific neutralizing antiserum. Furthermore, the HRV-31 sequence data generated here was distinct from all other HRV strains, indicating that cross-contamination had not occurred during RT-PCR. Based on the phylogenetic analysis of our data, HRV-31 is closely related to HRV-47 and HRV-32 clusters with HRV-9 (Fig. 1).
Vlasak et al. reported recently on the VP1 amino acid sequence for all HRV serotypes that utilize the low-density lipoprotein receptor family to bind cells (the minor receptor binding group HRVs) (20, 50). Their phylogenetic analyses are in excellent agreement with those reported here and support our data that HRV-31 clusters with HRV-47 as noted above. It is interesting that, as shown by our data and those reported previously, minor receptor-binding group HRV serotypes are distributed throughout the HRV-A species phylogenetic tree (Fig. 1 and reference 43). Indeed, HRV-25 (a major receptor-binding group virus) and HRV-62 (a minor receptor-binding group virus) are closely related pairs, with only 13 amino acid differences across all of VP1 (282 amino acid residues, 95.4% identity; Table 1). Despite this high level of amino acid identity in VP1, only a one-way heterologous cross-reactivity with antisera raised against these viruses was observed; i.e., antiserum to HRV-25 neutralized HRV-62 at an ∼10-fold-higher concentration than it neutralized HRV-25 (data not shown and references 7 and 8). Antiserum to HRV-62 did not cross-react with HRV-25 (data not shown).
Of the 13 amino acid residue differences between HRV-25 and HRV-62 in VP1, 4 fall within the region proposed by Vlasak et al. to be involved in receptor discrimination: Ser87 in HRV-62 to Lys in HRV-25, Lys89 to Asp, Asp90 to Glu, and Thr85 to Lys (in the neighboring VP1 loop) (50). Both viruses contain a Lys at 224 that is proposed to be essential for interaction with the minor receptor protein (50). If the hypothesis proposed in that study is correct, then conversion of one or more of these residues in HRV-25 to the corresponding HRV-62 amino acid should result in a virus capable of utilizing either ICAM-1 (intercellular adhesion molecule-1) or a member of the low-density-lipoprotein receptor family to bind to cells.
Several of the serotype pairs presented in Table 1 have been reported to share some degree of antigenic cross-reactivity (7, 8, 18). The sequencing of VP1 reported here indicated that HRV-8-HRV-95 and HRV-21-HRV-Hanks were closely related pairs, with amino acid identities of 98.6 and 97.2%, respectively. This raised the question of the serotypic uniqueness of HRV-95 and HRV-Hanks. Indeed, monospecific antisera raised against HRV-8 and HRV-21 neutralized, respectively, HRV-95 and HRV-Hanks at similar dilutions. These results suggest that HRV-95 was erroneously classified as a novel HRV serotype (18) and that the previously untyped HRV-Hanks is in fact, HRV-21.
Amino acid conservation in the VP1 drug-binding pocket and relationship to virus susceptibility to pleconaril.
X-ray crystallographic studies have shown that the pocket into which compounds such as pleconaril integrate is nestled within the eight-stranded antiparallel β-barrel sandwich of VP1 (27, 45). In most wild-type picornavirus structures, this pocket is occupied by a natural fatty acid ligand referred to as “pocket factor” (12, 26, 50). The integration of compounds such as pleconaril results in displacement of the pocket factor from the hydrophobic pocket, which in turn leads to measurable stabilization of the virus capsid and inhibition of capsid functions (1, 17, 30, 32, 39-41).
Overall, there is a strikingly high degree of amino acid identity within the drug-binding pockets of viruses within the two HRV species. Of the 25 amino acid residue positions in the drug-binding pocket of HRV-A and HRV-B species viruses, 18 and 24, respectively, are identical or differ by only a single amino acid across all of the respective genetic group members (Tables 2 and 3). The median amino acid identity in the drug-binding pocket of HRV-A and HRV-B viruses (80.0 and 88.0%, respectively) is significantly higher than the identity in VP1 overall (70.2 and 74.3%, respectively; P < 0.0001 [Mann-Whitney nonparametric test]). Moreover, drug susceptibility tracks remarkably well with pocket conservation. For any two viruses with identical pocket amino acid composition, their susceptibility to pleconaril differs by an average of only 2.1-fold (95% CI = 1.7 to 2.5) for HRV-A species members and by only 2.8-fold (95% CI = 2.3 to 3.4) for HRV-B species members (Fig. 5). This result was unexpected in that several factors could be involved in compound binding to and inhibition of virus replication beyond the primary amino acid sequence of the drug-binding pocket. These factors could include, for example, pocket shape dictated by amino acids in flanking beta strands, conformational changes induced by compound binding, or accessibility of the pocket to the compound (39, 45). Nevertheless, the data presented here indicate that primary amino acid sequence in the drug-binding pocket can be used to predict susceptibility of a virus isolate to pleconaril.
Genetic basis for naturally occurring phenotypic resistance to pleconaril.
All seven HRV-B serotypes that are naturally resistant to the antiviral effects of pleconaril in cell culture cluster tightly on the phylogenetic tree (Fig. 2). All share an amino acid pattern of Phe152 and Leu191 in the drug-binding pocket, which, when observed in HRV-B viruses, is positively correlated with naturally occurring resistance or weak susceptibility to pleconaril. No atomic structures are available for HRV-B viruses possessing a Phe152 and Leu191 amino acid residue pattern in VP1. However, the capsid structure of HRV-14, an HRV-B virus, has been determined to atomic resolution (3, 42). HRV-14 is susceptible to inhibition by pleconaril (EC50 = 0.16 μM) and differs from the naturally resistant serotypes HRV-5 and HRV-42 at only the 152 (Tyr in HRV-14) and 191 (Val) positions in the drug-binding pocket (Table 3).
In HRV-14 (and HRV-16, an HRV-A virus), the hydroxyl function of Tyr152 is hydrogen bonded to the backbone carbonyl oxygen of Met221 (oxygen-to-oxygen distance of 2.9 Å), enhancing rigidity in that region of the pocket. The absence of this hydrogen bond in viruses possessing a Phe152, when computationally modeled, does not result in a significant rearrangement of the benzyl side chain moiety either in the presence or in the absence of ligand. More conformational flexibility is observed in the Met221 side chain when its carbonyl hydrogen bond to Tyr152 is removed. An increase in the conformational freedom of Met221 may contribute to constriction of the drug-binding pocket diameter, which may in turn affect the binding affinity of pleconaril. However, it is not possible to predict or quantify this potential impact on drug binding.
In contrast, the Leu side chain at position 191 in HRV-B viruses is likely to play a critical role in determining drug susceptibility. Overall, the drug-binding pocket of HRV-14 is significantly narrower than that of HRV-16. This constriction of the HRV-14 pocket is attributed largely to the amino acid residues surrounding the dimethylphenoxy moiety of pleconaril: Trp102, Tyr128, Val191, and Met224 (Fig. 6). Modeling of a bulkier Leu in place of Val at position 191 results in a further narrowing of the drug-binding pocket in HRV-14, which would likely have a deleterious steric effect on the binding of pleconaril.
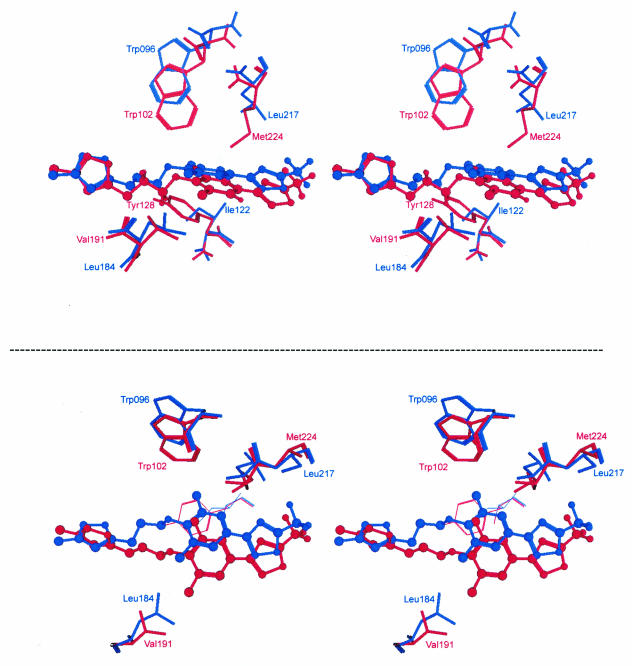
FIG. 6.
Amino acid residues proposed to affect binding of central ring system of pleconaril and to affect natural resistance to the drug. The crystal structure of pleconaril bound in HRV-14 (red) and HRV-16 (blue) is shown. Two stereo pair images are shown to emphasize the difference in amino acid composition in the top of the virus pockets (upper image) and the position of the dimethylphenoxy group of pleconaril (bottom image). The image was generated in MOE.
It is of interest to note that the majority of HRV-A viruses possess a Leu at the amino acid position corresponding to 191 in HRV-B viruses (Table 2). In HRV-16, the side chain of Trp96 (the corresponding amino acid to Trp102 in HRV-14) is shifted by 1.5 to 2.2 Å out from the center of the pocket relative to its position in HRV-14. In addition, the neighboring Met224 of HRV-14 is replaced by the smaller Leu217 side chain in HRV-16. Together, these two residue differences create significant additional space in the roof of the HRV-16 pocket (Fig. 6). The dimethylphenoxy moiety of pleconaril is shifted considerably in HRV-16 relative to HRV-14 (1.8 to 2.5 Å) in order to then accommodate the bulkier Leu side chain at position 184 in the bottom of the HRV-16 pocket (Fig. 6). This positional shift is tolerated because of the space afforded in the pocket roof by Trp96 and Leu217. Taken together, the crystallographic results suggest a plausible explanation for why Leu191 is associated with poor drug susceptibility in HRV-B viruses but not at the corresponding position in HRV-A viruses. Our data indicate that Phe152/Leu191 in VP1 of HRV-B viruses is diagnostic of naturally occurring weak susceptibility or full resistance to pleconaril. Further sequencing of naturally occurring, weakly susceptible or resistant clinical isolates will be required to fully establish this association. In addition, site-directed mutagenesis experiments using an infectious cDNA clone of HRV-14 to quantify the relative contribution of Phe152 and Leu191 in the naturally occurring drug resistance phenotype are ongoing.
Genetic comparison of naturally occurring and drug treatment-emergent resistance to pleconaril.
In the autumn of 2000, ViroPharma conducted two large phase 3 clinical studies to examine the safety and efficacy of pleconaril for treatment of community-acquired viral upper-respiratory-tract disease of picornavirus etiology. Virus isolation from patients was performed prior to pleconaril treatment (baseline) and after initiation of treatment (postbaseline [days 3 and 6]). Of patients with baseline drug-susceptible viruses, postbaseline nonsusceptible viruses were isolated from only 2.7% of pleconaril-treated patients (19). Sequencing of the VP1 protein-coding region of these isolates revealed that the majority were HRV-A viruses. Two positions in the drug-binding pocket were frequently associated with a reduction in drug susceptibility: (i) Ile98 (to Met or Phe) and (ii) Ile122 to Phe or Val122 to Leu (unpublished data). Interestingly, neither Met nor Phe occurs at amino acid position 98 in any prototype HRV-A virus or at the equivalent position in HRV-B viruses (position 104). The same is true for Phe122 (position 130 in HRV-B viruses). These results indicate that drug selection pressure resulted in the emergence of virus variants with amino acid sequence variations distinct from those observed in the naturally drug-resistant viruses. Virus variants with the drug treatment-emergent VP1 sequence phenotype (Ile at position 98 or 122) do exist at low levels in drug-susceptible, wild-type virus populations (unpublished results and reference 17). The fact that the VP1 sequence pattern of drug treatment-emergent viruses with reduced susceptibility to pleconaril is not observed among the prototypic HRVs may indicate that such variant viruses are at a competitive disadvantage or suffer a fitness liability (17).
The absence of Met or Phe at position 98 and Phe at 122 in prototype strains may serve as a useful molecular diagnostic marker for pleconaril treatment-emergent resistance should the drug be approved for widespread use in treatment of viral upper-respiratory-tract disease. Combined with the results discussed above, a resistance-monitoring program could be envisioned in which viruses with naturally occurring resistance (Phe152/Leu191 in the HRV-B group) can be readily distinguished from those with treatment-emergent resistance (often associated with an amino acid change at position 98 or 122 in the HRV-A group), if such viruses are able survive or compete in nature.
In summary, the data reported here contribute to the HRV sequence database and provide insight into the phylogenetic relationships among the many viruses of the HRV genus. These data may form the basis for possible molecular serotyping of HRVs, analogous to the serotyping reported for the human enterovirus genus (36, 37). Further, upon analysis of the drug-binding pocket in VP1, amino acid sequence patterns associated with naturally occurring virus resistance to pleconaril were identified that are distinct for those involved in drug treatment-emergent resistance. If confirmed through sequencing of clinical HRV isolates that are naturally resistant to pleconaril, these data could form the basis of a resistance-monitoring program should pleconaril be approved for broad use in the treatment of viral upper respiratory infection.
Acknowledgments
We thank Gillian Kunze and Maureen Kealey for technical assistance and Mark McKinlay for critical review of the manuscript.
REFERENCES
- 1.Andries, K., B. Dewindt, J. Snoeks, L. Wouters, H. Moereels, P. J. Lewi, and P. A. J. Janssen. 1990. Two groups of rhinoviruses revealed by a panel of antiviral compounds present sequence divergence and differential pathogenicity. J. Virol. 64:1117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andries, K., B. Dewindt, J. Snoeks, R. Willebrords, K. van Eemeren, R. Stokbroekx, and P. A. Janssen. 1992. In vitro activity of pirodavir (R77975), a substituted phenoxy-pyridazinamine with broad-spectrum antipicornaviral activity. Antimicrob. Agents Chemother. 36:100-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold, E., and M. G. Rossmann. 1990. Analysis of the structure of a common cold virus, human rhinovirus 14, refined at a resolution of 3.0 Å. J. Mol. Biol. 211:763-801. [DOI] [PubMed]
- 4.Bertino, J. S. 2002. Cost burden of viral respiratory infections: issues for formulary decision makers. Am. J. Med. 112:42S-49S. [DOI] [PubMed]
- 5.Blomqvist, S., C. Savolainen, L. Raman, M. Roivainen, and T. Hovi. 2002. Human rhinovirus 87 and enterovirus 68 represent a unique serotype with rhinovirus and enterovirus features. J. Clin. Microbiol. 40:4218-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conant, R. M., and V. V. Hamparian. 1968. Rhinoviruses: basis for a numbering system. II. Serologic characterization of prototype strains. J. Immunol. 100:114-119. [PubMed] [Google Scholar]
- 7.Cooney, M. K., J. A. Wise, G. E. Kenny, and J. P. Fox. 1975. Broad antigenic relationships among rhinovirus serotypes revealed by cross-immunization of rabbits with different serotypes. J. Immunol. 114:635-639. [PubMed] [Google Scholar]
- 8.Cooney, M. K., J. P. Fox, and G. E. Kenny. 1982. Antigenic groupings of 90 rhinovirus serotypes. Infect. Immun. 37:642-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denny, F. W., Jr. 1995. The clinical impact of human respiratory virus infections. Am. J. Respir. Crit. Care Med. 152:S4-S12. [DOI] [PubMed] [Google Scholar]
- 10.Dewindt, B., K. van Eemeren, and K. Andries. 1994. Antiviral capsid-binding compounds can inhibit the adsorption of minor receptor rhinoviruses. Antivir. Res. 25:67-72. [DOI] [PubMed] [Google Scholar]
- 11.Fendrick, A. M., A. S. Monto, B. Nightengale, and M. Sarnes. 2003. The economic burden of non-influenza-related viral respiratory tract infection in the United States. Arch. Intern. Med. 163:487-494. [DOI] [PubMed] [Google Scholar]
- 12.Filman, D. J., R. Syed, M. Chow, A. J. Macadam, P. D. Minor, and J. M. Hogle. 1989. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 8:1567-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox, M. P., M. J. Otto, and M. A. McKinlay. 1986. Prevention of rhinovirus and poliovirus uncoating by WIN 51711, a new antiviral drug. Antimicrob. Agents Chemother. 30:110-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzales, R., J. G. Bartlett, R. E. Besser, R. J. Cooper, J. M. Hickner, J. R. Hoffman, and M. A. Sande. 2001. Principles of appropriate antibiotic use for treatment of acute respiratory tract infections in adults: background, specific aims, and methods. Ann. Intern. Med. 134:479-486. [DOI] [PubMed] [Google Scholar]
- 15.Gonzales, R., J. F. Steiner, and M. A. Sande. 1997. Antibiotic prescribing for adults with colds, upper respiratory tract infections, and bronchitis by ambulatory care physicians. JAMA 278:901-904. [PubMed] [Google Scholar]
- 16.Greve, J. M., G. Davis, A. M. Meyer, C. P. Forte, S. C. Yost, C. W. Marlor, M. E. Kamarck, and A. McClelland. 1989. The major human rhinovirus receptor is ICAM-1. Cell 56:839-847. [DOI] [PubMed] [Google Scholar]
- 17.Groarke, J. M., and D. C. Pevear. 1999. Attenuated virulence of pleconaril-resistant coxsackievirus B3 variants. J. Infect. Dis. 179:1538-1541. [DOI] [PubMed] [Google Scholar]
- 18.Hamparian, V. V., R. J. Colonno, M. K. Cooney, E. C. Dick, J. M. Gwaltney, J. H. Hughes, W. S. Jordan, Jr., A. Z. Kapikian, W. J. Mogabgab, A. Monto, C. A. Phillips, R. R. Rueckert, J. H. Schieble, E. J. Stott, and D. A. J. Tyrrell. 1987. A collaborative report: rhinoviruses-extension of the numbering system from 89 to 100. Virology 159:191-192. [DOI] [PubMed] [Google Scholar]
- 19.Hayden, F. G., D. T. Herrington, T. L. Coats, K. Kim, E. C. Cooper, S. A. Villano, S. Liu, S. Hudson, D. C. Pevear, M. Collett, M. McKinlay, et al. 2003. Efficacy and safety of oral pleconaril for treatment of colds due to picornaviruses in adults: results of 2 double-blind, randomized, placebo-controlled trials. Clin. Infect. Dis. 36:1523-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofer, F., M. Gruenberger, H. Kowalski, H. Machat, M. Huettinger, E. Kuechler, and D. Blaas. 1994. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc. Natl. Acad. Sci. USA 91:1839-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hogle, J. M., M. Chow, and D. J. Filman. 1985. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science 229:1358-1365. [DOI] [PubMed] [Google Scholar]
- 22.Horsnell, C., R. E. Gama, P. J. Hughes, and G. Stanway. 1995. Molecular relationships between 21 human rhinovirus serotypes. J. Gen. Virol. 76:2549-2555. [DOI] [PubMed] [Google Scholar]
- 23.Ishiko, H., R. Miura, Y. Shimada, A. Hayashi, H. Nakajima, S. Yamazaki, and N. Takeda. 2002. Human rhinovirus 87 identified as human enterovirus 68 by VP4-based molecular diagnosis. Intervirology 45:136-141. [DOI] [PubMed] [Google Scholar]
- 24.Kapikian, A. Z., R. M. Conant, V. V. Hamparian, R. M. Chanock, P. J. Chapple, E. C. Dick, J. D. Fenters, J. M. Gwaltney, Jr., D. Hamre, J. C. Holper, W. S. Jordan, Jr., E. H. Lennette, J. L. Melnick, W. J. Mogabgab, M. A. Mufson, C. A. Phillips, J. H. Schieble, and D. A. J. Tyrrell. 1967. Rhinoviruses: a numbering system. Nature 213:761-763.4291698 [Google Scholar]
- 25.Kapikian, A. Z., R. M. Conant, V. V. Hamparian, R. M. Chanock, E. C. Dick, J. D. Fenters, J. M. Gwaltney, Jr., D. Hamre, W. S. Jordan, Jr., G. E. Kenny, E. H. Lennette, J. L. Melnick, W. J. Mogabgab, C. A. Phillips, J. H. Schieble, E. J. Stott, and D. A. J. Tyrrell. 1971. A collaborative report: rhinoviruses-extension of the numbering system. Virology 43:524-526.
- 26.Kim, S., T. J. Smith, M. S. Chapman, M. G. Rossmann, D. C. Pevear, F. J. Dutko, P. J. Felock, G. D. Diana, and M. A. McKinlay. 1989. The crystal structure of human rhinovirus serotype 1A HRV1A. J. Mol. Biol. 210:91-111. [DOI] [PubMed] [Google Scholar]
- 27.Kim, K. H., P. Willingmann, Z. X. Gong, M. J. Kremer, M. S. Chapman, I. Minor, M. A. Oliveira, M. G. Rossmann, K. Andries, G. D. Diana, F. J. Dutko, M. A. McKinlay, and D. C. Pevear. 1993. A comparison of the anti-rhinoviral drug binding pocket in HRV14 and HRV1A. J. Mol. Biol. 230:206-227. [DOI] [PubMed] [Google Scholar]
- 28.Mainous, A. G., III, W. J. Hueston, M. M. Love, M. E. Evans, and R. Finger. 2000. An evaluation of statewide strategies to reduce antibiotic overuse. Fam. Med. 32:22-29. [PubMed] [Google Scholar]
- 29.McIsaac, W. J., N. Levine, and V. Goel. 1998. Visits by adults to family physicians for the common cold. J. Fam. Pract. 47:366-369. [PubMed] [Google Scholar]
- 30.Moeremans, M., M. De Raeymaeker, G. Daneels, M. De Brabander, F. Aerts, C. Janssen, and K. Andries. 1992. Study of the parameters of binding of R 61837 to human rhinovirus 9 and immunobiochemical evidence of capsid-stabilizing activity of the compound. Antimicrob. Agents Chemother. 36:417-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori, J., and J. P. Clewley. 1994. Polymerase chain reaction and sequencing for typing rhinovirus RNA. J. Med. Virol. 44:323-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosser, A. G., and R. R. Rueckert. 1993. WIN 51711-dependent mutants of poliovirus type 3: evidence that virions decay after release from cells unless drug is present. J. Virol. 67:1246-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.National Center for Health Statistics, Centers for Disease Control and Prevention, Vital and Health Statistics. 1996. Current estimates from the National Health Interview Survey. [Online] http://www.cdc.gov/nchs/data/series/sr_10/sr10_200.pdf.
- 34.National Institute of Allergy and Infectious Diseases. 2001. The common cold. NIAID Fact Sheet, National Institute of Allergy and Infectious Diseases, National Institutes of Health, U.S. Department of Health and Human Services. [Online] http://www.niaid.nih.gov/factsheets/cold.htm.
- 35.Nyquist, A. C., R. Gonzales, J. F. Steiner, and M. A. Sande. 1998. Antibiotic prescribing for children with colds, upper respiratory tract infections, and bronchitis. JAMA 279:875-877. [DOI] [PubMed] [Google Scholar]
- 36.Oberste, M. S., K. Maher, D. R. Kilpatrick, M. R. Flemister, B. A. Brown, and M. A. Pallansch. 1999. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 37:1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oberste, M. S., K. Maher, D. R. Kilpatrick, and M. A. Pallansch. 1999. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 73:1941-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira, M. A., R. Zhao, W.-M. Lee, M. J. Kremer, I. Minor, R. R. Rueckert, G. D. Diana, D. C. Pevear, F. J. Dutko, M. A. McKinlay, and M. G. Rossmann. 1993. The structure of human rhinovirus 16. Structure 1:51-68. [DOI] [PubMed] [Google Scholar]
- 39.Pevear, D. C., M. J. Fancher, P. J. Felock, M. G. Rossmann, M. S. Miller, G. Diana, A. M. Treasurywala, M. A. McKinlay, and F. J. Dutko. 1989. Conformational change in the floor of the human rhinovirus canyon blocks adsorption to HeLa cell receptors. J. Virol. 63:2002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phelps, D. K., and C. B. Post. 1995. A novel basis for capsid stabilization by antiviral compounds. J. Mol. Biol. 254:544-551. [DOI] [PubMed] [Google Scholar]
- 41.Phelps, D. K., P. J. Rossky, and C. B. Post. 1998. Influence of an antiviral compound on the temperature dependence of viral protein flexibility and packing; a molecular dynamics study. J. Mol. Biol. 276:331-337. [DOI] [PubMed] [Google Scholar]
- 42.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H. J. Hecht, J. E. Johnson, G. Kamer, M. Luo, A. G. Mosser, R. R. Rueckert, B. Sherry, and G. Vriend. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145-153. [DOI] [PubMed] [Google Scholar]
- 43.Savolainen, C., S. Blomqvist, M. N. Mulders, and T. Hovi. 2002. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J. Gen. Virol. 83:333-340. [DOI] [PubMed] [Google Scholar]
- 44.Shepard, D. A., B. A. Heinz, and R. R. Rueckert. 1993. WIN 52035-2 inhibits both attachment and eclipse of human rhinovirus 14. J. Virol. 67:2245-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, T. J., M. J. Kremer, M. Luo, G. Vriend, E. Arnold, G. Kamer, M. G. Rossmann, M. A. McKinlay, G. D. Diana, and M. J. Otto. 1986. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science 233:1286-1293. [DOI] [PubMed] [Google Scholar]
- 46.Staunton, D. E., V. J. Merluzzi, R. Rothlein, R. Barton, S. D. Marlin, and T. A. Springer. 1989. A cell adhesion molecular, ICAM-1, is the major surface receptor for rhinoviruses. Cell 56:849-853. [DOI] [PubMed] [Google Scholar]
- 47.Steinman, M. A., C. S. Landefeld, and R. Gonzales. 2003. Predictors of broad-spectrum antibiotic prescribing for acute respiratory tract infections in adult primary care. JAMA 289:719-725. [DOI] [PubMed] [Google Scholar]
- 48.Stone, S., R. Gonzales, J. Maselli, and S. R. Lowenstein. 2000. Antibiotic prescribing for patients with colds, upper respiratory tract infections, and bronchitis: a national study of hospital-based emergency departments. Ann. Emerg. Med. 36:320-327. [DOI] [PubMed] [Google Scholar]
- 49.Uncapher, C. R., C. M. DeWitt, and R. J. Colonno. 1991. The major and minor group receptor families contain all but one human rhinovirus serotype. Virol. 180:814-817. [DOI] [PubMed] [Google Scholar]
- 50.Vlasak, M., S. Blomqvist, T. Hovi, E. Hewat, and D. Blaas. 2003. Sequence and structure of human rhinoviruses reveal the basis of receptor discrimination. J. Virol. 77:6923-6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeates, T. O., D. H. Jacobson, A. Martin, C. Wychowski, M. Girard, D. J. Filman, and J. M. Hogle. 1991. Three-dimensional structure of a mouse-adapted type 2/type 1 poliovirus chimera. EMBO J. 10:2331-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeichhardt, M. J. H., Otto, M. A. McKinlay, P. Willingmann, and K. O. Habermehl. 1987. Inhibition of poliovirus uncoating by disoxaril (WIN 51711). Virology 160:281-285. [DOI] [PubMed] [Google Scholar]