Summary
Programmed cell death is a gene-directed process involved in the development and homeostasis of multicellular organisms. The most common mode of programmed cell death is apoptosis, which is characterized by a stereotypical set of biochemical and morphological hallmarks. Here we report that Escherichia coli also exhibit characteristic markers of apoptosis – including phosphatidylserine exposure, chromosome condensation and DNA fragmentation – when faced with cell death-triggering stress, namely bactericidal antibiotic treatment. Notably, we also provide proteomic and genetic evidence for the ability of multifunctional RecA to bind peptide sequences that serve as substrates for eukaryotic caspases, and regulation of this phenotype by the protease, ClpXP, under conditions of cell death. Our findings illustrate that prokaryotic organisms possess mechanisms to dismantle and mark dying cells in response to diverse noxious stimuli, and suggest that elaborate, multilayered proteolytic regulation of these features may have evolved in eukaryotes to harness and exploit their deadly potential.
Introduction
Apoptosis is a genetically regulated form of programmed cell death (PCD) that is essential to the development and long-term viability of multicellular organisms (Kerr et al., 1972; Wyllie et al., 1980). Induction of apoptosis, which takes place in response to a variety of intra- and extracellular stimuli and stresses, typically involves the deployment of a family of conserved cysteine-dependent regulatory proteases with aspartic acid specificity, or caspases (Hengartner, 2000; Nicholson and Thornberry, 1997; Strasser et al., 2000); it should be noted, however, that apoptosis may occur in a caspase-independent manner involving catalytically-distinct proteases (Broker et al., 2005). In general, a commitment to apoptosis (whether caspase-dependent or caspase-independent) is characterized by a series of defined biochemical and morphological events that predispose, precede and accompany death. The cumulative effects of these physiological changes are designed to induce cell cycle arrest, halt DNA repair and homeostasis, inactivate apoptosis inhibitor proteins, facilitate ultrastructural modifications, and tag the dying cell, thereby achieving the deconstruction of biomolecular architecture, breakdown of cellular contents and marking for death (Danial and Korsmeyer, 2004).
This series of controlled events includes chromatin condensation, DNA fragmentation, and exposure of phosphatidylserine (PS) phospholipid on the outer membrane leaflet (Kerr et al., 1972; Wyllie, 1980). These physiological changes ultimately prevent harm from befalling neighboring cells that would occur during uncontrolled death, and together lead to recognition and phagocytosis of the dying cell by macrophages, or engulfment by nearby cells, to achieve this goal (Reddien and Horvitz, 2004; Ren and Savill, 1998). As such, the phenotypic hallmarks listed above routinely provide the basis for differentiation between apoptosis and other major forms of PCD. Accordingly, it has been proposed by the Nomenclature Committee on Cell Death that several of these biochemical features be examined to precisely determine which mode of cell death is induced by a given stress in a particular model (Kroemer et al., 2009).
Included among the diverse range of apoptosis-inducing triggers and apoptotic effector molecules are intrinsically-generated reactive oxygen species (ROS) (Buttke and Sandstrom, 1994). Typically, ROS are efficiently dealt with by endogenous oxidant remediation systems (Hockenbery et al., 1993; Tan et al., 1998). Levels that exceed defense capabilities, or sustained production of these ROS, however, are considered potent inducers of cell death (Chandra et al., 2000; Jacobson, 1996). In the case of mitochondria-mediated apoptosis involving ROS, an interdependent increase in mitochondrial respiratory rate and membrane potential (ΔΨ) during the initial phase (fueling ROS production) is followed by a ΔΨ collapse related to the opening of the permeability transition pore (Skulachev, 2006). Mitochondrial membrane depolarization is required for the critical release of cytochrome c into the cytoplasm (Li et al., 1997), activating the “classic” apoptosis program (Green and Reed, 1998).
In previous work, we have shown that treatment of Escherichia coli (E. coli) with bactericidal antibiotics induces the generation of ROS, via a common metabolic mechanism, which contribute to drug-induced killing (Dwyer et al., 2007; Kohanski et al., 2007; Kohanski et al., 2008). Considering the roles played by ROS in apoptotic cell death, and given that markers of apoptosis have been observed in unicellular eukaryotes (Carmona-Gutierrez et al., 2010; Madeo et al., 1997) under a variety of conditions including oxidative stress (Burhans et al., 2003; Jin and Reed, 2002; Madeo et al., 1999), we sought to determine whether we could detect physiological hallmarks of apoptosis in E. coli following exposure to cell death-inducing stresses.
In this work, we demonstrate that drug-induced bacterial cell death is indeed accompanied by DNA fragmentation, chromosomal condensation, extracellular exposure of PS, ΔΨ dissipation and loss of structural integrity, all markers of eukaryotic apoptosis. We also uncover novel roles for the multifunctional DNA recombinase, RecA, and the core bacterial protease, ClpXP, as critical effectors of the apoptosis-like phenotypes exhibited during the bacterial cell death process.
Results
Bactericidal antibiotics induce TUNEL-detectable DNA fragmentation
We treated cells with functionally unique representatives of the bactericidal β-lactam (ampicillin), fluoroquinolone (norfloxacin) and aminoglycoside (gentamicin) drug classes. These drugs have been shown to induce changes in cellular metabolism that promote ROS production (Dwyer et al., 2007; Kohanski et al., 2007). As experimental controls, we treated cells with the bacteriostatic antibiotic, spectinomycin, as well as chemotherapeutic [mitomycin C (MMC)] and anti-metabolite [5-fluorouracil (5-FU)] drugs; MMC and 5-FU were chosen as controls for their ability to induce apoptosis markers in model eukaryotic systems.
We observed that MMC efficiently induced cell death, while 5-FU treatment exhibited a more bacteriostatic effect on E. coli cells; this latter result is due to the ability of E. coli to efficiently export 5-FU. Generally speaking, cells treated with spectinomycin or 5-FU exhibited inhibited growth, while treatment with ampicillin, norfloxacin, gentamicin or MMC resulted in cell killing (Figure 1A).
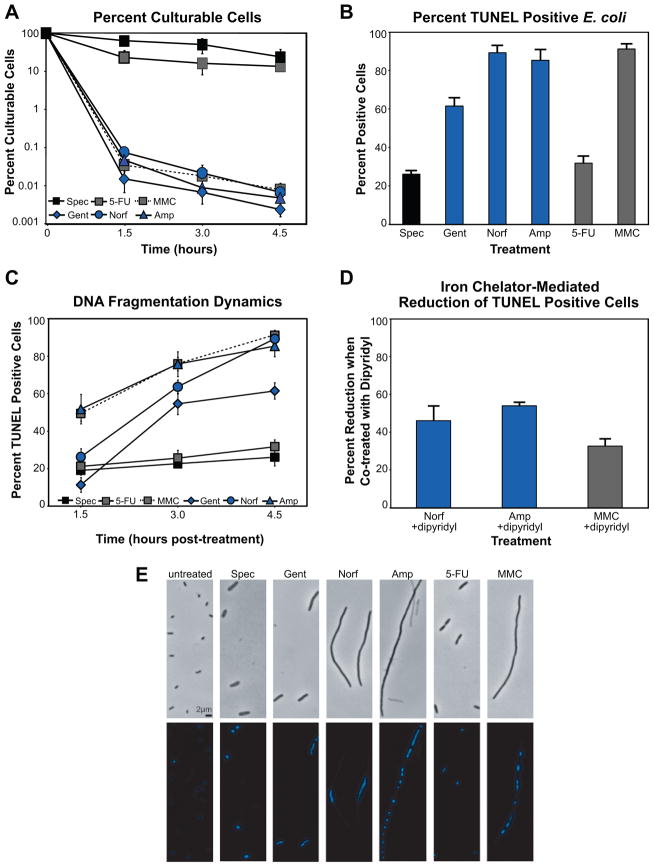
Figure 1. Cell death, DNA fragmentation and DNA condensation induced by bactericidal stress.
(A) % culturable (mean ± SD) E. coli cells following treatment with gentamicin, norfloxacin, ampicillin, spectinomycin, mitomycin C, or 5-fluorouracil. Samples are taken when indicated and viable cells counted after 16 hours growth on solid media. (B) % TUNEL positive E. coli (mean ± SD at 4.5 hours post-treatment). Values reflect the mean percentage of a treated population exceeding the fluorescence of 99% of untreated cells, at each timepoint. (C) DNA fragmentation dynamics. Graph depicts mean ± SD percent TUNEL positive cells. (D) DNA fragmentation reduction via hydroxyl radical inhibition. % reduction in TUNEL positive E. coli (mean ± SD at 4.5 hours post-treatment) when co-treated with the iron chelator, dipyridyl. (E) Effect of drug treatment on the structural state of the E. coli chromosome and cellular morphology. Shown are representative bright-field and fluorescent (Hoechst 33342) micrographs of untreated and drug-treated cells at 4.5 hours post-treatment.
To determine whether death-inducing stresses were capable of promoting DNA fragmentation of the E. coli chromosome, we performed terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) (Gavrieli et al., 1992) on bactericidally treated cells. We employed FITC-conjugated dUTP to directly label 3′-hydroxyl termini, and flow cytometry to detect TUNEL staining of individual cells. The percentage of TUNEL positive cells reported for a given perturbation reflects the mean percentage of a treated population exceeding the signal detected in 99% of untreated cells, at each timepoint.
As shown in Figures 1B and 1C, we routinely observed weak TUNEL staining of cells treated with spectinomycin or 5-FU. In contrast, treatment with norfloxacin, ampicillin, gentamicin, or MMC, as well as UV irradiation, all consistently yielded significant percentages of TUNEL positive cells (Figures 1B and S1, Tables S1 and S2). These data indicate that fragmentation of E. coli DNA occurs upon treatment with cell death-inducing agents.
In light of our previous work on ROS generation, we sought to determine the contribution of OH• to our TUNEL assay results. To accomplish this, we simultaneously treated E. coli with the most potent inducers of TUNEL positive staining in our study (norfloxacin, ampicillin, MMC) and the iron chelator, dipyridyl; this chelator has been used previously to block the generation of OH• via the Fenton reaction in both prokaryotic (Dwyer et al., 2007) and eukaryotic models (de Mello Filho and Meneghini, 1985). We found that inhibition of OH• formation reduced the number of cells exhibiting TUNEL positive staining, thereby implicating drug-induced ROS in this phenotype (Figure 1D and Table S1).
Bactericidal antibiotics induce chromosome condensation
To monitor the structural state of the bacterial chromosome following drug treatment, we employed the DNA-specific and conformation-sensitive dye, Hoechst 33342. Hoechst exhibits increased fluorescence intensity in the presence of condensed chromosomal material, and has been considered the only such stain that can be used with intact cells supravitally (Darzynkiewicz et al., 2004). Untreated E. coli exhibited light staining in our experiments (Figure 1E), which was expected given the known fluorescence properties of the dye. Following treatment with either spectinomycin or 5-FU, we observed highly ordered and focused, yet dim chromosomal staining in those cells where fluorescence could be detected.
In contrast, more dramatic staining was observed following treatment with those drugs that induced significant cell death. Treatment with MMC or ampicillin consistently resulted in unique patterns of intense fluorescence, and in both cases, several focal points were readily observed in the majority of intact cells, indicating that localized condensation of chromosomal material was occurring (Figure 1E). These treatments were also strong inducers of cell filamentation, a marker of cell division arrest typically mediated under stress conditions by SulA in E. coli (Huisman et al., 1984); ampicillin-induced filamentation was consistent with the previously reported ability of this β-lactam to stimulate the SOS response (Kohanski et al., 2007; Miller et al., 2004).
While we observed a similar spotting or banding pattern of fluorescence in a small number of norfloxacin-treated cells (data not shown), the vast majority exhibited a linear region of increased fluorescence geometrically centralized within the filamented cells (Figure 1E). In response to gentamicin treatment, we routinely observed a linear region of increased fluorescence that spanned the length of the relatively small and non-filamented cells.
Bactericidal antibiotics induce phosphatidylserine exposure at the outer membrane
Controlled exposure of PS on the outer leaflet of the plasma membrane of mammalian cells is a defining biochemical marker of apoptosis (Fadok et al., 1992). Using flow cytometry and fluorescently-labeled annexin V, a human anticoagulant which binds PS with high specificity (Martin et al., 1995), we were able to detect the extracellular exposure of PS in dying E. coli at single-cell resolution (Figure 2). The percentage of annexin V positive cells reported for a given perturbation reflects the mean percentage of a treated population exceeding the signal detected in 95% of untreated cells, at each timepoint.
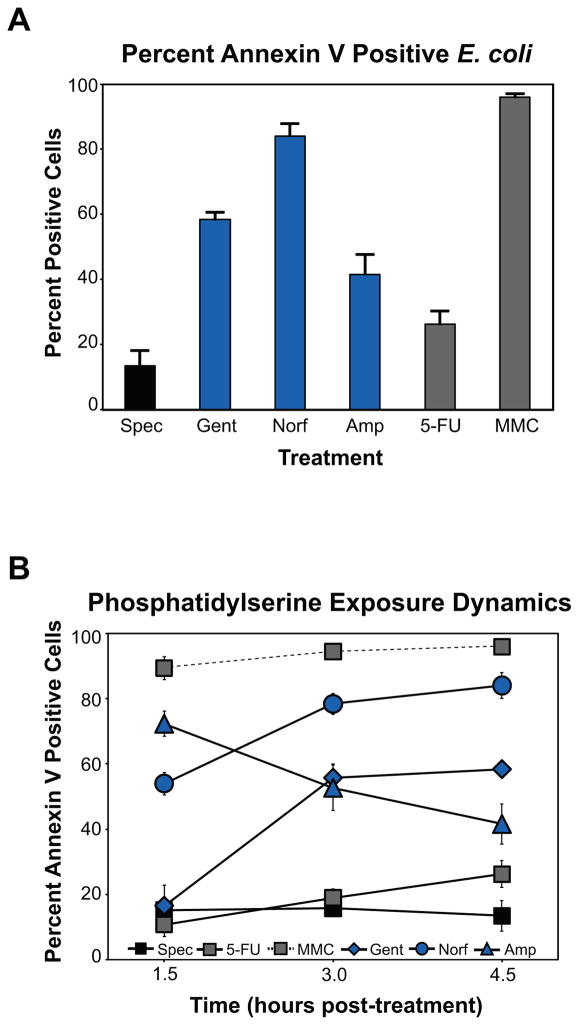
Figure 2. Phosphatidylserine exposure is induced by bactericidal stress.
(A) % annexin V labeled E. coli (mean ± SD at 4.5 hours post-treatment) following bactericidal treatment. Shown are results for cells treated with spectinomycin, gentamicin, norfloxacin, ampicillin, 5-FU, or MMC. Values reflect the mean percentage of a treated population exceeding the fluorescence of 95% of untreated cells, at each timepoint. (B) PS exposure dynamics. Graph depicts mean ± SD percent annexin V labeled cells.
As shown in Figure 2, we observed modest annexin V staining of spectinomycin- and 5-FU-treated E. coli throughout our experimental timecourse (Figure 2, Tables S1 and 2). With regard to the other drugs in our study, MMC treatment proved to be the most proficient at inducing detectable PS exposure, while norfloxacin was also a potent stimulator of PS translocation, with ~75% of the tested population labeled by annexin V by 3 hours post-treatment (Figures 2 and S2). Significant numbers (>50%) of annexin V-positive E. coli were also detected in gentamicin-treated (Figure 2) and UV-irradiated cultures by this same timepoint (Figure S1).
Treatment with ampicillin yielded the most dramatic PS exposure dynamics in our study (Figure 2), exhibiting a rapid surge and subsequent decline of percent cells with detectable PS. This result is very likely related to the breakdown of typical membrane biodynamics and lysis which accompany ampicillin’s primary mode of action. Notably, the majority of drug-treated, annexin V-positive cells in our study did not display a loss of membrane integrity [monitored by staining with the fluorescent intercalating dye, propidium iodide (PI), which requires structural permeabilization for detection], indicating that PS exposure was primarily detected at the outer membrane; in eukaryotic models, cells that are both annexin V- and PI-positive are considered to be dead. In our study, the exception was ampicillin treatment, where PI signal was increased at our later timepoints (Tables S1 and S2).
PS typically accounts for <1% of total membrane phospholipid in E. coli due to its rapid conversion to phosphatidylethanolamine by the membrane-bound enzyme, PS decarboxylase (Psd) (Kanfer and Kennedy, 1964). While psd is an essential gene, temperature-sensitive conditional mutants have been generated which afford for accumulation of PS at high concentrations (~50% of total phospholipid) (Hawrot and Kennedy, 1978). Perhaps more importantly, the ability of psd mutants to actively translocate PS between the inner and outer bacterial membranes has previously been described (Langley et al., 1982), thus providing precedence for the high level of PS staining we observed.
Further, these data suggest that drug-induced translocation of PS in E. coli occurs in an active manner, similar to apoptosis, in contrast to the spontaneous exposure of PS that takes place in other forms of cell death, including necrosis. This conclusion is particularly supported by our gentamicin results, where the inhibition of new protein synthesis following aminoglycoside treatment did not lead to a passive increase in annexin V-labeled cells over time.
Bactericidal antibiotics induce expression of a protein with caspase-like substrate specificity
Caspases are canonically regarded as the critical regulators of apoptotic PCD in metazoans (Thornberry and Lazebnik, 1998), and exert their control by proteolytic modification of the function of a variety of metabolic, structural and DNA repair protein targets (Cohen, 1997). Increasingly, non-apoptotic roles for caspases, in core cellular processes (i.e., proliferation and signaling), have also been identified, thereby placing these proteolytic regulators in unique positions of influence over homeostatic cell growth and survival (Lamkanfi et al., 2007). While distant, sequence-based relatives of caspase-like proteins (e.g., meta- and paracaspases) have been identified in silico in various unicellular organisms, bacterial orthologs have yet to be discovered based on DNA sequence (Vercammen et al., 2007). To this end, we attempted to determine experimentally if functional orthologs exist in E. coli by searching for bacterial proteins with the ability to bind synthetic caspase substrate peptides (Garcia-Calvo et al., 1998; Talanian et al., 1997) following treatments that induce bacterial cell death (Figure 3).
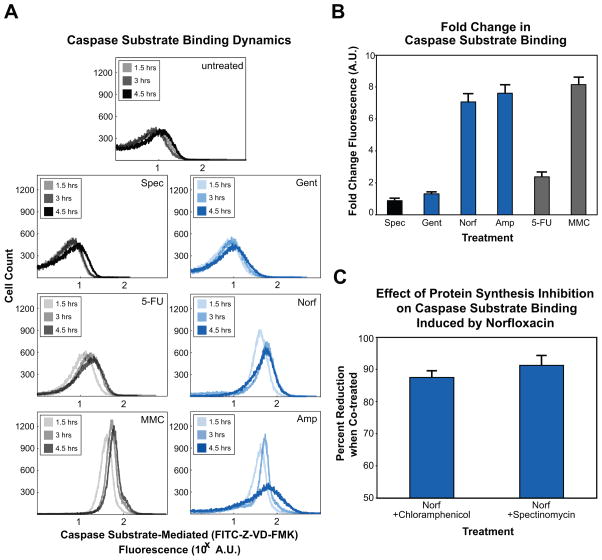
Figure 3. Bactericidal stress induces expression of a bacterial protein with affinity for caspase substrate peptides.
(A) Representative flow cytometer fluorescence timecourse histograms of untreated cells and cells treated with spectinomycin, gentamicin, norfloxacin, ampicillin, 5-FU, or MMC. Increased FITC-Z-VD-FMK fluorescence indicates increased binding of this pan-caspase inhibitor to a bacterial protein with similar substrate specificity, identified as RecA. (B) Fold change in FITC-Z-VD-FMK fluorescence (mean ± SD at 4.5 hours post-treatment), relative to untreated wildtype cells. (C) Percent reduction in FITC-Z-VD-FMK fluorescence (mean ± SD at 4.5 hours post-treatment) when co-treated with norfloxacin and one of the protein synthesis inhibitors, chloramphenicol or spectinomycin.
We employed a FITC-conjugated peptide pan-caspase inhibitor, Z-VD-FMK, to detect if lethal stress induced the expression of proteins that were capable of binding to caspase substrate peptides. Intracellular fluorescence, monitored by flow cytometry, is therefore indicative of stable binding of FITC-Z-VD-FMK to bacterial proteins with affinity for a general caspase substrate, and an increase in fluorescence reflects an increase in the concentration of these bacterial proteins.
Critically, we were unable to detect changes in FITC-Z-VD-FMK-mediated fluorescence in untreated cultures (Figure 3A). We did find, however, that treatment of E. coli with norfloxacin, ampicillin and MMC, as well as UV irradiation, each resulted in strong expression of a bacterial protein (or proteins) with caspase-like substrate specificity (Figures 3A, 3B and S1). Further, we observed an increase in mean fluorescence over the timecourse for these treatments, revealing that the concentration of any caspase-like protein was increasing in response to prolonged and potentially overwhelming drug-induced stress (Figures 3A and 3B, Tables S1 and S2).
Consistent with our other assays, treatment with 5-FU resulted in modest increases in caspase substrate-mediated fluorescence over our timecourse (Figures 3A and 3B). Perhaps most importantly, we were unable to detect an increase in fluorescence following treatment with either of the protein synthesis inhibitors used in our study, spectinomycin or gentamicin. These findings imply that an increase in FITC-Z-VD-FMK fluorescence is directly related to the expression of properly translated and folded peptides during periods of significant stress. To test this hypothesis, we performed follow-up experiments in which we simultaneously treated cultures with the antibiotics norfloxacin and spectinomycin, or norfloxacin and gentamicin (Figure 3C). In both cases, we were also unable to detect an increase in FITC-Z-VD-FMK-related fluorescence, thereby supporting the idea that new protein synthesis is necessary for this phenotype.
Proteomics analysis identifies RecA as a caspase substrate binding partner
To identify the bacterial protein (or proteins) binding to the caspase substrate, we designed a proteomics-based approach involving affinity chromatography and mass spectrometry of MMC-treated whole-cell extracts; MMC treatment was initially chosen as the perturbation for this screen because of the potency and rate with which it induced FITC-Z-VD-FMK binding. Briefly, isolation was performed using a non-fluorescent, biotin-conjugated variant (Z-VKD-biotin-FMK) of the fluorescent caspase substrate described above; the ability of the biotinylated substrate to reproduce our FITC substrate results was validated by flow cytometry prior to our proteomics screen using covalently linked FITC-streptavidin (Figure S3). Mass spectrometry was then used to identify peptides composing the single MMC treatment specific band (of approximately 40 kDa) following SDS-PAGE separation (Figure S4). Via this method, it was determined that RecA, a multifunctional regulator, was the most likely candidate for caspase substrate peptide binding. This finding was initially confirmed by SDS-PAGE and western blot of Z-VKD-biotin-FMK isolates, following both MMC and norfloxacin treatment, using an anti-RecA antibody (Figures 4A and S4).
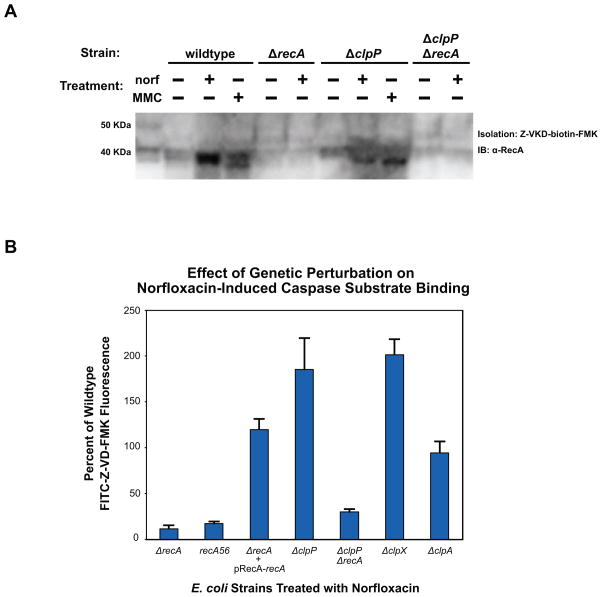
Figure 4. Influence of RecA and ClpXP on caspase substrate binding.
(A) RecA expression and caspase substrate binding assayed by western blot. Wildtype, ΔrecA, ΔclpP and ΔclpPΔrecA E. coli were treated with 125ng/mL norfloxacin or 0.5ug/mL MMC as shown, and samples taken at 4.5 hours post-treatment. Whole cell lysates were incubated with Z-VKD-biotin-FMK, then affinity purified with streptavidin and run on SDS-PAGE gels. Western blots were probed with a polyclonal E. coli RecA antibody. (B) % change in FITC-Z-VD-FMK fluorescence (mean ± SD at 4.5 hours post-treatment) exhibited by ΔrecA, ΔclpP, ΔclpPΔrecA, ΔclpX and ΔclpA E. coli when treated with 125ng/mL norfloxacin relative to similarly treated wildtype. Also shown is the effect of this treatment on ΔrecA cells expressing RecA from a rescue plasmid, and recA56 E. coli.
To validate that RecA exhibits shared peptide substrate specificity with eukaryotic caspases, we first attempted to detect FITC-Z-VD-FMK fluorescence in ΔrecA cells treated with norfloxacin and found that substrate binding was abolished in the absence of RecA expression (Figure 4B). Considering that DNA damage induces a conformational shift in RecA to its active form, we also attempted to detect FITC-Z-VD-FMK fluorescence in cells expressing a mutant allele of RecA (recA56) previously shown to be deficient in nearly all biological functions (Lauder and Kowalczykowski, 1993), including the conformational shift induced when RecA adopts its activated form, as well as SOS response induction, recombination, and stimulation of LexA autocleavage (Cox, 2007). Similar to our results in ΔrecA cells, we were unable to detect substrate binding in E. coli expressing inactive RecA56 following norfloxacin treatment (Figure 4B).
To further confirm this result, we transformed ΔrecA cells with a low-copy ‘rescue’ plasmid that ectopically restored RecA expression from its native promoter. We observed an increase in FITC-Z-VD-FMK fluorescence following norfloxacin treatment of ‘rescued’ cells that was consistent with our wildtype control (Figure 4B). Cumulatively, these data indicate that the bacterial regulatory protein, RecA, exhibits a specific affinity for caspase substrate signal sequences under conditions of overwhelming DNA stress, and that the binding of RecA to these substrate motifs requires that RecA be converted to its active form, which would occur upon recognition of significant cellular stress.
Caspase substrate binding by RecA is proteolytically modulated by ClpP
Considering the central role of proteolytic activity in the regulation of eukaryotic cell death programs, we next attempted to identify any potential role for the bacterial proteolytic machinery in the caspase substrate binding ability of RecA. In E. coli, proteolysis is achieved with high environmental plasticity by the concerted efforts of four families of energy-dependent proteases – Lon and FtsH single-component polypeptides, as well as ClpP and HslV complexes (Gottesman, 2003).
To assess whether these proteases may be playing a role in the appearance of apoptotic hallmarks, we screened single-gene knockouts of lon, ftsH, clpP and hslV using the fluorescent pan-caspase inhibitor substrate (FITC-Z-VD-FMK), following treatment with norfloxacin. We found that the absence of specific protease activity in Δlon, ΔftsH and ΔhslV cells yielded no change in FITC-Z-VD-FMK binding (data not shown). In contrast, the RecA-mediated ability of E. coli to bind FITC-Z-VD-FMK was increased approximately 175% in ΔclpP cells, compared to wildtype cells similarly treated with norfloxacin (Figure 4B, Tables S1 and S2). Together, these results suggest that ClpP acts as the peptidase which modulates the amount of RecA that is therefore capable of binding caspase substrate peptide sequences under conditions of DNA stress.
To provide further support for our ClpP result, we generated a ΔclpP ΔrecA double knockout strain to compare its behavior under terminal stress conditions to our ΔrecA single-gene knockout strain. Similar to our ΔrecA results, we observed a complete loss of FITC-Z-VD-FMK binding in our assay following norfloxacin treatment of ΔclpP ΔrecA cells, thereby reinforcing the critical role of RecA in this phenotype, and highlighting a link between RecA and ClpP in dying cells.
Next, in an attempt to the explore a possible proteolytic relationship between RecA and ClpP, we probed the Z-VKD-biotin-FMK-derived isolates of norfloxacin-treated ΔrecA, ΔclpP and ΔclpP ΔrecA cells, via western blot, using the anti-RecA antibody described above (Figures 4A and S3). As anticipated, we were unable to detect RecA in samples isolated from our ΔrecA and ΔclpP ΔrecA cellular controls. Interestingly, in wildtype isolates, we did observe differences in the number of RecA bands between norfloxacin- and MMC-treated cultures. Of note, the single RecA band observed in norfloxacin-treated wildtype isolates, and the “upper” band observed in MMC isolates, migrated similarly to the background band seen in untreated wildtype isolates, as well as the single RecA band detected in whole-cell lysates (including untreated, and norfloxacin- and MMC-treated samples). Perhaps more importantly, we also observed a stark difference in the pattern of RecA when comparing wildtype and ΔclpP isolates. Specifically, we detected a single RecA band in ΔclpP isolates that migrated like the “lower” band observed in MMC-treated wildtype isolates, yet differently than wildtype and ΔclpP whole-cell lysates, as well as the background band seen in untreated wildtype and ΔclpP isolates. These findings are supportive of the hypothesis that the differences observed in fluorescent caspase substrate binding between wildtype and ΔclpP are related to proteolytic regulation and modification of RecA by ClpP.
Lastly, in order to identify the ClpP protease complex responsible for this phenotype, we screened single-gene knockouts of the clpA and clpX ATPase chaperone genes to determine if either strain exhibited an increase in FITC-Z-VD-FMK binding. As shown in Figure 4B, we observed no increase in the fluorescence of norfloxacin-treated ΔclpA cells when compared to similarly treated wildtype cells. In ΔclpX cells, however, we did observe an increase in norfloxacin-induced FITC-Z-VD-FMK fluorescence that was in line with our ΔclpP results. This finding is consistent with previous work which has elucidated a core role for ClpXP during conditions of terminal DNA stress in E. coli and restoration of homeostasis following SOS response activation (Neher et al., 2006).
RecA, ClpP and the SOS response affect induction of apoptotic markers
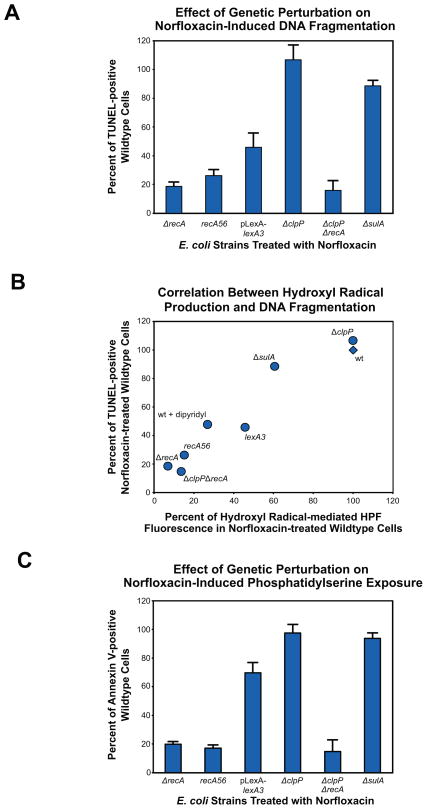
Given the critical role that RecA plays in the induction of the LexA-regulated bacterial SOS stress response, and the roles which RecA and ClpP play in the control of the SOS program (Friedberg et al., 2006; Neher et al., 2006), we next attempted to genetically link RecA, ClpP, and the SOS regulon to the display of apoptotic phenotypes by dying bacteria. To accomplish this, we assayed for DNA fragmentation and PS exposure in ΔrecA, ΔclpP and ΔclpPΔrecA cells, recA56 cells, as well as wildtype cells expressing an uncleavable mutant of the LexA repressor that will suppress SOS activity (Mount et al., 1972). Lastly, as a control for the effects of cell filamentation, we also assayed for these apoptotic phenotypes in a sulA knockout strain.
With respect to DNA fragmentation, we found that the absence of native RecA activity in both ΔrecA and recA56 cultures treated with norfloxacin resulted in the significant reduction of TUNEL positive cells (~80% and ~75% decrease, respectively) relative to similarly treated wildtype cells (Figure 5A, Tables S1 and S2). Comparable results were observed when ΔclpPΔrecA cells were treated with norfloxacin, whereas ΔclpP cultures were nearly indistinguishable from wildtype cultures. When the SOS response was suppressed by LexA3(Ind-) expression, we found that the number of TUNEL positive cells was reduced by approximately 60% relative to wildtype cells. Finally, we found the effect of the sulA knockout to be minimal on DNA fragmentation, routinely observing a decrease of approximately 10% when compared to wildtype cells. To further explore the genetic effects of these mutants on DNA fragmentation, we also measured how efficient these strains were at generating OH• via the common mechanism upon treatment with norfloxacin. We found there to be a strong correlation between the degree of DNA fragmentation and the efficiency of OH• production in these mutant strains (Figure 5B).
Figure 5. Influence of RecA, ClpP and the SOS response on DNA fragmentation, hydroxyl radical production and phosphatidylserine exposure.
(A) % change in DNA fragmentation detectable by TUNEL (mean ± SD at 4.5 hours post-treatment) exhibited by ΔrecA, ΔclpP, ΔclpPΔrecA and ΔsulA cells, recA56 cells and wildtype E. coli expressing the LexA3(Ind-) mutant repressor protein compared to similarly treated wildtype cells (125ng/mL norfloxacin). (B) Correlation between norfloxacin-induced DNA fragmentation and hydroxyl radical production. Data reflect % change in DNA fragmentation detectable by TUNEL and hydroxyl radical production detectable by the fluorescent dye, HPF (mean ± SD at 4.5 hours post-treatment), compared to similarly treated wildtype cells (125ng/mL norfloxacin). (C) % change in PS exposure detectable by annexin V (mean ± SD at 4.5 hours post-treatments) exhibited by above strains compared to similarly treated wildtype cells (125ng/mL norfloxacin).
Much like our DNA fragmentation findings, when we assessed the role of RecA in PS exposure, we found that annexin V labeling of the phospholipid was reduced throughout our timecourse by approximately 85% in the absence of RecA functionality (ΔrecA, ΔclpPΔrecA and recA56 cells) when compated to wildtype cells following norfloxacin treatment (Figure 5C and Supplemental Table 1). When examining the potential influence of SOS suppression on PS exposure, we found that LexA3(Ind-) expression resulted in significantly delayed PS accumulation. More specifically, the number of SOS suppressed cells that were annexin V positive was reduced by approximately 85% at our earliest timepoint, and by nearly 30% by the end of our timecourse. Next assaying ΔsulA cells, we found that inhibition of filamentation did affect accumulation of PS (~40% less than wildtype at 1.5 hours), but by the end of our timecourse ΔsulA cells were quite similar to wildtype. Lastly, we found that the absence of ClpP proteolytic activity resulted in an initially reduced number of PS exposing cells (~30% less than wildtype). By 3 hours post treatment, however, ΔclpP culutres were again nearly indistinguishable from wildtype cultures. Interestingly, these dynamics were similar to our observations related to SOS response activation when studying the effects of the clpP knockout on SOS induction (Figure S5); these results are discussed in the Supplemental Information.
In sum, these data provide evidence for the central role played by RecA in the appearance of canonical apoptotic cell death-accompanying phenotypes. These results also point to a critical role for the proteolytic activity of ClpP in this process, particularly with respect to PS exposure and SOS activity, and highlight the importance of an intact SOS response for the efficient induction and completion of apoptosis-resembling cell death.
Discussion
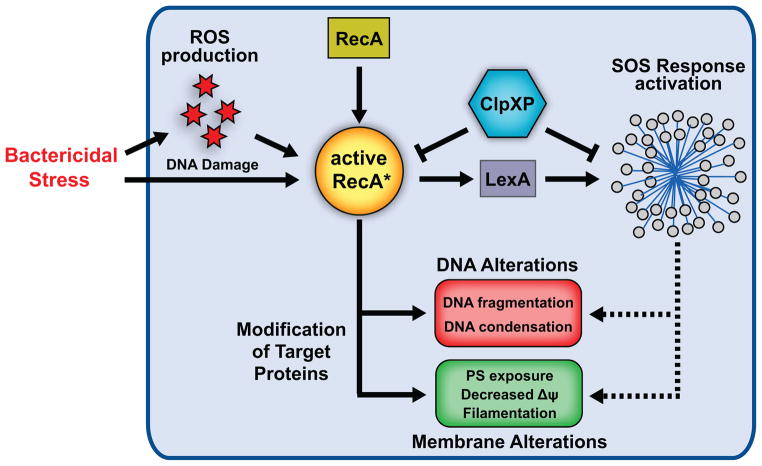
In eukaryotic organisms, genetically encoded cell death pathways are associated with fundamental changes in core cellular processes and alteration of critical biomolecules. In this study, we reveal that terminally stressed E. coli exhibit hallmarks of apoptotic PCD following treatment with bactericidal antibiotics, inducers of eukaryotic apoptosis, or UV irradiation. Our results shed light upon the physiological and biochemical changes that accompany (and presumably ensure) bacterial cell death, brought about by the interaction of varied drug molecules and their respective intracellular targets, which enable the dismantling of dying cells. As depicted in Figure 6, our findings reveal that this means of bacterial cell death involves ROS generation and is accompanied by DNA fragmentation, chromosomal condensation, PS exposure and membrane depolarization. Our model highlights critical participants in the exhibition of these phenotypes – namely, the multifunctional RecA, the stress-responsive ClpXP protease complex and broadly, the SOS stress response regulon – that functionally interact to elicit physiological modifications in a high-stress environment. These results provide evidence supporting the notion that prokaryotes possess the requisite biochemical machinery to facilitate their own termination once cell death has been triggered by functionally distinct antibacterial drugs.
Figure 6. Physiological and biochemical hallmarks of apoptosis exhibited by terminally-stressed E. coli.
Facing bactericidal stress, metabolic alterations fuel production of DNA-damaging reactive oxygen species (ROS), namely hydroxyl radicals (OH•), via the common mechanism of oxidative stress-related cell death. DNA damage induced by OH• alone, or in concert with the specific effects of the bactericidal stress, drives RecA to conformationally shift from its inactive to its active form. Following initial attempts to respond to this stress, namely SOS response network activation, E. coli exhibit several hallmarks of apoptosis which accompany cell death. These hallmarks include DNA fragmentation and condensation, and membrane alterations including PS exposure, decreased membrane potential ΔΨ, and cell division arrest [SulA-induced filamentation]. RecA plays a central role in the exhibition of these phenotypes, while the SOS response appears to play a downstream role in this process. Our findings also suggest that ClpXP affects the ability of RecA to induce these apoptotic phenotypes.
Eukaryotes encode several homologues of recA, including the essential recombinase Rad51 (Lin et al., 2006). Recent work has shown that Rad51 migrates to the mitochondria, and plays a critical role in the repair of damaged mitochondrial DNA, under conditions of ROS-induced DNA stress (Sage et al., 2010). When facing DNA stress, metazoans spend a finite amount of time in the repair phase before apoptosis is initiated, a switch designed to prevent carcinogenic mutation accumulation and energetic collapse. When DNA damage is induced by ROS or environmental insult, a eukaryote immediately activates DNA repair processes and arrests cell cycle progression. If the amount of cellular DNA damage is overwhelming (e.g., the number of lesions, the time required to repair the damage, or the energetic cost of repair), apoptosis is triggered and caspases are deployed to inactivate repair proteins like Rad51, thereby blocking any further attempts to repair the DNA damage and conserving ATP and NADH for programmed cell death (Skulachev, 2006).
Our findings suggest that broadly similar decision-making logic may exist in prokaryotes dealing with terminal stress. As noted, SOS response duration is controlled by regulated proteolysis of DNA repair proteins (Gottesman, 2003), which is related to the extent of DNA damage and may similarly serve as a bioenergetic throttle. The ClpXP protease, which we implicate here as a regulator of RecA (and some apoptotic phenotypes), has previously been shown to play a considerable role in reshaping the cellular proteome after DNA damage (Neher et al., 2006). In light of our current data, it appears likely that RecA, ClpXP and the SOS response act in a concerted manner to alter cellular behavior when cellular stress is overwhelming by modulating the function of target proteins involved in the appearance of apoptotic phenotypes and pushing the cell towards its death.
Considering that ΔrecA cells exhibit a significantly inhibited ability to induce apoptotic phenotypes, their heightened sensitivity to bactericidal treatments (Kohanski et al., 2007) presents a seemingly counterintuitive result. Intriguing parallels, however, can be found in the literature regarding highly conserved caspase 3 and caspase 2. Caspase 3 is regarded as the quintessential effector caspase and is responsible for the bulk of target protein modification during the demolition phase of apoptosis. In vivo and in vitro studies involving Casp3-null mice have revealed a critical role for this central regulator in both survival and the appearance of apoptotic phenotypes, as the loss of caspase 3 function results in decreased survival as well as diminished apoptotic features in derived cell types (Los et al., 1999). Critical subsequent findings revealed that Casp3−/− cells exhibit either fully decreased or delayed cleavage of caspase 3 substrates, suggesting that the disrupted ability of caspase 3 to modulate its array of targets is the likely cause of apoptosis defects during cell death. With regard to caspase 2, interestingly, two forms of the caspase, referred to as 2L and 2S, have been identified and found to play antagonistic roles when cells are faced with overwhelming DNA stress (Wang et al., 1994). Specifically, caspase-2L is considered to be an inducer of cell death, whereas 2S has been shown to suppress cell death, a function attributed to its potential ability to detect DNA damage in the nucleus. More recent results revealing that caspase 2 plays non-apoptotic roles in the eukaryotic DNA damage response, repair of DNA damage, and cell cycle arrest under conditions of genotoxic stress (including ROS accumulation), support this notion (Kumar, 2009).
As such, the same questions which have been raised about these caspases, including whether the diverse set of functions and connection to cell death can be related to the modification of a particular set of common substrates, or to the potential context-specific pleiotropy of this regulator, can similarly be raised about RecA in our bacterial model. Plausible explanations for our RecA results can therefore be derived from hypotheses attempting to account for the seemingly counterintuitive activities of caspase 3 and caspase 2. With respect to caspase 3, it may be that the role of RecA is so centralized and significant during cell death, that the diminished ability to regulate the phenotypes of cell death via modulation of target proteins cannot overcome the functional essentiality of RecA and the fundamental physiological defects which accompany the loss of RecA. In light of what is known about caspase 2, it may be that the immediate protective functions of RecA (i.e., detection of DNA damage, activation of the SOS response, modification of repair proteins) are essential for regulating cellular defenses, while any unexpected lack of survival may be the result of compensation in ΔrecA cells, perhaps the result of regulatory or functional redundancy, or the specific conditions leading to cell death (e.g., ROS production).
It has been widely postulated that genetic regulation of a cell death program arose in eukaryotes to manage the inherent toxicity of acquired aerobic metabolism (Ameisen, 1996; Blackstone and Green, 1999; Koonin and Aravind, 2002; Kroemer, 1997), following endosymbiosis of the prokaryotic protomitochondria billions of years ago (Gray et al., 1999; Margulis, 1996). In eukaryotes, this cell death program is most often manifest physiologically as apoptosis, and the mitochondria serves as the core mediator of death (Green and Reed, 1998). Considering that the human homologue of the ClpP protease is a mitochondrial protein (de Sagarra et al., 1999), that Rad51 will localize to the mitochondria under DNA stress conditions, and that quinolone antibiotics have been shown to induce apoptotic hallmarks in mammalian cells by a mitochondrial-dependent and caspase-independent mechanism (Boya et al., 2003), this work may shed light on novel participants in the eukaryotic apoptosis cascade.
Perhaps more importantly, this work may provide additional insight into the evolution of both apoptosis and the eukaryotic mitochondria. Our findings appear to lend support to the theory that unicellular organisms have long harbored the biochemical ability to insure their own demise. (Ameisen, 2002). This theory further proposes that evolution of multicellular organisms was accompanied by the development of tight regulatory layers to control these abilities, rather than a sudden and simultaneous materialization of both ability and control. It is therefore plausible that complex genetic regulation of the apoptotic program arose in eukaryotes to manage and tune these physiological and biochemical hallmarks evident in bacteria exposed to cell death-triggering stresses, thereby taking advantage of their deadly potential when necessary.
Experimental Procedures
Cell strains and growth analysis
We compared the growth and phenotypic changes associated with treatment of wildtype MG1655 E. coli (ATCC# 700926) with spectinomycin (400μg/mL), 5-fluorouracil (500μg/mL), mitomycin C (5μg/mL), ampicillin (5μg/mL), gentamicin (10μg/mL), and norfloxacin (750ng/mL) to untreated cultures (Sigma, Fisher, Acros). The respective drug concentrations were chosen for the ability to induce comparable cell death. We also studied the growth and phenotypic changes of a number of recA-related knockout strains treated with a lower concentration of norfloxacin (125ng/mL). For details on growth conditions (including UV irradiation experiments), please see the Supplemental Information (SI).
Analysis of DNA fragmentation
For DNA fragmentation experiments, cells were grown as described in SI for terminal deoxynucleotide transferase dUTP nick end labeling (TUNEL). Labeling of DNA fragments was performed using the Apo-Direct Kit (BD Bioscience), which employs FITC-conjugated deoxyuridine triphosphate (FITC-dUTP). See SI for details of collection and staining.
For detection, samples were collected using either a FACSCalibur or a FACS Aria II flow cytometer (Becton Dickinson). The FACSCalibur is equipped with a 488-nm argon laser for excitation, and 530 +/− 15nm (FL1, for FITC fluorescence) and 585 +/− 21nm (FL2, for propidium iodide fluorescence) emission filters, and used at a low flow rate. The following PMT voltage settings were used: E00 (FSC, primary collection parameter), 300 (SSC), and 800 (FL1). The FACS Aria II is equipped with a 488-nm argon laser for excitation, and 515 +/− 15nm (with 495nm long-pass mirror for FITC fluorescence) and 610 +/− 20nm (with 600nm long-pass mirror for propidium iodide fluorescence) emission filters, and used with a 70 μm nozzle at 70 PSI. The following PMT voltages were used: 353 (FSC), 271 (SSC), 675 (FITC), and 500 (PI). At least 100,000 cells were collected for each sample. Percent positive cells were determined using MATLAB software and reflect the number of TUNEL positive cells exceeding the fluorescence of 99% of untreated cells.
To determine the contribution of hydroxyl radicals to TUNEL positive labeling, we co-treated cells with the iron chelator, dipyridyl (500μM). Sample preparation, labeling and analysis were as described above.
Analysis of DNA structure
For microscopy experiments, cells were grown as described in SI. Fluorescent staining of DNA was performed using the DNA specific stain, Hoechst 33342 (Invitrogen), according to kit instructions. Counterstaining was performed using PI.
After staining, cells were analyzed using a Nikon Eclipse 80i microscope with a 20x objective (200x magnification), outfitted with a CoolSnap HQ CCD camera (Roper Scientific), operated with IPLab software (Scanalytics). For fluorescence images, cells were analyzed using UV/488nm dual excitation and emission was measured using standard DAPI and PI filters.
Analysis of phosphatidylserine exposure
For phosphatidylserine (PS) exposure experiments, cells were grown as described in SI. Annexin V labeling of cells was performed using FITC- and AlexaFluor488-conjugated annexin V (Invitrogen). PI was used as a counterstain for identification of dead cells. See SI for details of collection and labeling.
For detection, samples were collected using either a FACSCalibur or FACS Aria II flow cytometer as described above. A FL1 PMT voltage of 750 was used with the FACSCalibur, while a FITC PMT voltage of 520 was used with the FACS Aria II. Percent positive cells were determined using MATLAB software and reflect the number of annexin V positive cells exceeding the fluorescence of 95% of untreated cells. To monitor PS exposure by microscopy, we used the Nikon Eclipse 80i microscope described above.
Detection of a bacterial protein with caspase-like substrate affinity
We used a FITC-conjugated, pan-caspase inhibitor peptide, Z-VD-FMK (ApoStat, R&D Systems), to detect expression of bacterial proteins that could bind peptides encoding caspase substrate sequences. Cultures were grown as described in SI.
Experiments can be performed directly in culture medium at 37°C, and require 30 minutes incubation. See SI for further detail on procedure. Samples were collected using either a FACSCalibur or FACS Aria II flow cytometer as described above. A FL1 PMT voltage of 750 was used with the FACSCalibur, while a FITC PMT voltage of 590 was used with the FACS Aria II.
To identify the bacterial protein or proteins binding to the FITC-labeled caspase substrate, we used a biotinylated variant, Z-VKD-biotin-FMK (R&D Systems). To confirm system suitability, we used a streptavidin-FITC detection solution (R&D Systems). See SI for further detail on procedure. Samples were collected using a FACSCalibur flow cytometer as described above, with an FL1 PMT voltage of 750.
Identification of a bacterial protein with caspase-like substrate specificity
Isolation of the caspase substrate-binding protein was performed by Blue Sky Biotech. The project outline was as follows: (a) two 1L cultures of wild type E. coli strain MG1655 were grown (+/− MMC); (b) enrichment conditions using streptavidin-agarose (Promega) chromatography and Z-VKD-biotin-FMK were determined with a small-scale lysate; (c) enrichment using optimal conditions was utilized to prepare elution fractions for tryptic mass spectrometry (MS) analysis; and (d) enriched fractions were snap-frozen for tryptic MS analysis. This method is described in full in SI.
Detection of RecA in Z-VKD-biotin-FMK isolates by western blot
Isolation of RecA and western blot analysis was performed by Blue Sky Biotech. The project outline was as follows: (a) ten 50mL cultures of each E. coli strain and condition tested were grown; (b) cell lysates were collected, clarified and batch incubated with Z-VKD-biotin-FMK; (c) SDS-PAGE was performed on isolated samples; and (d) following transfer, samples were analyzed by western blot using a polyclonal anti-RecA antibody (Abcam, AB63797). As a control, whole cell extracts were also analyzed for RecA expression.
Blots were probed with the anti-RecA primary antibody at 1:2,000 dilution, then a peroxidase-conjugated secondary antibody (Jackson) at 1:20,000 dilution. Detection was performed using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher). This method and related data are described in the SI.
Detection of hydroxyl radical production following norfloxacin treatment
To assess hydroxyl radicals production in response to norfloxacin (125ng/mL) treatment, we employed the fluorescent reporter dye 3′(p-hydroxyphenyl) fluorescein (HPF, Invitrogen) at a final concentration of 5 μM. HPF was added to 25mL cultures at inoculation. Samples were collected using a FACS Aria II flow cytometer as described above. A FITC PMT voltage of 600 was used and flow data were analyzed as described above.
Supplementary Material
Highlights.
Hallmarks of apoptosis accompany bacterial cell death induced by antibiotics.
Ultrastructural alterations induced by bactericidal drugs include phosphatidylserine exposure.
The multifunctional regulator RecA exhibits affinity for caspase substrate sequences.
RecA and the ClpXP protease are identified as critical regulators of these apoptotic phenotypes.
Acknowledgments
We thank Kim McCall and members of the Collins lab for their suggestions, and Scott Gridley for his technical assistance. This work was supported by the NIH (DP1 OD003644), NSF (EMSW21-RTG) and HHMI.
References
- Ameisen JC. The origin of programmed cell death. Science. 1996;272:1278–1279. doi: 10.1126/science.272.5266.1278. [DOI] [PubMed] [Google Scholar]
- Ameisen JC. On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell Death Differ. 2002;9:367–393. doi: 10.1038/sj.cdd.4400950. [DOI] [PubMed] [Google Scholar]
- Blackstone NW, Green DR. The evolution of a mechanism of cell suicide. Bioessays. 1999;21:84–88. doi: 10.1002/(SICI)1521-1878(199901)21:1<84::AID-BIES11>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Boya P, Andreau K, Poncet D, Zamzami N, Perfettini JL, Metivier D, Ojcius DM, Jaattela M, Kroemer G. Lysosomal membrane permeabilization induces cell death in a mitochondrion-dependent fashion. J Exp Med. 2003;197:1323–1334. doi: 10.1084/jem.20021952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broker LE, Kruyt FA, Giaccone G. Cell death independent of caspases: a review. Clin Cancer Res. 2005;11:3155–3162. doi: 10.1158/1078-0432.CCR-04-2223. [DOI] [PubMed] [Google Scholar]
- Burhans WC, Weinberger M, Marchetti MA, Ramachandran L, D’Urso G, Huberman JA. Apoptosis-like yeast cell death in response to DNA damage and replication defects. Mutat Res. 2003;532:227–243. doi: 10.1016/j.mrfmmm.2003.08.019. [DOI] [PubMed] [Google Scholar]
- Buttke TM, Sandstrom PA. Oxidative stress as a mediator of apoptosis. Immunol Today. 1994;15:7–10. doi: 10.1016/0167-5699(94)90018-3. [DOI] [PubMed] [Google Scholar]
- Carmona-Gutierrez D, Eisenberg T, Buttner S, Meisinger C, Kroemer G, Madeo F. Apoptosis in yeast: triggers, pathways, subroutines. Cell Death Differ. 2010;17:763–773. doi: 10.1038/cdd.2009.219. [DOI] [PubMed] [Google Scholar]
- Chandra J, Samali A, Orrenius S. Triggering and modulation of apoptosis by oxidative stress. Free Radic Biol Med. 2000;29:323–333. doi: 10.1016/s0891-5849(00)00302-6. [DOI] [PubMed] [Google Scholar]
- Cohen GM. Caspases: the executioners of apoptosis. Biochem J. 1997;326(Pt 1):1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM. Regulation of bacterial RecA protein function. Crit Rev Biochem Mol Biol. 2007;42:41–63. doi: 10.1080/10409230701260258. [DOI] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Crissman H, Jacobberger JW. Cytometry of the cell cycle: cycling through history. Cytometry A. 2004;58:21–32. doi: 10.1002/cyto.a.20003. [DOI] [PubMed] [Google Scholar]
- de Mello Filho AC, Meneghini R. Protection of mammalian cells by o-phenanthroline from lethal and DNA-damaging effects produced by active oxygen species. Biochim Biophys Acta. 1985;847:82–89. doi: 10.1016/0167-4889(85)90156-9. [DOI] [PubMed] [Google Scholar]
- de Sagarra MR, Mayo I, Marco S, Rodriguez-Vilarino S, Oliva J, Carrascosa JL, Casta n JG. Mitochondrial localization and oligomeric structure of HClpP, the human homologue of E. coli ClpP. J Mol Biol. 1999;292:819–825. doi: 10.1006/jmbi.1999.3121. [DOI] [PubMed] [Google Scholar]
- Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol Syst Biol. 2007;3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok VA, Voelker DR, Campbell PA, Cohen JJ, Bratton DL, Henson PM. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J Immunol. 1992;148:2207–2216. [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2. Washington, D.C: ASM Press; 2006. [Google Scholar]
- Garcia-Calvo M, Peterson EP, Leiting B, Ruel R, Nicholson DW, Thornberry NA. Inhibition of human caspases by peptide-based and macromolecular inhibitors. J Biol Chem. 1998;273:32608–32613. doi: 10.1074/jbc.273.49.32608. [DOI] [PubMed] [Google Scholar]
- Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- Hawrot E, Kennedy EP. Phospholipid composition and membrane function in phosphatidylserine decarboxylase mutants of Escherichia coli. J Biol Chem. 1978;253:8213–8220. [PubMed] [Google Scholar]
- Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- Hockenbery DM, Oltvai ZN, Yin XM, Milliman CL, Korsmeyer SJ. Bcl-2 functions in an antioxidant pathway to prevent apoptosis. Cell. 1993;75:241–251. doi: 10.1016/0092-8674(93)80066-n. [DOI] [PubMed] [Google Scholar]
- Huisman O, D’Ari R, Gottesman S. Cell-division control in Escherichia coli: specific induction of the SOS function SfiA protein is sufficient to block septation. Proc Natl Acad Sci U S A. 1984;81:4490–4494. doi: 10.1073/pnas.81.14.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MD. Reactive oxygen species and programmed cell death. Trends Biochem Sci. 1996;21:83–86. [PubMed] [Google Scholar]
- Jin C, Reed JC. Yeast and apoptosis. Nat Rev Mol Cell Biol. 2002;3:453–459. doi: 10.1038/nrm832. [DOI] [PubMed] [Google Scholar]
- Kanfer J, Kennedy EP. Metabolism and Function of Bacterial Lipids. Ii. Biosynthesis of Phospholipids in Escherichia Coli. J Biol Chem. 1964;239:1720–1726. [PubMed] [Google Scholar]
- Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135:679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Aravind L. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ. 2002;9:394–404. doi: 10.1038/sj.cdd.4400991. [DOI] [PubMed] [Google Scholar]
- Kroemer G. Mitochondrial implication in apoptosis. Towards an endosymbiont hypothesis of apoptosis evolution. Cell Death Differ. 1997;4:443–456. doi: 10.1038/sj.cdd.4400266. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. Caspase 2 in apoptosis, the DNA damage response and tumour suppression: enigma no more? Nat Rev Cancer. 2009;9:897–903. doi: 10.1038/nrc2745. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Festjens N, Declercq W, Vanden Berghe T, Vandenabeele P. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 2007;14:44–55. doi: 10.1038/sj.cdd.4402047. [DOI] [PubMed] [Google Scholar]
- Langley KE, Hawrot E, Kennedy EP. Membrane assembly: movement of phosphatidylserine between the cytoplasmic and outer membranes of Escherichia coli. J Bacteriol. 1982;152:1033–1041. doi: 10.1128/jb.152.3.1033-1041.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauder SD, Kowalczykowski SC. Negative co-dominant inhibition of recA protein function. Biochemical properties of the recA1, recA13 and recA56 proteins and the effect of recA56 protein on the activities of the wild-type recA protein function in vitro. J Mol Biol. 1993;234:72–86. doi: 10.1006/jmbi.1993.1564. [DOI] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Lin Z, Kong H, Nei M, Ma H. Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. Proc Natl Acad Sci U S A. 2006;103:10328–10333. doi: 10.1073/pnas.0604232103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los M, Wesselborg S, Schulze-Osthoff K. The role of caspases in development, immunity, and apoptotic signal transduction: lessons from knockout mice. Immunity. 1999;10:629–639. doi: 10.1016/s1074-7613(00)80062-x. [DOI] [PubMed] [Google Scholar]
- Madeo F, Frohlich E, Frohlich KU. A yeast mutant showing diagnostic markers of early and late apoptosis. J Cell Biol. 1997;139:729–734. doi: 10.1083/jcb.139.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Frohlich E, Ligr M, Grey M, Sigrist SJ, Wolf DH, Frohlich KU. Oxygen stress: a regulator of apoptosis in yeast. J Cell Biol. 1999;145:757–767. doi: 10.1083/jcb.145.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L. Archaeal-eubacterial mergers in the origin of Eukarya: phylogenetic classification of life. Proc Natl Acad Sci U S A. 1996;93:1071–1076. doi: 10.1073/pnas.93.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SJ, Reutelingsperger CP, McGahon AJ, Rader JA, van Schie RC, LaFace DM, Green DR. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J Exp Med. 1995;182:1545–1556. doi: 10.1084/jem.182.5.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C, Thomsen LE, Gaggero C, Mosseri R, Ingmer H, Cohen SN. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science. 2004;305:1629–1631. doi: 10.1126/science.1101630. [DOI] [PubMed] [Google Scholar]
- Mount DW, Low KB, Edmiston SJ. Dominant mutations (lex) in Escherichia coli K-12 which affect radiation sensitivity and frequency of ultraviolet lght-induced mutations. J Bacteriol. 1972;112:886–893. doi: 10.1128/jb.112.2.886-893.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher SB, Villen J, Oakes EC, Bakalarski CE, Sauer RT, Gygi SP, Baker TA. Proteomic profiling of ClpXP substrates after DNA damage reveals extensive instability within SOS regulon. Mol Cell. 2006;22:193–204. doi: 10.1016/j.molcel.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Nicholson DW, Thornberry NA. Caspases: killer proteases. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Horvitz HR. The engulfment process of programmed cell death in caenorhabditis elegans. Annu Rev Cell Dev Biol. 2004;20:193–221. doi: 10.1146/annurev.cellbio.20.022003.114619. [DOI] [PubMed] [Google Scholar]
- Ren Y, Savill J. Apoptosis: the importance of being eaten. Cell Death Differ. 1998;5:563–568. doi: 10.1038/sj.cdd.4400407. [DOI] [PubMed] [Google Scholar]
- Sage JM, Gildemeister OS, Knight KL. Discovery of a novel function for human Rad51: maintenance of the mitochondrial genome. J Biol Chem. 2010;285:18984–18990. doi: 10.1074/jbc.M109.099846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skulachev VP. Bioenergetic aspects of apoptosis, necrosis and mitoptosis. Apoptosis. 2006;11:473–485. doi: 10.1007/s10495-006-5881-9. [DOI] [PubMed] [Google Scholar]
- Strasser A, O’Connor L, Dixit VM. Apoptosis signaling. Annu Rev Biochem. 2000;69:217–245. doi: 10.1146/annurev.biochem.69.1.217. [DOI] [PubMed] [Google Scholar]
- Talanian RV, Quinlan C, Trautz S, Hackett MC, Mankovich JA, Banach D, Ghayur T, Brady KD, Wong WW. Substrate specificities of caspase family proteases. J Biol Chem. 1997;272:9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- Tan S, Sagara Y, Liu Y, Maher P, Schubert D. The regulation of reactive oxygen species production during programmed cell death. J Cell Biol. 1998;141:1423–1432. doi: 10.1083/jcb.141.6.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornberry NA, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- Vercammen D, Declercq W, Vandenabeele P, Van Breusegem F. Are metacaspases caspases? J Cell Biol. 2007;179:375–380. doi: 10.1083/jcb.200705193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Miura M, Bergeron L, Zhu H, Yuan J. Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell. 1994;78:739–750. doi: 10.1016/s0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- Wyllie AH. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature. 1980;284:555–556. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- Wyllie AH, Kerr JF, Currie AR. Cell death: the significance of apoptosis. Int Rev Cytol. 1980;68:251–306. doi: 10.1016/s0074-7696(08)62312-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.