Abstract
Replication of nearly all RNA viruses depends on a virus-encoded RNA-dependent RNA polymerase (RdRp). Our earlier work found that purified recombinant hepatitis C virus (HCV) RdRp (NS5B) was able to initiate RNA synthesis de novo by using purine (A and G) but not pyrimidine (C and U) nucleotides (G. Luo et al., J. Virol. 74:851-863, 2000). For most human RNA viruses, the initiation nucleotides of both positive- and negative-strand RNAs were found to be either an adenylate (A) or guanylate (G). To determine the nucleotide used for initiation and control of HCV RNA replication, a genetic mutagenesis analysis of the nucleotides at the very 5′ and 3′ ends of HCV RNAs was performed by using a cell-based HCV replicon replication system. Either a G or an A at the 5′ end of HCV genomic RNA was able to efficiently induce cell colony formation, whereas a nucleotide C at the 5′ end dramatically reduced the efficiency of cell colony formation. Likewise, the 3′-end nucleotide U-to-C mutation did not significantly affect the efficiency of cell colony formation. In contrast, a U-to-G mutation at the 3′ end caused a remarkable decrease in cell colony formation, and a U-to-A mutation resulted in a complete abolition of cell colony formation. Sequence analysis of the HCV replicon RNAs recovered from G418-resistant Huh7 cells revealed several interesting findings. First, the 5′-end nucleotide G of the replicon RNA was changed to an A upon multiple rounds of replication. Second, the nucleotide A at the 5′ end was stably maintained among all replicon RNAs isolated from Huh7 cells transfected with an RNA with a 5′-end A. Third, initiation of HCV RNA replication with a CTP resulted in a >10-fold reduction in the levels of HCV RNAs, suggesting that initiation of RNA replication with CTP was very inefficient. Fourth, the 3′-end nucleotide U-to-C and -G mutations were all reverted back to a wild-type nucleotide U. In addition, extra U and UU residues were identified at the 3′ ends of revertants recovered from Huh7 cells transfected with an RNA with a nucleotide G at the 3′ end. We also determined the 5′-end nucleotide of positive-strand RNA of some clinical HCV isolates. Either G or A was identified at the 5′ end of HCV RNA genome depending on the specific HCV isolate. Collectively, these findings demonstrate that replication of positive-strand HCV RNA was preferentially initiated with purine nucleotides (ATP and GTP), whereas the negative-strand HCV RNA replication is invariably initiated with an ATP.
Hepatitis C virus (HCV) is an enveloped RNA virus of the genus Hepacivirus of the Flaviviridae family (16, 46). It causes chronic infections of approximately 4 million people in the United States and 170 million people worldwide (10, 55). The majority (∼85%) of individuals with HCV infection develop chronic hepatitis, which leads to cirrhosis (10 to 20%) and hepatocellular carcinoma (1 to 5%). HCV infection results in 10,000 to 12,000 deaths each year in the United States alone, a figure expected to triple within the next 10 to 20 years if no effective intervention is developed (10). Currently, there is no specific antiviral therapy for treating HCV infection.
HCV has a single positive-strand RNA genome that contains a single open reading frame encoding a large viral polyprotein of 3,010 to 3,040 amino acids (12, 45). Upon translation, the viral polyprotein is proteolytically processed into structural and nonstructural viral proteins. The structural proteins C (core), E1 and E2 (envelope), and p7 are processed from the N-terminal third of the polyprotein by cellular signal peptidases (19, 29, 45). The nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) result from cleavages at the NS2-NS3 junction by the NS2-3 metalloprotease and at the downstream sites of the NS3 protein by the NS3 serine protease (2, 13, 17, 20). Recently, a number of studies have demonstrated that subgenomic HCV replicon RNAs encoding the nonstructural proteins NS3 to NS5B were able to replicate in a human hepatoma cell line, Huh7 (5, 18, 21, 31, 37). Findings from these studies suggest that the nonstructural viral proteins NS3, NS4A, NS4B, NS5A, and NS5B are sufficient for HCV RNA replication in the cell.
Replication of nearly all RNA viruses depends on a virus-encoded RNA-dependent RNA polymerase (RdRp) in association with other viral and/or cellular proteins (3, 9, 23, 27). Sequence comparison studies revealed that the HCV NS5B protein contains functional motifs characteristic for all known RNA polymerases (26, 38, 41). The RdRp activity of NS5B was experimentally demonstrated by using purified recombinant NS5B in an in vitro assay for RNA synthesis (4). Further biochemical characterization of recombinant NS5B has revealed that HCV RdRp catalyzed in vitro RNA polymerization on both HCV-specific and nonviral RNA templates by either using an oligonucleotide primer (primer-dependent RNA synthesis) or by extending the 3′ end of the RNA template itself (self-priming or copy-back) (15, 22, 30, 33, 34, 57). Primer-dependent RNA synthesis was reported as a mechanism of RNA replication for members of the Picornaviridae family, in which the viral protein VPg was used to initiate RNA transcription and replication (43). Copy-back initiation of RNA synthesis was mainly observed for in vitro RNA synthesis catalyzed by purified viral RdRps (4, 30, 35, 60). However, we and others have found that purified HCV NS5B expressed in either insect cells or Escherichia coli was able to initiate RNA synthesis de novo (24, 36, 39, 40, 62). De novo initiation of RNA synthesis by HCV RdRp was observed when a full-length HCV RNA genome (39), RNAs derived from the HCV 3′-untranslated region (3′UTR) (40, 62), or even nonviral RNA and RNA homopolymers were used as templates (24, 36). De novo RNA synthesis has been also demonstrated for many other viral RNA polymerases (1, 42, 49, 50). Although replication of picornavirus requires primer-dependent RNA synthesis, de novo initiation of RNA synthesis is the mechanism commonly used for RNA replication by most positive-, negative-, and double-strand RNA viruses (1, 11, 23, 36, 52). In the case of HCV, de novo initiation is the most likely mechanism for HCV RNA replication in vivo since the copy-back or self-priming RNA synthesis is unable to produce authentic viral RNA with precise 5′ and 3′ ends.
Our earlier work found that purified recombinant HCV RdRp was able to initiate RNA synthesis de novo with purine (A and G) but not pyrimidine (C and U) nucleotides when homopolymer RNAs were used as templates (36). In addition, the HCV RdRp exhibited a higher Km for nucleotide substrates ATP and GTP than for CTP and UTP during in vitro RNA synthesis (36). These findings suggest that HCV RdRp possesses a propensity to selectively utilize purine nucleotides for initiation of RNA replication. Consistent with these findings, the first nucleotide at the 5′ end of positive- and negative-strand HCV RNAs was reported to be a G and an A, respectively, suggesting that GTP and ATP might be utilized for the initiation of de novo RNA synthesis in vivo (7, 23, 56). However, the role of the initiation nucleotides in HCV RNA replication in vivo has not been experimentally determined.
In the present study, we performed a molecular genetic analysis of the presumed initiation nucleotides of HCV RNAs at the very 5′ and 3′ ends to determine their roles in the control of HCV RNA replication. The initiation of the positive-strand HCV RNA replication was investigated by varying the 5′-end nucleotide with G, A, and C, respectively. The replicon RNAs with a 5′-end nucleotide G or A were able to efficiently induce cell colony formation, whereas a nucleotide C at the 5′ end dramatically reduced the efficiency of cell colony formation. However, sequence analysis revealed that the 5′-end nucleotide G of the HCV replicons recovered from cell lines was changed to a nucleotide A, whereas the 5′-end nucleotide A was stably maintained in all replicon RNAs recovered from different cell lines. These findings demonstrate that an ATP was preferentially and stably utilized by HCV replicase to initiate positive-strand HCV RNA replication in the cell. In addition, two replicon RNAs recovered from the 5′UTR-C RNA-transfected Huh7 cells were found to contain a C at the very 5′ end, which resulted in >10-fold-lower levels of HCV RNA replication. To determine the initiation of the negative-strand HCV RNA replication, we mutated the 3′-end nucleotide of the HCV replicon RNA from a U to a C, G, or A, respectively. A U-to-C mutation at the 3′ end did not significantly affect the efficiency of cell colony formation. However, a U-to-G mutation remarkably decreased the frequency of cell colony formation, and a U-to-A mutation resulted in a complete abolition of cell colony formation. The 5′-end nucleotide of HCV genomic RNA derived from various genotypes of clinical HCV isolates was also determined by RACE (rapid amplification of cDNA ends) reverse transcription-PCR (RT-PCR). Both nucleotides G and A were identified at the 5′ end of HCV genome varying from isolate to isolate. Collectively, these findings demonstrate that replication of HCV RNA is preferentially initiated with purine nucleotides in vivo.
MATERIALS AND METHODS
Cell culture.
A human hepatoma cell line, Huh7, was maintained in Dulbecco modified essential medium (DMEM; Invitrogen) supplemented with nonessential amino acids and 10% fetal bovine serum (FBS; Mediatech). Huh7 cells harboring an HCV replicon were selected and maintained by addition of G418 sulfate (0.5 mg/ml) to DMEM containing 10% FBS.
DNA construction.
As described in our earlier study (37), a subgenomic HCV replicon cDNA, pBR322/I377/NS3-3′/S1179I was constructed by PCR and cloning with synthetic oligonucleotides based on the published sequences (31). The original HCV replicon RNA contains a G at the 5′ end and a U at the 3′ end (37). Mutations of the nucleotides at the very 5′ and 3′ ends were introduced by PCR with synthetic oligonucleotide primers (Table 1). To mutate the 5′-end nucleotide from G to A, the 5′UTR cDNA was amplified by PCR with pBR322/I377/NS3-3′/S1179I as a template and oligonucleotides 5′UTR-A and Asc3 as primers (Table 1). The PCR DNA fragment was digested with ClaI and AscI and inserted to the vector pBR322/I377/NS3-3′/S1179I similarly digested by ClaI and AscI. The resulted DNA construct was designated pBR322/I377/NS3-3′/S1179I/5′UTR-A. The 5′ end nucleotide G to C and U mutations were introduced in the same way as the 5′ end G-to-A mutation except that oligonucleotides 5′UTR-C and 5′UTR-U (Table 1) were used as primers. The resulted DNA constructs were designated pBR322/I377/NS3-3′/S1179I/5′UTR-C and pBR322/I377/NS3-3′/S1179I/5′UTR-U, respectively.
TABLE 1.
Oligonucleotides used for PCR-directed mutagenesis and RT-PCR amplificationa
| Oligonucleotide | Sequence (5′ to 3′) |
|---|---|
| 5′UTR-A | CCATCGATAATACGACTCACTATAACCAGCCCCCGATTGGGGGCG |
| 5′UTR-C | CCATCGATAATACGACTCACTATACCCAGCCCCCGATTGGGGGCG |
| 5′UTR-U | CCATCGATAATACGACTCACTATATCCAGCCCCCGATTGGGGGCG |
| Asc3 | ATGGCGCGCCCTTTGGTTTTTCTTTGAGGTTTAGGATTC |
| Oligo/EcoRI | GCGGAATTCGGGGGCCGGAACCT |
| AfeI/M | GCCGCTTCTGCTTTCGTAGGC |
| AfeI/PacI | CCTTAATTAAGCGCTTGATCTGCAGAGAGGCCAG |
| MfeI/5 | CTGGAAGACACTGAGACACC |
| FseI/PacI | CCTTAATTAAGGCCGGCCTTGATCTGCAGAGAGGC |
| NsiI/3670/5 | ATGTATGTCGGCTGACCTGGAG |
| NsiI/3670/3 | GCCGACATACATGCCATGATGTATTTG |
| NsiI/7110/5 | ATGTATCTGGCAAAAGGGTGTACTATC |
| NsiI/7110/3 | GCCAGATACATCGTGCGCGACTGACAC |
| NsiI/AseI | GCGCGCATTAATGCATCTTGATCTGCAGAGAGGC |
| Oligo-BsrGI | CCACATTGGTGTACATTTG |
| P7485 | ACATCGGGCCAGAAGTGTCCGCGCTAGG |
| P7741 | CTGTAGGGGTAGGCATCTATC |
| P7896 | GCTCCATCTTAGCCCTAGTC |
| 5′RACE outer | GCTGATGGCGATGAATGAACACTG |
| 5′RACE inner | CGCGGATCCGAACACTGCGTTTGCTGGCTTTGATG |
| 206 | GACTGTTGTGGCCTGCAGGGCCGAATT |
| 192 | TTGAATTCGGCCCTGCAGGCCACAACAGTC |
Nucleotide mutations are underlined. Oligonucleotides were synthesized by Integrated DNA Technology.
Mutations of the very 3′-end nucleotide were also introduced by PCR with synthetic oligonucleotides as primers (Table 1). To create a U-to-C mutation at the very 3′ end, an AfeI restriction enzyme site was introduced, and cleavage of this enzyme will result in an RNA molecule with a precise 3′-end C upon T7 RNA polymerase transcription (Fig. 1). The unique internal AfeI site at nucleotide 4197 of the replicon was destroyed by a silent T-to-C mutation at nucleotide 4200 with oligonucleotides AfeI/M and EcoRI (Table 1) as primers in PCR. The PCR DNA was digested with EcoRI and then inserted into the vector pBR322/I377/NS3-3′/S1179I that was cut by AfeI and EcoRI. The resulting construct was designated pBR322/I377/NS3-3′/S1179I/ΔAfeI. To introduce a unique AfeI site into the 3′ end of the replicon pBR322/I377/NS3-3′/S1179I/ΔAfeI, the DNA fragment between MfeI and PvuI sites was amplified by PCR with the oligonucleotides MfeI/5 and AfeI/PacI (Table 1) as primers and pBR322/I377/NS3-3′/S1179I as a template. The PCR DNA fragment was cut by MfeI and PacI and ligated to pBR322/I377/NS3-3′/S1179I/ΔAfeI similarly digested by MfeI and PvuI (compatible cohesive end with PacI), resulting in a construct designated pBR322/I377/NS3-3′/S1179I/3′UTR-C. The 3′-end U-to-G mutation was also introduced by PCR with oligonucleotides MfeI/5 and FseI/PacI as primers (Table 1). The PCR DNA was digested with MfeI and PacI and inserted into the vector pBR322/I377/NS3-3′/S1179I between the MfeI and PvuI sites, resulting in pBR322/I377/NS3-3′/S1179I/3′UTR-G that produces a replicon RNA with the 3′-end U-to-G mutation. To mutate the 3′-end nucleotide from a U to an A, an NsiI restriction enzyme site was created at the 3′ end, producing a construct designated pBR322/I377/NS3-3′/S1179I/3′UTR-A. Two internal NsiI sites at positions 3670 and 7110 were destroyed by PCR-directed mutagenesis with the oligonucleotides NsiI/3670/5 and NsiI/3670/3 or the oligonucleotides NsiI/7110/5 and NsiI/7110/3 (Table 1). A construct with the 5′UTR-A and 3′UTR-C double mutations was made by replacement of the DNA fragment between the ClaI and AscI sites of pBR322/I377/NS3-3′/S1179I/3′UTR-C with the one derived from pBR322/I377/NS3-3′/S1179I/5′UTR-A. This construct was designated pBR322/I377/NS3-3′/S1179I/5′UTR-A/3′UTR-C. All mutations introduced to the HCV replicon cDNA were confirmed by DNA sequence analysis (Elim Biopharmaceuticals, Hayward, Calif.).
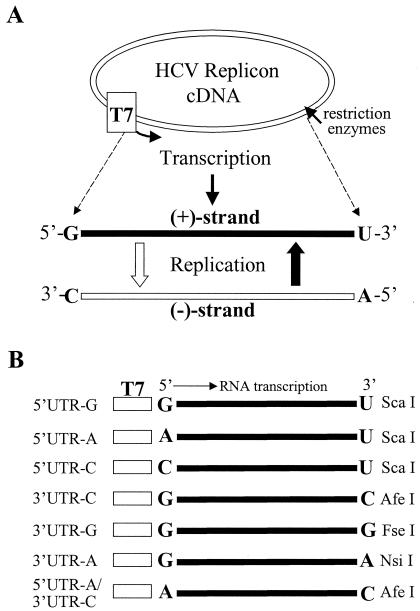
FIG. 1.
(A) Diagram of the in vitro transcription and replication cycle of HCV replicon RNA. The HCV replicon RNAs were transcribed by a T7 RNA polymerase from DNA vectors digested with restriction enzymes as indicated on the right side of panel B. The in vitro-transcribed RNA was transfected into Huh7 cells. Upon replication, the positive-strand HCV RNA is converted to the complementary negative strand, which in turn serves as a template for synthesis of more positive-strand RNA. The 5′- and 3′-end nucleotides of both positive- and negative-strand RNAs are highlighted in boldface letters. (B) Schematic presentation of the in vitro-transcribed HCV replicon RNAs. The nucleotides at the 5′ and 3′ ends of the replicon RNAs are highlighted in boldface letters. The T7 promoter is indicated by an open box. The restriction enzyme used to linearize each cDNA clone of the replicon RNA is shown on the right side. The replicon was named after the 5′- or 3′-end nucleotide, as shown on the left side. The original HCV replicon RNA contains a G at the 5′ end and a U at the 3′ end (37).
Preparation of RNA transcripts.
Subgenomic HCV replicon RNAs were transcribed in vitro by a T7 RNA polymerase from the above-described DNA constructs linearized by restriction enzyme digestion by using large-scale RNA production kits (Promega) (Fig. 1). Restriction enzymes used for linearization of the DNA constructs are indicated on the right side of Fig. 1B. Digestion of DNAs with restriction enzymes ScaI and AfeI resulted in blunt ends, whereas cleavage by FseI and NsiI produced an overhang at the 3′ end of the DNA constructs. Therefore, DNAs digested by FseI and NsiI were further treated with Klenow fragment of DNA polymerase I to remove the 3′-end overhangs prior to T7 RNA polymerase transcription. The in vitro-transcribed RNA molecules were named after the mutation as shown on the left side of Fig. 1B. For instance, the replicon RNAs containing a 5′-end nucleotide G, A, or C are designated 5′UTR-G, 5′UTR-A, and 5′UTR-C, respectively. Likewise, the replicon RNAs with a 3′-end nucleotide C, G, or A are designated 3′UTR-C, 3′UTR-G, and 3′UTR-A, respectively (Fig. 1B). After extensive treatment with RNase-free DNase I, the in vitro-transcribed RNAs were purified by phenol-chloroform extraction, ethanol precipitation, and centrifugation through Sephadex G-50 columns (Roche). The RNA concentration was determined by using a spectrophotometer.
RNA transfection and selection of G418-resistant Huh7 cells.
The subgenomic HCV replicon RNAs were transfected into Huh7 cells by electroporation with a GenePulser system (Bio-Rad). Briefly, 2 μg of the in vitro-transcribed and purified replicon RNAs were electroporated into 8 × 106 Huh7 cells in 0.4 ml (2 × 107 cells/ml) of ice-cold phosphate-buffered saline buffer. The replicon-transfected Huh7 cells were seeded in 100-mm dishes at different cell densities and incubated with DMEM containing 10% FBS. At 24 h posttransfection, the cell culture medium was replaced with DMEM containing 10% FBS and 0.5 mg of G418 sulfate/ml. The medium was changed every 3 to 4 days. After an ∼4-week selection with G418 sulfate, individual cell colonies were transferred into new dishes for amplification and further characterization. Cell colonies in other dishes were fixed and stained with a solution containing 0.01% crystal violet and 19% methanol.
RNA extraction and quantitation of HCV RNA by RPA.
Total RNA was extracted from HCV replicon-harboring cell lines and serum samples derived from either HCV patients or a chimpanzee with TRIzol reagent (Invitrogen) and collected by isopropanol precipitation. The levels of positive- and negative-strand HCV RNAs were determined by RNase protection assay (RPA) with [α-32P]UTP-labeled HCV-specific RNA probes. For measurement of positive-strand RNA, a negative-strand [(−)] 3′UTR RNA probe was transcribed from HindIII-digested pUC19/T7(−)3′UTR by a T7 RNA polymerase and labeled with α32P[UTP]. For detection of negative-strand RNA, a positive-strand [(+)] 5′UTR RNA probe was synthesized by in vitro T7 RNA transcription from pUC19/T7(+)5′UTR linearized with EcoRI. The level of β-actin RNA was used as a control to normalize the amount of total RNA in each sample. The RNA probe for β-actin RNA was transcribed by a T7 RNA polymerase from the DNA pTRI-β-actin-125-Human (Ambion) and labeled with [α-32P]UTP. A total of 15 μg of total cellular RNA was used in an RPA for hybridization with 5 × 104 cpm of [α-32P]UTP-labeled β-actin probe and 105 cpm of either (−)3′UTR or (+)5′UTR RNA probe. RPA was carried out by using an RPA III kit (Ambion). After digestion with RNase A/T1, RNA products were analyzed by electrophoresis in a 6% polyacrylamide-7.7 M urea gel. The levels of RNAs were then determined by quantitation with a PhosphorImager (Molecular Dynamics).
RT-PCR and sequence analysis.
The nucleotides of the 5′, 3′, or the 5′ and 3′ ends of the subgenomic HCV replicon RNAs either extracted from G418-resistant Huh7 cells or in vitro transcribed by a T7 RNA polymerase were determined by RT-PCR and sequence analysis. The 5′UTR RNAs of both positive- and negative-strand RNAs of HCV replicons were reverse transcribed and amplified by RNA ligase-mediated RACE (RLM-RACE; Ambion) (54). A 45-base RNA adapter (5′-GCUGAUGGCGAUGAAUGAACACUGCGUUUGCUGGCUUUGAUGAAA-3′; Dharmacon) was ligated to the 5′ ends of the replicon RNAs by using T4 RNA ligase (NEB). To determine the 5′-end nucleotide, the positive-strand HCV RNA was reverse transcribed by using SuperScript II RNase H− reverse transcriptase (Invitrogen) with oligo-BsrGI (Table 1) as a primer, which is complementary to nucleotides 2020 to 2038 of the replicon. The cDNA of the positive-strand 5′UTR was subsequently amplified by PCR with oligonucleotides Asc3 and 5′RACE outer/inner primers (Table 1). The PCR-amplified 5′UTR cDNAs were used for direct DNA sequencing (Elim Biopharmaceuticals) and/or cloned into a pCR2.1 vector by using a TA cloning kit (Invitrogen). To determine the 3′-end nucleotide, the negative-strand HCV RNA was reverse transcribed by using the primer P7485 (Table 1), which was derived from nucleotides 7485 to 7512 of the replicon. The conserved 98 nucleotides of the negative-strand 5′UTR cDNA was amplified by PCR with oligonucleotide P7896 (nucleotides 7896 to 7915) and 5′RACE outer/inner primers (Table 1). PCR DNA was cloned into a pCR2.1 vector. The 3′-end nucleotides of the in vitro T7 transcripts were determined by RT-PCR by using a procedure described previously (25). Briefly, the synthetic oligonucleotide 206 (Table 1) was modified by the addition of a cordycepin (Sigma) at the 3′ end and phosphorylation at the 5′ end. Modified oligonucleotide 206 was then ligated to the 3′ end of the in vitro T7 transcripts (25). The cDNA of the 3′UTR was amplified by RT-PCR with oligonucleotides 192 and P7741 as primers (Table 1) (25). The nucleotides at both the 5′ and 3′ ends were determined by DNA sequence analysis (Elim Biopharmaceuticals).
RESULTS
Approaches.
The findings from our previous studies suggest that purified recombinant HCV RdRp preferentially utilize purine (ATP and GTP) but not pyrimidine (CTP and UTP) nucleotides for the initiation of de novo RNA synthesis in vitro (36). The question arose which nucleotide is preferentially utilized for the initiation of HCV RNA replication in vivo and, furthermore, how HCV RNA replication is controlled by the initiation nucleotides. To address these questions, we used an HCV replicon replication system to examine the effects of mutations of the initiation nucleotides on replication of the positive- and negative-strand HCV RNAs (5, 31). The 5′-end nucleotide of the subgenomic HCV replicon I377-NS3-3′/S1179I was mutated from a G to an A, C, or U, and the 3′ end nucleotide U was changed to a C, G, or A, respectively (Fig. 1). Upon replication, the 3′-end nucleotide will be copied to become the complementary 5′-end nucleotide of the negative-strand HCV RNA. Therefore, mutations of the 3′-end nucleotide will reveal their effects on the initiation of negative-strand RNA replication (Fig. 1A). The effects of mutations of the 5′- and 3′-end nucleotides on HCV RNA replication were initially examined for the ability of the replicon RNAs to induce cell colony formation. Cell colony formation directly correlates with replication of the subgenomic HCV replicon RNA in Huh7 cells (5, 31). The levels of both positive- and negative-strand RNAs of HCV replicons in Huh7 cells resistant to G418 sulfate were then determined by RPA (36, 37). The nucleotides at the 5′ and 3′ ends of the replicon RNAs isolated from G418-resistant Huh7 cells were determined by RT-PCR and sequence analysis (54).
Confirmation of the 5′-end nucleotide identity of the in vitro-transcribed replicon RNAs by sequence analysis.
The HCV replicon 5′UTR-A, 5′UTR-C and 5′UTR-U RNAs were synthesized to much lower levels by a T7 RNA polymerase compared to the replicon 5′UTR-G (data not shown). T7 RNA polymerase is known to preferentially initiate in vitro RNA transcription with a guanylate (G). The question arose as to whether the in vitro T7 transcripts had correct nucleotides at their 5′ ends when initiated with nucleotide A, C, or U. The 5′-end nucleotide of the in vitro T7 transcripts was then determined by RLM-RACE and sequence analysis. The results clearly show that the 5′-end nucleotides G, A, and C of the HCV replicons were correctly transcribed by a T7 RNA polymerase (see Fig. 8, 5′UTR-A and 5′UTR-C). To further confirm the homogeneity of the 5′-end nucleotide of the replicon 5′UTR-A and 5′UTR-C, cDNAs of the T7 transcripts were amplified by RT-PCR and cloned into pCR2.1 vector. Sequence analysis of eight individual clones for each cDNA derived from either 5′UTR-A or 5′UTR-C verified that both 5′UTR-A and 5′UTR-C are homogeneous as to the 5′-end expected nucleotides (data not shown). However, the 5′-end nucleotide U of the 5′UTR-U RNA was not transcribed. Instead, the T7 RNA polymerase skipped the first nucleotide U and initiated RNA transcription from the second nucleotide C (data not shown). As a result, the replicon 5′UTR-U RNA has the first nucleotide U deletion and a C at the 5′ end. Upon transfection into Huh7 cells, the 5′UTR-U mutant RNA was unable to induce cell colony formation (data not shown). The expected 3′-end nucleotides of the replicons 3′UTR-G and 3′UTR-A were also confirmed by using a procedure described previously (25) (see Materials and Methods; data not shown).
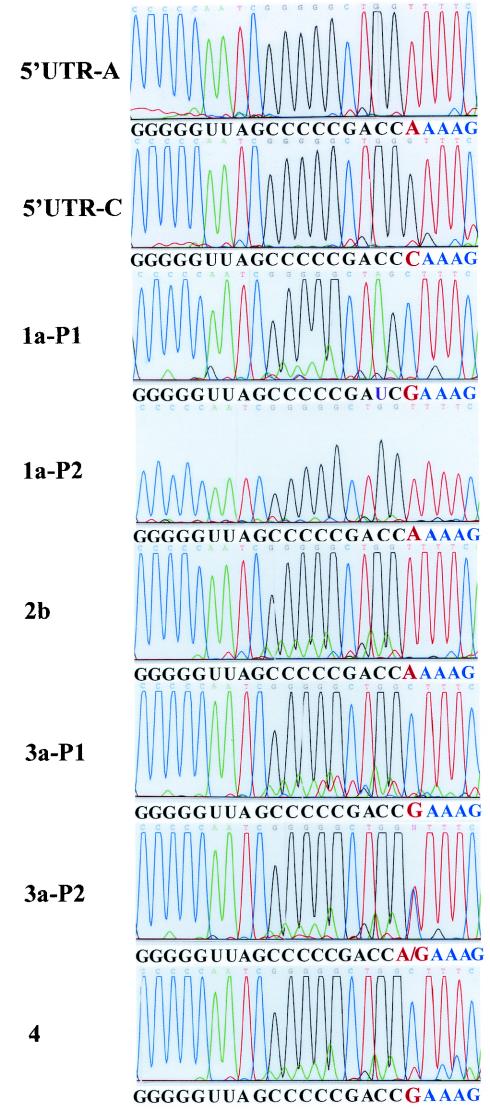
FIG. 8.
Determination of the 5′-end nucleotide of T7 RNA transcripts and HCV genomic RNAs derived from clinical HCV isolates. The 5′UTR-A and 5′UTR-C RNAs were in vitro transcribed by a T7 RNA polymerase. HCV RNAs were extracted with TRIzol reagent from serum samples of patients infected with different genotypes of HCV. The 5′-end nucleotide of T7 transcripts and HCV genomic RNAs were determined by RLM-RACE as described in Fig. 7A legend. Nucleotide sequences of each chromogram are complementary to HCV genomic RNA (in black and red) and adapter RNA sequence (in blue), as highlighted underneath each chromogram. Nucleotides in red are the 5′-end nucleotides of HCV RNAs. The highlighted sequence underneath each chromogram is in the order from 3′ to 5′ ends. HCV genotypes are indicated by 1a, 2b, 3a, and 4. P1 and P2 stand for HCV RNAs derived from patient 1 and patient 2, who were infected with the same genotype. A mutation from C to U at position 3 from the 5′ end was identified in one clinical isolate (1a-P1) and is highlighted by a purple letter U.
Effects of the 5′-end nucleotide mutations on cell colony formation.
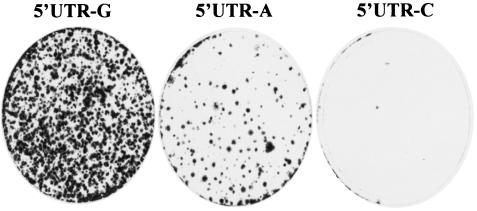
To determine the effects of different nucleotides at the 5′ end of the subgenomic replicon on cell colony formation as a result of RNA replication, the in vitro-transcribed replicon RNAs were transfected into Huh7 cells by electroporation. Replication of the HCV replicon RNA in Huh7 cells will result in expression of a selectable marker, neomycin phosphotransferase (Neo), which renders Huh7 cells resistant to G418 sulfate (5, 31). After an ∼4-week selection with G418 sulfate, Huh7 cell colonies resulted from HCV replicon replication were stained with a crystal violet solution. The ability of HCV RNAs to replicate in Huh7 cells was measured by the efficiency of cell colony formation. The results are shown in Fig. 2. The replicon 5′UTR-A RNA was replicated in Huh7 cells, although it resulted in ∼10-fold-lower efficiency in cell colony formation than that of the replicon 5′UTR-G (Fig. 2). Likewise, the efficiency of cell colony formation induced by the replicon 5′UTR-C was remarkably reduced compared to the replicon 5′UTR-G (Fig. 2). The lower efficiency of cell colony formation induced by the replicons 5′UTR-A and 5′UTR-C was confirmed by three independent experiments. However, it was difficult to precisely determine the efficiency of cell colony formation between the replicons 5′UTR-G, 5′UTR-A, and 5′UTR-C since the numbers of cell colonies varied between experiments. Overall, there was a preference of the 5′-end nucleotide of the HCV replicon RNA in the order of G, A, and C for cell colony formation (Fig. 2 and data not shown).
FIG. 2.
Effects of the 5′-end nucleotide mutations on cell colony formation. Then, 2 μg each of the in vitro-transcribed RNAs was transfected into 8 × 106 Huh7 cells by electroporation. The RNA-transfected Huh7 cells were incubated with DMEM containing 10% FBS. After 24 h of incubation at 37°C and 5% CO2, cell culture medium was replaced by DMEM containing 10% FBS and 0.5 mg of G418 sulfate/ml. The medium was changed twice a week. After an ∼4-week selection with G418, cell clones were fixed, stained by a solution containing 0.01% crystal violet and 19% methanol, and photographed.
Effects of the 5′-end nucleotide mutations on the levels of positive- and negative-strand replicon RNAs in the cell.
The questions arose whether cell colony formation was the result of HCV RNA replication and whether different nucleotides at the 5′ end affect the levels of HCV RNA replication. We performed an RPA to determine the levels of both positive- and negative-strand RNAs present in Huh7 cells resistant to G418 sulfate. The positive-strand RNA was detected by using a radiolabeled (−)3′UTR RNA probe, whereas the negative-strand RNA was measured by using a radiolabeled (+)5′UTR RNA probe (37). As shown in Fig. 3, the levels of positive-strand replicon RNAs present in Huh7 cell clones induced by the 5′UTR-A RNA were comparable to those of the 5′UTR-G replicon (Fig. 3A). However, it appears that the levels of negative-strand RNA in cell colonies resulted from the 5′UTR-A are ∼2-fold higher than those in cells transfected with the 5′UTR-G (Fig. 3A). Conversely, two of three cell colonies that resulted from the replication of the replicon 5′UTR-C had 10- and 14-fold-lower levels of both positive- and negative-strand RNAs (Fig. 3B, numbers 1 and 2) compared to those present in the third cell colony (Fig. 3B, number 3). Sequence analysis revealed that the two low-replicating RNAs contain a C at the 5′ end. In contrast, the third cell colony had similar levels of positive- and negative-strand RNAs to those of the replicon 5′UTR-A (Fig. 3B, number 3). The 5′-end nucleotide of the replicon RNA extracted from the third cell clone was actually changed from a C to an A, as shown by sequence analysis (see Fig. 7B). These results demonstrate that the initiation of positive-strand RNA replication with ATP is much more efficient than RNA replication initiated with a CTP in cell culture.
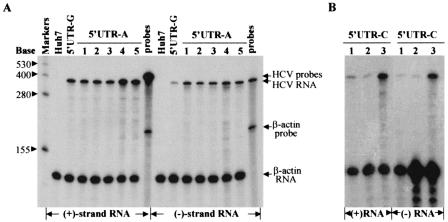
FIG. 3.
RPA determination of the positive-strand and negative-strand RNAs of HCV replicons isolated from different Huh7 cell lines. (A) Detection of both positive- and negative-strand RNAs of HCV replicons isolated from Huh7 cell colonies resulted from replication of the 5′UTR-A RNA. Total cellular RNA was extracted with TRIzol reagent from Huh7 cells. A total of 15 μg of total RNA was used in an RPA for hybridization with 5 × 104 cpm of [α-32P]UTP-labeled β-actin RNA probe (Ambion) and either 105 cpm of [α-32P]UTP-labeled (−)3′UTR (for the detection of positive-strand RNA) or (+)5′UTR RNA probe (for the detection of negative-strand RNA). After RNase A/T1 digestion, RNA products were analyzed in a 6% polyacrylamide-7.7 M urea gel. The RNA levels were determined by quantitation with a PhosphorImager (Molecular Dynamics). The sizes of the RNA markers are indicated on the left, and arrows on the right highlight the RNA products. Numbers on the top indicate different cell colonies that resulted from replication of the 5′UTR-A RNA. Huh7, RNA extracted from regular Huh7 cells as a negative control; 5′UTR-G, RNA extracted from a Huh7 cell line resulted from transfection with the 5′UTR-G RNA (wild type). (B) Detection of both positive-strand and negative-strand RNAs of HCV replicons recovered from Huh7 cells transfected with the 5′UTR-C RNA. Total RNA was extracted from Huh7 cell colonies that resulted from transfection with the 5′UTR-C RNA. Otherwise, detection was the same as described for panel A. Positive- and negative-strand RNAs are indicated at the bottom.
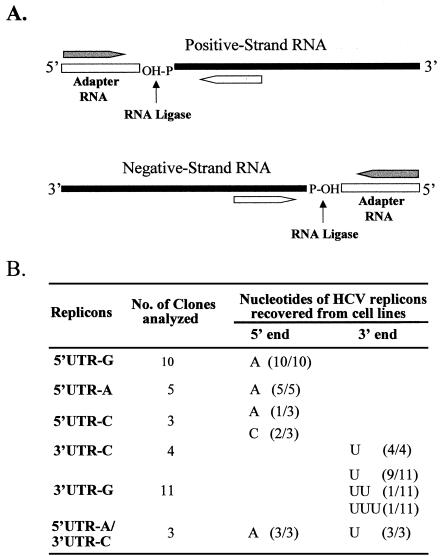
FIG. 7.
Determination of nucleotides at the 5′ and 3′ ends of HCV replicon RNAs recovered from G418-resistant Huh7 cells. (A) Schematic of the RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) (Ambion). An adapter RNA (open bar) was ligated to the 5′ end of either the positive-strand (for determination of the 5′-end nucleotide) or the negative-strand (for determination of the 3′-end nucleotide) HCV replicon RNA isolated from different cell lines. The cDNAs of both positive and negative strands of HCV RNA were reverse transcribed by using HCV-specific primers and amplified by PCR with 5′RACE outer/inner primers (gray arrow) and HCV-specific primers (open arrow). The 5′-end nucleotides of both positive and negative strands of HCV RNA were determined by DNA sequence analysis. (B) 5′- and 3′-end nucleotides of HCV RNA determined by RLM-RACE. Letters indicate nucleotides at the 5′ and 3′ ends as shown on the top. The number indicates the number of cell clones from which HCV replicon RNAs were analyzed. Numbers in parentheses indicate the frequency of the nucleotides determined. The HCV replicon RNAs given on the left column were the ones transcribed in vitro by a T7 RNA polymerase.
Effects of mutations of the 3′-end nucleotide on cell colony formation.
Sequence analysis and comparison studies revealed that the 3′-end nucleotide of the HCV RNA genome is an invariant U among different HCV isolates. Upon replication, the 3′-end nucleotide U is converted to a complementary A at the 5′ end of negative-strand RNA, which is the presumed nucleotide for initiation of negative-strand RNA replication (Fig. 1A). To determine the nucleotide preferentially utilized for initiation of negative-strand RNA replication, we performed a genetic analysis of the 3′-end nucleotide of HCV RNA. Mutation of the 3′-end nucleotide from U to C did not significantly affect the efficiency of cell colony formation, whereas a U-to-G mutation at the 3′ end dramatically decreased the efficiency of cell colony formation (Fig. 4). Furthermore, when the 3′-end nucleotide was changed from a U to an A, it resulted in a complete abolition of cell colony formation (Fig. 4). These results suggest that HCV RNAs prefer a pyrimidine but not purine nucleotide at the 3′ end for efficient replication in cell culture.
FIG. 4.
Effects of the 3′-end nucleotide mutations on cell colony formation. The in vitro-transcribed replicon RNAs contain nucleotide mutations from a U (3′UTR-U) to a C (3′UTR-C), to a G (3′UTR-G), and to an A (3′UTR-A) at the 3′ end, respectively. Otherwise, transfection and selection were done in the same way as described as for Fig. 2. The 3′UTR-U is a wild-type replicon RNA.
Effects of mutations of the 3′-end nucleotide on the levels of positive- and negative-strand replicon RNAs in the cell.
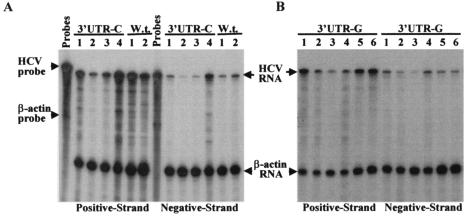
To further determine whether cell colony formation induced by the 3′-end mutant replicons was a consequence of RNA replication and whether mutations of the 3′-end nucleotide affected HCV RNA replication, the levels of both positive- and negative-strand RNAs of HCV replicons extracted from Huh7 cells were determined by RPA (Fig. 5). Both positive- and negative-strand RNAs of the HCV replicon were detected, confirming that G418-resistant cell clones were indeed the result of HCV RNA replication. However, the levels of both positive- and negative-strand RNAs varied by up to sixfold between different cell lines. This observation suggests that cell colony formation induced by HCV replicon replication might not be an ideal way to quantitate HCV RNA replication, since it is complicated by the virally adapted mutations and cellular coevolution. It appeared that there was no significant pattern in difference of the levels of HCV RNAs among cell clones induced by the replicons 3′UTR-U (wild type), 3′UTR-C, and 3′UTR-G (Fig. 5).
FIG. 5.
Determination of the positive- and negative-strand RNAs of the subgenomic HCV replicons isolated from Huh7 cells transfected with the 3′UTR-C and 3′UTR-G RNAs. (A) Quantitation of replicon RNAs derived from the 3′UTR-C RNA-transfected Huh7 cells. RPA was essentially the same as described for Fig. 3 except that total RNAs were extracted from different Huh7 cell lines as a result of replication of the 3′UTR-C RNA and selection with G418 sulfate. W.t. is the 5′UTR-G RNA (Fig. 1). (B) Quantitation of replicon RNAs isolated from different Huh7 cell lines that resulted from replication of the 3′UTR-G RNA by RPA. The positive- and negative-strand RNAs are indicated at the bottom. Numbers indicate replicon RNAs derived from different cell lines.
Effects of double mutations from a G to an A at the 5′ end and a U to a C at the 3′ end on HCV RNA replication.
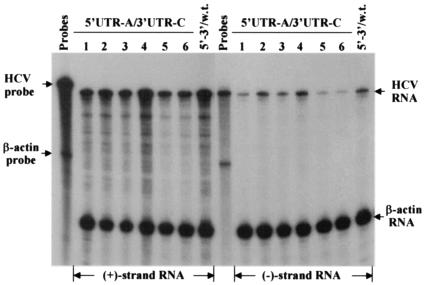
Findings from the experiments described above demonstrate that HCV replicon RNAs could tolerate nucleotide variation at both the 5′ and 3′ ends with respect to the induction of cell colony formation. To determine whether the replicon RNA was able to tolerate combined mutations at both the 5′ and the 3′ ends, we made a construct to produce a replicon RNA with an A at the 5′ end and a C at the 3′ end. The replicon RNA with double mutations induced cell colony formation with an efficiency similar to that of the 5′UTR-A RNA (data not shown). The levels of both positive- and negative-strand RNAs of the mutant replicon were also comparable to those of the wild-type replicon, although the RNA levels vary between different cell lines (Fig. 6).
FIG. 6.
Determination of the positive- and negative-strand RNAs of HCV replicon RNAs isolated from Huh7 cells transfected with the 5′UTR-A/3′UTR-C RNA. Total RNAs were extracted from six different Huh7 cell lines resulted from replication of the transfected 5′UTR-A/3′UTR-C RNA. RPA was done in the same way as described in the Fig. 3 legend. 5′-3′/w.t., RNA extracted from an Huh7 cell line resulting from replication of the transfected wild-type replicon RNA (5′UTR-G RNA, see Fig. 1B). Replicon RNAs derived from different cell lines are indicated by numbers on the top. The positive- and negative-strand RNAs are indicated at the bottom.
Determination of the 5′-end nucleotides of both positive- and negative-strand HCV RNA by RT-PCR and sequence analysis.
To determine the stability of the mutations introduced into the 5′ and 3′ ends, the replicon RNAs isolated from G418-resistant Huh7 cell lines were subject to RT-PCR and sequence analysis. Individual Huh7 cell clones resulted from replication of wild-type and mutant HCV replicon RNAs were passaged approximately 7 to 10 times. Total RNAs were then extracted from replicon-harboring Huh7 cells. The 5′UTR cDNAs of both the positive- and negative-strand RNAs were amplified by RLM-RACE, and the 5′-end nucleotide identity was determined by sequence analysis. The results are summarized in Fig. 7.
The 5′-end nucleotide sequence of a total of 10 replicon RNAs isolated from the 5′UTR-G RNA-transfected Huh7 cells was determined. Strikingly, all 10 replicon RNAs contained nucleotide A rather than G at the 5′ end (Fig. 7). This finding suggests that replication of HCV RNA in cell culture was preferentially initiated with ATP as a result of G418 selection. Consistent with this finding, the 5′-end nucleotide A of the replicon 5′UTR-A was stably maintained even after numerous rounds of RNA replication. All five replicon RNAs isolated from Huh7 cells transfected with the 5′UTR-A RNA contain a nucleotide A at the 5′ end, indicating that ATP is favorable and stable for initiation of HCV RNA replication in the cell culture system. In addition, one replicon RNA recovered from the 5′UTR-C RNA-transfected Huh7 cells was found to have the 5′-end nucleotide changed to an A. Interestingly, the nucleotide C was retained at the 5′ end of two other replicon RNAs isolated from Huh7 cells transfected with the same 5′UTR-C RNA. How the 5′-end nucleotide C was stably maintained in HCV RNA replication was not clear. Also, HCV RNA extracted from a series of serum samples derived from a chimpanzee inoculated with a homogeneous infectious HCV RNA (53) was found to contain a nucleotide C at the 5′ end (Z. Cai et al., unpublished results). Whether the initiation nucleotide of HCV RNA controls the level of HCV RNA replication in vivo remains to be determined.
Reversion of the 3′-end nucleotide mutations was analyzed by determination of the 5′-end nucleotide of the negative-strand HCV RNA. The (−)5′UTR cDNA was amplified by RLM-RACE. Sequence analysis revealed that the 3′-end nucleotide mutations were all reverted back to a wild-type nucleotide U from the mutated nucleotides C and G (Fig. 7). In addition, two revertants recovered from the 3′UTR-G RNA-transfected Huh7 cells have one and two extra U residues at the 3′ end, respectively. A previous study found that some clinical isolates of HCV genotype 1b also contained extra U residues at the 3′ end (56). We do not know, however, how the extra U residues were added to the 3′ end of the HCV RNA during RNA replication. One possible explanation is that ATP is a component of the HCV replication complex, and a nucleotide G at the 3′ end might affect the affinity of the binding of the HCV replication complex to the RNA template during negative-strand RNA replication. As a result, extra ATP was mistakenly added during the initiation of negative-strand RNA replication.
Determination of the 5′-end nucleotides of HCV genomic RNAs derived from various clinical isolates.
To confirm the biological relevance of our findings derived from cell culture replication of subgenomic HCV replicon RNAs, we sought to determine the 5′-end nucleotides of HCV RNA genomes of different clinical isolates. HCV RNAs of genotypes 1, 2, 3, and 4 were used in the present study for the determination of the initiation nucleotide of positive-strand HCV RNA by RLM-RACE (see Materials and Methods). The results are shown in Fig. 8. Both nucleotides A and G were found at the 5′ ends of HCV RNAs of different clinical isolates, confirming that both ATP and GTP are used by the HCV replicase in vivo to initiate positive-strand RNA replication. However, there is no correlation between the initiation nucleotide (ATP or GTP) and different HCV genotypes. The initiation nucleotide of the genomic HCV RNA varies between different HCV isolates even within the same genotype (Fig. 8). Interestingly, polymorphic nucleotides A and G were found at the 5′ end of HCV RNA derived from the same patient (Fig. 8, 3a-P2). Further analyses of HCV RNA derived from this patient by cDNA cloning and DNA sequence analysis revealed that the mixture of HCV RNA consists of 60% of RNA with a 5′-end nucleotide G and 40% of RNA with a 5′-end nucleotide A. It will be interesting to determine how the initiation of HCV RNA replication is controlled in vivo by selective utilization of ATP and GTP.
DISCUSSION
Various lines of evidence derived from in vitro biochemical studies suggest that ATP and GTP are favorable nucleotides used by recombinant HCV RdRp for the initiation of de novo RNA synthesis (36, 61, 62). Our earlier study revealed that purified recombinant HCV RdRp exhibited a preference for purine nucleotides in de novo RNA synthesis (36). Others reported that HCV RdRp preferred GTP as the initiation nucleotide in the in vitro RNA synthesis (62). Consistent with these findings, GTP was found to stimulate the in vitro RdRp activity by up to 100-fold (32). The role of GTP in stimulation of the RdRp activity was further supported by identification of a GTP-specific binding site on the surface of the enzyme ∼30 Å away from the active site. The GTP binding pocket lies at the interface between fingers and thumb domains of NS5B. It was speculated that binding of a GTP to the specific GTP binding pocket might trigger a conformational rearrangement of the enzyme to allow alternative interactions between the two domains, which therein renders efficient initiation of RNA synthesis (6).
However, findings derived from our genetic analysis of HCV RNA replication here demonstrate that replication of both positive- and negative-strand RNAs of a subgenomic HCV RNA replicon was selectively initiated with ATP rather than GTP in cell culture. The 5′-end nucleotide GTP was present in HCV replicon RNA transcribed by a T7 RNA polymerase in vitro. Upon transfection into Huh7 cells and multiple rounds of RNA replication, however, the 5′-end GTP of the HCV replicon RNA was changed to an ATP, as revealed by sequence analysis (Fig. 7). This finding indicates that ATP was preferentially utilized by HCV replicase for initiation of the positive-strand HCV RNA replication in cell culture. This conclusion was further supported by the finding that a nucleotide A at the 5′ end was stably maintained in the replicon RNAs recovered from Huh7 cells after many passages. The nucleotide A was invariantly found at the 5′ ends of all five replicon RNAs isolated from Huh7 cells transfected with RNAs containing an ATP at the 5′ end (Fig. 7). CTP was also found at the 5′ end of the 5′UTR-C RNA upon multiple rounds of replication, suggesting that CTP was used for the initiation of HCV RNA replication in cell culture. However, the 5′-end nucleotide C caused a dramatic reduction in cell colony formation (Fig. 2). Also, the levels of both positive- and negative-strand RNAs of the replicon RNAs with a nucleotide C at the 5′ end were >10-fold lower than those of replicon RNAs initiated with an ATP (Fig. 3B). The nucleotide C at the 5′ end could also revert back to a nucleotide A (Fig. 3 and 7). Reversion of the 5′-end nucleotides G and C to an A suggests that ATP is favorable and more stable for initiation of HCV RNA replication in cell culture (Fig. 7). These findings are superimposed by those derived from the mutagenesis analysis of the 3′-end nucleotide of the HCV replicon RNA. Mutations of the 3′-end nucleotide of HCV RNA were expected to affect the initiation of the negative-strand RNA replication. Our results clearly show that pyrimidine nucleotides (U and C) at the 3′ end were favorable for efficient cell colony formation (Fig. 4). The analysis of nucleotide reversion further revealed that the 3′-end nucleotide U-to-C and -G mutations were all reverted back to a wild-type nucleotide U at the 3′ end. Considering that the 3′-end nucleotide U became the initiation nucleotide A of the negative-strand RNA, reversion of the 3′-end nucleotide mutations clearly indicates that replication of the negative-strand HCV RNA was preferentially initiated with ATP. These findings are consistent with those derived from a transient HCV replicon replication system (59). The 3′-end nucleotide mutations of the replicon RNA was found to revert back to the wild-type nucleotide U as early as 1 day posttransfection (59). Taken together, these findings demonstrate that ATP was selectively utilized by HCV replicase as the initiation nucleotide for efficient and stable replication of both positive- and negative-strand RNAs of HCV in cell culture.
The question arose why the replicon 5′UTR-A RNA resulted in lower efficiency in cell colony formation compared to that of the 5′UTR-G RNA (Fig. 2). The exact reason for the discrepancy between cell colony formation and nucleotide preference of ATP for initiation of RNA replication in cell culture is not known. One possible interpretation is that higher levels of positive-strand RNA replication initiated with ATP might be lethal to the cell. It has been recently reported that HCV propagation in B lymphocytes could cause a significant level of apoptosis (51). Cell colony formation induced by the HCV replicon depends on the replication of HCV RNA, as well as on cell proliferation. Only cells that were able to tolerate HCV RNA replication would be selected to form cell colonies. Therefore, cell colony formation might not be an ideal assay system for determination of the efficiency of HCV RNA replication. A transient HCV replicon replication system is probably a better approach in this regard (58).
Findings derived from sequence analysis of HCV RNAs of clinical isolates demonstrate that both ATP and GTP were used by HCV replicase in vivo for initiation of positive-strand RNA replication (Fig. 8). Also, sequence analysis revealed that HCV RNA derived from a chimpanzee, which was intrahepatically inoculated with an infectious HCV RNA (53), contained a nucleotide C at the 5′ end, suggesting that CTP was also utilized for the initiation of positive-strand RNA replication (Cai et al., unpublished). The discrepancy in selection of the initiation nucleotide between HCV RNA replication in cell culture and in vivo might be due to the difference in sequence and/or structure of the HCV replicase between different HCV isolates. Alternatively, the HCV propagation sites (e.g., liver versus spleen) and/or the nucleotide pool in the cell where HCV RNA is replicated could also determine the selection of nucleotides used for initiation of HCV RNA replication. Further investigation is needed to determine whether specific amino acid and/or structure of the HCV replicase discriminate the incoming nucleotide for efficient initiation of HCV RNA replication. Furthermore, it will be interesting to see whether HCV RNA replication is also controlled by different initiation nucleotides in vivo.
The question also arises how initiation of HCV RNA replication in cell culture differs from the one catalyzed by the purified recombinant RdRp in vitro as to the selection of nucleotides for initiation of de novo RNA synthesis. In vivo, replication of most RNA viruses is a highly orchestrated process involving multiple proteins through protein-RNA and protein-protein interactions. The virus-encoded RdRp is a central component of the replication complex that consists of multiple viral and cellular proteins and viral RNA. In the absence of other viral and cellular proteins, viral RdRp by itself often lacks template specificity (27). For instance, purified recombinant HCV RdRp was able to copy HCV-specific RNA, nonviral RNA, or even DNA templates in vitro (24, 36, 39, 40). However, the replication complex associated with the intracellular membrane specifically catalyzes replication of HCV RNA. Therefore, HCV RdRp assembled in the replication complex in the cell may be different in both structure and function from purified RdRp in vitro.
The underlying molecular mechanism of the initiation of HCV RNA replication with different nucleotides remains enigmatic. The preference of ATP and GTP as initiation nucleotides in vivo is probably due to the fact that the selective nucleotide is a component of the viral initiation complex assembled prior to RNA replication like the transcription initiation complex of the polymerase II polymerase (14, 48). In addition, ATP and GTP may provide energy for the formation of the first phosphodiester bond during initiation of HCV RNA replication. It was previously reported that the β-λ imido analogue of ATP was unable to initiate but was able to elongate RNA transcription catalyzed by the RdRp of vesicular stomatitis virus in vitro, suggesting that a hydrolyzable form of the β-λ phosphodiester bond in ATP might be required for initiation of vesicular stomatitis virus RNA transcription (52). A hydrolyzable ATP was also found to be an essential component of the transcription initiation complex of the RNA polymerase II, a DNA-dependent RNA polymerase (8, 44). Whether a hydrolyzable ATP is required for synthesis of the first phosphodiester bond during initiation of HCV RNA replication remains to be determined.
Mutations of the nucleotides at the 5′ and 3′ ends of the HCV replicon RNA might also influence the function of the promoters for synthesis of the positive- and negative-strand RNAs. The nucleotide at the very 3′ end is likely a part of the promoter sequence required for RNA replication, and therein mutations of the initiation nucleotides may impair promoter function. This possibility may explain why a G-to-C mutation at the 5′ end and U-to-G or -A mutations at the 3′ end resulted in a dramatic decrease or a complete abolition of cell colony formation (Fig. 2 and 4). However, it will be difficult to determine the role of the 3′-end nucleotide in the function of the promoter since recognition of the promoter and initiation of RNA synthesis by the viral replication complex are tightly coupled.
Identification of the initiation nucleotide required for efficient HCV RNA replication provides a novel target for rational design and screen of nucleoside and nucleotide analogue inhibitors. Initiation of RNA synthesis is likely a rate-limiting step in HCV RNA replication. Therefore, inhibitors targeting the initiation of HCV RNA replication will be more potent and efficacious than elongation inhibitors for the inhibition of HCV replication in vivo. An HBV inhibitor, Entacavir, blocking the initiation of the viral genome replication exhibited much greater efficacy than the one (lamivudine or 3TC) inhibiting the elongation step (28, 47). It remains to be determined whether ATP analogues are more potent than other nucleoside analogues in inhibition of the initiation of HCV RNA replication in vitro and in the cell.
Acknowledgments
We thank Ralf Bartenschlager for providing a Huh7 cell line used in this study and Robert Geraghty for critical reading of the manuscript.
This study was supported by NIH grants R01CA93712 and R01AI51592.
REFERENCES
- 1.Ackermann, M., and R. Padmanabhan. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 276:39926-39937. [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1993. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J. Virol. 67:3835-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartholomeusz, A., and P. Thompson. 1999. Flaviviridae polymerase and RNA replication. J. Viral Hepat. 6:261-270. [DOI] [PubMed] [Google Scholar]
- 4.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 5.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1975. [DOI] [PubMed] [Google Scholar]
- 6.Bressanelli, S., L. Tomei, F. A. Rey, and R. De Francesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukh, J., R. H. Purcell, and R. H. Miller. 1992. Sequence analysis of the 5′ noncoding region of hepatitis C virus. Proc. Natl. Acad. Sci. USA 89:4942-4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunick, D., R. Zandomeni, S. Ackerman, and R. Weinmann. 1982. Mechanism of RNA polymerase II-specific initiation of transcription in vitro: ATP requirement and uncapped runoff transcripts. Cell 29:877-886. [DOI] [PubMed] [Google Scholar]
- 9.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. 1998. Recommendations for prevention and control of hepatitis C virus (HCV) infection and HCV-related chronic disease. Morb. Mortal. Wkly. Rep. 47:1-39. [Google Scholar]
- 11.Chen, D., and J. T. Patton. 2000. De novo synthesis of minus strand RNA by the rotavirus RNA polymerase in a cell-free system involves a novel mechanism of initiation. RNA 6:1455-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 13.De Francesco, R., and C. Steinkuhler. 2000. Structure and function of the hepatitis C virus NS3-NS4A serine proteinase. Curr. Top. Microbiol. Immunol. 242:149-169. [DOI] [PubMed] [Google Scholar]
- 14.Dvir, A., K. P. Garrett, C. Chalut, J. M. Egly, J. W. Conaway, and R. C. Conaway. 1996. A role for ATP and TFIIH in activation of the RNA polymerase II preinitiation complex prior to transcription initiation. J. Biol. Chem. 271:7245-7248. [DOI] [PubMed] [Google Scholar]
- 15.Ferrari, E., J. Wright-Minogue, J. W. Fang, B. M. Baroudy, J. Y. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francki, R. I. B., C. M. Fauquet, D. L. Knudson, and F. Brown. 1991. Classification and nomenclature of viruses. Fifth report of the International Committee on Taxonomy of Viruses. Arch. Virol. 1991(Suppl. S2):223-233.
- 17.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. A second hepatitis C virus-encoded proteinase. Proc. Natl. Acad. Sci. USA 90:10583-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hijikata, M., N. Kato, Y. Ootsuyama, M. Nakagawa, and K. Shimotohno. 1991. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl. Acad. Sci. USA 88:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hijikata, M., H. Mizushima, T. Akagi, S. Mori, N. Kakiuchi, N. Kato, T. Tanaka, K. Kimura, and K. Shimotohno. 1993. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J. Virol. 67:4665-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishii, K., Y. Tanaka, C. C. Yap, H. Aizaki, Y. Matsuura, and T. Miyamura. 1999. Expression of hepatitis C virus NS5B protein: characterization of its RNA polymerase activity and RNA binding. Hepatology 29:1227-1235. [DOI] [PubMed] [Google Scholar]
- 23.Kao, C. C., P. Singh, and D. J. Ecker. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:251-260. [DOI] [PubMed] [Google Scholar]
- 24.Kao, C. C., X. Yang, A. Kline, Q. M. Wang, D. Barket, and B. A. Heinz. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74:11121-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197-2206. [DOI] [PubMed] [Google Scholar]
- 27.Lai, M. M. 1998. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology 244:1-12. [DOI] [PubMed] [Google Scholar]
- 28.Levine, S., D. Hernandez, G. Yamanaka, S. Zhang, R. Rose, S. Weinheimer, and R. J. Colonno. 2002. Efficacies of entecavir against lamivudine-resistant hepatitis B virus replication and recombinant polymerases in vitro. Antimicrob. Agents Chemother. 46:2525-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, C., B. D. Lindenbach, B. M. Pragai, D. W. McCourt, and C. M. Rice. 1994. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J. Virol. 68:5063-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 32.Lohmann, V., H. Overton, and R. Bartenschlager. 1999. Selective stimulation of hepatitis C virus and pestivirus NS5B RNA polymerase activity by GTP. J. Biol. Chem. 274:10807-10815. [DOI] [PubMed] [Google Scholar]
- 33.Lohmann, V., A. Roos, F. Korner, J. O. Koch, and R. Bartenschlager. 1998. Biochemical and kinetic analyses of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology 249:108-118. [DOI] [PubMed] [Google Scholar]
- 34.Lohmann, V., A. Roos, F. Korner, J. O. Koch, and R. Bartenschlager. 2000. Biochemical and structural analysis of the NS5B RNA-dependent RNA polymerase of the hepatitis C virus. J. Viral Hepat. 7:167-174. [DOI] [PubMed] [Google Scholar]
- 35.Lubinski, J. M., G. Kaplan, V. R. Racaniello, and A. Dasgupta. 1986. Mechanism of in vitro synthesis of covalently linked dimeric RNA molecules by the poliovirus replicase. J. Virol. 58:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo, G., S. Xin, and Z. Cai. 2003. Role of the 5′-proximal stem-loop structure of the 5′ untranslated region in replication and translation of hepatitis C virus RNA. J. Virol. 77:3312-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, R. H., and R. H. Purcell. 1990. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc. Natl. Acad. Sci. USA 87:2057-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh, J. W., T. Ito, and M. M. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oh, J. W., G. T. Sheu, and M. M. Lai. 2000. Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J. Biol. Chem. 275:17710-17717. [DOI] [PubMed] [Google Scholar]
- 41.O'Reilly, E. K., and C. C. Kao. 1998. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology 252:287-303. [DOI] [PubMed] [Google Scholar]
- 42.Osman, T. A., and K. W. Buck. 1996. Complete replication in vitro of tobacco mosaic virus RNA by a template-dependent, membrane-bound RNA polymerase. J. Virol. 70:6227-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paul, A. V., J. H. van Boom, D. Filippov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 44.Rappaport, J., and R. Weinmann. 1987. Purine triphosphate beta-gamma bond hydrolysis requirements for RNA polymerase II transcription initiation and elongation. J. Biol. Chem. 262:17510-17515. [PubMed] [Google Scholar]
- 45.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 46.Rice, C. M. 1996. Flaviviridae: the viruses and their replication. Lippincott-Raven, Philadelphia, Pa.
- 47.Seifer, M., R. K. Hamatake, R. J. Colonno, and D. N. Standring. 1998. In vitro inhibition of hepadnavirus polymerases by the triphosphates of BMS-200475 and lobucavir. Antimicrob. Agents Chemother. 42:3200-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Serizawa, H., Z. Tsuchihashi, and K. Mizumoto. 1997. The RNA polymerase II preinitiation complex formed in the presence of ATP. Nucleic Acids Res. 25:4079-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh, R. N., and T. W. Dreher. 1997. Turnip yellow mosaic virus RNA-dependent RNA polymerase: initiation of minus strand synthesis in vitro. Virology 233:430-439. [DOI] [PubMed] [Google Scholar]
- 50.Song, C., and A. E. Simon. 1994. RNA-dependent RNA polymerase from plants infected with turnip crinkle virus can transcribe (+)- and (−)-strands of virus-associated RNAs. Proc. Natl. Acad. Sci. USA 91:8792-8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sung, V. M., S. Shimodaira, A. L. Doughty, G. R. Picchio, H. Can, T. S. Yen, K. L. Lindsay, A. M. Levine, and M. M. Lai. 2003. Establishment of B-cell lymphoma cell lines persistently infected with hepatitis C virus in vivo and in vitro: the apoptotic effects of virus infection. J. Virol. 77:2134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Testa, D., and A. K. Banerjee. 1979. Initiation of RNA synthesis in vitro by vesicular stomatitis virus. Role of ATP. J. Biol. Chem. 254:2053-2058. [PubMed] [Google Scholar]
- 53.Thomson, M., M. Nascimbeni, S. Gonzales, K. K. Murthy, B. Rehermann, and T. J. Liang. 2001. Emergence of a distinct pattern of viral mutations in chimpanzees infected with a homogeneous inoculum of hepatitis C virus. Gastroenterology 121:1226-1233. [DOI] [PubMed] [Google Scholar]
- 54.Trowbridge, R., and E. J. Gowans. 1998. Identification of novel sequences at the 5′ terminus of the hepatitis C virus genome. J. Viral Hepat. 5:95-98. [DOI] [PubMed] [Google Scholar]
- 55.World Health Organization. 1998. W. H. O. concerns hepatitis C. Lancet 351:1415.
- 56.Yamada, N., K. Tanihara, A. Takada, T. Yorihuzi, M. Tsutsumi, H. Shimomura, T. Tsuji, and T. Date. 1996. Genetic organization and diversity of the 3′ noncoding region of the hepatitis C virus genome. Virology 223:255-261. [DOI] [PubMed] [Google Scholar]
- 57.Yamashita, T., S. Kaneko, Y. Shirota, W. Qin, T. Nomura, K. Kobayashi, and S. Murakami. 1998. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 273:15479-15486. [DOI] [PubMed] [Google Scholar]
- 58.Yi, M., and S. M. Lemon. 2003. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 77:3557-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yi, M., and S. M. Lemon. 2003. Structure-function analysis of the 3′ stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA 9:331-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Young, D. C., D. M. Tuschall, and J. B. Flanegan. 1985. Poliovirus RNA-dependent RNA polymerase and host cell protein synthesize product RNA twice the size of poliovirion RNA in vitro. J. Virol. 54:256-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhong, W., E. Ferrari, C. A. Lesburg, D. Maag, S. K. Ghosh, C. E. Cameron, J. Y. Lau, and Z. Hong. 2000. Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:9134-9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhong, W., A. S. Uss, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]