Abstract
Alteration in hepatitis B virus (HBV) secretion efficiency may have pathological consequences. Naturally occurring mutations that regulate virion secretion have not been defined. We recently identified HBV genomes displaying high (4B), substantially reduced (3.4), or negative (4C) virion secretion. In the present study, the underlying mutations were mapped. A T552C point mutation in the 4B genome was responsible for its enhanced virion secretion, whereas a G510A mutation in 3.4 and G660C in 4C impaired virus secretion. The three point mutations generate M133T, G119E, and R169P substitutions in the S domains of viral envelope proteins, respectively, without modifying the coding capacity of the overlapping polymerase gene. The mutated residues are predicted to lie in the luminal side of the endoplasmic reticulum (ER) or to be embedded in the ER membrane and thus are not involved in contact with core particles during envelopment. Of the two mutations inhibitory of virion secretion, G510A greatly reduced small envelope protein (hepatitis B surface antigen [HBsAg]) levels both inside cells and in culture medium, whereas G660C specifically abolished HBsAg secretion. Surprisingly, a T484G mutation in the 4B genome, generating an I110M substitution in the S domain, could also reduce HBsAg secretion and block virion secretion. However, its inhibitory effect was suppressed in the 4B genome by the T552C mutation, the enhancer of virion secretion. T552C can also override the inhibitory G510A mutation, but not the G660C mutation. These findings suggest a hierarchy in the regulation of virion secretion and a close link between defective virion secretion and impaired HBsAg formation or secretion.
The replication cycle of the hepatitis B virus (HBV) family consists of pregenomic RNA packaging into core protein particles, minus-strand DNA synthesis, RNA pregenome degradation, and plus-strand DNA synthesis (8). Subsequent interaction of core particles with viral envelope proteins anchored on the endoplasmic reticulum (ER) leads to envelopment and virion secretion. Virion formation is linked to the degree of genome maturation, with preferential secretion of core particles containing double-stranded DNA genomes (23, 26, 30). It apparently requires interaction between exposed residues on the core protein and the cytosolic part of viral envelope proteins, although membrane association of core particles may occur independently of viral envelope proteins (19). Residues in the core protein critical for virion formation, although scattered throughout the linear core protein sequence, occupy a ringlike groove around the base of the spike or a small area at the capsid surface (25). Some of these residues have been found mutated in naturally occurring HBV variants, causing reduced virion secretion or secretion of enveloped particles containing single-stranded viral genomes (16, 31).
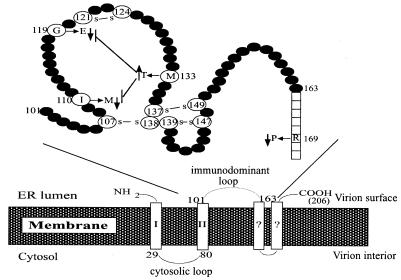
The three envelope proteins (small, middle, and large) are products of alternative translational initiation; hence the middle envelope protein contains 55 extra residues at the N terminus compared to the small envelope protein, whereas the large envelope protein has 108 to 119 more residues than the middle envelope protein. The middle envelope protein is dispensable for virion formation (7). The small envelope protein, historically referred to as hepatitis B surface antigen (HBsAg), is expressed at extremely high levels and may be secreted alone as subviral particles. In addition, it is a component of virions and may be the morphogenic factor. The large envelope protein is not secreted when expressed alone and inhibits the secretion of the small envelope protein (5, 24). This feature may recruit the small envelope protein for virion formation. When anchored on the ER, the entire pre-S domain of the large envelope protein is exposed in the cytosol. A linear sequence in this domain (residues 103 to 124) has been implicated in HBV envelopment and secretion (2, 17). On the other hand, the S domain sequence required for virion secretion is less clear. According to the proposed topology of the small envelope protein, most of the C terminus of HBsAg is buried in the membrane (Fig. 1). Residues 29 to 79 form the major cytoplasmic loop and therefore interact with core particles (Fig. 1). Residues 101 to 163, which are located inside the ER lumen, resurface on secreted virions and are thus called the immunodominant loop. This region contains eight cysteine residues that may participate in the formation of intramolecular as well as intermolecular disulfide bonds (20) (Fig. 1). These events may be important for oligomerization of HBsAg, leading to its secretion. The major antigenic epitope in the immunodominant loop is called the α determinant and is composed of residues 124 to 147.
FIG. 1.
Proposed topology of HBV small envelope protein. The lower panel depicts the 206-amino-acid small envelope protein traversing the ER membrane, probably four times. The major cytosolic loop (residues 29 to 80) and luminal loop (residues 101 to 163) are shown. In secreted virions, the luminal loop turns up on the viral surface and is called the immunodominant loop. Its detailed structure, as described by Wallace and Carman (29), is shown above. We have identified in this study four naturally occurring mutations in the S domain that enhance or impair virion secretion. Note that the M133T mutation can suppress the inhibitory effect of G119E and I110M mutations on virion secretion.
Two research groups have performed deletional analysis of the cytosolic component of the envelope protein. While the C-terminal sequence was found to be essential for the formation of subviral particles and hence also mandatory for virion formation, residues 29 to 59 were dispensable for hepatitis delta virus virion formation (11) yet critical for the production of HBV virions (18). Considering that liver injury associated with HBV infection is mediated in large part by the host immune response, the secretion efficiency of an HBV strain may greatly affect viral pathogenicity. In support of this view, an HBV genome associated with fulminant hepatitis was found to be impaired in virion secretion (12). During the characterization of naturally occurring core promoter mutants of genotype A (22), we identified clones with a high-virion-secretion phenotype (4B and 5.2) as well as those with impaired virion secretion (3.4 and 4C) (Table 1). In the present study, the mutations responsible were mapped to the S gene. Surprisingly, clone 4B harbored not only a mutation enhancing virion secretion but also an inhibitory mutation, although its effect was suppressed in the presence of the enhancer. A close correlation between alteration in virion secretion and change in HBsAg expression or secretion was observed.
TABLE 1.
Properties of HBV clones used in this study
| Clone | Presence or level of:
|
||||
|---|---|---|---|---|---|
| Core promoter mutations | Genome replication | Virion secretion | HBeAg secretion | HBsAg secretion | |
| 3.4 | + | +++ | +/−a | ++++ | +/− |
| 4B | + | ++++ | +++ | ++ | ++++ |
| 4C | + | ++++ | +/− | ++ | − |
| 5.2 | − | + | ++ | ++++ | ++++ |
| 6.2 | − | + | + | ++++ | ++++ |
| 7.2 | − | + | +/− | +++ | ++++ |
| 7.4 | − | + | + | ++++ | ++++ |
+/−, borderline positive.
MATERIALS AND METHODS
Constructs.
The parental clones for this study, 3.4, 4B, 4C, 5.2, 5.4, 6.2, 7.2, and 7.4 (Table 1), have been described (22). These are full-length clones of genotype A HBV amplified from serum samples of hepatitis B e antigen (HBeAg)-positive French patients through high-fidelity PCR. Clones 3.4, 4B, and 4C originated from low-viremia samples (4B and 4C were from the same patient), whereas the other clones were obtained from highly viremic samples. Clone 5.4 is defective in viral DNA replication due to a single nucleotide deletion in the core gene and thus served as a negative control. Clone 6.2 is considered to have a wild-type genome since it was isolated from a patient with high viremia titer (and hence likely at early HBeAg phase of infection) and contained wild-type sequences in both the core promoter region and the S gene. Moreover, the genome replication, virion secretion, and HBsAg/HBeAg secretion phenotypes of 6.2 are typical of those of wild-type clones. Mutations that alter virion secretion were mapped through the construction of chimeric clones, which was facilitated by the AvrII, EcoRV, RsrII, and ApaI sites that are unique on the HBV genome but absent on the pUC18 vector (Fig. 2). When partial replacement of a restriction fragment was intended, the chimeric sequence encompassing the restriction fragment was generated by overlap extension PCR (Expand high-fidelity PCR system; Roche) and subsequently digested with the two restriction enzymes. This methodology was also employed for the creation of site-specific mutants. Each construct was sequenced to confirm its chimeric nature, and complete sequencing of the insert was performed when the restriction fragment was derived from PCR products. The hybrid genomes were converted into tandem dimers in the EcoRI site of the pUC18 vector as described previously (1, 22). Once the secretion determinant was mapped to the AvrII-EcoRV restriction fragment, 1.5-mer genomes of HBV clones 3.4, 4B, and later 6.2 were generated in the pBluescript vector. This allowed us to create additional mutant constructs simply by replacement of the AvrII-EcoRV fragment, since both AvrII and EcoRV cut only once on the 1.5-mer genome. To make the 1.5-mer HBV genome, the 2.2-kb EcoRV-EcoRI fragment of the HBV genome was first cloned into the SmaI-EcoRI sites of the pBluescript vector. Next, the 2.6-kb EcoRI-ApaI HBV fragment was inserted into the EcoRI-XhoI sites of the initial construct (the ApaI end of HBV was rendered blunt by T4 DNA polymerase and deoxynucleoside triphosphate [dNTP], while the XhoI end of the initial construct was flushed by Klenow fragments in the presence of dNTP).
FIG. 2.
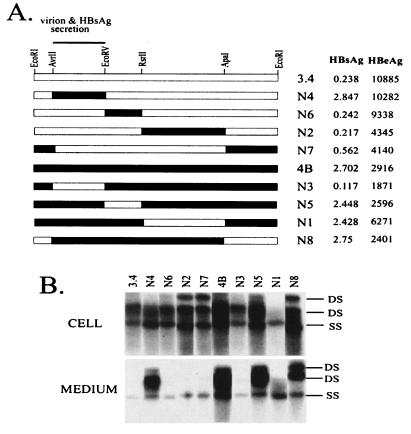
Mapping of the virion secretion determinant to the AvrII-EcoRV restriction fragment. (A) Cartoon view of the constructs. Blank box, 3.4 sequence; filled box, 4B sequence. The AvrII-EcoRV restriction fragment determines the low-virion-secretion and low-HBsAg-secretion phenotypes of clone 3.4. Dimers of HBV constructs (10 μg) were transfected into Huh7 cells grown in 6-cm dishes. Cells and culture supernatant were harvested 5 days later. Shown on the right are the HBsAg and HBeAg values from 1:5-diluted culture medium, with values from nontransfected cells subtracted. (B) HBV DNA replication and virion secretion analyzed from the same experiment. Cell, intracellular HBV DNA isolated from core particles; medium, extracellular HBV DNA associated with Dane or core particles; DS, double-stranded DNA (relaxed circular or linear); SS, single-stranded DNA. For extracellular particles, DS corresponds to Dane particles while SS segregates with naked core particles (22).
Transfection and analysis of DNA replication and virion secretion.
The dimers and 1.5-mers of the HBV genome were purified by a commercial kit (Marligen), further extracted with phenol and chloroform-isoamyl alcohol, precipitated with ethanol, and dissolved in Tris-EDTA buffer. DNA was transfected into the Huh7 human hepatoma cell line by the calcium phosphate method (Promega) using 10 μg of dimer DNA and 2 μg of luciferase DNA per 6-cm-diameter dish of cells. Cells were harvested 5 days posttransfection (1, 22). Later experiments employed the Lipofectamine transfection method (Mirus), which gave much higher transfection efficiency. The transIT-LT1 reagent from Mirus (4 μl) was mixed with serum-free minimal essential medium (150 μl), vortexed, and incubated at room temperature for 15 min. Following addition of 1.5 μg of DNA and a further incubation for15 min, the complex was added in dropwise fashion to cells seeded in six-well plates (at 40 to 80% confluence). Medium (2 ml/well) was replaced 4 to 16 h later, and cells were harvested 5 days later. The cell pellet was used for the luciferase assay as well as for the extraction of core particle-associated HBV DNA (22, 28). Extracellular viral particles were concentrated from 1.8 ml of culture supernatant by ultracentrifugation through 10 and 20% sucrose cushions, at 39,000 rpm (Sorvall TH-641 rotor) and 4°C for 18 h. Following nuclease treatment and proteinase K digestion, DNA was extracted. Viral DNA derived from both intracellular and extracellular viral particles was separated by 1% agarose gel, omitting ethidium bromide during electrophoresis. As a result, the double- and single-stranded viral genomes ran as sharp bands close to each other (22). DNA was transferred to a GeneScreen Plus membrane (Perkin-Elmer) and hybridized with a highly pure HBV DNA probe. Densitometric analysis was performed with National Institutes of Health IMAGE, version 1.61, software.
When separation of virions from naked core particles was needed, core particles and virions that had been pelleted down through 10 and 20% sucrose cushions were subjected to ultracentrifugation in a CsCl gradient at 46,000 rpm (Sorvall AH-650 rotor) for 48 h (22). Fractions of 400 μl were taken from the top, weighed to determine density values, and dialyzed against TEN (10 mM Tris, pH 8.0, 1 mM EDTA, 150 mM NaCl) buffer. Following nuclease treatment and proteinase K digestion, HBV DNA was analyzed by Southern blotting.
Measurement of HBeAg and HBsAg.
A small fraction of the culture supernatant (3 to 40 μl) was diluted with phosphate-buffered saline to 200 μl for the detection of HBsAg (Auszyme kit; Abbott) and HBeAg (EBK kit; DiaSorin). In later experiments, we also used 1/100 to 1/10 of the cell lysate to measure intracellular HBsAg.
RESULTS
The AvrII-EcoRV restriction fragment of the HBV genome controls differential virion secretion between clones 3.4 and 4B.
Both 3.4 and 4B are core promoter mutants. Clone 3.4 has a slightly lower replication capacity than 4B (Table 1). It secretes much higher levels of HBeAg than 4B but very low levels of HBsAg. This clone is also characterized by a mutated pre-S2 initiation codon (ATA). The presence of four unique restriction sites in the HBV genome in addition to the EcoRI site enabled us to divide the genome into four segments (Fig. 2A). Individual restriction fragments of the 3.4 genome were replaced with cognate sequences from clone 4B, generating chimeric constructs N2, N4, N6, and N7. In the reverse approach, the four fragments of the 4B genome were swapped with sequence derived from 3.4 (constructs N1, N3, N5, and N8). Such a two-way exchange strategy allowed reliable mapping of the secretion determinant. Tandem dimers of the eight chimeric constructs were transfected in parallel with the parental clones into Huh7 cells. A dimer of clone 5.4 which was defective in genome replication due to a single nucleotide deletion in the core gene (22) was routinely used as a negative control for the transfection experiments. No HBV DNA was detected in lysate of 5.4-transfected cells or in the corresponding culture supernatant (data not shown). Virions in the culture supernatant were concentrated by ultracentrifugation through a sucrose cushion before DNA extraction and Southern blot analysis. Although this method also precipitates naked core particles (which are artifacts of human hepatoma cells), we have previously established that they contained single-stranded viral genomes, rather than the double-stranded DNA associated with virions (22). Clone N1 showed limited HBV signals both intracellularly and extracellularly, as well as the lowest luciferase level (Fig. 2B). Results from the other seven constructs clearly implicated the AvrII-EcoRV fragment as the sole determinant for differential virion secretion. Thus, all four of the constructs with a 3.4 sequence in this region (N2, N3, N6, and N7) displayed weak signals of double-stranded DNA in the culture supernatant, in contrast to constructs with 4B-derived AvrII-EcoRV fragments (N4, N5, and N8). This fragment, which codes for most of the S domain (residues 9 to 226), is also responsible for reduced HBsAg secretion by clone 3.4. In contrast, the RsrII-ApaI fragment, encompassing core promoter and precore/core coding sequence, determines HBeAg expression as anticipated.
Secretion phenotypes of 3.4 and 4B are interconvertible through exchange of genomic fragment 520-730.
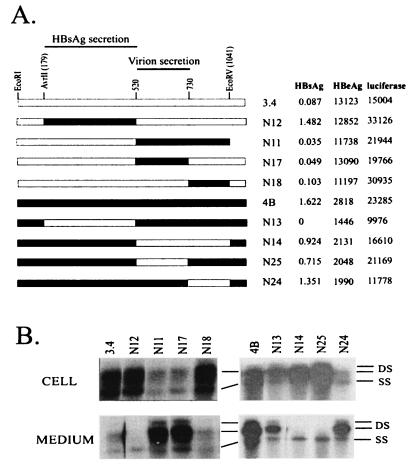
Conversion of a dimer to a monomeric form during restriction fragment replacement necessitates dimer making for each new construct. Considering that the S gene is represented only once in the pregenomic RNA, we generated 1.5-mer versions of 3.4 and 4B genomes that survived AvrII/EcoRV double digestion (see Materials and Methods). When transfected into Huh7 cells, each 1.5-mer reproduced the replication, virion secretion, and HBsAg secretion properties of its dimeric version (data not shown). Replacing the 179 (AvrII)-to-520 region of the 3.4 genome with 4B restored HBsAg secretion to the level of clone 4B yet still failed to rescue the virion secretion defect (N12; Fig. 3). On the other hand, substitution of fragment 520-1041 (EcoRV) rescued virion secretion without restoring extracellular HBsAg levels (N11). Reciprocal experiments with 4B-based constructs corroborated assignment of a determinant(s) for HBsAg production or secretion to 179-520 and virion secretion to 520-1041 (N13 and N14; Fig. 3).
FIG. 3.
Exchange of genomic fragment 520-730 switches the virion secretion phenotypes between clones 3.4 and 4B. (A) Cartoon view of the constructs. The low-HBsAg-secretion phenotype of 3.4 is controlled by the 179-to-520 region, whereas efficiency of virion secretion is reciprocated by exchange of the 520-to-730 region between 3.4 and 4B. The experimental conditions are similar to those described for Fig. 2, except that 1.5-mer genomes were used. The HBsAg, HBeAg, and luciferase values are shown to the right (with 3.4-based constructs from one transfection experiment and 4B-based constructs from another experiment). (B) HBV DNA replication inside transfected cells and virion secretion into the culture medium. DS, double-stranded DNA; SS, single-stranded DNA.
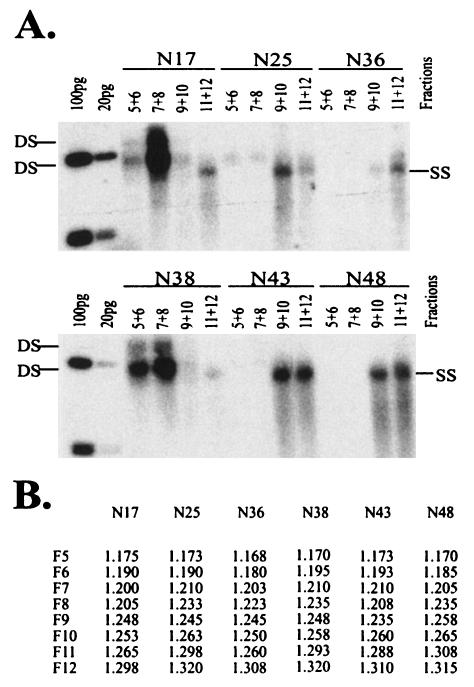
Four more constructs were used to further define the mutations that affect virion secretion. Replacing the 520-to-730, but not the 731-to-1041, segment of the 3.4 genome with the 4B sequence restored efficient virion secretion (N17 and N18; Fig. 3). Conversely, replacement of that particular 4B sequence by 3.4 impaired virion secretion (N25 and N24). To reaffirm that single- and double-stranded DNAs derived from extracellular particles correspond to core particles and virions, respectively, culture supernatant of N17- and N25-transfected cells was subject to centrifugation through a CsCl gradient. Southern blot analysis of gradient fractions confirmed efficient virion secretion by the N17 construct in contrast to that by N25 (Fig. 4).
FIG. 4.
Separation of extracellular virions from core particles by CsCl gradient. Virions and core particles were first concentrated from culture supernatant by centrifugation through 10 and 20% sucrose. Next, the two types of particles were separated by centrifugation through a CsCl gradient. Thirteen or 14 fractions were collected from the top, weighed, and dialyzed. DNA was extracted from fractions 5 to 12 and analyzed by Southern blotting using pooled samples (fractions 5+6, 7+8, 9+10, and 11+12). (A) Southern blots showing efficient virion secretion by the N17 and N38 constructs but not the other four constructs. Irrespective of the constructs used, double-stranded DNA is always associated with Dane particles (fractions 5 to 8), while single-stranded DNA is present in core particles (fractions 9 to 12). (B) Buoyant densities of these fractions.
The T552C mutation in the 4B genome is responsible for the high-secretion phenotype.
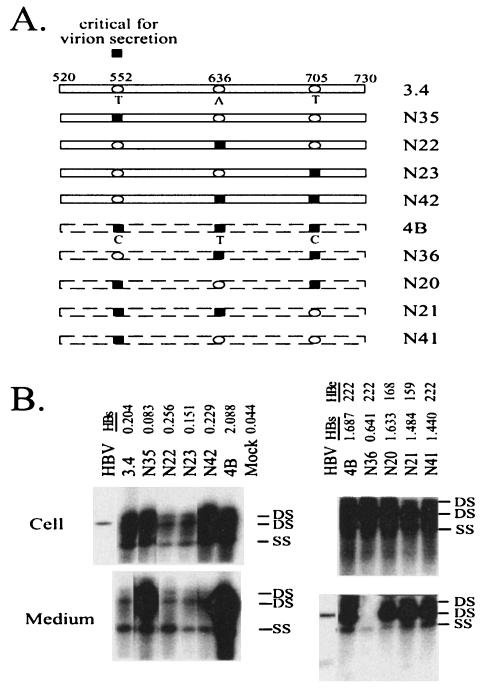
Alignment of genomic sequence of 3.4 with that of 4B revealed three nucleotide substitutions within 520-730: 552T, 636A, and 705T versus 552C, 636T, and 705C. The 3.4 sequence but not the 4B sequence conforms to consensus genotype A (21). The triple mutation caused M133T, Y161F, and V184A substitutions in HBsAg but was silent in the overlapping polymerase gene (Table 2). It is noteworthy that F161 is the consensus sequence for genotypes C, D, and E, while A184 is the consensus sequence for genotype E and also present in some genotype C strains (21). Therefore only M133T (or T552C at the nucleotide level) is a true mutation. To determine which divergent position(s) was critical for the observed secretion phenotype, we generated site-directed mutants from both 3.4 and 4B backgrounds. Reversion of nucleotide 636 or 705 or both to the wild type failed to perturb the secretion efficiency of clone 4B (N20, N21, and N41; Fig. 5). On the other hand, a C-to-T back mutation at position 552 abolished virion secretion (N36; Fig. 4 and 5). Thus, the T552C mutation at the nucleotide level, or the M133T substitution at the envelope protein level, is necessary and sufficient for the high-secretion phenotype of clone 4B. Consistent with this, introducing the T552C mutation, but not mutations at 636 or 705 or both, enhanced virion secretion of the 3.4 clone (compare N35 with N22, N23, and N42; Fig. 5).
TABLE 2.
Mutations in the S genes of HBV genomes and their impact on protein translation
| Clone(s) | Mutationa in:
|
||
|---|---|---|---|
| Nucleotide | S domain | Polymerase | |
| 3.4 | T176C | F8L | I363T |
| G510A | G119E | Silent | |
| T777C | I208T | Silent | |
| 4B and 4C | A273G | N40S | Silent |
| T287G | S45A | I400S | |
| T484G | I110M | S466A | |
| A496G | Silent | N470D | |
| T552C | M133T | Silent | |
| A636T | Y161F* | Silent | |
| T705C | V184A* | Silent | |
| G779T | V209L | R564L | |
| T784G | S210R | S566A | |
| 4C (not 4B) | G660C | R169P | Silent |
| 5.2 | T216C | L21S | Silent |
| T766A | S204R | S560T | |
Boldface, mutations that modulate virus secretion; *, wild-type sequence in some other genotypes.
FIG. 5.
The T552C mutation is responsible for efficient virion secretion by 4B and can confer a high-secretion phenotype to 3.4. (A) Cartoon view of the constructs. Clones 3.4 and 4B differ at three positions within the 520-to-730 region: 552, 636, and 705. Open ovals, wild-type 3.4 sequences; black squares, mutated 4B sequences. Eight site-directed mutants were prepared and tested. (B) HBV 1.5-mer DNA (1.5 μg) was transfected into Huh7 cells grown in six-well plates with the Mirus reagents and harvested 5 days later. HBsAg and HBeAg from culture supernatant were measured after 40- and 33-fold dilutions, respectively. HBV DNA replication inside transfected cells and virion secretion into the culture medium were analyzed by Southern blotting. DS, double-stranded DNA; SS, single-stranded DNA; HBV, a mixture of the EcoRI-linearized HBV genome (3.2 kb) and EcoRI/RsrII double-cut HBV DNA (1.7 and 1.5 kb).
HBV clones with altered virion secretion often harbor missense mutations in the S gene.
The 0.8-kb AvrII-EcoRV fragment covers most of the S gene but does not encompass the pre-S regions (Fig. 6). Although the enhanced secretion capacity of clone 4B could be attributed to the M133T mutation, the defect in clone 3.4 could not have been imposed by a wild-type sequence in this region. However, 3.4 contained substitutions in the S domain (F8L, G119E, and I208T; Fig. 6) owing to the T176C, G510A, and T777C mutations in the viral genome (Table 2), which might account for the impairment of virion secretion. To explain the unusual results shown in Fig. 3, we hypothesized that both the 3.4 and 4B genomes harbored secretion-inhibitory mutations, possibly in the same region, which were nevertheless suppressible by the M133T mutation (T552C at the nucleotide level).
FIG. 6.
Comparison of S domain sequences among HBV genomes with different virion secretion efficiencies. WT, wild-type consensus sequence of genotype A (21). Such a wild-type sequence is found in clones 6.2, 7.2, and 7.4. For other clones, only mutated residues are given. The landmarks for fragment exchange (AvrII and EcoRV sites, nucleotide positions 520 and 730) are indicated. Underlined is a stretch of amino acid residues specified by fragment 520-730, the determinant for differential virion secretion between 3.4 and 4B.
We sequenced several clones with wild-type sequences in the core promoter region (22) to find one with a wild-type S gene sequence for construction of chimeras. In addition, clones with abnormal secretion phenotypes were sequenced to evaluate the significance of mutations in the S gene (Table 1). Two clones with normal secretion profile (6.2 and 7.4) and a clone with defective virion secretion (7.2) were found to have wild-type nucleotide sequences in this region. Thus, the mutation responsible for impaired virion secretion of clone 7.2 must lie elsewhere in the viral genome. A clone with high virion secretion efficiency (5.2) harbored two amino acid substitutions (L21S and S204R) (Fig. 6). Clone 4C, which was derived from the same individual as clone 4B yet was defective in virion secretion and HBsAg expression (22), contained an additional R169P substitution in the S domain owing to a G660C mutation (Table 2 and Fig. 6). Otherwise the entire pre-S region and promoter region were found to be identical to those of the 4B genome.
Both 3.4 and 4B genomes harbor mutations inhibitory of virion secretion at the 179-to-520 region.
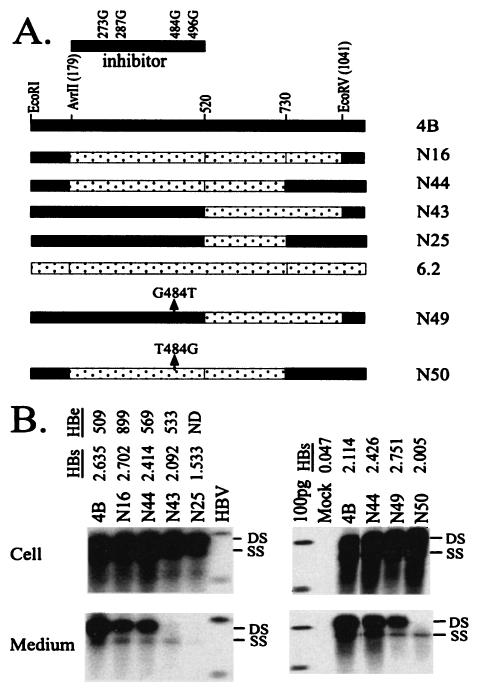
To understand why a wild-type 520-730 sequence was associated with defective virion secretion in the 4B background (N25; Fig. 3), we replaced the entire AvrII-EcoRV fragment of clone 4B with the wild-type sequence. Interestingly, this construct retained one-half of the virion secretion efficiency of 4B (N16; Fig. 7). This result not only confirms enhancement of virion secretion by the T552C mutation (by twofold) but also argues against the wild-type fragment 520-730 sequence as an inhibitor of virion secretion. Experiments with additional chimeric constructs implicated the 179-to-520 region in clone 4B as an inhibitor of virion secretion when the 520-to-730 region is wild type (N43 and N44; Fig. 7). Four nucleotide changes (A273G, T287G, T484G, and A496G) were found in this part of the 4B genome, resulting in three amino acid substitutions in the S domain (N40S, S45A, and I110M) (Table 2 and Fig. 6). To determine whether mutations in the cytosolic loop (N40S and S45A) or the immunodominant loop (I110M) inhibited virion secretion, we mutated genomic position 484, part of the 110th codon of the S gene. Indeed, a G-to-T back mutation at this position of the N43 genome restored virion secretion (N49; Fig. 7), whereas a T484G mutation in N44 abolished virion secretion (N50). Thus, the T484G mutation in the HBV genome, corresponding to the I110M substitution in the S gene, blocked virion secretion.
FIG. 7.
Identification of a point mutation in clone 4B that inhibits virion secretion. (A) Cartoon view of the constructs. The inhibitor of virion secretion is located in the AvrII (179-to-520) region, which contains four nucleotide changes. Since the T552C mutation can suppress the inhibitory mutation, constructs in this series all had the wild-type fragment 520-730 sequence. Construct N49 was derived from N43 by reversion of position 484 back to T; construct N50 differs from N44 by a T484G mutation. (B) HBV DNA replication inside transfected cells and virion secretion into the culture medium. DS, double-stranded DNA; SS, single-stranded DNA; HBV, 3.2-, 1.7-, and 1.4-kb HBV DNA. Also shown are HBsAg and HBeAg values from culture supernatants of transfected Huh7 cells (diluted 13-fold for results shown on the left and 25-fold for HBsAg values shown on the right).
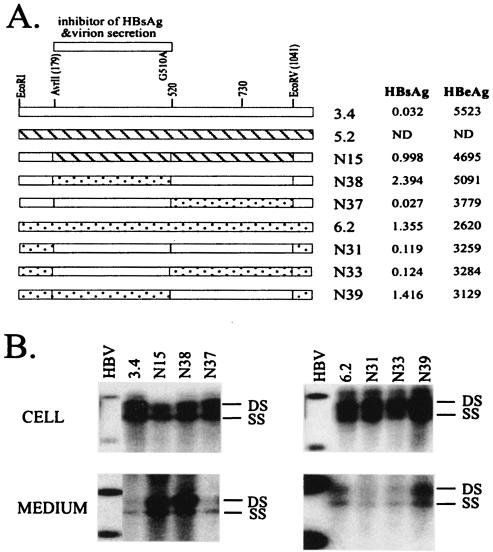
Replacement of the AvrII-EcoRV fragment of 3.4 genome with the sequence from clone 5.2 (containing the wild-type 520-to-730 region) significantly improved virion secretion (N15; Fig. 8), reinforcing the hypothesis that a wild-type fragment 520-730 sequence was not detrimental to virion secretion. Replacement of fragment 179-520, but not 520-1041, of the 3.4 genome with the wild-type (6.2) sequence improved both virion secretion and HBsAg expression, thus implicating the single point mutation within 179-520, G510A, as responsible for reduced HBsAg expression and virion secretion (N37 and N38; Fig. 4 and 8). This point mutation produced a G119E substitution in the S domain but was silent in the polymerase gene (Table 2). The role of the G510A mutation was validated in the reverse experiment, where its introduction to clone 6.2 impaired both HBsAg expression and virion secretion (N31, N33, and N39; Fig. 8).
FIG. 8.
Mapping the mutation in clone 3.4 that inhibits HBsAg and virion secretion. (A) Cartoon view of the constructs. The 179-to-520 region of clone 3.4, which contains a single G510A point mutation, inhibits both HBsAg and virion secretion. Experiments were performed with both 3.4-based constructs (N15, N38, and N37) and 6.2-based constructs (N31, N33, and N39). Shown on the right are HBsAg and HBeAg values from culture supernatants of transfected Huh7 cells diluted 13-fold, following subtraction of values for mock-transfected cells. (B) HBV DNA replication inside transfected cells and virion secretion into the culture medium. DS, double-stranded DNA; SS, single-stranded DNA; HBV, 3.2-, 1.7-, and 1.4-kb HBV DNA.
The presence of a mutation(s) inhibitory to virion secretion within the 179-to-520 regions of both the 3.4 and 4B genomes (positions 510 and 484, respectively) explains why the inhibitory mutation of clone 3.4 was erroneously mapped to the 520-to-730 region with 3.4/4B chimeras (Fig. 3). These findings, taken together, lead us to believe that the T552C mutation of 4B can override the secretion-inhibiting mutation present in the same genome (T484G) or the G510A mutation present in the 3.4 HBV strain.
A single point mutation in the 4C genome impaired secretion of both viral and subviral particles.
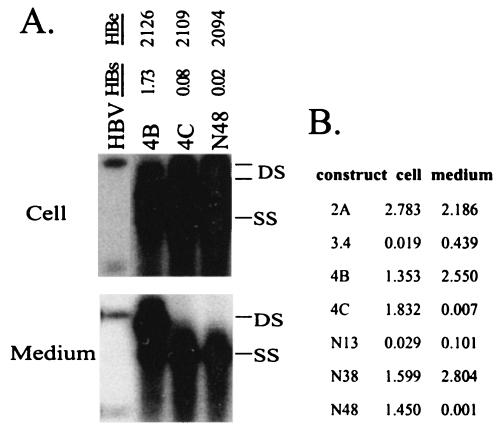
To determine whether the unique R169P mutation present in clone 4C was responsible for its lack of virion secretion, this mutation was introduced into 4B by replacement of the AvrII-EcoRV fragment. The resultant construct, N48, behaved similarly to clone 4C in having undetectable levels of virions and HBsAg in culture supernatant (Fig. 4 and 9A). Surprisingly, normal levels of HBsAg were detected in cell lysate, suggesting a specific block in HBsAg secretion (Fig. 9B). In contrast, 3.4 and its derivative (N13) produced low HBsAg levels both inside and outside transfected Huh7 cells.
FIG. 9.
A single extra nucleotide in clone 4C is responsible for its impaired HBsAg secretion and virion secretion. (A) Clone 4C and construct N48 are defective in virion secretion. N48 is a 4B-based construct with the AvrII-EcoRV fragment derived from 4C. It differs from 4B by a single G660C mutation. HBsAg and HBeAg were measured after 1:40 and 1:10 dilutions, respectively, and values from mock-transfected cells were subtracted. DS, double-stranded DNA; SS, single-stranded DNA. (B) HBsAg expression and secretion by the mutants. HBsAg from 1:25-diluted culture supernatant as well as 1/25 of the lysate of Huh7 cells grown in six-well plates was measured 5 days following transfection. Note that cells transfected with 3.4 and N13 displayed low HBsAg levels both in cell lysate and culture medium, whereas 4C and N48 had a specific block in HBsAg secretion.
The efficiency of virion secretion is correlated with the efficiency of HBsAg secretion.
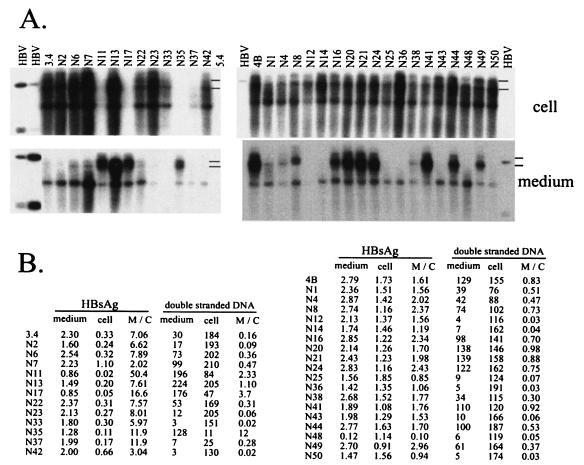
To search for a possible link between the efficiency of virion secretion and HBsAg secretion in other constructs, we tested the majority of constructs in two series: those with high HBsAg levels (derived from 4B or 6.2; Fig. 10, right) and those with low HBsAg levels (based on 3.4; Fig. 10, left). Both secreted HBsAg and intracellular HBsAg were measured, and ratios of extracellular/intracellular (M/C) signals were calculated. Meanwhile, levels of double-stranded viral DNA in both cell-associated core particles and extracellular particles were quantified by densitometric analysis, and the M/C ratios were obtained. Interestingly, there was a general correlation between the efficiency of HBsAg secretion and virion secretion (Fig. 10B). For example, 3.4-based constructs with enhanced virion secretion (N11, N17, and N35) had higher HBsAg M/C ratios than 3.4. Similarly, impaired virion secretion in 4B-based constructs was accompanied by reduced HBsAg secretion (N14, N25, and N36), although the reduction was modest. Finally, gain of virion secretion in N49 compared with N43 was correlated with an increased HBsAg M/C ratio, whereas loss of virion secretion in N50 in comparison with N44 was associated with a reduced M/C ratio. Thus, mutations identified in this study modulate the secretion of both viral and subviral particles, although the impact on virion secretion was much severer.
FIG. 10.
Quantitative analysis of virion secretion as well as HBsAg expression and secretion. (A) Southern blot analysis of intracellular HBV DNA (top) and extracellular HBV DNA (bottom). Constructs with low HBsAg expression were grouped on the left, while those with high HBsAg expression were analyzed in a separate experiment (right). Positions of double-stranded DNA are indicated. (B) Quantitative analysis. For 3.4-based constructs (left), HBsAg was analyzed from 40 μl of culture supernatant and 20 μl (1/10) of cell lysate. For 4B- and 6.2-based constructs, HBsAg was measured from 4 μl of culture supernatant and 2 μl (1/100) of cell lysate. Double-stranded HBV DNA in the Southern blots was quantified by densitometric analysis of X-ray films (with unsaturated signals). Values from a same-size spot lacking specific HBV signals were subtracted.
DISCUSSION
Based on these results, we conclude that virion secretion is markedly reduced by the G510A mutation derived from 3.4 and abolished by the T484G mutation from the 4B genome, as well as the G660C mutation from 4C. In contrast, the T552C mutation from 4B enhances virion secretion and suppresses the activity of both the G510A mutation from 3.4 and the T484G mutation from 4B. Currently the cis effect of T484G, G510A, T552C, and G660C mutations on virion secretion has not been excluded experimentally, but these mutations most likely function in trans through amino acid changes in the S domains of viral envelope proteins: I110M, G119E, M133T, and R169P, respectively (Table 2). The S gene overlaps with the coding sequence for the reverse transcriptase (RT) domain of the viral polymerase. However, these mutations do not induce amino acid changes in the RT domain except T484G, which causes S466A substitution in the polymerase (Table 2). Mutations in the polymerase inhibit virion secretion when they block viral DNA replication at an early step, such as minus-strand DNA synthesis (30). In this regard, efficient synthesis of double-stranded DNA was observed for all the constructs studied here.
Although parental clones harboring these mutations, 3.4, 4B, and 4C, were obtained by PCR (22), the high-fidelity PCR system that we used in the previous study contained the proofreading enzyme to greatly reduce PCR errors. We are confident that the secretion-modifying mutations identified in this study are truly naturally occurring rather than artifacts of PCR. For example, the I110M and M133T mutations are present in clones 4B, 4C, and 8.22 and the M133T mutation has been detected in HBsAg-negative patients at extremely high frequency (9). The G119E mutation is present in clone 1B, which has a full-length sequence identical to that of 3.4 (22). This mutation is also present in one of the genotype A clones compiled by Norder and colleagues (21). All of the four point mutations (M133T, I110M, G119E, and R169P) can be found in the GenBank deposit of HBV sequences.
From the topological point of view, residues 110, 119, and 133 are all located in the first loop of the immunodominant region (with residue 133 inside α determinant), while residue 169 is predicted to be membrane associated (Fig. 1). Therefore, the mutations probably do not affect the interaction of envelope proteins with core particles, which requires the cytosolic loop (residues 29 to 80) of the S domain. The S domain is present in all the three types of envelope proteins: large, middle, and small. At present we do not know whether the S domain mutations mentioned above influence virion secretion through the small or large envelope protein and whether the mutant proteins are dominant over wild-type envelope proteins when coexpressed. In this regard, artificial mutations in the cytosolic loop of either the large or small envelope protein inhibited virion secretion, although a mutated large envelope protein did not impair virion secretion completely (18).
How could the I110M, G119E, and R169P mutations inhibit virion secretion? Analysis of HBsAg expression and secretion patterns has provided clues to answer this question. The R169P mutation did not affect intracellular levels of HBsAg but completely blocked its secretion (N48; Fig. 9B). Thus, a defect in virion secretion may be a secondary event. The subcellular compartment where the R169P mutant protein is trapped remains to be determined. Concerted reduction in the secretion of HBsAg and virions has also been reported for an HBV strain implicated in fulminant hepatitis, although several mutations other than R169P were implicated (12). Similar to the R169P mutation, the I110M mutation also impaired HBsAg secretion; however, the effect was very mild (N43, N44, N49, and N50; Fig. 10B).
The G119E mutation, in contrast, strikingly reduced HBsAg levels in both cell lysate and culture supernatant, thus arguing against a specific defect in the secretion of subviral particles. In fact, the reduction was even more pronounced in cell lysate than in culture medium (Fig. 9B and 10B), suggesting enhanced HBsAg secretion. Another naturally occurring HBV variant with reduced virion secretion was found to express similarly reduced levels of HBsAg (10). The remarkable effect of a single missense mutation in the S gene on its expression (or degradation) is surprising. Presumably, low HBsAg levels of the G119E mutant may have been artifacts of impaired recognition of the mutant by the antibodies used in the assay. Residue 119 is located in the first extracellular loop of HBsAg (Fig. 1), where substitution in numerous residues, including those at nearby positions such as 116, 118, 120, and 122, could alter the α determinant and reduce HBsAg antigenicity (3, 4, 15, 27, 29). The nonconservative change from glycine to glutamic acid increases the negative charge and may induce a major conformational change considering its proximity to C121, the cysteine residue involved in an intramolecular disulfide bond with C124 (Fig. 1). Nevertheless, the strong secretion efficiencies but low HBsAg titers of the N11 and N17 constructs (Fig. 3) suggest that a secondary mutation in the first extracellular loop (M133T) can rescue the virion secretion defect without correcting structural changes brought about by the G119E substitution.
How could the M133T (T552C) mutation suppress the inhibitory effect of the G119E (G510A) mutation from 3.4 as well as the I110M (T484G) mutation of the 4B genome, but not the R169P (G660C) mutation from 4C? In this regard, residues 110, 119, and 133 are all clustered in the loop formed by the Cys107-Cys138 disulfide bond, whereas residue 169 is more distally located in a membrane-bound segment (Fig. 1). It is tempting to suggest that the suppressive effect of the M133T mutation is linked to its proximity to residues 110 and 119. Results for constructs N35 and N36 suggest that the M133T mutation can enhance HBsAg secretion, which may be intricately linked to its ability to enhance virion secretion (Fig. 10B). At any rate, the functional interaction among these mutations underline the danger of predicting virion secretion phenotype based on isolated studies of individual point mutations. Analogous to our findings, the naturally occurring I97L mutation in the core protein causes “immature” secretion of virions containing single-stranded genomes (31), which is nevertheless suppressed by other coevolving mutations in the core protein, such as P5T and P130T (6, 32).
While the manuscript was under review, Kalinina and colleagues reported that a virion secretion defect of the fulminant hepatitis virus strain could be rescued by coexpressing the wild-type small, but not the large, envelope protein (13). They also found that the G145R immune escape mutation, which is present in the fulminant hepatitis strain, reduced virion secretion by about fourfold (14). These observations and our present findings highlight the important role of mutations in the immunodominant loop of small envelope protein in modulating virion secretion.
Acknowledgments
This work was supported by grants CA35711, AA20169, p20RR15578, CA095490, AI054535, and DK062857 from the National Institutes of Health and from Life Span Research Funds. It was also supported by an Alpha Omega Alpha Student Research Fellowship (to N.K.), Brain Korea 21 Project for Medical Science (to S.H.A.), and Undergraduate Teaching and Research Assistantship from Brown University (to G.B.). J.L. was a Liver Scholar from the American Liver Foundation.
REFERENCES
- 1.Ahn, S., A. Kramvis, S. Kawai, H. Spangenberg, J. Li, G. Kimbi, M. Kew, J. Wands, and S. Tong. 2003. Sequence variation upstream of precore translation initiation codon reduces hepatitis B virus e antigen production. Gastroenterology 125:1370-1378. [DOI] [PubMed] [Google Scholar]
- 2.Bruss, V. 1997. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J. Virol. 71:9350-9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carman, W., A. Nanetti, P. Karyiannis, J. Waters, G. Manzillo, E. Tanzi, A. Zuckerman, and H. Thomas. 1990. Vaccine-induced escape mutant of hepatitis B virus. Lancet 336:325-329. [DOI] [PubMed] [Google Scholar]
- 4.Chiou, H., T. Lee, J. Kuo, Y. Mau, and M. Ho. 1997. Altered antigenicity of ‘a’ determinant variants of hepatitis B virus. J. Gen. Virol. 78:2639-2645. [DOI] [PubMed] [Google Scholar]
- 5.Chisari, F., P. Filippi, A. McLachlan, D. Milich, M. Riggs, S. Lee, R. Palmiter, C. Pinkert, and R. Brinste. 1986. Expression of hepatitis B virus large envelope polypeptide inhibits hepatitis B surface antigen secretion in transgenic mice. J. Virol. 60:880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chua, P., Y. Wen, and C. Shih. 2003. Coexistence of two distinct secretion mutations (PT5 and I97L) in hepatitis B virus core produces a wild-type pattern of secretion. J. Virol. 77:7673-7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernholz, D., P. Galle, M. Stemler, M. Brunetto, F. Bonino, and H. Will. 1993. Infectious hepatitis B virus variant defective in pre-S2 protein expression in a chronic carrier. Virology 194:137-148. [DOI] [PubMed] [Google Scholar]
- 8.Ganem, D., and R. Schneider. 2001. Hepadnaviridae: the viruses and their replication, p. 2939-2970. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 9.Hou, J., Z. Wang, J. Cheng, Y. Lin, G. Lau, J. Sun, F. Zhou, J. Waters, P. Karayiannis, and K. Luo. 2001. Prevalence of naturally occurring surface gene variants of hepatitis B virus in nonimmunized surface antigen-negative Chinese carriers. Hepatology 34:1027-1034. [DOI] [PubMed] [Google Scholar]
- 10.Jeantet, D., I. Chemin, B. Mandrand, F. Zoulim, C. Trepo, and A. Kay. 2002. Characterization of two hepatitis B virus populations isolated from a hepatitis B surface antigen-negative patient. Hepatology 35:1215-1224. [DOI] [PubMed] [Google Scholar]
- 11.Jenna, S., and C. Sureau. 1998. Effect of mutations in the small envelope protein of hepatitis B virus on assembly and secretion of hepatitis delta virus. Virology 251:176-186. [DOI] [PubMed] [Google Scholar]
- 12.Kalinina, T., A. Riu, L. Fischer, H. Will, and M. Sterneck. 2001. A dominant hepatitis B virus population defective in virus secretion because of several S-gene mutations from a patient with fulminant hepatitis. Hepatology 34:385-394. [DOI] [PubMed] [Google Scholar]
- 13.Kalinina, T., A. Riu, L. Fischer, T. Santantonio, H. Will, and M. Sterneck. 2003. Selection of a secretion-incompetent mutant in the serum of a patient with severe hepatitis B. Gastroenterology 125:1077-1084. [DOI] [PubMed] [Google Scholar]
- 14.Kalinina, T., A. Iwanski, H. Will, and M. Sterneck. 2003. Deficiency in virion secretion and decreased stability of the hepatitis B virus immune escape mutant G145R. Hepatology 38:1274-1281. [DOI] [PubMed] [Google Scholar]
- 15.Kim, K., K. Lee, H. Chang, S. Ahn, S. Tong, Y. Yoon, B. Seong, S. Kim, and K. Han. 2003. Evolution of hepatitis B virus sequence from a liver transplant recipient with rapid breakthrough despite hepatitis B immune globulin prophylaxis and lamivudine therapy. J. Med. Virol. 71:367-375. [DOI] [PubMed] [Google Scholar]
- 16.Le Pogam, S., T. Yuan, G. Sahu, S. Chatterjee, and C. Shih. 2000. Low-level secretion of human hepatitis B virus virions caused by two independent, naturally occurring mutations (P5T and L60V) in the capsid protein. J. Virol. 74:9099-9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Seyec, J., P. Chouteau, I. Cannie, C. Guguen-Guillouzo, and P. Gripon. 1998. Role of the pre-S2 domain of the large envelope protein in hepatitis B virus assembly and infectivity. J. Virol. 72:5573-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loffler-Mary, H., J. Dumortier, C. Klentsch-Zimmer, and R. Prange. 2000. Hepatitis B virus assembly is sensitive to changes in the cytosolic S loop of the envelope proteins. Virology 270:358-367. [DOI] [PubMed] [Google Scholar]
- 19.Mabit, H., and H. Schaller. 2000. Intracellular hepadnavirus nucleocapsids are selected for secretion by envelope protein-independent membrane binding. J. Virol. 74:11472-11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangold, C. M., and R. E. Streeck. 1993. Mutational analysis of the cysteine residues in the hepatitis B virus small envelope protein. J. Virol. 67:4588-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norder, H., B. Hammas, S. Lee, K. Bile, A. Courouce, I. Mushahwar, and L. Magnius. 1993. Genetic relatedness of hepatitis B viral strains of diverse geographical origin and natural variations in the primary structure of the surface antigen. J. Gen. Virol. 74:1341-1348. [DOI] [PubMed] [Google Scholar]
- 22.Parekh, S., F. Zoulim, S. Ahn, A. Tsai, J. Li, S. Kawai, N. Khan, C. Trepo, J. Wands, and S. Tong. 2003. Genome replication, virion secretion, and e antigen expression of naturally occurring hepatitis B virus core promoter mutants. J. Virol. 77:6601-6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perlman, D., and J. Hu. 2003. Duck hepatitis B virus virion secretion requires a double-stranded DNA genome. J. Virol. 77:2287-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Persing, D., H. Varmus, and D. Ganem. 1986. Inhibition of secretion of hepatitis B surface antigen by a related presurface polypeptide. Science 234:1388-1391. [DOI] [PubMed] [Google Scholar]
- 25.Ponsel, D., and V. Bruss. 2003. Mapping of amino acid side chains on the surface of hepatitis B virus capsids required for envelopment and virion formation. J. Virol. 77:416-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Summers, J., and W. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 27.Tang, J., C. Yeh, T. Chen, S. Hsieh, C. Chu, and Y. Liaw. 1998. Emergence of an S gene mutant during thymosin a1 therapy in a patient with chronic hepatitis B. J. Infect. Dis. 178:866-869. [DOI] [PubMed] [Google Scholar]
- 28.Tong, S., J. Li, L. Vtvitski, and C. Trepo. 1992. Replication capacities of natural and artificial precore stop codon mutants of hepatitis B virus: relevance of pregenome encapsidation signal. Virology 191:237-245. [DOI] [PubMed] [Google Scholar]
- 29.Wallace, W., and W. Carman. 1997. Surface variation of HBV: scientific and medical relevance. Viral Hepat. Rev. 3:5-16. [Google Scholar]
- 30.Wei, Y., J. Tavis, and D. Ganem. 1996. Relationship between viral DNA synthesis and virion envelopment in hepatitis B viruses. J. Virol. 70:6455-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan, T., G. Sahu, W. Whitehead, R. Greenberg, and C. Shih. 1999. The mechanism of an immature secretion phenotype of a highly frequent naturally occurring missense mutation at codon 97 of human hepatitis B virus core antigen. J. Virol. 73:5731-5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan, T., and C. Shih. 2000. A frequent, naturally occurring mutation (P130T) of human hepatitis B virus core antigen is compensatory for immature secretion phenotype of another frequent variant (I97L). J. Virol. 74:4929-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]