Abstract
Fourteen isolates of Rice yellow mottle virus (RYMV) were selected as representative of the genetic variability of the virus in Africa from a total set of 320 isolates serologically typed or partially sequenced. The 14 isolates were fully sequenced and analyzed together with two previously reported sequences. RYMV had a genomic organization similar to that of Cocksfoot mottle sobemovirus. The average nucleotide diversity among the 16 isolates of RYMV was 7%, and the maximum diversity between any two isolates was 10%. A strong conservative selection was apparent on both synonymous and nonsynonymous substitutions, through the amino acid replacement pattern, on the genome size, and through the limited number of indel events. Furthermore, there was a lack of positive selection on single amino acid sites and no evidence of recombination events. RYMV diversity had a pronounced and characteristic geographic structure. The branching order of the clades correlated with the geographic origin of the isolates along an east-to-west transect across Africa, and there was a marked decrease in nucleotide diversity moving westward across the continent. The insertion-deletion polymorphism was related to virus phylogeny. There was a partial phylogenetic incongruence between the coat protein gene and the rest of the genome. Overall, our results support the hypothesis that RYMV originated in East Africa and then dispersed and differentiated gradually from the east to the west of the continent.
Rice yellow mottle virus (RYMV) is one of the most important emergent plant viruses and poses a threat to rice cultivation in Africa (41). First reported in Kenya in 1966 (4), RYMV has since been detected in most rice-growing countries in Africa (1) and recently in Central Africa (45) but not outside the continent. The natural host range of RYMV is mostly restricted to grasses of the Oryzeae tribe (4). It is transmitted by several species of beetles (Coleoptera), most of which belong to the Chrysomelidae family (4), and is not seed borne (13, 19). Additional means of biotic transmission by rats and other vertebrates have been reported (38). There are conflicting observations on the role of abiotic transmission by mechanical means during cultural practices, through soil residues, or by irrigation water (2). A highly conserved RNA satellite, not involved in pathogenicity, is often associated with RYMV (33).
RYMV is an RNA virus belonging to the genus Sobemovirus (46). Based on organizational differences in the central part of the genome (encoding the virus polyprotein), the sobemoviruses were subdivided into Southern cowpea mosaic virus (SCPMV)-like and Cocksfoot mottle virus (CfMV)-like types (42, 46). First reported to have an SCPMV-like organization (28), RYMV is now suspected to be of the CfMV type (12). Its genome harbors four open reading frames (ORFs) (42). ORF1, which is located at the 5′ end of the genome, encodes a small protein involved in virus movement (7) and in suppressing gene silencing (47). ORF2, which encodes the central polyprotein, has two overlapping ORFs. ORF2a encodes a serine protease and a viral genome-linked protein. ORF2b, which is translated through a −1 ribosomal frameshift mechanism as a fusion protein, encodes the RNA-dependent RNA polymerase. The coat protein gene (ORF4) is expressed by a subgenomic RNA at the 3′ end of the genome. The requirement of encapsidation for long-distance movement has been suggested (8). Additionally, the genome comprises three noncoding regions (NCR) at the 5′ (5′ NCR) and 3′ (3′ NCR) ends and between ORF1 and ORF2.
RNA viruses have a potential for much genetic variability due to the intrinsically high mutation rate associated with the RNA-dependent RNA polymerase, their high rates of replication, and their large population sizes (9, 10, 22, 34). RYMV variability was first apparent from the detection of several serotypes in immunological studies with polyclonal and monoclonal antibodies (13, 20, 23, 29). Moreover, sequencing of the coat protein gene revealed genetic variation within each serotype (11). These studies suggested that the strains followed a geographic distribution with a split between East and West African strains (3, 32, 45).
Comparative analysis of gene sequence data and geographic information can elucidate the origin and spread of viruses as well as the evolutionary processes that underlie their genetic diversity. Accordingly, we sequenced the full genome of 14 isolates selected as representatives of the genetic variability as well as of the geographic distribution of RYMV from a set of 320 isolates that had been serologically typed or partially sequenced. These 14 sequences, together with two previously reported ones (28), were analyzed. First, we identified the major evolutionary constraints operating on the genome. Phylogenetic analyses were then made to determine the genetic relationships between the isolates. Last, phylogeographic studies were conducted to assess the links between geographic and genetic distances. Altogether, these analyses suggested that (i) RYMV evolution operated under a conservative selection, in the absence of adaptation or recombination events, (ii) RYMV dispersed and differentiated gradually from the east to the west of Africa, and (iii) RYMV originated and evolved in wild graminaceous species and only recently infected cultivated rice.
MATERIALS AND METHODS
Isolate selection.
Altogether, 320 RYMV isolates from cultivated rice and wild graminaceous species have been collected over the past 15 years in 13 countries from all agro-ecological zones where rice is cultivated in Africa (Burkina-Faso, Cameroon, Chad, Côte d'Ivoire, Ghana, Guinea, Kenya, Madagascar, Mali, Nigeria, Sierra-Leone, Tanzania, and Togo). This collection is fully representative of the geographic distribution of RYMV in Africa and is one of the most comprehensive to have been used for plant virus diversity studies. All the field isolates were inoculated on the susceptible Oryzae sativa cultivar IR64 to increase virus concentration and to allow direct sequencing of reverse transcription-PCR products. We checked that no modification of the sequences occurred after passages on this host (S. Fatogoma and D. Fargette, unpublished data). The isolates were typed with discriminant monoclonal antibodies and assigned to one of the five recorded serotypes (29). The coat protein gene of 145 isolates representative of the five serotypes was then sequenced. Molecular typing confirmed the serotyping and further identified several variants within each of the serotypes (11). Fourteen isolates were selected for analysis to represent the genetic diversity, i.e., one or two isolates of each serotype and variant, and their genomes were fully sequenced. Details of the 14 isolates, name, country of origin, host species, year of isolation, and sequence accession number, are shown in Table 1. Additionally, we included isolates CIb and Nia, whose genomes had been previously sequenced (28).
TABLE 1.
Origins and references of isolates of RYMV used in the study
| Isolatea | Region | Country of origin | Original host plant | Yr of isolation | Accession no. |
|---|---|---|---|---|---|
| CIa | West Africa | Côte d'Ivoire | O. sativa | 1985 | AJ608206 |
| CIb | West Africa | Côte d'Ivoire | O. sativa | 1985 | L20893 |
| CI4 | West Africa | Côte d'Ivoire | O. sativa | 1995 | AJ608207 |
| CI63 | West Africa | Côte d'Ivoire | O. sativa | 1997 | AJ608208 |
| Ma10 | West Africa | Mali | O. sativa | 1996 | AJ608209 |
| Ma77 | West Africa | Mali | Oryzae barthii | 2000 | AJ608210 |
| Mg1 | East Africa | Madagascar | O. sativa | 1990 | AJ608211 |
| Mg2 | East Africa | Madagascar | O. sativa | 1990 | AJ608212 |
| Nia | Central Africa | Nigeria | O. sativa | 1985 | U23142 |
| Ni1 | Central Africa | Nigeria | O. sativa | 1985 | AJ608213 |
| Ni2 | Central Africa | Nigeria | O. sativa | 1985 | AJ608214 |
| SL4 | West Africa | Sierra Leone | O. sativa | 1985 | AJ608215 |
| Tz3 | East Africa | Tanzania | O. sativa | 1997 | AJ608216 |
| Tz5 | East Africa | Tanzania | O. sativa | 1997 | AJ608217 |
| Tz8 | East Africa | Tanzania | O. sativa | 1996 | AJ608218 |
| Tz11 | East Africa | Tanzania | O. sativa | 2001 | AJ608219 |
All sequences were deposited in the EMBL, except isolates CIb and Nia, which were deposited in GenBank.
Genome sequencing.
Nucleotide sequences of the entire genome of each isolate were determined by using at least two overlapping sequences for all regions. Four genome fragments were transcribed and amplified by reverse transcription-PCR after extracting total RNA from leaves according to the method of Pinel et al. (32). Sequencing was performed by using the Taq terminator sequencing kit (Applied Biosystems) and analyzed on an Applied Biosystems 373A sequencer. Two readings per base (in the 3′-to-5′ and 5′-to-3′ directions) led to sequence accuracy >99.9%. Sequences were assembled by Seqman (DNASTAR). Four pairs of primers were used for amplification. Nucleotide numbering is that of isolate CIa. The primers are as follows: AS (nucleotides [nt] 2 to 22), 5′-CAATTGAAGCTAGGAAAGGAG-3′; AAS (nt 982 to 1000), 5′-ACCCCAGGATTTACTCTTT-3′; BS (nt 892 to 915), 5′-CTCGGGGTTACGTGGTTGATGTTT-3′; BAS (nt 2401 to 2424), 5′-ACTTCGCCGGTTTCGCAGAGGATT-3′; CS (nt 2138 to 2157), 5′-CATGCTGGGAAAAGTGTCTG-3′; CAS (nt 3597 to 3616), 5′-GGCCAGGTGTTAGAAGATAG-3′; DS (nt 3442 to 3457), 5′-CAAAGATGGCCAGGAA-3′; DAS (nt 4430 to 4452), 5′-CTCCCCCACCCATCCCGAGAATT-3′.
Sequence analyses.
The following analyses were conducted on the 16 sequences.
(i) Sequence alignment.
The sequences were aligned using CLUSTAL W with default parameters (43). The alignment was corrected by hand in some inappropriate gaps that were not multiples of 3 nt in coding regions to maintain the alignment of the encoded amino acids.
(ii) Sequence diversity.
The diversity index (π), which is the average number of nucleotide differences per site between two sequences (26), was calculated along the whole 16 genome sequences by using a 100-nt sliding window with a 25-nt step. The value assigned to the nucleotide was that of the window midpoint. Additionally, for each ORF, πa, the average number of nucleotide substitutions per nonsynonymous site, πs, the average number of nucleotide substitutions per synonymous site, and their ratio (ω = πa/πs) were calculated (27). All diversity indices were calculated by using DNAsp version 3.5 (36).
(iii) Search for positive selection.
A search for positive selection was performed on each of the four ORFs (excluding the overlapping parts) with the 16 sequences. More extensive samples of 36 and 48 sequences of ORF4 representatives of the full corpus of 145 sequences were also tested. The pattern of selection was inferred through ω values corresponding, respectively, to negative selection (ω < 1), neutral evolution (ω = 1), and positive selection (ω >1) (50). Estimations of ω were performed within the maximum-likelihood (ML) framework which used codon-based models of sequence evolution that account for phylogenetic structure, biases in codon usage, and the transition/transversion ratio (51). Efficient determination of sites under positive selection only required implementation of six models of codon substitution: M0, M1, M2, M3, M7, and M8 (50). Null models M0, M1, and M7 did not allow the existence of positively selected sites because ω values were fixed or estimated between 0 and 1 boundaries, whereas models M2, M3, and M8 allowed positive selection with parameters that estimate ω to be >1. The significance of positive selection was then evaluated through a likelihood ratio test between null models and those which allowed positive selection. Models M0 and M1 are both nested within M2 and M3, M2 is nested within M3, and M7 is nested within M8. Once positively selected sites were found, a Bayesian approach was used to infer the most likely value for each site. Models were implemented by using the CODEML program of the PAML package, version 3.1 (49).
(iv) Residue substitution.
Residue substitution was estimated by using the most parsimonious series of substitutions that could give rise to the residue differences observed in the alignment given the current phylogenetic tree relationships. This was applied on the whole genome to calculate the transition-to-transversion ratio and to assess, on each ORF, the amino acid replacements according to their physical properties determined by their pairwise physicochemical distances (15).
(v) Phylogenetic analyses.
Phylogenetic relationships between the isolates were determined by three methods: the ML with the transition/transversion ratio estimated through ML, the maximum-parsimony (MP) method, and a distance method where the nucleotide pairwise distances were corrected by using the Kimura two-parameter methods and trees were reconstructed by the neighbor-joining (NJ) method. The full heuristic search option was used, and the significance of the internal branches was evaluated by using 1,000 bootstrap replications for MP and NJ analyses and 100 replications for ML analyses. All analyses were implemented with PAUP, version 4.0 (40).
(vi) Bremer support index.
MACLADE 4 (21) was used to calculate, under the most parsimonious hypothesis, the Bremer support index of the branches (number of nucleotide substitutions necessary to break a node).
(vii) Search for recombination.
The aligned sequences were checked for incongruent relationships that might have resulted from recombination by using a distance method implemented in PHYLPRO (48) and a nucleotide substitution distribution method implemented in GENECOV (39).
(viii) Phylogeographic studies.
The genetic distances between the isolates were expressed in matrices of pairwise nucleotide divergence percentages (26). We applied the log10 (d + 100) transformation of the geographic distances (d; in kilometers) to test the relationships with the genetic distances. Correlations between genetic and geographic distances (after logarithmic transformation) were assessed with the Mantel test (24) implemented in GENETIX 4.02 (5).
(ix) Genetic distances between clades.
The average number of substitutions per site between two populations (27) was calculated by using DNAsp, version 3.5 (36), to assess the genetic distances between the clades.
RESULTS
Genomic studies.
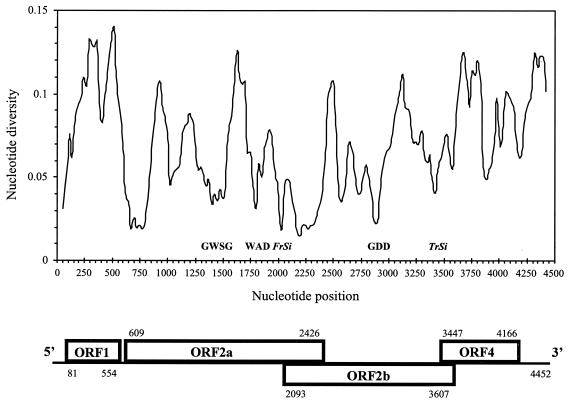
The genomes of the 14 RYMV isolates had CfMV-like overlapping ORF2a and ORF2b in their central part instead of the long continuous SCPMV-like ORF2 reported earlier (Fig. 1). The comparison to the first two fully sequenced genomes (which had suggested a SCPMV-like genomic organization) showed the presence of an extra nucleotide U at position 2244. This occurred in a region prone to sequencing errors because of gel compression when sequencing was made manually. Resequencing these two isolates confirmed the sequence error and clearly established that RYMV had a CfMV-like genomic organization. Without this erroneous extra nucleotide, a stop codon appeared at position 2424 which resulted in the ORF2a-ORF2b CfMV-like overlap instead of the translation of a long ORF2 and a nested ORF3 of the SCPMV type.
FIG. 1.
Genomic organization of RYMV (bottom) and nucleotide diversity index along the genome (top). The diversity index (π), average number of nucleotide differences per site between two sequences, was calculated along the genome by using a sliding window of 100 nt moved by steps of 25 nt after alignment of the 16 sequences. The value is assigned to the nucleotide at the midpoint of the window. Key conserved amino acid motifs and frameshift (FrSi) or transcription (TrSi) signals are indicated (see text for details).
The average nucleotide diversity among the 16 isolates of RYMV was 7%, and the maximum diversity between any two isolates was 10% (Table 2). Peaks of variability of >10% were apparent within each ORF (Fig. 1). By contrast, lower variability in the rest of the genome illustrated the extent and frequency of the genomic constraints. They operated both on synonymous and nonsynonymous sites. The first 60 nt of the 5′ NCR, which included sites involved in the translation initiation region of ORF1, were highly conserved. There were eight other fully conserved stretches of nucleotides over 25 nt long in the genome, located mostly in ORF2a and ORF2b. One included the frameshift signal made of the heptanucleotide UUUAAAC (nt 1979 to 1985) and a predicted 32-nt-long stem-loop structure located 7 nt downstream from the heptamer. This motif was conserved in CfMV and SCPMV as well. Another conserved nucleotide motif was ACAAA (nt 3441 to 3445) in the putative transcription site of the subgenomic RNA. Low diversity at the beginning of the 3′ NCR possibly reflected a conserved secondary structure as suggested by MFOLD analysis (25) and by the presence of a large majority of compensatory substitutions (data not shown). There were two strictly constant regions of 74 amino acids (aa) (nt 987 to 1208) and 92 aa (nt 1287 to 1562) within ORF2a. The motifs GWSG (nt 1455 to 1466) and WAD (nt 1776 to 1784), characteristic of the serine protease and of the viral genome-linked protein, respectively, were found (42). Within ORF2b, a stretch of 94 aa (nt 2660 to 2941) as well as other motifs, such as GDD, characteristic of the polymerase (nt 3146 to 3154) were fully conserved. The overlapping ORF2a/ORF2b regions were characterized by an overall low nucleotide diversity, a likely consequence of the combined constraints on each ORF translated in different frames. There were several highly constrained regions with unidentified functions, with the most constant one being located at the beginning of ORF2a (11 variable positions within a stretch of 161 nt).
TABLE 2.
Diversity indices (average and maximum) calculated on the total (π), synonymous (πs), and nonsynonymous (πa) sites and the ratio (ω = πa/πs) for the four ORFs after alignment of the 16 RYMV sequences
| Genome part | Diversity index for:
|
ω | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total sites
|
Synonymous sites
|
Nonsynonymous sites
|
||||||||
| NSa | Avg π | Maximum π | NS | Avg πs | Maximum πs | NS | Avg πa | Maximum πa | ||
| Full genome | 4,462 | 0.070 | 0.103 | |||||||
| ORF1 | 471 | 0.105 | 0.167 | 112 | 0.290 | 0.587 | 359 | 0.048 | 0.095 | 0.164 |
| ORF2a | 1,815 | 0.054 | 0.075 | 462 | 0.161 | 0.243 | 1,353 | 0.017 | 0.028 | 0.104 |
| ORF2b | 1,509 | 0.056 | 0.080 | 357 | 0.185 | 0.305 | 1,152 | 0.017 | 0.025 | 0.090 |
| ORF4 | 717 | 0.083 | 0.143 | 184 | 0.216 | 0.387 | 533 | 0.036 | 0.057 | 0.166 |
| 3′ NCR | 289 | 0.107 | 0.235 | |||||||
NS, number of sites.
Nucleotide diversity was variable among ORFs. ORF1 and ORF4 were the most variable, whereas ORF2a and ORF2b were the most conserved. The average variation in synonymous sites ranged from 16 to 29%, and the maximum variation between two isolates ranged from 24 to 59%, an indication of the extent of divergence which developed during RYMV evolution. The nonsynonymous diversity (πa) was 6 to 11 times less than the synonymous diversity and ranged between 2 and 5%, with the maximum between any two isolates being 9% (Table 2). This indicated that the conservative selection pressure operated mostly at the protein level. Patterns of amino acid changes also provided information on the selection pressure which acted on proteins. With RYMV, the physicochemical properties of the amino acids as defined by Grantham (15) were conserved in most replacements, no matter which the ORF (data not shown). However, selection pressure also occurred at synonymous sites, as apparent in the conservation of the various nucleotide signals and secondary structures reported above, and in the differences in πs between ORFs (Table 2). The nucleotide diversity of the 3′ NCR was similar to that of the ORF1 total diversity index, which was further indication that the conservative pressure also operated in NCR. Nucleotide substitution by transition was more frequent than by transversion, with a transition/transversion ratio of 7.2.
None of the models used to assess diversifying selection detected sites under positive selection within the first three ORFs. By contrast, M3 and M8 detected a single site under positive selection in ORF4 (Table 3). Analyses of the 36 and 48 sequences of ORF4 gave the same results. Bayesian analyses assigned this site to threonine218 (nt 4095 to 4097) with posterior probabilities of 0.98 (model M3) and 0.92 (model M8). However, the likelihood ratio tests indicated that the models detecting positive selection were not the most likely ones (Table 3). A conservative conclusion is that there is no site under positive selection in the RYMV genome, even within ORF4.
TABLE 3.
ML analysis of the evolution of RYMV coat protein with models allowing ω to vary across amino acid sitesa
| Modelb | ML
|
Likelihood ratio test | P valuef | ||
|---|---|---|---|---|---|
| ωc | ω maxd | Fe | |||
| M0 | 0.12 | M0 vs M2 | <0.001 | ||
| M1 | 0.25 | M1 vs M2 | <0.001 | ||
| M2 | 0.12 | 0.30 | 0.27 | M0 vs M3 | <0.001 |
| M3 | 0.12 | 1.93 | 0.01 | M1 vs M3 | <0.001 |
| M7 | 0.12 | M2 vs M3 | 0.17 | ||
| M8 | 0.12 | 1.98 | 0.01 | M7 vs M8 | 0.36 |
Results were obtained with the CODEML program implemented in PAML.
Models M0, M1, and M7 assume that no amino acid sites are under diversifying selection, whereas M2, M3, and M8 allow sites with a ω of >1.
Mean value of ω over the whole ORF.
Maximum ω category.
Estimated frequency of sites belonging to the maximum ω category.
Probability that twice the difference between the log likelihoods is smaller than a χ2 distribution.
Phylogenetic studies.
The complete genome sequences of CfMV and SCPMV were too distantly related to RYMV (30 and 21% nucleotide identity, respectively) to be used as out-groups in the phylogenetic analyses of the RYMV isolates. Consequently, preliminary phylogenetic analyses were conducted on the ORF2b sequences, the most conserved region among sobemoviruses, with CfMV and SCPMV as out-groups (59 and 52% nucleotide identity with RYMV, respectively) to identify the most basal RYMV isolates. This indicated that Tz11 and Tz3 were the most basal isolates. This was confirmed with midpoint rooting on the complete genome, which assigned the root to the midpoint of the two longest paths between the two terminal taxa in the tree. Then the in-group (RYMV) was rooted at the branch (Tz3 or Tz11) where the out-group (CfMV or SCPMV) joined the in-group tree (37). Tz11 was used as the out-group for the phylogenetic analyses with the full sequences, although similar topologies were obtained with Tz3. The phylogenetic relationships between the 16 isolates were reconstructed by using the ML, MP, and NJ methods. Whatever the method, the general topology of the trees was similar and showed the same branching order of the clades. The trees were distinctive in that the branching order reflected the geographic origin of the isolates along an east-to-west transect across Africa (Fig. 2a). The most basal isolates originated from eastern Tanzania. There were successive branchings of other East African and Madagascar isolates, whereas isolates from West Central Africa constituted a monophyletic group split into two sister groups comprising isolates from West and Central Africa (Fig. 2a). West African isolates split into further groups westward.
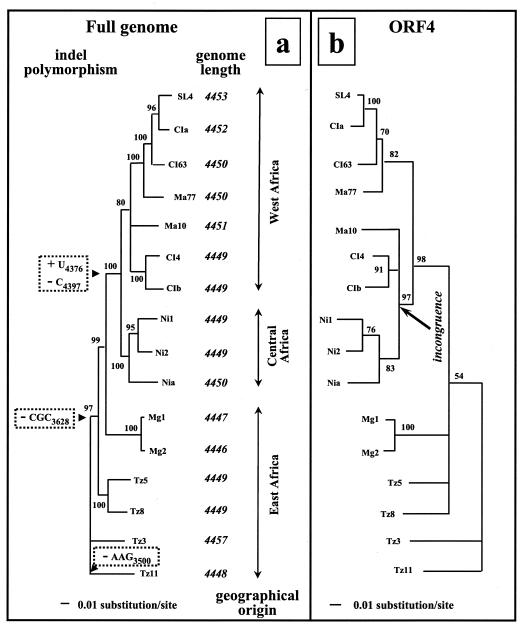
FIG. 2.
Phylogenetic trees of 16 representative isolates of RYMV calculated from the complete genome (a) and from the ORF4 sequences (b). These trees were constructed by using the ML method. The numbers at each node indicate the percentage of bootstrap support (values of >70% are shown for the full genome; values of >50% are shown for ORF4). Phylogenetically constrained insertion-deletion events are indicated at the corresponding node at the left of panel a. The geographic origins and sizes of the genomes of the 16 isolates are shown at the right of panel a. The incongruence between full-genome and ORF4 tree topologies is indicated by an arrow in panel b.
The genome size of RYMV was highly conserved and ranged from 4,447 to 4,457 nt (Fig. 2a). There were very few insertion-deletion events, and only two were in coding regions. A coding indel polymorphism occurred in the N-terminal part of the coat protein. The first indel was in the overlapping ORF2b-ORF4 region, and both events involved basic amino acids. Coding indel events were related to virus phylogeny. The basal isolate Tz3 had both a lysine19 (AAG; nt 3498 to 3500) and an arginine60 (CGC; nt 3626 to 3628), Tz11 lacked the lysine19 located within the putative bipartite nuclear targeting motif (28), and all other isolates lacked the arginine60. The deletion of the arginine60 in all isolates except the most basal ones suggested that it occurred in one of their common ancestors at the most internal node (Fig. 2a). At least two single-nucleotide indels were also determined phylogenetically. An insertion of U4376 and a deletion of C4397 were shared by all the isolates from West and Central Africa, which suggested that it occurred in the ancestor of the West Central African monophyletic group (Fig. 2a).
The phylogenetic analyses were conducted on each individual ORF and on the 3′ NCR with the ML, MP, and NJ methods. Compared to the full genome, there was a loss of phylogenetic resolution in trees based on ORF2a and ORF2b sequences, which were the most conserved regions. Due to the lower number of informative sites, some groups separated in the full-length sequence analysis were collapsed within the same group. Minor incongruences were observed in ORF1, ORF2a, and ORF2b. However, the only incongruence which broke the relation between the branching order and the east to west origins of the isolates was through the coat protein gene (ORF4) phylogenetic analysis (Fig. 2b). With the full genome (Fig. 2a), isolates Ma10, CIb, and CI4 from Mali and Côte d'Ivoire in West Africa belonged to a monophyletic group with all other isolates from West Africa ((CIb, CI4), Ma10, (Ma77, (CI63, (CIa, SL4)))). By contrast, with ORF4 (Fig. 2b), the West African isolates CIb, CI4, and Ma10 formed a sister group of Central African isolates Nia, Ni1, and Ni2 within a paraphyletic group (((Ni1, Ni2), Nia), (CIb, CI4), Ma10). This was apparent whatever the methods used and whether they were applied on nucleotide or amino acid sequences (data not shown). This paraphyletic grouping of isolates from Central Africa with some isolates from West Africa from savanna ecologies on the basis of ORF4, previously referred to as savanna strains (30), was also observed with the extended samples of 36 and 48 sequences of ORF4.
The partial incongruence in ORF4 was not a signal of ancient interstrain recombination between ORF4 and other parts of the genome, as no recombination events were detected by using PHYLPRO or GENECOV (data not shown). MP analysis also indicated that the substitutions supporting the node of the paraphyletic group (((Ni1, Ni2), Nia), (CIb, CI4), Ma10) were spread along the coat protein gene and not gathered within any given region, as expected after a recombination event. Neither was ORF4 incongruence due to convergent evolution on the CP, as no adaptive selection was detected through the study of ω values (see above). Actually, the corresponding Bremer support index, the number of nucleotide substitutions necessary to break the node of the paraphyletic group, was only 4. The Bremer support index of the node supporting the isolates from West Africa was 6 when calculated on the full genome. Then a nonuniform distribution of a few substitutions among some lineages across the genome could explain this incongruence. Practically, this incongruence indicates that ORF4 sequencing is a reliable typing tool to assign the RYMV isolates to the main clades but that it is inappropriate to assess the phylogenetic relationships between some clades.
Phylogeographic studies.
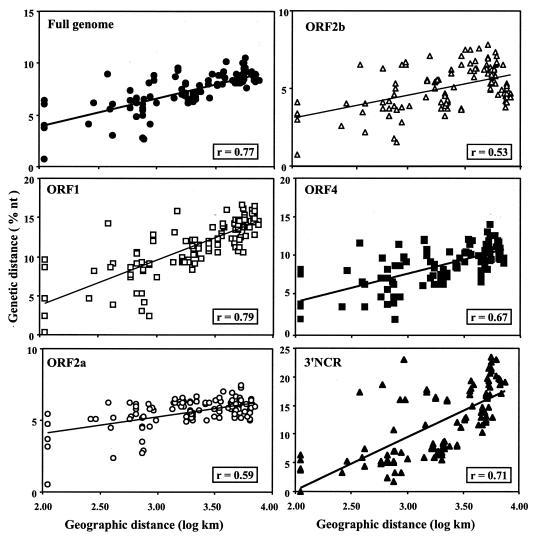
There was a close relationship between the geographic distance (after logarithmic transformation) and the genetic distance between isolates (Fig. 3). The more apart the origin of the isolates, the greater was their genetic distance. This was verified on the full genome and also with each individual ORF and with the 3′ NCR (correlation coefficients [r] ranged between 0.53 and 0.79; P < 0.001). In no instance did isolates separated by a long distance have a low genetic divergence. This was true also with the 145 isolates partially sequenced (data not shown).
FIG. 3.
Pairwise comparisons and corresponding linear regression lines of geographic distance (in kilometers, after logarithmic transformation) and genetic distance (percent divergence in nucleotides) between each of the 16 isolates calculated on the full genome, on each ORF, and on the 3′ NCR.
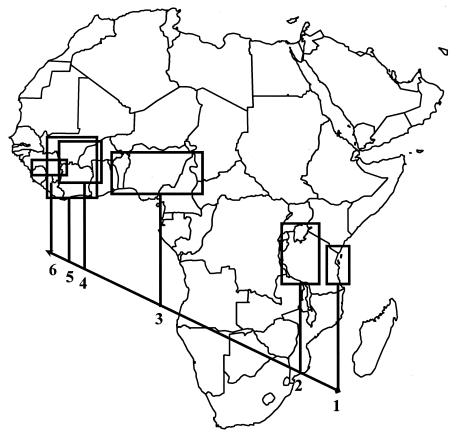
Considering the branching order of the ML phylogram, six clades were defined which followed an east-to-west orientation across mainland Africa. Madagascar was not included in the analysis because the very few isolates available for study were not representative of the country. Each of the isolates partially sequenced was assigned to one of the six clades by phylogenetic analysis. Subsequently, the area of each of the clades was derived from the locations where the corresponding isolates were collected (Fig. 4). Clade 1 (reference isolates Tz3 and Tz11; previously referred to as strains S5 and S6) gathered isolates found exclusively in eastern Tanzania, in the Eastern Arc mountains, and on Pemba island (close to Zanzibar). Clade 2 (Tz5 and Tz8; previously strain S4) included isolates from the west and north of Tanzania and from Kenya. Clade 3 (Nia, Ni1, and Ni2; previously S1/AC) included isolates from Cameroon, Chad, Nigeria, and Togo in Central Africa from 16° to 1°E. The most intensive surveys were conducted in West Africa. They revealed that Clades 4, 5, and 6 had partially overlapping distributions but also showed a westward orientation, as terminal clades 5 and 6 spread over more western regions than clade 4. Isolates of clade 4 (CI4, CIb, and Ma10; previously S1/AO) occurred exclusively in the savanna regions in Côte d'Ivoire, Mali, and Burkina-Faso from 7° to 16°N and from 1° to 8°W. Isolates from clade 5 (Ma77 and CI63; previously S2) had a more western geographic distribution than that of clade 4, as they originated from 3° to 10°W and from 5° to 16°N. These isolates were found in savanna regions of Mali, Burkina-Faso, and Côte d'Ivoire and further south in the forest regions of Côte d'Ivoire, Ghana, and Guinea. Clade 6 (SL4 and CIa; previously S3) had a restricted distribution and had the most westward location (13°W).
FIG. 4.
Geographic distribution of the six clades in mainland Africa. The six clades were defined by analysis of the complete genomic sequences of representative isolates of RYMV by using the ML method. Each isolate partially sequenced was assigned to one of the six clades by phylogenetic analyses. The geographic area of each clade was derived from the locations where these isolates were collected.
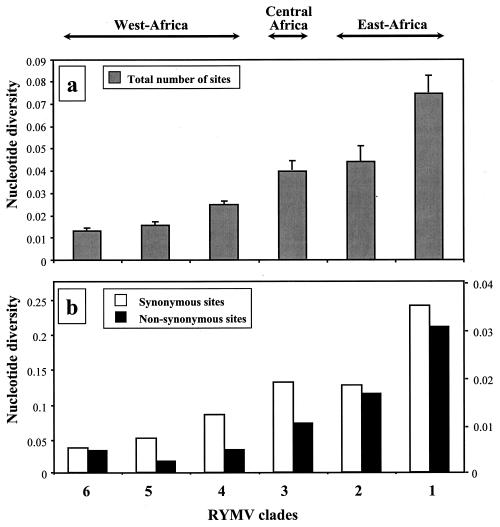
From the isolates partially sequenced, 10 isolates representative of the genetic diversity of each clade were selected, except for clades 2 and 6 where only 7 isolates were available. The diversity index (π) was calculated on the total number of sites and also by distinguishing the synonymous (πs) and nonsynonymous (πa) sites. Nucleotide diversity gradually decreased moving westward, being the greatest in the eastern clade 1 and minimal in the most western clades 5 and 6 (Fig. 5). This decrease was observed whether the diversity index was calculated on the total nucleotide substitutions (Fig. 5a) or on synonymous or nonsynonymous substitutions (Fig. 5b). Genetic distances among clades were calculated from the number of substitutions per site among the corresponding isolates. Altogether, they were consistent with the general topology of the phylogenetic tree with an increase in genetic distances between clades from west to east. The genetic distance of the most western clade 6 to other clades increased from 3.5% with clade 5 up to 11% with the most eastern clade 1.
FIG. 5.
Total nucleotide diversity index and standard error calculated for 10 representative isolates of each clade oriented along an east-to-west transect across Africa (a) and diversity indices calculated for synonymous and nonsynonymous sites (left and right scales, respectively) (b).
DISCUSSION
A strong conservative selection operates on most animal and plant viruses (9, 14). With RYMV, a pronounced purifying selection was apparent in all coding and noncoding regions and through the high conservation of the genome size. The genetic stability of RYMV was also supported by the following observations. The first isolate studied by Bakker in 1966 (4) had a sequence very similar to that of neighboring isolates collected 30 years later in the Lake Victoria region (3). Some isolates from Mali collected at intervals of 10 years from locations dozens of kilometers apart were identical. Moreover, the sequences of several isolates were conserved after serial passages in various cultivars (our unpublished data). Altogether, the study of the genomic constraints and the experimental evidence suggested a low rate of change of RYMV. Similarly, a low rate of change of Turnip yellow mosaic virus was inferred from biogeographical evidence (6).
The average nucleotide diversity among the 16 isolates of RYMV was 7%, and the maximum diversity between any two isolates was 10%. RYMV diversity showed a pronounced and characteristic spatial structure. The branching order of the clades correlated with the geographic origin of the isolates along an east-to-west transect across Africa and was associated with a marked decrease in the nucleotide diversity westward. The indel polymorphism and the nucleotide substitution patterns were related. There was a close relationship between genetic and geographic distances. In no instances were two distant isolates (from sites >100 km apart) genetically close (<1%). This was apparent not only with the 16 isolates fully sequenced but also with the 145 isolates partially sequenced (data not shown). This relationship between geographic and genetic distances and the associated marked geographic structure would not be apparent if a long-distance spread had occurred. Overall, this indicated that dispersion of the virus during its evolutionary history was gradual. There was no evidence that differences in geographic distribution of the strains along an east-to-west transect across Africa had any sort of selective basis such as adaptation to particular hosts, vectors, or agro-ecologies. This was supported by analyses of the sequences which showed that no single amino acid sites were under positive selection. Considering that these analyses are highly conservative (50, 51), we conclude that adaptation did not play a major role in RYMV evolution. Altogether, RYMV evolutionary history showed features markedly different from those of other plant viruses subjected to similar analyses of full sequences of several representative isolates. In particular, reassortment was found to be critical for Cucumber mosaic virus (35), recombination and long-distance transport by humans were involved for Turnip mosaic virus (44), and adaptive selection was suspected for Potato leafroll virus (17).
Full-genome analyses excluded earlier hypotheses (30), based on coat protein gene sequences, of an adaptation to savanna regions of some isolates from Central Africa (clade 3) and West Africa (clade 4). Actually, conflicts among data partitions appear to be the rule rather than the exception (16, 37). The partial incongruence between ORF4 and the rest of the genome was not due to obvious recombination or positive selection events. Considering the few informative sites in ORF4 supporting the paraphyletic group, the most conservative conclusion is that the partial incongruence is due to a nonuniform distribution of a few substitutions along lineages across the genome. Practically, this incongruence indicates that sequencing the coat protein gene, the most widely used gene in phylogenetic studies of plant viruses, may lead to erroneous evolutionary assumptions. However, for RYMV at least, the coat protein gene is a valid typing tool to assign the isolates to the main clades without ambiguity and to assess intraclade diversity, but it is inappropriate to determine the phylogenetic relationships between some of the clades.
Information on the evolutionary history of viruses can be inferred from the analyses of their spatial genetic structures (14). Our results suggested an origin of RYMV in East Africa, a gradual dispersion and differentiation westward across the continent, and a genetic isolation by distance. Strain interaction may reinforce genetic isolation of the clades. In particular, we found that isolates from clade 5 dominated when coinoculated with isolates from clade 4 (30). This was observed in cultivated rice (30) and in wild grasses (our unpublished data). These interactions may explain the wider distribution of clade 5 over clade 4 in West Africa and the lack of double infection. Genetic isolation of clades due to strain interaction was also suggested for Wheat streak mosaic virus (18).
These results provide information on the conditions of emergence and on the tempo of evolution of RYMV. A recent and substantial long-range virus dissemination by vectors or humans from Kenya since 1966 to the rest of Africa is most unlikely as phylogeographic relationships would not have been preserved if long-distance dispersal had occurred. This is consistent with the lack of efficient biotic and abiotic long-range means of dispersal for RYMV, as there is no seed transmission, a short retention time in the vector, and a low flight ability of the beetle vector. Coevolution between RYMV and cultivated rice is unlikely also. Two species of rice are cultivated in Africa, Oryzae glaberrima and O. sativa. Both are susceptible to RYMV. O. glaberrima was domesticated in West Africa c. 2,500 years ago (31). O. sativa was introduced to Africa from Asia, where RYMV is absent, a few hundred years ago. RYMV evolution characterized by a higher diversity in East Africa does not fit with the longer history of cultivated rice in West Africa. Overall, this suggests that the observed evolutionary history of RYMV developed in primary hosts and colonized cultivated rice only later. The primary hosts are likely to be wild grass species, as the present host range of the virus is limited to the gramineaceous species.
Acknowledgments
We thank A. Ghesquière, J.-F. Guégan, B. D. Harrison, J.-P. Hugot, B. Lafay, J. Maley, J.-C. Pintaud, and J. M. Thresh for helpful discussion and constructive criticism of the manuscript and J. Aribi for technical assistance.
REFERENCES
- 1.Abo, M., A. Sy, and M. Alegbejo. 1998. Rice yellow mottle virus in Africa: evolution, distribution, economic significance and sustainable rice production and management strategies. J. Sust. Agric. 11:85-111. [Google Scholar]
- 2.Abo, M., M. Alegbejo, A. Sy, and S. Misari. 2000. An overview of the mode of transmission, host plants and methods of detection of rice yellow mottle virus. J. Sust. Agric. 17:19-36. [Google Scholar]
- 3.Abubakar, Z., F. Ali, A. Pinel, O. Traoré, P. N′Guessan, J.-L. Notteghem, F. Kimmins, G. Konaté, and D. Fargette. 2003. Phylogeography of Rice yellow mottle virus in Africa. J. Gen. Virol. 84:733-743. [DOI] [PubMed] [Google Scholar]
- 4.Bakker, W. 1974. Characterisation and ecological aspects of rice yellow mottle virus in Kenya. Agric. Res. Rep. (Wageningen) 829:1-152. [Google Scholar]
- 5.Belkhir, K., P. Borsa, L. Chikhi, N. Raufaste, and F. Bonhomme. 2002. GENETIX 4.04, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier, France.
- 6.Blok, J., A. Mackenzie, P. Guy, and A. Gibbs. 1987. Nucleotide sequence comparisons of turnip yellow mosaic virus isolates from Australia and Europe. Arch. Virol. 97:283-295. [DOI] [PubMed] [Google Scholar]
- 7.Bonneau, C., C. Brugidou, L. Chen, R. Beachy, and C. Fauquet. 1998. Expression of the rice yellow mottle virus P1 protein in vitro and in vivo and its involvement in virus spread. Virology 244:79-86. [DOI] [PubMed] [Google Scholar]
- 8.Brugidou, C., C. Holt, M. Ngon, S. Zhang, R. Beachy, and C. Fauquet. 1995. Synthesis of an infectious full-length cDNA clone of rice yellow mottle virus and mutagenesis of the coat protein. Virology 206:108-115. [DOI] [PubMed] [Google Scholar]
- 9.Domingo, E., K. Biebricher, M. Eigen, and J. Holland. 2001. Quasispecies and RNA virus evolution: principles and consequences. Landes Bioscience, Georgetown, Tex.
- 10.Drake, J., and J. Holland. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 96:13910-13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fargette, D., A. Pinel, H. Halimi, C. Brugidou, C. Fauquet, and M. van Regenmortel. 2002. Comparison of molecular and immunological typing of the isolates of Rice yellow mottle virus. Arch. Virol. 147:583-596. [DOI] [PubMed] [Google Scholar]
- 12.Fargette, D., A. Pinel, P. N′Guessan, O. Traoré, Z. Abubakar, C. Brugidou, E. Hébrard, and G. Konaté. 2002. Rice yellow mottle virus and its RNA satellite: genomic organisation, diversity and evolution, p. 10. In Proceedings of the 12th International Congress of Virology. EDK, Paris, France.
- 13.Fauquet, C., and J.-C. Thouvenel. 1977. Isolation of the rice yellow mottle virus in Ivory Coast. Plant Dis. Rep. 61:443-446. [Google Scholar]
- 14.Garcia-Arenal, F., A. Fraile, and J. Malpica. 2001. Variability and genetic structure of plant virus populations. Annu. Rev. Phytopathol. 39:157-186. [DOI] [PubMed] [Google Scholar]
- 15.Grantham, R. 1974. Amino acid difference formula to help explain protein evolution. Science 185:862-864. [DOI] [PubMed] [Google Scholar]
- 16.Graur, D., and W. Li. 2000. Fundamentals of molecular evolution, 2nd ed. Sinauer Associates, Sunderland, Mass.
- 17.Guyader, S., and D. Giblot Ducray. 2002. Sequence analysis of Potato leafroll virus isolates reveals genetic stability, major evolutionary events and differential selection pressure between overlapping reading frame products. J. Gen. Virol. 83:1799-1807. [DOI] [PubMed] [Google Scholar]
- 18.Hall, J., R. French, G. Hei, J. Morris, and D. Stenger. 2001. Three distinct mechanisms facilitate genetic isolation of sympatric wheat streak mosaic virus lineages. Virology 282:230-236. [DOI] [PubMed] [Google Scholar]
- 19.Konaté, G., S. Sarra, and O. Traoré. 2001. Rice yellow mottle is seed-borne but not seed transmitted in rice seeds. Eur. J. Plant Pathol. 107:361-364. [Google Scholar]
- 20.Konaté, G., O. Traoré, and M. Coulibaly. 1997. Characterisation of rice yellow mottle virus isolates in Sudano-Sahelian areas. Arch. Virol. 142:1117-1124. [DOI] [PubMed] [Google Scholar]
- 21.Madison, R., and W. Madison. 2000. MACLADE 4. Sinauer Associates, Sunderland, Mass.
- 22.Malpica, J., A. Fraile, I. Moreno, C. Obies, J. Drake, and F. Garcia-Arenal. 2002. The rate and character of spontaneous mutation in an RNA virus. Genetics 162:1505-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mansour, A., and K. Baillis. 1994. Serological relationships among rice yellow mottle virus isolates. Ann. Appl. Biol. 125:133-140. [Google Scholar]
- 24.Mantel, N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27:209-220. [PubMed] [Google Scholar]
- 25.Mathews, D., J. Sabina, M. Zuker, and D. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 26.Nei, M. 1987. Molecular evolutionary genetics. Columbia University, New York, N.Y.
- 27.Nei, M., and T. Gojobori. 1986. Simple method for estimating the number of synonymous and non-synonymous substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 28.Ngon, A. Y., C. Ritzenthaler, C. Brugidou, C. Fauquet, and R. Beachy. 1994. Nucleotide sequence and genome characterization of rice yellow mottle virus RNA. J. Gen. Virol. 75:249-257. [DOI] [PubMed] [Google Scholar]
- 29.N’Guessan, P., A. Pinel, M. Caruana, R. Frutos, A. Sy, A. Ghesquière, and D. Fargette. 2000. Evidence of the presence of two serotypes of rice yellow mottle sobemovirus in Côte d'Ivoire. Eur. J. Plant Pathol. 106:167-178. [Google Scholar]
- 30.N′Guessan, P., A. Pinel, A. Sy, A. Ghesquière, and D. Fargette. 2001. Distribution, pathogeny and interactions of two strains of Rice yellow mottle virus in forested and savannah zones of West-Africa. Plant Dis. 85:59-64. [DOI] [PubMed] [Google Scholar]
- 31.Oka, H. 1988. Origin of cultivated rice. Development in crop science 14. Japan Scientific Societies Press, Elsevier, Tokyo, Japan.
- 32.Pinel, A., P. N′Guessan, M. Bousalem, and D. Fargette. 2000. Molecular variability of geographically distinct isolates of Rice yellow mottle virus in Africa. Arch. Virol. 145:1621-1638. [DOI] [PubMed] [Google Scholar]
- 33.Pinel, A., O. Traoré, Z. Abubakar, G. Konaté, and D. Fargette. 2003. Molecular epidemiology of the RNA satellite of the Rice yellow mottle virus. Arch. Virol. 148:1721-1733. [DOI] [PubMed] [Google Scholar]
- 34.Roossinck, M. 1997. Mechanisms of plant virus evolution. Annu. Rev. Phytopathol. 35:191-209. [DOI] [PubMed] [Google Scholar]
- 35.Roossinck, M. 2002. Evolutionary history of Cucumber mosaic virus deduced by phylogenetic analyses. J. Virol. 76:3382-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rozas, J., and R. Rozas. 1999. DNAsp version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174-175. [DOI] [PubMed] [Google Scholar]
- 37.Sanderson, M., and H. Shaffer. 2002. Troubleshooting molecular phylogenetic analyses. Annu. Rev. Ecol. Syst. 33:49-72. [Google Scholar]
- 38.Sara, S., and D. Peters. 2003. Rice yellow mottle virus is transmitted by cows, donkeys, and grass rats in irrigated rice crops. Plant Dis. 87:804-808. [DOI] [PubMed] [Google Scholar]
- 39.Sawyer, S. 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6:526-538. [DOI] [PubMed] [Google Scholar]
- 40.Swofford, D. 2000. PAUP: phylogenetic analysis using parsimony, version 4. Sinauer Associates, Sunderland, Mass.
- 41.Sy, A., J. Hughes, and A. Diallo. 2001. Rice yellow mottle virus (RYMV): economic importance, diagnosis and management strategies. West Africa Rice Development Association, Bouaké, Côte d'Ivoire.
- 42.Tamm, T., and E. Truve. 2000. Sobemoviruses. J. Virol. 74:6231-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson, J., D. Higgins, and T. Gibson. 1994. CLUSTAL W. Improving the sensitivity of the progressive multiple sequence alignment through sequence weighting, positions gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tominura, K., A. Gibbs, C. Jenner, J. Walsh, and K. Ohshima. 2003. The phylogeny of Turnip mosaic virus; comparisons of thirty-eight genomic sequences reveal an Eurasian origin and a recent emergence in east Asia. Mol. Ecol. 12:2099-2111. [DOI] [PubMed] [Google Scholar]
- 45.Traoré, O., A. Pinel, D. Fargette, and G. Konaté. 2001. First report and characterization of Rice yellow mottle virus in Central Africa. Plant Dis. 85:920. [DOI] [PubMed] [Google Scholar]
- 46.Van Regenmortel, M., C. Fauquet, D. Bishop, E. Carstens, M. Estes, S. Lemon, J. Maniloff, M. Mayo, D. McGeoch, C. Pringle, and R. Wickner. 2000. Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, New York, N.Y.
- 47.Voinnet, O., Y. Pinto, and D. Baulcombe. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96:14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiller, G. 1998. Phylogenetic profiles: a graphical method for detecting genetic recombination in homologous methods. Mol. Biol. Evol. 15:326-335. [DOI] [PubMed] [Google Scholar]
- 49.Yang, Z. 1997. PAML: a program package for the phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555-556. [DOI] [PubMed] [Google Scholar]
- 50.Yang, Z., and J. Bielawski. 2000. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15:496-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, Z., R. Nielsen, N. Goldman, and A. Pedersen. 2000. Codon-substitution models for heterogenous selection pressure at amino-acid sites. Genetics 155:431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]