Abstract
Cytochrome c oxidase is the terminal enzyme of the mitochondrial electron transport chain, without which oxidative metabolism cannot be carried to completion. It is one of only four unique, bigenomic proteins in mammalian cells. The holoenzyme is made up of three mitochondrial-encoded and ten nuclear-encoded subunits in a 1:1 stoichiometry. The ten nuclear subunit genes are located in nine different chromosomes. The coordinated regulation of such a multisubunit, multichromosomal, bigenomic enzyme poses a challenge. It is especially so for neurons, whose mitochondria are widely distributed in extensive dendritic and axonal processes, resulting in the separation of the mitochondria from the nuclear genome by great distances. Neuronal activity dictates COX activity that reflects protein amount, which, in turn, is regulated at the transcriptional level. All 13 COX transcripts are up- and downregulated by neuronal activity. The ten nuclear COX transcripts and those for Tfam and Tfbms important for mitochondrial COX transcripts are transcribed in the same transcription factory. Bigenomic regulation of all 13 transcripts is mediated by nuclear respiratory factors 1 and 2 (NRF-1 and NRF-2). NRF-1, in addition, also regulates critical neurochemicals of glutamatergic synaptic transmission, thereby ensuring the tight coupling of energy metabolism and neuronal activity at the molecular level in neurons.
12.1 Cytochrome c Oxidase
Cytochrome c oxidase (COX, cytochrome aa3, ferrocytochrome c oxygen oxidoreductase, complex IV, E.C. 1.9.3.1) is the terminal enzyme of the mitochondrial electron transport chain. It catalyzes the oxidation of its substrate, cytochrome c, and the reduction of molecular oxygen to water. It assists in the pumping of protons from the matrical to the cytosolic side of the inner mitochondrial membrane, setting up the electrochemical proton gradient that drives the synthesis of ATP from ADP and phosphate by ATP synthase (complex V). Inactivation of COX by cyanide, azide, or carbon monoxide is incompatible with life, as oxidative metabolism cannot be carried to completion, and no ATP can be generated from mitochondria. Thus, highly oxidative organs such as the heart, liver, kidney, skeletal muscles, and especially the brain, are critically dependent on COX for their normal functioning and survival.
COX is one of the most ancient enzymes known, parts of it evolved more than a billion years ago. It is one of only 4 bigenomic proteins in mammalian cells: complexes I, III, IV, and V of the electron transport chain, each of which has subunits from either the nuclear or the mitochondrial genome, and none of which is encoded entirely by a single genome. This implies that a) the mitochondrial genome retains its control of key subunits of the electron transport chain through evolution; and b) the two genomes have to work closely together to ensure proper functioning of the oxidative phosphorylation machinery. COX holoenzyme has 13 subunits with 1:1 stoichiometry (Kadenbach et al 1983). The largest three subunits (COX I, II, III) are encoded in the maternally inherited mitochondrial genome, and the remaining ten (COX IV, Va,b, VIa,b,c, VIIa,b,c, and VIII) are nuclear-encoded in nine different chromosomes. To form a functional holoenzyme, precise coordination between the two genomes is necessary. The mechanism of regulating such a complex, multisubunit, bigenomic enzyme appears daunting and poorly understood until recent years, when the regulatory machinery was beginning to be revealed.
12.2 Cytochrome c Oxidase as a Metabolic Marker for Neurons
Neurons are ideal cells for investigating the regulation of COX. Unlike glial cells that can fare quite well under anaerobic condition, neurons are dependent almost entirely on oxidative metabolism for their function and survival. Being postmitotic, they do not undergo constant turnover and rebirth, and hence their metabolic activity reflects primarily their constitutive functional demands. Of all the ATP-demanding functions of neurons, such as the synthesis of proteins and other molecules, turnover of transmitters and receptors, active anterograde and retrograde transport of proteins and organelles, and active transport of ions against their concentration and electrical gradients, the first two consume relatively little energy, the third one accounts for only a minor fraction of the energy, but the last one is by far the most energy-demanding function of neurons, especially with regard to repolarizing the membrane after excitatory depolarizing activity (Wong-Riley 1989; 2010; Attwell and Laughlin 2001). Indeed, it is neuronal activity that controls energy expenditure, and not vice versa (Lowry 1975; Wong-Riley 1989). The control is precise, such that energy is not generated until energy is spent. As different compartments of a single neuron require varying amounts of energy, the entire neuron is not metabolically homogeneous. Dendrites, being the major receptive sites of incoming depolarizing input and whose membranes have to be constantly repolarized, consume the bulk of energy, whereas axonal trunks, especially myelinated axons, consume very little energy (Wong-Riley 1989). Energy demand of cell bodies is largely dependent on the magnitude and frequency of excitatory input they receive, and that of axon terminals reflects how tonically active they are (Wong-Riley, 1989).
Can cytochrome c oxidase serve as a sensitive and reliable metabolic marker for neurons? At the biochemical, histochemical, immunohistochemical, cytochemical, and molecular levels, this proves to be the case, and they correlate well with the neuron’s functional activity (Wong-Riley 1979; Wong-Riley et al 1989a,b; Hevner et al 1995; Hevner and Wong-Riley 1989; 1991). In altering the functional demands of neurons, such as with tetrodotoxin (TTX)-induced impulse blockade or with KCl-mediated depolarizing stimulation, the levels of cytochrome c oxidase in affected neurons are down- or up-regulated accordingly (Wong-Riley and Carroll 1984; Hevner and Wong-Riley 1990; DeYoe et al 1995; Zhang and Wong-Riley 1999). Such alterations exist not only in the activity of the enzyme, but also in its protein and mRNA amount, indicating that the activity reflects mainly the protein amount, which, in turn, is regulated mainly at the transcriptional level (Wong-Riley et al. 1998a).
12.3 The Challenge of Bigenomic Coordination in Neurons
As COX subunit transcripts and proteins are of two genomic sources, this poses a unique challenge for neurons. For, unlike most cells, neurons have mitochondria-laden dendrites as well as axons that can extend far from the cell bodies, hence the nuclear and the mitochondrial genomes can be separated by great distances. Do the nuclear transcripts migrate from the cell body to distal dendrites for local translation, or do they stay within the cell body? To answer this question, in situ hybridization was done at both the light and electron microscopic (EM) levels. It was found that, indeed, the mitochondrial mRNAs are located within the mitochondria that are distributed throughout the cell bodies, dendrites, and axons, but the nuclear transcripts are restricted only to the cell bodies, indicating that translation of nuclear transcripts occurs only in the cell bodies (Hevner and Wong-Riley 1991; Wong-Riley et al 1997). How, then, can the nuclear-encoded subunit proteins get to distal dendrites, where they are most needed? Do they take the intra- or the extra-mitochondrial route in their transit from the cell body to distal processes?
The answer for at least one of the nuclear-encoded subunits, COXIV, is that the precursor protein, which contains the mitochondrial-targeting presequence, is translated in the cell body, incorporated into the mitochondria within the cell body, and is translocated intra-mitochondrially to distal processes (Liu and Wong-Riley 1994). There, it can remain as precursor protein, or be processed into the mature form to be incorporated into the holoenzyme with the other nuclear- and mitochondrial subunit polypeptides. Thus, neurons have devised a mechanism by which the precursor proteins are not immediately processed upon entry into the mitochondria, as in the case of yeast and rat hepatocytes (Mori et al 1981; Reid et al 1982), but rather, can form a precursor pool in dendrites and axons until such time when additional energy demand triggers further processing into their mature forms. This mechanism ensures conservation of energy and bypasses the need for constant shuttling of individual mitochondrion back to the cell body for a fresh supply of precursor proteins each time a new stock of holoenzyme is called for.
12.4 Bigenomic Coordination of Cytochrome c Oxidase in Response to Changing Neuronal Activity
Are all 13 subunits of cytochrome c oxidase coordinately or disparately regulated by neuronal activity? Both in vivo and in vitro approaches have been used to probe this question. In vitro, all 13 COX subunit transcripts are significantly upregulated after 5 h of depolarizing stimulation, and they are all downregulated by TTX blockade (Liang et al 2006). However, the levels of the three mitochondrial-encoded transcripts fall earlier than those of the ten nuclear-encoded ones (2 versus 4 days). By the 6th day after inactivation, all 13 transcripts are downregulated to about the same extent (to ~ 20% of controls). Likewise, in vivo sensory deprivation with retinal impulse blockade or enucleation induces an earlier and more severe downregulation of the mitochondrial- than the nuclear-encoded subunit transcripts (Hevner and Wong-Riley 1993; Liang et al 2006). This implies that the mitochondrial genome exerts a greater control over the activity and amount of the enzyme in neurons. The merit of such a mechanism includes: a) mitochondria in distal dendrites and axon terminals are strategically located at the “business” ends of neurons, where they can sense local energy demand and adjust the supply of holoenzymes accordingly; b) the mitochondrial genome is responsible for the largest three subunits that form the catalytic core of the enzyme, although the ten nuclear-encoded subunits also play important roles in energy metabolism (Kadenbach et al 2000); and c) as stated above, there is a reservoir of nuclear-encoded subunit proteins in distal neuronal processes, so the downregulation of these subunits may be delayed. Ultimately, however, all 13 subunits are up- or downregulated by neuronal activity (Liang et al 2006).
12.4.1 Synthesis Versus Degradation of Bigenomic Transcripts
Are activity-induced changes in COX transcripts due to RNA synthesis rate or stability, or both? The answer came from an experiment in which primary neurons in culture were stimulated with 20 mM KCl for 5 h (Zhang and Wong-Riley 2000a). It was found that the synthesis rate of both the mitochondrial-encoded Cox2 and the nuclear-encoded Cox4 transcripts are increased significantly after 3 h of depolarizing stimulation. The rate of Cox2 remains higher than controls at 4 and 5 h of stimulation, but that of Cox4 returns to control levels after 3 h. The degradation rate was monitored by 3Huridine pulse-chase labeling, and it revealed a half-life of 84 min for Cox2 and 50 min for Cox4 mRNA. With KCl stimulation, the half life of Cox2 transcripts remains relatively constant, whereas that of Cox4 increases to 102 min. These data indicate that the mitochondrial transcripts are regulated mainly at the transcriptional level, but that the nuclear transcripts are regulated at both the synthetic and degradative levels, and that both are tightly governed by neuronal activity (Zhang and Wong-Riley 2000a).
12.5 Transcription Factors as Bigenomic Coordinators
The bigenomic nature of COX imposes a special need for transcriptional coordination between the two genomes. Is there a transcription factor or factors that may serve such a role? Two factors have been proposed to mediate nuclear-mitochondrial interactions. They are nuclear respiratory factors 1 and 2 (NRF-1 and NRF-2) (Scarpulla 2008).
12.5.1 Role of Nuclear Respiratory Factor 2
NRF-2 is the human homologue of the murine GA-binding protein (GABP) (Evans and Scarpulla 1990; Thompson et al 1991; Virbasius et al 1993a). It belongs to the Ets (E26 transformation-specific) family of transcription factors, recognizing the consensus sequence (C/A)GGA(A/T)(A/G) (LaMarco et al 1991; Thompson et al 1991; Virbasius and Scarpulla 1991). NRF-2 is a heteromeric protein made up of mainly α and β subunits (β1, β2), and γ1 and γ2 are splice variants of the β subunit. The α subunit has the Ets domain and is required for DNA binding. The β subunit contains four Notch-ankyrin repeats that mediate dimerization with the α subunit, but is incapable of binding DNA alone. The β subunit also has the transactivating domain and the nuclear localizing signal. Heterodimerization of α and β is required for stabilization and specificity of α-DNA binding (De la Brousse et al 1994; Batchelor et al 1998). The homodimerization domain of the β subunit enables the formation of α2β2 heterotetramer that binds to tandem repeats of NRF-2 in target gene promoters (Scarpulla 2002). DNA binding and in vitro studies have implicated the transactivational activity of NRF-2 and its regulatory role in the expression of a number of subunits of respiratory chain enzymes, especially some of the nuclear-encoded COX subunits (rat and mouse subunits IV and Vb, human subunit Vb, bovine subunit VIIaL (Virbasius and Scarpulla 1991; Carter et al 1992; Virbasius et al 1993a; Carter and Avadhani 1994; Scarpulla 1997), and human COX VIaL (Ongwijitwat and Wong-Riley 2004). In addition, NRF-2 also regulates genes that encode mitochondrial transcription factors A and B (TFAM, TFB1M, and TFB2M) (Virbasius and Scarpulla 1994; Gleyzer et al 2005), which are nuclear-derived and function inside the mitochondria as regulators of mtDNA transcription and replication (Fisher and Clayton 1988; Falkenberg et al 2002). NRF-2 also regulates three of the four human succinic dehydrogenase subunit genes, as well as genes for human TOMM20, mitochondrial transcription termination factor (mTERF), RNA polymerase POLRMT, and the B subunit of the DNA Polγ, among others (Au and Scheffler 1998; Blesa et al 2007; Scarpulla 2008; Bruni et al 2010). Knockout of NRF-2/GABP is embryonically lethal before implantation, attesting to the essential role of this transcription factor in embryogenesis; whereas the heterozygous nulls appear normal (Ristevski et al 2004). Thus, NRF-2 potentially links the nucleus and mitochondria by regulating COX-related gene expression in the two genomes (Scarpulla 2008).
12.5.1.1 NRF-2 Itself Responds to Changes in Neuronal Activity
What is known about the significance of NRF-2 in neurons? Remarkably, the pattern of NRF-2’s distribution in the primate visual cortex is virtually identical to that of COX (Nie and Wong-Riley 1999), a unique feature not shared by any other transcription factors studied thus far. NRF-2 is also more strongly expressed in cell types that have higher COX activity than those with lower activity (Wong-Riley et al 2005). Moreover, NRF-2 itself responds to monocular impulse blockade by downregulating its protein and message levels in deprived cortical columns and neurons in which COX activity is suppressed (Nie and Wong-Riley 1999; Guo et al 2000; Wong-Riley et al 2005). In response to KCl depolarization in cultured primary neurons, NRF-2 protein is upregulated prior to the upregulation of COX subunit message and activity (Zhang and Wong-Riley 2000b), and both α and β subunits of NRF-2 respond to increased neuronal activity by translocating from the cytoplasm to the nucleus, where they associate primarily with euchromatin to activate their target genes (Yang et al 2004).
12.5.1.2 NRF-2 Regulates All 13 Cytochrome c Oxidase Subunit Genes in Neurons
To determine if NRF-2 regulates all COX subunit genes in neurons, in vitro electrophoretic mobility shift (EMSA) and supershift assays, in vivo chromatin immunoprecipitation (ChIP) assays, and promoter mutational analysis were performed. It was found that, indeed, NRF-2 functionally regulates all ten nuclear-encoded subunit genes of COX (Ongwijitwat and Wong-Riley 2005). Moreover, functional silencing of NRF-2 with small hairpin interference RNA (shRNA) significantly reduces the expression of all 10 nuclear-encoded COX subunit genes, as well as of Tfam and Tfb1m, which regulate the expression of the three mitochondrial-encoded COX subunit genes (Ongwijitwat et al 2006). As discussed above, the role of NRF-2 in directly regulating Tfam and Tfbms has been established (Virbasius and Scarpulla 1994; Gleyzer et al 2005). These findings, then, are consistent with NRF-2’s proposed role as a transcriptional activator of COX and that its own expression is regulated by neuronal activity (reviewed in Wong-Riley et al 2008).
12.5.2 Role of Nuclear Respiratory Factor 1
NRF-1 was first discovered as a transcriptional regulator of the somatic cytochrome c, the substrate for COX (Evans and Scarpulla 1989). NRF-1 also activates other genes whose products function within the mitochondria, such as a few of the nuclear-encoded COX subunits (Vb and VIa in humans, Vb and VIc in rats, and VIIaL in cows), specific nuclear-encoded subunits of complexes I, II, III and V, mitochondrial RNA processing (MRP) RNA, as well as 5-aminolevulinate synthase, which is important for regulating the supply of heme to the cytochromes and other hemoproteins (reviewed in Kelly and Scarpulla 2004). Together with NRF-2, NRF-1 also activates Tfam and the Tfbms (Virbasius and Scarpulla 1994; Gleyzer et al 2005). Thus, NRF-1 is another potential coordinator of mitochondrial- and nuclear-encoded subunits of COX.
NRF-1, unlike NRF-2, is a single-gene product whose gene is mapped to human chromosome 7 (7q31) (Gopalakrishnan and Scarpulla 1995) and is ~104-kb long (Huo and Scarpulla 1999). The DNA-binding domain is at the amino terminus and is highly conserved, whereas the transactivation domain is at the carboxy terminus, which is quite divergent among the species (Virbasius et al 1993b; Gugneja et al 1996). NRF-1 binds the palindromic consensus sequence (T/C)GCGCA(T/C)GCGC(A/G) (Evans and Scarpulla 1990; Virbasius et al. 1993b; Scarpulla 1997). However, the GCA core is found to be invariant, whereas the flanking GC-rich sequences can be somewhat variable (Dhar et al 2008). Phosphorylation of NRF-1 greatly enhances its DNA-binding and transactivational activity (Gugneja and Scarpulla 1997). Homozygous NRF-1 knockout mice are embryonically lethal at E3.5 to E6.5, and the blastocysts have greatly reduced mtDNA levels (Huo and Scarpulla 2001). This is consistent with the key role of NRF-1 in the maintenance of mtDNA and respiratory chain function during early embryogenesis. On the other hand, heterozygous mice developed normally, and no apparent deficits have been detected. Interestingly, mutations in a homologue of NRF-1 in the zebrafish, known as Not really finished, cause a progressive degeneration of photoreceptors and other cells in the retina, optic tectum, and the brain (Becker et al 1998).
12.5.2.1 NRF-1 Itself Responds to Changes in Neuronal Activity
What is the role of NRF-1 in neurons? Does it respond to changes in neuronal activity? By means of light and EM immunohistochemistry, western blotting, and real-time quantitative PCR, it was found that both NRF-1 protein and mRNA are present in mammalian visual cortical neurons, and that both are regulated by neuronal activity (Liang and Wong-Riley 2006; Yang et al 2006). In vitro impulse blockade with TTX and in vivo monocular enucleation lead to a significant downregulation of NRF-1 mRNA and protein in deprived neurons after 6 or 7 days of deprivation (Liang and Wong-Riley 2006). On the other hand, depolarizing stimulation with KCl progressively upregulates both NRF-1 message and protein in a time-dependent manner, increasing above controls after 1 h and remaining high at 3, 5, and 7 h (Yang et al 2006). NRF-1 message increases in both the nucleus and the cytoplasm of stimulated neurons, and EM quantification of immunogold particles is consistent with an activity-induced cytoplasmic-to-nuclear translocation of NRF-1 protein. Levels of NRF-1 mRNA and protein progressively decline when the stimulation is withdrawn, with the former reaching basal levels by 5 h and the latter by 7 h (Yang et al 2006). Thus, NRF-1 upregulates swiftly to functional stimulation but declines more slowly with functional impulse blockade in neurons. These findings are consistent with an activity dependency of the synthesis, distribution, and possibly stability of NRF-1 mRNA and protein in neurons, and that the regulation is primarily at the transcriptional level.
12.5.2.2 NRF-1 Regulates All 13 Cytochrome c Oxidase Subunit Genes in Neurons
Does NRF-1 regulate all 13 COX subunit genes? Its indirect activation of the three mitochondrial-encoded subunit genes via TFAM and TFBMs is already known (Virbasius and Scarpulla 1994; Gleyzer et al 2005). In silico analysis of the rat genome revealed the typical NRF-1 binding motif in only three of the ten nuclear-encoded COX subunit promoters: COX5b, 6a1, and 6c (Ongwijitwat and Wong-Riley 2005). These three have been reported previously in humans and rats (Bachman et al 1996; Ongwijitwat and Wong-Riley 2004; Evans and Scarpulla 1990). In silico analysis has revealed another NRF-1 binding site on the bovine Cox7a2 promoter (Seelan et al 1996). However, after using multiple approaches, such as EMSA, supershift, ChIP, and promoter mutational analysis, it was found that NRF-1 functionally regulates all ten nuclear subunits of COX in neurons (Dhar et al 2008). The reason that in silico analysis failed to detect those sites is that the classical NRF-1 cis motif can actually vary slightly with respect to the sequence of GCs, as long as the GCA core remains intact (Dhar et al 2008). Silencing of NRF-1 with shRNA significantly downregulates all ten nuclear COX mRNAs, as well as messages for TFAM, TFB1M, TFB2M, SURF1 (surfeit 1), VDAC (voltage-dependent anion channel), and TOM20 (transporter of outer mitochondrial membrane) (Dhar et al 2008), the last five are known target genes of NRF-1 (Scarpulla 2002; Kelly and Scarpulla 2004; Gleyzer et al 2005). The extent of reduction ranges from ~35% to 70% (P < 0.05 – 0.01). Thus, both NRF-1 and NRF-2 prove to be key bigenomic coordinators for transcriptional regulation of all COX subunit genes in neurons.
12.6 Transcriptional Coactivators: Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1α (PGC-1α)
In recent years, an important transcriptional coactivator of NRF-1 and NRF-2 has been identified as the peroxisome proliferator-activated receptor gamma coactivator 1 (PGC-1 or PGC-1α) (Wu et al 1999). It was first cloned from a brown fat cDNA library to be induced by cold exposure in both the brown fat and skeletal muscles of mice (Puigserver et al 1998). Now it is known that it belongs to a family of regulated coactivators, which include PGC-1α, PGC-1β, and PGC-1α-related coactivator (PRC) (reviewed in Scarpulla 2008). As a coactivator, PGC-1α does not bind DNA directly, but rather, in response to appropriate signals in a tissue-specific manner, such as cold exposure in brown adipose tissue and muscle, fasting in the heart, and prolonged physical exercise in skeletal muscles, it interacts with nuclear receptors and transcription factors to activate genes involved in energy and nutrient homeostasis (Puigserver et al 1998; Lehman et al 2000; Goto et al 2000; Baar et al 2002). These factors include peroxisome proliferator-activated receptor gamma (PPARγ) and alpha (PPARα), thyroid hormone receptor, estrogen-related receptor (ERRα), glucocorticoid receptor, mineralocorticoid receptor, myocyte enhancer factor 2C, Ying Yang 1 (YY1), as well as NRF-1 and NRF-2 (Puigserver et al 1998; Wu et al 1999; Knutti and Kralli 2001; Scarpulla 2011). The target genes of these factors include those that encode for the uncoupling proteins (UCPs), subunits of mitochondrial electron transport chain complexes, TFAM, and TFBMs, among others (Puigserver et al 1998; Knutti and Kralli 2001; Scarpulla 2008, 2011). Thus, PGC-1α plays a key role in adaptive thermogenesis, glucose and fatty acid metabolism, skeletal muscle fiber type switching, heart development, and mitochondrial biogenesis (Puigserver et al 1998; Knutti and Kralli 2001; Scarpulla 2011). Surprisingly, PGC-1α knockout mice are viable, but exhibit multisystem abnormalities, decreased mitochondrial function, defective thermogenic response, and lesions in the striatum and cerebral cortex (Lin et al 2004; Leone et al 2005). Over-expression of PGC-1α increases mitochondrial content, induces the expression of genes involved in energy production and transduction pathways, and protects cultured cells from oxidative stress-induced death (Lehman et al 2000; St-Pierre et al 2006; Scarpulla 2011). However, cardiac-specific over-expression of PGC-1α in transgenic mice can lead to uncontrolled mitochondrial proliferation and dilated cardiomyopathy (Lehman et al 2000).
12.6.1 PGC-1α Responds to Changes in Neuronal Activity
In neurons, PGC-1α is localized mainly to nuclear euchromatin and cytoplasmic free ribosomes (Meng et al 2007). Depolarizing stimulation for 0.5 h significantly increases PGC-1α in both the nucleus and the cytoplasm (Meng et al 2007). The level is sustained up to 3 h of stimulation, but decreases from 5 h onward and returns to baseline level by 10 h. Thus, PGC-1α responds very early to increased neuronal activity (earlier than either NRF-1 or NRF-2) by upregulating its own synthesis in the cytoplasm and having the protein being translocated to the nucleus for gene activation. When neuronal activity is reduced by impulse blockade in vitro or sensory deprivation in vivo, the levels of PGC-1α mRNA and proteins are significantly downregulated earlier than those of NRF-1 and NRF-2 (Liang and Wong-Riley 2006). Neuronal activity, therefore, directly regulates PGC-1α, and PGC-1α is likely to be a critical sensor of activity-dependent energy demand in neurons.
12.6.2 Regulation of PGC-1α in Neurons
Depolarizing activation of PGC-1α in neurons is found to be mediated by p38 mitogen-activated protein kinase (MAPK) and calcium channels (Liang et al 2010). Stimulation upregulates PGC-1α mRNA and protein levels in 0.5 and 1 h, respectively, but both p38 MAPK and phosphorylated p38 MAPK levels are increased after only 15 min. Such upregulation is suppressed by 30 min of pretreatment with SB203580 (a blocker of p38 MAPK that also blocks the upregulation of PGC-1α by KCl) or with nifedipine (a Ca2+ channel blocker). Furthermore, a knockdown of p38 MAPK with shRNA significantly suppresses both PGC-1α mRNAs and proteins (Liang et al 2010). Thus, both p38 MAPK and Ca2+ are critical in mediating signaling in depolarization-induced activation of PGC-1α in neurons.
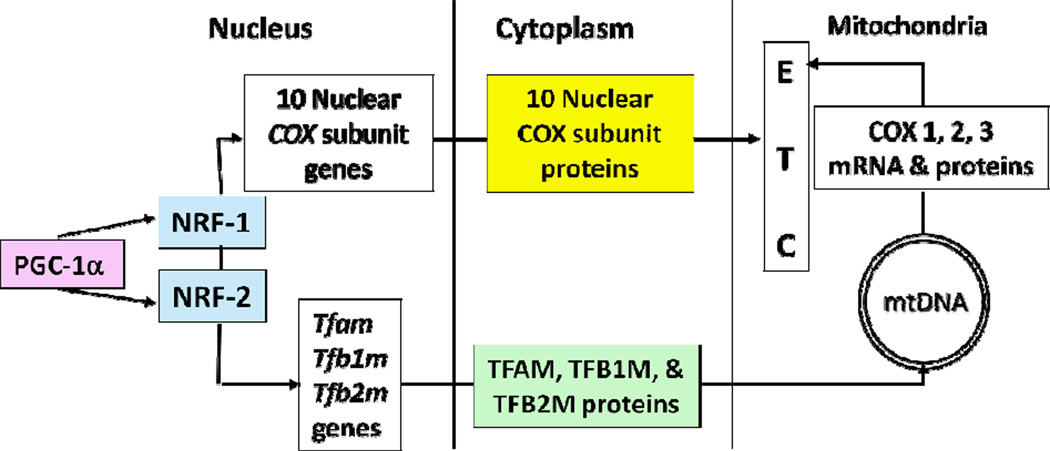
Taken together, PGC-1α is an early sensor of changes in neuronal activity, and it recruits NRF-1 and NRF-2 (among other factors) to regulate the expression of target genes, such as COX, important in energy metabolism that is tightly coupled to neuronal activity. Such a chain of events regulates not only the three mitochondrial-encoded COX subunits via TFAM and TFBMs, but also all ten of the nuclear-encoded COX subunit genes in neurons (Fig. 1).
Fig. 12.1.
Schematic diagram depicting that PGC-1α coordinates the induction of NRF-1 and NRF-2 in regulating the transcription of all ten nuclear-encoded COX subunit genes, as well as the genes for Tfam, Tfb1m, and Tfb2m in the nucleus. Translation in the cytoplasm leads to the generation of the respective proteins, all of which enter into the mitochondria. Within the mitochondrion, the three mitochondrial-encoded COX genes are transcribed aided by the TFs and translated into polypeptides. Together, the nuclear- and mitochondrial-encoded COX subunits form the holoenzyme that is complex IV of the electron transport chain (ETC).
12.7 Is There a Transcription Factory for the 13 Genomic Loci Involved in the Bigenomic Transcription of COX in Neurons?
Transcription factories have been described as dynamic but discrete loci in the nucleus that actively transcribe related genes. These sites are thought to contain several RNA polymerase II molecules, relevant transcription factors, and loops of chromatin-containing genes to be transcribed together (Jackson et al 1998; Osborne et al 2004; Zhou et al 2006). To demonstrate such long-range interactions among related genes, chromosome conformation capture (3C) has been developed (Dekker et al 2002; Miele and Dekker 2009). This technique converts chromatin conformation and physical interactions in vivo into specific ligation products demonstrable with polymerase chain reaction. Interactions between loci from the same chromosome or from two different chromosomes have been described (Spilianakis and Flavell 2004; Ling et al 2006; Lomvardas et al 2006; Schoenfelder et al 2010). However, to demonstrate interactions among ten genomic loci of the ten nuclear-encoded COX subunit genes located in nine different chromosomes poses a distinct challenge.
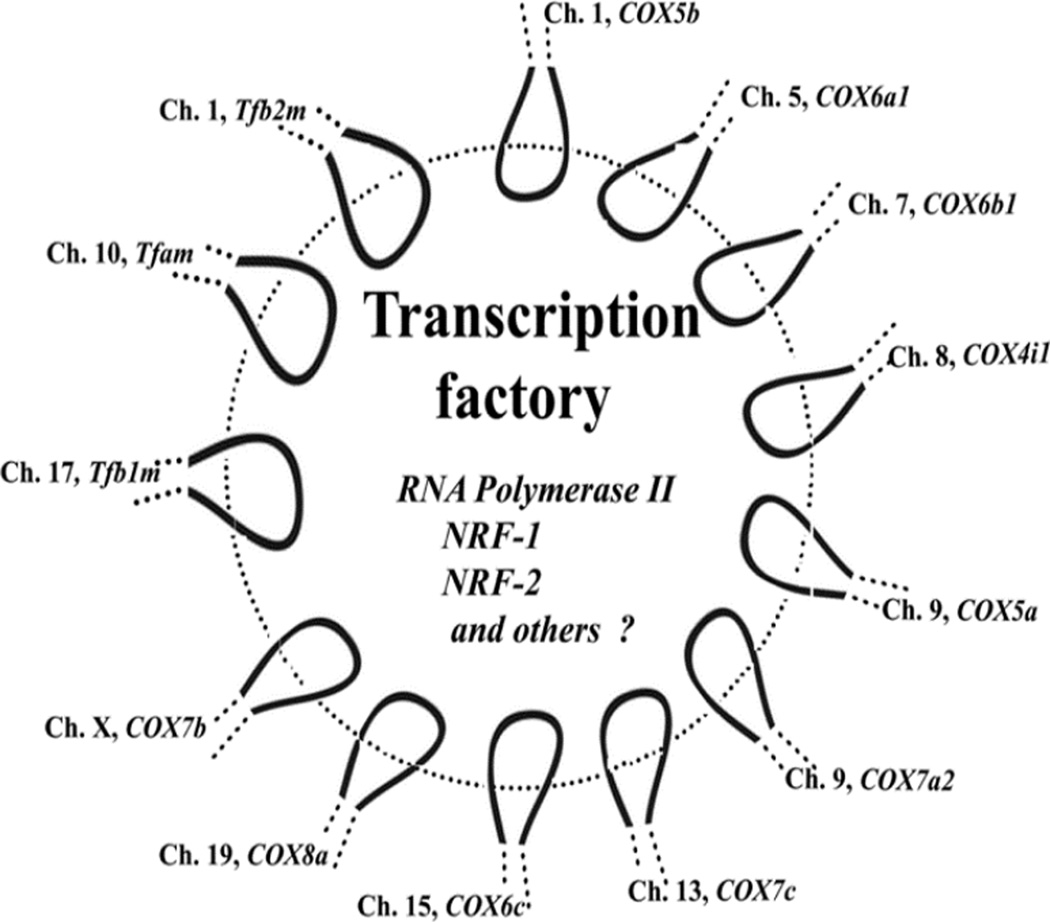
By means of 3C, it was found that not only do these ten genomic loci interact in the same transcription factory, but that genes from three chromosomes encoding Tfam, Tfb1m, and Tfb2m that are critical for the transcription of the three mitochondrial-encoded COX subunit genes all occupy common intranuclear sites in the murine neuronal nuclei (Dhar et al 2009a). Moreover, interactions between COX subunit and Tf genes are upregulated by depolarizing stimulation and downregulated by impulse blockade in primary neurons in culture (Dhar et al 2009a). No doubt, such “transcription factories” are dynamic entities regulated by the energy demand of neurons. Taken together, there is indeed an exquisite mechanism in place for a coordinated and synchronized transcriptional regulation of the multisubunit, multichromosomal, bigenomic COX enzyme in neurons (Fig. 2).
Fig. 12.2.
Schematic rendition of a dynamic transcription factory in which the loops of 13 genomic loci for the ten nuclear-encoded COX subunit genes and genes for Tfam, Tfb1m, and Tfb2m are cotranscribed, with the aid of RNA polymerase II, NRF-1, NRF-2, and possibly other transcription factors and coactivators. (Reproduced with permission from Dhar et al 2009a).
12.8 Tight Coupling Between Neuronal Activity and Energy Metabolism at the Transcriptional Level
The tight coupling between neuronal activity and energy metabolism has been well established at the cellular level (Wong-Riley 1989; Wong-Riley et al 1998a; 2008). As discussed above, repolarization of membrane potentials after depolarizing stimulation consumes the bulk of energy in neurons (Wong-Riley 1989). The more excitatory input a neuron receives, the greater its energy demand.
12.8.1 Glutamatergic System in Neurons
The main depolarizing agent in the brain is glutamate, a major and the most prevalent excitatory neurotransmitter (Fonnum 1984; Streit 1984). Its action is mediated by two major types of receptors, N-methyl-D-aspartate (NMDA) and non-NMDA (reviewed in Nakanishi 1992). NMDA receptor is an ionotropic, ligand-gated calcium channel with voltage-dependent magnesium block, and it is made up of the ubiquitous and obligatory NR1 (GluN1) subunit in a heterotetrameric complex with one or more of NR2 (NR2A-D or GluN2A-D) and/or NR3 (NR3A-B or GluN3A-B) subunits (Orrego and Villanueva 1993; Mori and Mishina 1995; Dingledine et al 1999; Salussolia et al 2011). Among the NR subunits, NR2B is important in synaptic signaling, long-term potentiation, learning and memory, as well as involvement in a number of human neurological disorders (Loftis and Janowsky 2003; Babb et al 2005). Within the non-NMDA receptor category, the AMPA (α-amino-3-hydroxyl-5-methyl-4-isoxazolpropionic acid) type is ionotropic, ligand-gated, mediates fast synaptic transmission, and is made up of GluR1–4 (GluR-A-D or GluA1–4) subunits in various combinations (Keinänen et al 1990). These subunits each undergoes RNA editing and alternative splicing, yielding either the flip or flop variants (Sommer et al 1990; Lomeli et al 1994). AMPA receptors play a critical role in synaptogenesis, neural circuitry formation, and synaptic plasticity (Tanaka et al 2000; Palmer et al 2005). Subunit 2 (GluR2, GluR-B, or GluA2) is of special interest because the glutamine residue in its transmembrane segment 2 is mRNA-edited to the positively charged arginine residue, and it is the only subunit that impedes Ca2+ entry into neurons (Verdoorn et al 1991). AMPA receptors that contain the GluR2 subunit are impermeable to Ca2+, show simple outward rectification, and are insensitive to blockage by external polyamines (Hollmann et al 1991; Burnashev et al 1992; Washburn et al 1997). Thus, GluR2 is dominant in determining the functional properties of heteromeric AMPA receptors (Jonas et al 1994; Tanaka et al 2000). The downregulation of GluR2 mRNA has been implicated in enhanced neurotoxicity with increased Ca2+ permeability in the affected neurons (reviewed in Pellegrini-Giampietro et al 1997).
In the CNS, regions rich in COX also have higher levels of glutamatergic and NMDA receptor-mediated synapses. When these regions are deprived of their excitatory input, the levels of both COX and NMDA receptor NR1 are downregulated (Wong-Riley et al 1998b,c). The expressions of GluR2 mRNA and proteins are also governed by neuronal activity (Wong-Riley and Jacobs 2002; Bai and Wong-Riley 2003). If excitatory glutamatergic neurotransmission goes hand in hand with COX expression, the question naturally arises as to whether the coupling between these two activities exists at the molecular level? That is, can the same transcription factor or factors regulate COX as well as neurochemicals of glutamatergic neurotransmission?
12.8.2 Does NRF-1 Coordinate the Transcriptional Regulation of Neurochemicals and Cytochrome c Oxidase in Neurons?
NRF-1 is a natural candidate for such an inquiry. Its role in regulating all 13 COX subunit genes has been well defined (see above). A consensus recognition sequence for NRF-1 has been reported for the GC-rich proximal promoter of the rat Gria2 (for GluR2) gene, but it has not been rigorously tested (Myers et al 1998). Whether NRF-1 regulates the other AMPA (Gria) and any of the NMDA receptor (Grin) subunit genes was entirely unknown. By means of in silico analyses, in vitro EMSA and supershift assays, in vivo ChIP, promoter mutational analyses, and shRNA, NRF-1 was found to functionally regulate Grin1 and Grin2b, but not the other, subunits of the NMDA receptor genes, and Gria2, but not the other, subunits of the AMPA receptor genes (Dhar and Wong-Riley 2009; Dhar et al 2009b). The transcripts are upregulated by KCl depolarizing stimulation and downregulated by TTX impulse blockade in cultured primary neurons. However, silencing of NRF-1 blocks the upregulation, and over-expression of NRF-1 rescues the downregulation, of Grin1, Grin2b, Gria2, as well as COX subunit transcripts in neurons (Dhar and Wong-Riley 2009; Dhar et al 2009b). NRF-1-binding sites on these genes are also highly conserved among rats, mice, and humans. As discussed above, NR1 is an essential subunit, NR2B is critical for a number of basic structural and functional attributes of the NMDA receptors, and GluR2 is an important regulatory subunit of AMPA receptors.
NRF-1 also functionally regulates neuronal nitric oxide synthase (Nos1) (Dhar et al 2009c), which links NMDA receptor transmission to the cGMP second messenger cascade (Garthwaite 1991). This regulation is specific, as NRF-1 does not control the expressions of either inducible NOS (iNOS or Nos2) or endothelial NOS (eNOS or Nos3) (Dhar et al 2009c). Silencing NRF-1 not only downregulates Nos1 mRNA and proteins, but also the transcripts of guanylyl cyclase, a downstream target of the nitric oxide pathway (Dhar et al 2009c).
12.8.3 Is There a Transcription Factory for Cytochrome c Oxidase and Genes of Glutamatergic Neurotransmission?
To verify that there is coordinated transcription of COX and those of neurochemicals coregulated by NRF-1, chromosome confirmation capture was utilized. Indeed, interactions were found among genomic loci for COX, Grin1, Grin2b, Gria2, and Nos1 in neurons, but not in C2C12 muscle cells, indicating that such a “factory” is neuron-specific (Dhar and Wong-Riley 2010). COX subunit genes also do not interact with Grin3a, Gria4, or Nos3, genes that are not regulated by NRF-1, nor with genes for calreticulin, a non-mitochondrial protein (Dhar and Wong-Riley 2010). Depolarizing stimulation increases the interaction frequencies between COX and neurochemical genes, whereas TTX impulse blockade or KCN inhibition of COX downregulates such interactions in neurons (Dhar and Wong-Riley 2010). Hence, these data are consistent with coordinated transcription of COX and specific glutamatergic neurochemical genes in the same transcription factory in neurons.
12.8.4 NRF-1 Coregulates NR2B and Its Transport Motor KIF17 in Neurons
More recently, NRF-1 was found to also regulate the expression of the kinesin superfamily protein KIF17 (Dhar and Wong-Riley 2011), which transports NR2B along microtubules specifically from the cell body to the dendrites, where it forms part of the NMDA receptor complex (Setou et al 2000). Interestingly, Kif17 is not regulated by NRF-2, and NRF-1 does not regulate other Kif transcripts, such as Kif1a (Dhar and Wong-Riley 2011). This is a clear example of how the same transcription factor regulates the expression of both the motor and its specific synaptic cargo in neurons.
12.8.5 Molecular Coupler(s) of Energy Metabolism and Neuronal Activity
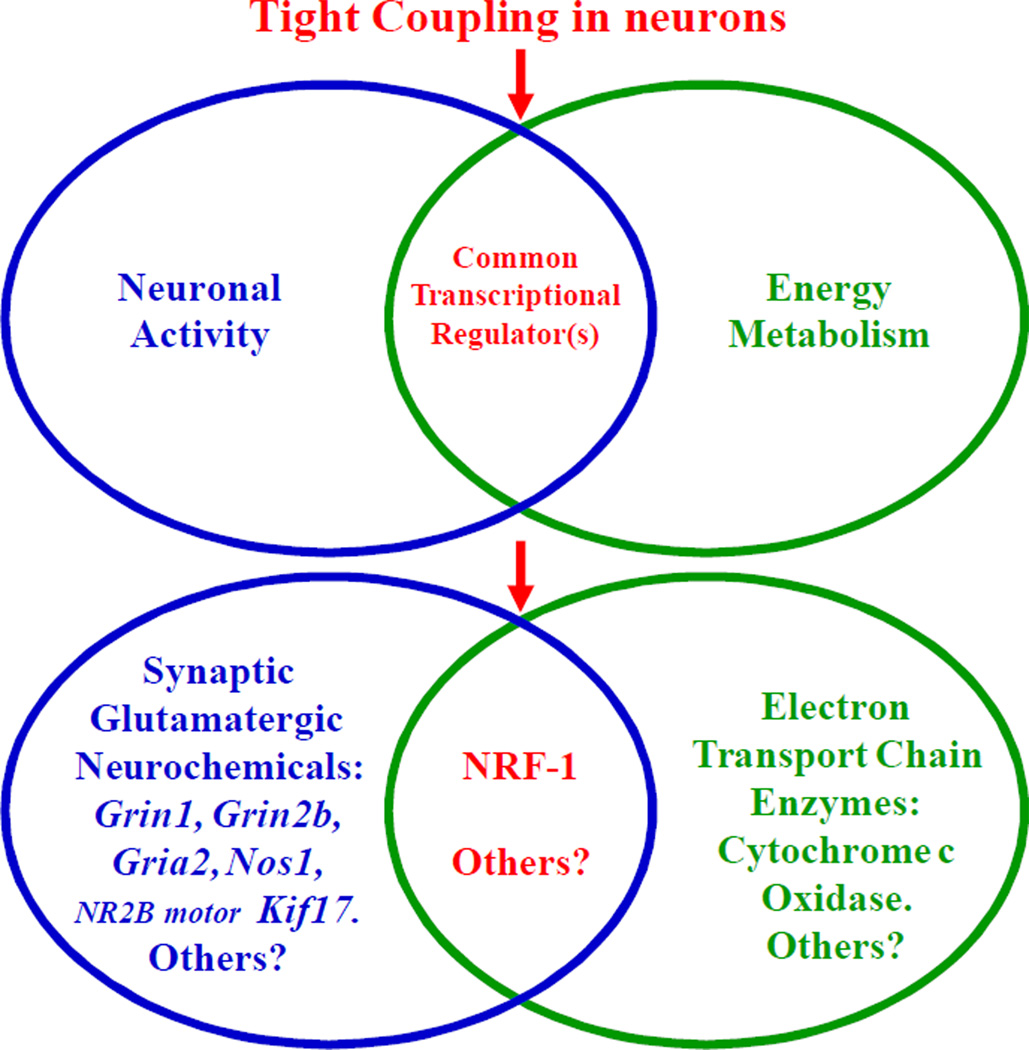
NRF-1 plays the heretofore unrecognized and unappreciated role of dually coordinating the expressions of neurochemicals of glutamatergic neurotransmission and agents of energy metabolism (COX). This coordinated expression ensures that energy production precisely matches energy utilization and thereby mediates the tight coupling between neuronal activity and energy metabolism at the molecular level (Fig. 3). Whether NRF-2 and/or other transcription factors also participate in this coupling remains to be explored. For example, it is unknown if NRF-2 coregulates glutamatergic neurochemicals together with NRF-1 in a complementary, concurrent, or a combination of complementary and concurrent manner. Whether NRF-1 and NRF-2 interact as they activate their common target genes is also not known at this time. However, silencing each of them with shRNA does not affect the expression of the other (Ongwijitwat et al 2006; Dhar et al 2008), suggesting that the two may function independently of each other in neurons.
Fig. 12.3.
A Venn diagram illustrating the tight coupling between neuronal activity and energy metabolism at the molecular level by having the same transcription factor, NRF-1, coregulating genes for critical glutamatergic neurochemicals (Grin1, Grin2b, Gria2, Nos1, and NR2B motor Kif17) as well as all 13 subunits of COX. Such coupling ensures that energy production exquisitely matches energy demand of neuronal activity.
12.9 Conclusions
Cytochrome c oxidase is one of the most ancient enzymes known. Its critical roles in the complete oxidation of carbohydrates, amino acids, and fatty acids and in the generation of a proton gradient necessary for ATP synthesis within the mitochondria are well recognized. The absolute dependence of neurons on COX for their proper functioning and survival is without question. However, only in recent years has the transcriptional regulation of this multisubunit, multichromosomal, bigenomic enzyme been extensively explored in neurons. NRF-1 and NRF-2 are proven bigenomic transcriptional coordinators of all 13 COX subunit transcripts from the two genomes, and both of them are under strict regulation of neuronal activity. NRF-1, in addition, regulates a number of neurochemicals crucial for glutamatergic neurotransmission. Thus, NRF-1 is the first transcription factor known to mediate the tight coupling between neuronal activity and energy metabolism at the molecular level. Other transcription factors, such as NRF-2, and coactivators, such as PGC-1α, may well participate in the coupling process to ensure the exquisite matching of energy production with energy demand of synaptic transmission in neurons.
Acknowledgments
The author expresses her deep appreciation for the valuable contributions from her past and present colleagues in her laboratory as well as in the relevant fields of bioenergetics and neuroscience. She also appreciates the continuous support from the National Institutes of Health. The current support is from NIH grant EY018441.
References
- Attwell D, Laughlin SB. An energy budget for signaling in the grey matter of the brain. J Cereb Blood Flow Metab. 2001;21:1133–1145. doi: 10.1097/00004647-200110000-00001. [DOI] [PubMed] [Google Scholar]
- Au HC, Scheffler IE. Promoter analysis of the human succinate dehydrogenase iron-protein gene – both nuclear respiratory factors NRF-1 and NRF-2 are required. Eur J Biochem. 1998;251:164–174. doi: 10.1046/j.1432-1327.1998.2510164.x. [DOI] [PubMed] [Google Scholar]
- Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, Holloszy JO. Adaptations of skeletal muscle to exercise: Rapid increase in the transcriptional coactivator PGC-1. Faseb J. 2002;16:1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- Babb TL, Mikuni N, Najm I, Wylie C, Olive M, Dollar C, MacLennan H. Pre- and postnatal expression of NMDA receptors 1 and 2B subunit proteins in the normal rat cortex. Epilepsy Res. 2005;64:23–30. doi: 10.1016/j.eplepsyres.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Bachman NJ, Yang TL, Dasen JS, Ernst RE, Lomax MI. Phylogenetic footprinting of the human cytochrome c oxidase subunit VB promoter. Arch Biochem Biophys. 1996;333:152–162. doi: 10.1006/abbi.1996.0376. [DOI] [PubMed] [Google Scholar]
- Bai X, Wong-Riley MTT. Neuronal activity regulates protein and gene expressions of GluR2 in postnatal rat visual cortical neurons in culture. J Neurocytol. 2003;32:71–78. doi: 10.1023/a:1027380315902. [DOI] [PubMed] [Google Scholar]
- Batchelor AH, Piper DE, De la Brousse FC, McKnight SL, Wolberger C. The structure of GABPalpha/beta: An ETS domain-ankyrin repeat heterodimer bound to DNA. Science. 1998;279:1037–1041. doi: 10.1126/science.279.5353.1037. [DOI] [PubMed] [Google Scholar]
- Becker TS, Burgess SM, Amsterdam AH, Allende ML, Hopkins N. Not really finished is crucial for development of the zebrafish outer retina and encodes a transcription factor highly homologous to human nuclear respiratory factor-1 and avian initiation binding repressor. Development. 1998;125:4369–4378. doi: 10.1242/dev.125.22.4369. [DOI] [PubMed] [Google Scholar]
- Blesa JR, Prieto-Ruiz JA, Hernández JM, Hernández-Yago J. NRF-2 transcription factor is required for human TOMM20 gene expression. Gene. 2007;391:198–208. doi: 10.1016/j.gene.2006.12.024. [DOI] [PubMed] [Google Scholar]
- Bruni F, Polosa PL, Gadaleta MN, Cantatore P, Roberti M. Nuclear respiratory factor 2 induces the expression of many but not all human proteins acting in mitochondrial DNA transcription and replication. J Biol Chem. 2010;285:3939–3948. doi: 10.1074/jbc.M109.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnashev N, Monyer H, Seeburg PH, Sakmann B. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron. 1992;8:189–198. doi: 10.1016/0896-6273(92)90120-3. [DOI] [PubMed] [Google Scholar]
- Carter RS, Avadhani NG. Cooperative binding of GA-binding protein transcription factors to duplicated transcription initiation region repeats of cytochrome c oxidase subunit IV gene. J Biol Chem. 1994;269:4381–4387. [PubMed] [Google Scholar]
- Carter RS, Bhat NK, Basu A, Avadhani NG. The basal promoter elements of murine cytochrome c oxidase subunit IV gene consist of tandemly duplicated ETS motifs that bind to GABP-related transcription factors. J Biol Chem. 1992;267:23418–23426. [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- De la Brousse FC, Birkenmeier EH, King DS, Rowe LB, McKnight SL. Molecular and genetic characterization of GABPβ. Genes Dev. 1994;8:1853–1865. doi: 10.1101/gad.8.15.1853. [DOI] [PubMed] [Google Scholar]
- DeYoe EA, Trusk TC, Wong-Riley MTT. Activity correlates of cytochrome oxidase-defined compartments in granular and supragranular layers of primary visual cortex of the macaque monkey. Vis Neurosci. 1995;12:629–639. doi: 10.1017/s0952523800008920. [DOI] [PubMed] [Google Scholar]
- Dhar SS, Wong-Riley MTT. Coupling of energy metabolism and synaptic transmission at the transcriptional level: Role of nuclear respiratory factor 1 in regulating both cytochrome c oxidase and NMDA glutamate receptor subunit genes. J Neurosci. 2009;29:483–492. doi: 10.1523/JNEUROSCI.3704-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Wong-Riley MTT. Chromosome conformation capture of transcriptional interactions between cytochrome coxidase genes and genes of glutamatergic synaptic transmission in neurons. J Neurochem. 2010;115:676–683. doi: 10.1111/j.1471-4159.2010.06956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Wong-Riley MTT. The kinesin superfamily protein KIF17 is regulated by the same transcription factor (NRF-1) as its cargo NR2B in neurons. BBA – Mol Cell Res. 2011;1813:403–411. doi: 10.1016/j.bbamcr.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Ongwijitwat S, Wong-Riley MTT. Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. J Biol Chem. 2008;283:3120–3129. doi: 10.1074/jbc.M707587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Ongwijitwat S, Wong-Riley MTT. Chromosome conformation capture of all 13 genomic loci in the transcriptional regulation of the multi-subunit bigenomic cytochrome c oxidase in neurons. J Biol Chem. 2009a;284:18644–18650. doi: 10.1074/jbc.M109.019976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Liang HL, Wong-Riley MTT. Nuclear respiratory factor 1 co-regulates AMPA glutamate receptor subunit 2 and cytochrome c oxidase: Tight coupling of glutamatergic transmission and energy metabolism in neurons. J Neurochem. 2009b;108:1595–1606. doi: 10.1111/j.1471-4159.2009.05929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Liang HL, Wong-Riley MTT. Transcriptional coupling of synaptic transmission and energy metabolism: Role of nuclear respiratory factor 1 in co-regulating neuronal nitric oxide synthase and cytochrome coxidase genes in neurons. BBA – Mol Cell Res. 2009c;1793:1604–1613. doi: 10.1016/j.bbamcr.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- Evans MJ, Scarpulla RC. Interaction of nuclear factors with multiple sites in the somatic cytochrome c promoter. Characterization of upstream NRF-1, ATF, and intron Sp1 recognition sequences. J Biol Chem. 1989;264:14361–14368. [PubMed] [Google Scholar]
- Evans MJ, Scarpulla RC. NRF-1: A trans-activator of nuclear-encoded respiratory genes in animal cells. Genes Dev. 1990;4:1023–1034. doi: 10.1101/gad.4.6.1023. [DOI] [PubMed] [Google Scholar]
- Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- Fisher RP, Clayton DA. Purification and characterization of human mitochondrial transcription factor 1. Mol Cell Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonnum F. Glutamate: A neurotransmitter in mammalian brain. J Neurochem. 1984;42:1–11. doi: 10.1111/j.1471-4159.1984.tb09689.x. [DOI] [PubMed] [Google Scholar]
- Garthwaite J. Glutamate, nitric oxide and cell-cell signaling in the nervous system. Trends Neurosci. 1991;14:60–67. doi: 10.1016/0166-2236(91)90022-m. = [DOI] [PubMed] [Google Scholar]
- Gleyzer N, Vercauteren K, Scarpulla RC. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol Cell Biol. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan L, Scarpulla RC. Structure, expression, and chromosomal assignment of the human gene encoding nuclear respiratory factor 1. J Biol Chem. 1995;270:18019–18025. doi: 10.1074/jbc.270.30.18019. [DOI] [PubMed] [Google Scholar]
- Goto M, Terada S, Kato M, Katoh M, Yokozeki T, Tabata I, Shimokawa T. cDNA cloning and mRNA analysis of PGC-1 in epitrochlearis muscle in swimming-exercised rats. Biochem Biophys Res Commun. 2000;274:350–354. doi: 10.1006/bbrc.2000.3134. [DOI] [PubMed] [Google Scholar]
- Gugneja S, Scarpulla RC. Serine phosphorylation within a concise amino-terminal domain in nuclear respiratory factor 1 enhances DNA binding. J Biol Chem. 1997;272:18732–18739. doi: 10.1074/jbc.272.30.18732. [DOI] [PubMed] [Google Scholar]
- Gugneja S, Virbasius CM, Scarpulla RC. Nuclear respiratory factors 1 and 2 utilize similar glutamine-containing clusters of hydrophobic residues to activate transcription. Mol Cell Biol. 1996;16:5708–5716. doi: 10.1128/mcb.16.10.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo AL, Nie F, Wong-Riley MTT. Human brain nuclear respiratory factor (NRF) 2α cDNA: Isolation, subcloning, sequencing and in situ hybridization of transcripts in normal and visually deprived macaque visual system. J Comp Neurol. 2000;417:221–232. doi: 10.1002/(sici)1096-9861(20000207)417:2<221::aid-cne7>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Wong-Riley MTT. Brain cytochrome oxidase: Purification, antibody production, and immunohistochemical/histochemical correlations in the CNS. J Neurosci. 1989;9:3884–3898. doi: 10.1523/JNEUROSCI.09-11-03884.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Wong-Riley MTT. Regulation of cytochrome oxidase protein levels by functional activity in the macaque monkey visual system. J Neurosci. 1990;10:1331–1340. doi: 10.1523/JNEUROSCI.10-04-01331.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Wong-Riley MTT. Neuronal expression of nucleaer and mitochondrial genes for cytochrome oxidase (CO) subunits analyzed by in situ hybridization; comparison with CO activity and protein. J Neurosci. 1991;11:1942–1958. doi: 10.1523/JNEUROSCI.11-07-01942.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Wong-Riley MTT. Mitochondrial and nuclear gene expression for cytochrome oxidase subunits are disproportionately regulated by functional activity in neurons. J Neurosci. 1993;13:1805–1819. doi: 10.1523/JNEUROSCI.13-05-01805.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Liu S, Wong-Riley MTT. A metabolic map of cytochrome oxidase in the rat brain: Histochemical, densitometric and biochemical studies. Neurosci. 1995;65:313–342. doi: 10.1016/0306-4522(94)00514-6. [DOI] [PubMed] [Google Scholar]
- Hollman M, Hartley M, Heinemann S. Ca2+ permeability of KA-AMPA-gated glutamate receptor channels depends on subunit composition. Science. 1991;252:851–853. doi: 10.1126/science.1709304. [DOI] [PubMed] [Google Scholar]
- Huo L, Scarpulla RC. Multiple 5’-untranslated exons in the nuclear respiratory factor 1 gene span 47 kb and contribute to transcript heterogeneity and translational efficiency. Gene. 1999;233:213–224. doi: 10.1016/s0378-1119(99)00135-3. [DOI] [PubMed] [Google Scholar]
- Huo L, Scarpulla RC. Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol Cell Biol. 2001;21:644–654. doi: 10.1128/MCB.21.2.644-654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Iborra FJ, Manders EM, Cook PR. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HaLa nuclei. Mol Biol Cell. 1998;9:1523–1536. doi: 10.1091/mbc.9.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas P, Racca C, Sakmann B, Seeburg PH, Monyer H. Differences in Ca2+ permeability of AMPA-type glutamate receptor channels in neocortical neurons caused by differential GluR-B subunit expression. Neuron. 1994;12:1281–1289. doi: 10.1016/0896-6273(94)90444-8. [DOI] [PubMed] [Google Scholar]
- Kadenbach B, Jaraush S, Hartmann R, Merle P. Separation of mammalian cytochrome c oxidase into 13 polypeptides by a sodium dodecyl sulfate- gel electrophoresis procedure. Anal Biochem. 1983;129:517–521. doi: 10.1016/0003-2697(83)90586-9. [DOI] [PubMed] [Google Scholar]
- Kadenbach B, Huttermann M, Arnold S, Lee I, Bender E. Mitochondrial energy metabolism is regulated via nuclear-coded subunits of cytochrome c oxidase. Free Radic Biol Med. 2000;29:211–221. doi: 10.1016/s0891-5849(00)00305-1. [DOI] [PubMed] [Google Scholar]
- Keinänen K, Wisden W, Sommer B, Werner P, Herb A, Verdoorn TA, Sakmann B, Seeburg PH. A family of AMPA-selective glutamate receptors. Science. 1990;249:556–560. doi: 10.1126/science.2166337. [DOI] [PubMed] [Google Scholar]
- Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- Knutti D, Kralli A. PGC-1, a versatile coactivator. Trends Endocrinol Metab. 2001;12:360–365. doi: 10.1016/s1043-2760(01)00457-x. [DOI] [PubMed] [Google Scholar]
- LaMarco K, Thompson CC, Byers BP, Walton EM, McKnight SL. Identification of Ets- and Notch-related subunits in GA-binding protein. Science. 1991;253:789–792. doi: 10.1126/science.1876836. [DOI] [PubMed] [Google Scholar]
- Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, Chen Z, Holloszy JO, Medeiros DM, Schmidt RE, Saffitz JE, Abel ED, Semenkovich CF, Kelly DP. PGC-1alpha deficiency causes multi-system energy metabolic derangements: Muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Wong-Riley MTT. Activity-dependent regulation of nuclear respiratory factor-1, nuclear respiratory factor-2, and peroxisome proliferators-activated receptor gamma coactivator-1 in neurons. NeuroReport. 2006;17:401–405. doi: 10.1097/01.wnr.0000204980.98876.11. [DOI] [PubMed] [Google Scholar]
- Liang HL, Ongwijitwat S, Wong-Riley MTT. Bigenomic functional regulation of all 13 cytochrome c oxidase subunit transcripts in rat neurons in vitro and in vivo . Neurosci. 2006;140:177–190. doi: 10.1016/j.neuroscience.2006.01.056. [DOI] [PubMed] [Google Scholar]
- Liang HL, Dhar SS, Wong-Riley MTT. p38 Mitogen-activated protein kinase and calcium channels mediate signaling in depolarization-induced activation of peroxisome proliferator-activated receptor gamma coactivator-1α in neurons. J Neurosci Res. 2010;88:640–649. doi: 10.1002/jnr.22222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D, Spiegelman BM. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- Liu S, Wong-Riley M. Nuclear-encoded mitochondrial precursor protein: Intramitochondrial delivery to dendrites and axon terminals of neurons and regulation by neuronal activity. J Neurosci. 1994;14:5338–5351. doi: 10.1523/JNEUROSCI.14-09-05338.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftis JM, Janowsky A. The N-methyl-D-aspartate receptor subunit NR2B: localization, functional properties, regulation, and clinical implications. Pharmacol Ther. 2003;97:55–85. doi: 10.1016/s0163-7258(02)00302-9. [DOI] [PubMed] [Google Scholar]
- Lomali H, Mosbacher J, Melcher T, Höger T, Geiger JRP, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Lowry OH. Energy metabolism in brain and its control. In: Ingvar DH, Lassen NA, editors. Brain Work: The Coupling of Function, Metabolism, and Blood Flow in the Brain, Alfred Benzon Synposium VIII. New York: Academic Press; 1975. pp. 48–64. [Google Scholar]
- Meng H, Liang HL, Wong-Riley M. Quantitative immune-electron microscopic analysis of depolarization-induced expression of PGC-1alpha in cultured rat visual cortical neurons. Brain Res. 2007;1175:10–16. doi: 10.1016/j.brainres.2007.07.063. [DOI] [PubMed] [Google Scholar]
- Miele A, Dekker J. Mapping cis- and trans-chromatin interaction networks using chromosome conformation capture (3C) Methods Mol Biol. 2009;464:105–121. doi: 10.1007/978-1-60327-461-6_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Mishina M. Structure and function of the NMDA receptor channel. Neuropharm. 1995;34:1219–1237. doi: 10.1016/0028-3908(95)00109-j. [DOI] [PubMed] [Google Scholar]
- Mori M, Morita T, Ikeda F, Amaya Y, Tatibana M, Cohen PP. Synthesis, intracellular transport, and processing of the precursor for mitochondrial ornithine transcarbamylase and carbamoylphosphate synthetase I in isolated hepatocytes. Proc Natl Acad Sci USA. 1981;78:6056–6060. doi: 10.1073/pnas.78.10.6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SJ, Peters J, Huang Y, Comer MB, Barthel F, Dingledine R. Transcriptional regulation of the GluR2 gene: Neural-specific expression, multiple promoters, and regulatory elements. J Neurosci. 1998;18:6723–6739. doi: 10.1523/JNEUROSCI.18-17-06723.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S. Molecular diversity of glutamate receptors and implications for brain function. Science. 1992;258:597–603. doi: 10.1126/science.1329206. [DOI] [PubMed] [Google Scholar]
- Nie F, Wong-Riley M. Nuclear respiratory factor-2 subunit protein: Correlation with cytochrome oxidase and regulation by functional activity in the monkey primary visual cortex. J Comp Neurol. 1999;404:310–320. doi: 10.1002/(sici)1096-9861(19990215)404:3<310::aid-cne3>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Ongwijitwat S, Wong-Riley MTT. Functional analysis of the rat cytochrome c oxidase subunit 6A1 promoter in primary neurons. Gene. 2004;337:163–171. doi: 10.1016/j.gene.2004.04.024. [DOI] [PubMed] [Google Scholar]
- Ongwijitwat S, Wong-Riley MTT. Is nuclear respiratory factor 2 a master transcriptional coordinator for all ten nuclear-encoded cytochrome c oxidase subunits in neurons? Gene. 2005;360:65–77. doi: 10.1016/j.gene.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Ongwijitwat S, Liang HL, Graboyes EM, Wong-Riley MTT. Nuclear respiratory factor 2 senses changing cellular energy demands and its silencing down-regulates cytochrome oxidase and other target gene mRNAs. Gene. 2006;374:39–49. doi: 10.1016/j.gene.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Orrego F, Villanueva S. The chemical nature of the main central excitatory transmitter: A critical appraisal based upon release studies and synaptic vesicle localization. Neurosci. 1993;56:539–555. doi: 10.1016/0306-4522(93)90355-j. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Palmer CL, Lin W, Hastie PGR, Toward M, Korolchuk VI, Burbidge SA, Banting G, Collingridge GL, Isaac JTR, Henley JM. Hippocalcin functions as a calcium sensor in hippocampal LTD. Neuron. 2005;47:487–494. doi: 10.1016/j.neuron.2005.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini-Giampietro DE, Gorter JA, Bennett MV, Zukin RS. The GluR2 (GluR-B) hypothesis: Ca2+-permeable AMPA receptors in neurologic disorders. Trends Neurosci. 1997;20:464–470. doi: 10.1016/s0166-2236(97)01100-4. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Reid GA, Yonetani T, Schatz G. Import of proteins into mitochondria: Import and maturation of the mitochondrial intermembrane space enzyme cytochemistry b2 and cytochrome c peroxidase in intact yeast cells. J Biol Chem. 1982;257:13068–13074. [PubMed] [Google Scholar]
- Ristevski S, O’Leary DA, Thornell AP, Owen MJ, Kola I, Hertzog PJ. The ETS transcription factor GABPalpha is essential for early embryogenesis. Mol Cell Biol. 2004;24:5844–5849. doi: 10.1128/MCB.24.13.5844-5849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salussolia CL, Prodromou ML, Borker P, Wollmuth LP. Arrangement of subunits in functional NMDA receptors. J Neurosci. 2011;31:11295–11304. doi: 10.1523/JNEUROSCI.5612-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC. Nuclear control of respiratory chain expression in mammalian cells. J Bioenerg Biomemb. 1997;29:109–119. doi: 10.1023/a:1022681828846. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biophys Acta. 2002;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- Scapulla RC. Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1 related coactivator. Ann N Y Acad Sci. 2008;1147:321–334. doi: 10.1196/annals.1427.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scapulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta. 2011;1813:1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelan RS, Gopalakrishnan L, Scarpulla RC, Grossman LI. Cytochrome c oxidase subunit VIIa liver isoform. Characterization and identification of promoter elements in the bovine gene. J Biol Chem. 1996;271:2112–2120. doi: 10.1074/jbc.271.4.2112. [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setou M, Nakagawa T, Seog DH, Hirokawa N. Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 2000;288:1796–1802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- Sommer B, Keinänen K, Verdoorn TA, Wisden W, Burnashev N, Herb A, Köhler M, Takagi T, Sakmann B, Seeburg PH. Flip and Flop: A cell-specific functional switch in glutamate-operated channels of the CNS. Science. 1990;249:1580–1585. doi: 10.1126/science.1699275. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Streit P. Glutamate and aspartate as transmitter candidate for systems of the cerebral cortex. In: Jones EG, Peters A, editors. Cerebral Cortex, vol 2, Functional Properties of Cortical Cells. New York: Plenum Press; 1984. pp. 119–143. [Google Scholar]
- Tanaka H, Grooms SY, Bennett MVL, Zukin RS. The AMPAR subunit GluR2: Still front and center-stage. Brain Res. 2000;886:190–207. doi: 10.1016/s0006-8993(00)02951-6. [DOI] [PubMed] [Google Scholar]
- Thompson CC, Brown TA, McKnight SL. Convergence of Ets- and Notch-related structural motifs in a heteromeric DNA binding complex. Science. 1991;253:762–768. doi: 10.1126/science.1876833. [DOI] [PubMed] [Google Scholar]
- Verdoorn TA, Burnashev N, Monyer H, Seeburg PH, Sakmann B. Structural determinants of ion flow through recombinant glutamate receptor channels. Science. 1991;252:1715–1718. doi: 10.1126/science.1710829. [DOI] [PubMed] [Google Scholar]
- Virbasius JV, Scarpulla RC. Transcriptional activation through ETS domain binding sites in the cytochrome c oxidase subunit IV gene. Mol Cell Biol. 1991;11:5631–5638. doi: 10.1128/mcb.11.11.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virbasius JV, Scarpulla RC. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: A potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci USA. 1994;91:1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virbasius JV, Virbasius CA, Scarpulla RC. Identity of GABP with NRF-2: A multisubunit activator of cytochrome oxidase expression, reveals a cellular role for an ETS domain activator of viral promoters. Gene Dev. 1993a;7:380–392. doi: 10.1101/gad.7.3.380. [DOI] [PubMed] [Google Scholar]
- Virbasius CA, Virbasius JV, Scarpulla RC. NRF-1, an activator involved in nuclear-mitochondrial interaction, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Gene Dev. 1993b;7:2431–2445. doi: 10.1101/gad.7.12a.2431. [DOI] [PubMed] [Google Scholar]
- Washburn MS, Numberger M, Zhang S, Dingledine R. Differential dependence on GluR2 expression of three characteristic features of AMPA receptors. J Neurosci. 1997;17:9393–9406. doi: 10.1523/JNEUROSCI.17-24-09393.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong-Riley M. Changes in the visual system of monocularly sutured or enucleated cats demonstrable with cytochrome oxidase histochemistry. Brain Res. 1979;171:11–28. doi: 10.1016/0006-8993(79)90728-5. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MTT. Cytochrome oxidase: An endogenous metabolic marker for neuronal activity. Trends Neurosci. 1989;12:94–101. doi: 10.1016/0166-2236(89)90165-3. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MTT. Energy metabolism of the visual system. Eye and Brain. 2010;2:99–116. doi: 10.2147/EB.S9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong-Riley M, Carroll EW. Effect of impulse blockage on cytochrome oxidase activity in monkey visual system. Nature. 1984;307:262–264. doi: 10.1038/307262a0. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MTT, Jacobs P. AMPA glutamate receptor subunit 2 in normal and visually deprived macaque visual cortex. Vis Neurosci. 2002;19:563–573. doi: 10.1017/s0952523802195022. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MTT, Tripathi SC, Trusk TC, Hoppe DA. Effect of retinal impulse blockage on cytochrome oxidase-rich zones in the macaque striate cortex: I. Quantitative electron-microscopic (EM) analysis of neurons. Vis Neurosci. 1989a;2:483–497. doi: 10.1017/s0952523800012384. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MTT, Trusk TC, Tripathi SC, Hoppe DA. Effect of retinal impulse blockage on cytochrome oxidase-rich zones in the macaque striate cortex: II. Quantitative electron-microscopic (EM) analysis of neuropil. Vis Neurosci. 1989b;2:499–514. doi: 10.1017/s0952523800012396. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MTT, Mullen MA, Huang Z, Guyer C. Brain cytochrome oxidase subunit complementary DNAs: Isolation, subcloning, sequencing, light and electron microscopic in situ hybridization of transcripts, and regulation by neuronal activity. Neurosci. 1997;76:1035–1055. doi: 10.1016/s0306-4522(96)00410-1. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MTT, Nie F, Hevner RF, Liu S. Brain cytochrome oxidase. In: Gonzalez-Lima F, editor. Cytochrome Oxidase in Neuronal Metabolism and Alzheimer’s Disease. New York: Plenum Press; 1998a. pp. 1–53. [Google Scholar]
- Wong-Riley M, Anderson B, Liebl W, Huang Z. Neurochemical organization of the macaque striate cortex: Correlation of cytochrome oxidase with Na+K+ATPase, NADPH-diaphorase, nitric oxide synthase, and N-methyl-D-aspartate receptor subunit 1. Neurosci. 1998b;83:1025–1045. doi: 10.1016/s0306-4522(97)00432-6. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MT, Huang Z, Liebl W, Nie F, Xu H, Zhang C. Neurochemical organization of the macaque retina: Effect of TTX on levels and gene expression of cytochrome oxidase and nitric oxide synthase and on the immunoreactivity of Na+K+ATPase and NMDA receptor subunit 1. Vision Res. 1998c;38:1455–1477. doi: 10.1016/s0042-6989(98)00001-7. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MTT, Yang SJ, Liang HL, Ning G, Jacobs P. Quantitative immuno-electron microscopic analysis of nuclear respiratory factor 2 alpha and beta subunits: Normal distribution and activity-dependent regulation in mammalian visual cortex. Visual Neurosci. 2005;22:1–18. doi: 10.1017/S0952523805221016. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MTT, Liang HL, Ongwijitwat S. Activity-dependent bigenomic transcriptional regulation of cytochrome c oxidase in neurons. In: Dudek SM, editor. Transcriptional Regulation by Neuronal Activity: To the Nucleus and Back. New York: Springer Science; 2008. pp. 209–228. Chapter 11. [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Yang SJ, Liang HL, Ning G, Wong-Riley MTT. Ultrasturctural study of depolarization-induced translocation of NRF-2 transcription factor in cultured rat visual cortical neurons. Eur J Neurosci. 2004;19:1153–1162. doi: 10.1111/j.1460-9568.2004.03250.x. [DOI] [PubMed] [Google Scholar]
- Yang SJ, Liang HL, Wong-Riley MTT. Activity-dependent transcriptional regulation of nuclear respiratory factor-1 in cultured rat visual cortical neurons. Neurosci. 2006;141:1181–1192. doi: 10.1016/j.neuroscience.2006.04.063. [DOI] [PubMed] [Google Scholar]
- Zhang C, Wong-Riley M. Expression and regulation of NMDA receptor subunit R1 and neuronal nitric oxide synthase in cortical neuronal cultures: Correlation with cytochrome oxidase. J Neurocytol. 1999;28:525–539. doi: 10.1023/a:1007053204929. [DOI] [PubMed] [Google Scholar]
- Zhang C, Wong-Riley MTT. Synthesis and degradation of cytochrome oxidase subunit mRNA in neurons: Differential bigenomic regulation by neuronal activity. J Neurosci Res. 2000a;60:338–344. doi: 10.1002/(SICI)1097-4547(20000501)60:3<338::AID-JNR8>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Zhang C, Wong-Riley M. Depolarization stimulation upregulates GA-binding protein in neurons: A transcription factor involved in the bigenomic expression of cytochrome oxidase subunits. Eur J Neurosci. 2000b;12:1013–1023. doi: 10.1046/j.1460-9568.2000.00997.x. [DOI] [PubMed] [Google Scholar]
- Zhou GL, Xin L, Song W, Di LJ, Liu G, Wu XS, Liu DP, Liang CC. Active chromatin hub of the mouse alpha-globin locus forms in a transcription factory of clustered house-keeping genes. Mol Cell Biol. 2006;26:5096–5105. doi: 10.1128/MCB.02454-05. [DOI] [PMC free article] [PubMed] [Google Scholar]