Abstract
A prevailing model for virus membrane fusion proteins has been that the hydrophobic fusion peptide is hidden in the prefusion conformation, becomes exposed once the fusion reaction is triggered, and then either inserts into target membranes or is rapidly inactivated. This model is in general agreement with the structure and mechanism of class I fusion proteins, such as the influenza virus hemagglutinin. We here describe studies of the class II fusion protein E1 from the alphavirus Semliki Forest virus (SFV). SFV fusion is triggered by low pH, which releases E1 from its heterodimeric interaction with the E2 protein and induces the formation of a stable E1 homotrimer. The exposure and target membrane interaction of the E1 fusion peptide (residues 83 to 100) were followed using a monoclonal antibody (MAb E1f) mapping to E1 residues 85 to 95. In agreement with the known structure of SFV and other alphaviruses, the fusion peptide was shielded in native SFV particles and exposed when E1-E2 dimer dissociation was triggered by acidic pH. In contrast, the fusion peptide on purified E1 ectodomains (E1*) was fully accessible at neutral pH. Functional assays showed that MAb E1f binding at neutral pH prevented subsequent low-pH-triggered E1* interaction with target membranes and trimerization. E1* was not inactivated by low pH when treated either in the absence of target membranes or in the presence of fusion-inactive cholesterol-deficient liposomes. Thus, the membrane insertion of the E1 fusion peptide is regulated by additional low-pH-dependent steps after exposure, perhaps involving an E1-cholesterol interaction.
Enveloped viruses catalyze a membrane fusion reaction between the virus and cellular membranes in order to infect the cell. This fusion event is mediated by a viral membrane glycoprotein through its dual interaction with the virus membrane and the cellular target membrane. The region of the virus fusion protein that inserts directly into the target membrane is termed the fusion peptide. Although they have not been rigorously identified in many viruses, in general fusion peptides are N-terminal or internal regions of ∼16 to 36 residues that are relatively hydrophobic in nature and are highly conserved within virus families (42). Since the interaction of the fusion peptide with the target membrane is critical to the fusion reaction, enveloped viruses must appropriately regulate their fusion proteins such that the fusion peptide-membrane interaction occurs only at the proper time and place.
The mechanism and regulation of viral fusion proteins are best understood for the influenza virus hemagglutinin (HA) protein, the prototype of the class I viral fusion proteins (reviewed in references 10 and 33). Class I fusion proteins are native trimers whose fusion can be triggered by endocytic low pH, as in the case of HA, or by receptor and/or coreceptor interactions. Fusion involves the refolding of the fusion protein to a six-helix bundle with a central coiled-coil and the fusion peptide and transmembrane domain at the same end of the rod-shaped molecule. As illustrated by the example of the influenza virus HA, interaction of class I fusion peptides with target membranes appears to have a relatively simple regulatory mechanism. The N-terminal HA fusion peptide is buried in an ionizable pocket in the native trimer interface (7), and its exposure is one of the first events to occur upon low-pH treatment (34, 43). Once exposed, the HA fusion peptide inserts into target membranes or becomes inactivated (41). Indicative of its hydrophobic nature, the fusion peptide will insert into a variety of target membranes, and in their absence it will bind detergent or aggregate with the fusion peptide of other HA molecules to form rosettes (29, 32). Exposure of the HA fusion peptide is thus a key regulatory element of the HA fusion reaction, because exposure is closely coupled to membrane insertion. Given the structural and functional similarities between the pre- and postfusion conformations of influenza virus HA and those of the other class I fusion proteins, this general scheme of fusion peptide regulation appears conserved in fusion proteins of this type.
Recent structural work on the fusion glycoproteins from the alphavirus Semliki Forest virus (SFV) and the flaviviruses tick-borne encephalitis virus (TBE) and dengue virus illustrates that they are very similar to each other and yet strikingly different from the class I fusion proteins, leading to their definition as class II viral membrane fusion proteins (23, 26, 28). In their native forms, both the alphavirus E1 protein and the flavivirus E protein have elongated structures that lie tangential to the virus membrane, and unlike the class I proteins both are composed primarily of β-sheets. The crystal structures of the ectodomains of these proteins show that they contain three domains. The internal fusion peptide is found in a loop at the tip of domain II at one end of the molecule, and the ectodomain C terminus connects to the stem and transmembrane region at the opposite end. In the virus particle the fusion protein is dimeric, forming an E-E homodimer in the case of flaviviruses and an E1-E2 heterodimer in the case of alphaviruses. Fusion of both alpha- and flaviviruses is triggered by low pH, which leads to the dissociation of the dimer and the formation of a highly stable homotrimer (HT) of the fusion protein (16, 17).
Based on hydrophobicity, sequence conservation, and the structure of the loop at the tip of domain II, the SFV fusion peptide has been placed within residues 83 to 100 of E1 (11, 23). Site-directed mutagenesis of single residues in this region showed that a number of mutations alter the pH dependence of fusion and that several mutations reduce the stability of the heterodimer (9, 20, 24). Importantly, replacement of glycine 91 with aspartate (G91D) completely blocks fusion and infection, and although the G91D mutant is able to undergo initial low-pH-dependent conformational changes, it does not form the HT (20). Proteolysis studies of the ectodomain HT showed that removal of a region of domain II including the fusion peptide released a truncated HT from the target membrane (14). Thus, the E1 fusion peptide region appears to be critical for fusion and target membrane interaction, and it is also implicated in formation of the HT. Analogous to the shielding of the fusion peptide in the influenza virus HA trimer interface, the fusion peptides of both alpha- and flaviviruses are masked by the dimer interaction in the native virion and become exposed and insert into the target membrane during fusion (16, 17). However, while the dissociation of the E1-E2 dimer, and thus exposure of the fusion peptide, is a critical first step in the alphavirus fusion reaction (31), several characteristics suggest that membrane insertion and regulation of the SFV fusion peptide may be more complex than those of the model described above.
The SFV fusion reaction is strongly dependent on the presence of cholesterol and sphingolipid in the target membrane, and cholesterol has been shown to be required in vivo for efficient SFV infection (reviewed in reference 17). Unlike the more nonspecific interaction of influenza virus or HA with target membranes, low-pH-triggered membrane binding of either SFV or the SFV E1 ectodomain (termed E1*) is dependent on the presence of cholesterol in the target bilayer. This suggests that the E1-membrane interaction requires additional specificity beyond simple hydrophobic interactions. Moreover, while the regulation of the viral E1 protein by its interaction with E2 is clear, studies have shown that the purified E1* ectodomain is monomeric rather than dimeric (21). Nonetheless, the E1* protein is fully competent to bind target liposomes and trimerize, with the same requirements as whole virus for cholesterol and low pH (1, 21). Together, these data suggest that there is a level of regulation built into the E1 fusion protein beyond its interaction with E2.
Here we have used a monoclonal antibody (MAb) termed MAb E1f to follow the regulation and function of the SFV fusion peptide. The epitope for MAb E1f was previously mapped to residues 85 to 95 of the fusion peptide (15), and our studies have shown that this epitope is masked when E1* inserts into cholesterol-containing target membranes at low pH (1). Here we demonstrate that regulation of the SFV fusion peptide-membrane interaction is very different from the influenza virus paradigm. While E2 plays an initial regulatory role in exposing the fusion peptide to solution, additional events are required for the membrane insertion of the fusion peptide. In the absence of cholesterol-containing target membranes, E1* appears insensitive to low pH.
(Some of the data in this paper are from a thesis submitted by D.L.G. in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Sue Golding Graduate Division of Medical Sciences, Albert Einstein College of Medicine, Yeshiva University.)
MATERIALS AND METHODS
Virus and cells.
The SFV used in these experiments was a well-characterized plaque-purified wild-type isolate (1) and was propagated in BHK-21 cells. Virus was radiolabeled with [35S]methionine and gradient purified as previously described (1).
COS-7 cell expression vectors and transfection.
For expression in COS-7 cells, the vectors pL2-SFV-88 and pL2-SFV-Δ83-92 were used, encoding the wild-type structural proteins and the structural proteins with a deletion of residues 83 to 92 of E1, respectively. These vectors were constructed and used for transient transfection in COS-7 cells as previously reported (24).
Expression analysis of viral proteins in infected BHK cells and transfected COS-7 cells.
COS-7 cells transfected 2 days prior with the wild type or Δ83-92 construct were preincubated for 15 min at 37°C in methionine-cysteine-deficient medium and then pulse-labeled with [35S]methionine-cysteine for 60 min, and the cell lysates were analyzed by immunoprecipitation and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described below.
Immunoprecipitation and SDS-gel analysis.
A polyclonal rabbit antibody against purified E1 and E2 proteins was prepared as previously described (19). MAb E1-1 recognizes both the acid and neutral conformations of E1, and MAb E1a-1 is specific for the acid conformation of E1 (19). Using an antibody-resistant virus mutant, the E1a-1 epitope was previously mapped to the vicinity of E1 G157 in domain I (2). MAb E2-1 recognizes both the acid and neutral conformations of E2 (19). MAbs E1f and E1n are specific for the E1 protein, and PepScan analysis localized the E1f epitope to a region between residues 85 to 95 of E1 with the sequence YPFMWGGAYCF (15). Immunoprecipitation was performed as previously described (19), except that for the experiments shown in Fig. 2 the reaction was performed in the absence of detergent. After several washes to remove unbound antibody, the samples were washed with buffers containing 1.0% Triton X-100, so that only those proteins that specifically bound antibody during the immunoprecipitation were recovered. Samples were analyzed by SDS-PAGE using 11% acrylamide gels and quantitated by PhosphorImager analysis with ImageQuant version 1.2 software (Molecular Dynamics, Sunnyvale, Calif.).
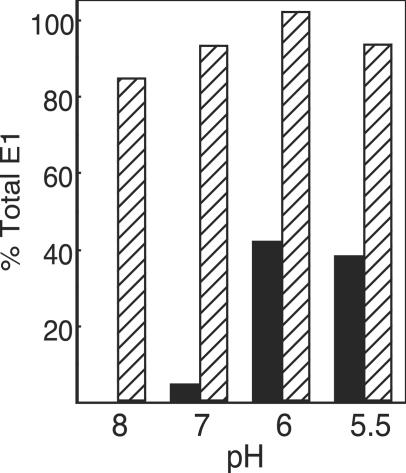
FIG. 2.
pH dependence of exposure of the MAb E1f epitope on virus particles or E1* ectodomains. Purified radiolabeled ectodomains (hatched bars) or intact virions (black bars) were treated at the indicated pH for 5 min at 37°C and then adjusted to pH 8.0. The samples were immunoprecipitated for 1 h on ice with a polyclonal antibody against the SFV glycoproteins or with MAb E1f and analyzed by SDS-PAGE. The amounts of E1 and E1* directly immunoprecipitated by MAb E1f were quantitated by phosphorimaging and graphed as a percentage of the total E1 or E1* immunoprecipitated by the polyclonal antibody. The data shown are representative of three experiments.
Preparation of E1 ectodomain.
Water-soluble ectodomain forms of the E1 and E2 transmembrane proteins (termed E1* and E2*) were prepared by limited proteolysis as previously described (18, 21). In brief, a mixture of 35S-labeled SFV (∼107 cpm) and purified unlabeled SFV (100 μg of protein) was digested with subtilisin (100 μg/ml) in phosphate-buffered saline containing 0.9 mM CaCl2, 0.5 mM MgCl2, and 0.5% Triton X-114 for 60 to 90 min on ice and purified by detergent phase separation and concanavalin A chromatography.
Liposomes.
Liposomes were prepared by extrusion as previously described (5) using phosphatidylcholine (from egg yolk)-phosphatidylethanolamine (derived from egg phosphatidylcholine by transphosphatidylation)-sphingomyelin (bovine brain)-cholesterol at an equimolar ratio of total phospholipids to cholesterol of 1:1:1:3. All phospholipids were purchased from Avanti Polar Lipids (Alabaster, Ala.), and cholesterol was from Steraloids (Wilton, N.H.).
Formation and assay of the E1* HT.
[35S]methionine-labeled ectodomains were preincubated with 1 mM liposomes for 5 min at 37°C, adjusted to pH 5.5 by addition of a precalibrated volume of 0.5 N acetic acid, incubated further as indicated in each figure legend, and then neutralized by addition of 0.5 N NaOH. The concentration of E1* in the reaction mixture was approximately 1 to 3 μg/ml. To maintain the HT and allow its quantitation by PAGE, samples were solubilized in SDS-sample buffer at 30°C for 3 min.
Sucrose density gradient flotation analysis.
Binding of ectodomains to liposomes was detected by cofloatation of protein with liposomes on sucrose step gradients as previously described (1, 21). Samples were adjusted to a final concentration of 40% sucrose, layered into the bottom of an ultracentrifuge tube, and then overlaid with a 25% sucrose step and a 5% sucrose step. The samples were centrifuged for 3 h at 54,000 rpm at 4°C in a TLS55 rotor. Seven fractions of 300 μl each were then collected from the gradients. The top four fractions and the bottom three fractions from each gradient were pooled for subsequent analysis. In control experiments, 3H-labeled liposomes were recovered in the top four fractions.
RESULTS
MAb E1f epitope mapping.
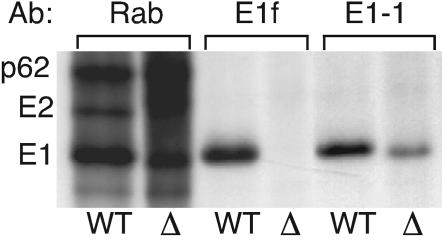
The epitope of MAb E1f was originally mapped by peptide scanning to residues 85 to 95 within the E1 fusion peptide loop (15). To confirm this result with the intact E1 protein, we used a previously described mutant with a deletion of E1 residues 83 to 92 (24). Constructs for either the wild-type structural proteins or the structural proteins containing the E1 Δ83-92 deletion were transiently expressed in COS cells. The cells were pulse-labeled, and the viral proteins in the cell lysates were immunoprecipitated with a polyclonal rabbit antiserum, MAb E1f, or a MAb recognizing an unrelated E1 epitope (E1-1). Both the wild-type and Δ83-92 envelope proteins were expressed well in COS cells, as shown by the total p62, E2, and E1 proteins precipitated by the polyclonal antiserum (Fig. 1, Rab lanes). The MAb E1f antibody efficiently precipitated wild-type E1, but not the Δ83-92 mutant E1. Extended exposure of the gel showed absolutely no signal from the MAb E1f immunoprecipitation of the Δ83-92 mutant (data not shown). In contrast, the control MAb E1-1 immunoprecipitated both wild-type and mutant E1 with similar efficiency.
FIG. 1.
Mapping the MAb E1f epitope on SFV E1. COS-7 cells were transfected with constructs expressing the structural proteins from wild-type virus (WT) or the E1Δ83-92 deletion mutant (Δ). Two days posttransfection the cells were pulse-labeled for 60 min with 200 μCi of [35S]methionine-cysteine/ml. Aliquots of the cell lysates were immunoprecipitated with a polyclonal antibody against the SFV glycoproteins (Rab), MAb E1f, or MAb E1-1 and analyzed by SDS-PAGE. The positions of p62, E2, and E1 are indicated.
Previous studies showed that the Δ83-92 E1 protein does not interact correctly with p62 and is not transported out of the endoplasmic reticulum (24). To confirm that MAb E1f failed to recognize the mutant because of the missing residues and not because of aberrant E1 folding or localization, we used virus-infected BHK cells treated with tunicamycin. This inhibitor of N-linked glycosylation traps the WT E1 and p62 proteins in the endoplasmic reticulum and causes misfolding, aggregation, and interchain disulfide bonds (25). MAb E1f showed comparable reactivity with E1 from either control or tunicamycin-treated cells, with ∼25% of the total E1 precipitated from each sample (data not shown). Thus, both the peptide and deletion analyses were consistent with placement of the E1f epitope within E1 residues 85 to 95.
Recognition of neutral- or low-pH-treated virus or ectodomains.
We next used MAb E1f to follow the solvent accessibility of the fusion peptide in virus particles and isolated ectodomains under native and low-pH-triggered conditions. Purified radiolabeled virus was incubated at pH 8.0, 7.0, 6.0, or 5.5 for 5 min at 37°C and adjusted to neutral pH. Samples were then incubated with antibodies in the absence of detergent in order to preserve particle structure and dimer interactions. A detergent wash was used as a final step so that only proteins that were specifically antibody bound in the intact virus particle were recovered. For comparison, a polyclonal antibody was used to quantitatively precipitate the total E2 and E1 in each sample. The MAb E1f showed negligible reactivity toward the fusion peptide of viral E1 at pH 8.0 or 7.0, while after treatment of virus particles at pH 6.0 or 5.5 approximately 40% of the total E1 protein was recognized by the antibody (Fig. 2). Parallel immunoprecipitation with the acid-specific MAb E1a-1 showed no reactivity with neutral pH virus and efficient recognition of E1 after treatment at low pH, while a nonspecific MAb showed no reactivity with any of the samples (data not shown). Our MAb E1f data are in agreement with previous Biocore data demonstrating that the MAb E1f epitope is hidden on whole virus at neutral pH and exposed at pH 6.5 and below (15), and they are also in agreement with the observed shielding of the E1 fusion peptide by the E2 protein in reconstructions of the native virus (23, 44).
Under the same conditions, the monomeric E1* ectodomain showed a strikingly different pattern of reactivity with MAb E1f. Greater than 80% of the E1* protein reacted with MAb E1f at pH 8.0, with only small increases at lower pH values (Fig. 2). In contrast, the acid-specific MAb E1a-1 did not recognize E1* even after treatment at pH 5.5 (data not shown), in agreement with previous observations that in the absence of membranes this acid-induced E1* conformational change does not occur (19) (see Fig. 4). The difference in reactivity with these two MAbs illustrates that exposure of the fusion peptide is pH independent and occurs before other acid-induced conformational changes. These results are consistent with the location of the fusion peptide on a loop at the tip of domain II in the native structure of the ectodomain (23). However, the full accessibility of the MAb E1f epitope in the native E1 ectodomain is surprising, given the generally hydrophobic nature of the amino acids in this epitope and the biochemical data demonstrating that this epitope inserts into target membranes at low pH (1).
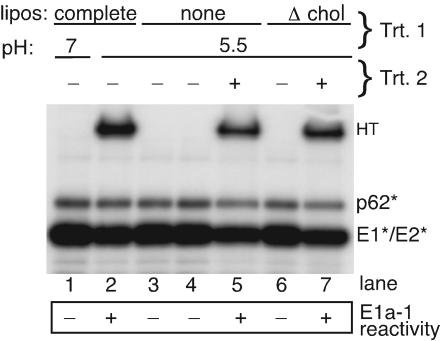
FIG. 4.
E1* is not inactivated by low-pH treatment in the absence of fusion-competent target membranes. Radiolabeled ectodomains were mixed with complete liposomes, buffer alone, or liposomes lacking cholesterol (Δ chol) and treated at pH 7.0 or 5.5 for 5 min at 37°C as indicated in treatment 1 (Trt. 1). All samples except for lane 4 were then adjusted to neutral pH. The mixtures were then assayed for HT formation by SDS-PAGE, tested for acid-specific epitope exposure by immunoprecipitation with MAb E1a-1 (results are in the box below the gel), or used for a second round of treatment where indicated (Trt. 2). For treatment 2 (lanes 5 and 7), the pretreated protein was mixed with cholesterol-containing liposomes (1 mM), treated again at pH 5.5 for 5 min at 37°C, and then assayed for HT formation and MAb E1a-1 reactivity. Data shown are a representative example of two experiments.
Functional effects of MAb E1f binding on E1* activity.
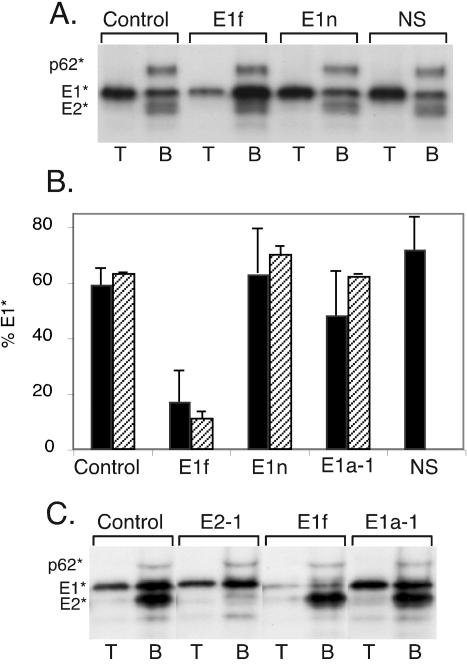
We then tested the biological effects of MAb E1f binding to E1*. Purified E1* and E2* were incubated at 4°C for 1 h with no antibody (control) or with MAb E1f, MAb E1n (which recognizes an unrelated E1 epitope [15]), the acid-conformation-specific MAb E1a-1, or a nonspecific isotype-matched MAb. Target liposomes were then added, and the samples were treated at pH 5.5 for 10 min and then adjusted to neutral pH. Each sample was analyzed for protein-liposome association by sucrose gradient floatation. As shown in the control sample (Fig. 3A) and in previous studies (1, 21), treatment at low pH in the presence of cholesterol-containing target membranes caused the majority of the E1* protein (∼60% of total) to associate with the liposomes and float at the top of the gradient, while E2*, p62*, and the remaining E1* did not bind membranes and remained in the bottom of the gradient. Similar results were observed when the samples were treated with MAb E1n or the nonspecific antibody (Fig. 3A) or with MAb E1a-1 (data not shown). However, in the MAb E1f-treated sample the majority of the E1* protein was found at the bottom of the gradient. Quantitation of results from multiple liposome floatation experiments (Fig. 3B) showed that MAb E1f reduced the interaction of E1* protein with target membranes by approximately 66%. Parallel samples were tested for HT formation by SDS-PAGE (Fig. 3B). MAb E1f reduced the trimerization of E1* from approximately 60% of the total E1* to about 11%, while the other antibodies had no effect.
FIG. 3.
Functional effects of MAb E1f binding on E1* membrane association and HT formation. (A) Radiolabeled ectodomains were incubated for 1 h at 4°C with control buffer, MAb E1f, MAb E1n, or a nonspecific isotype-matched MAb (NS) at 15 μg/ml. Samples were then mixed with liposomes (1 mM), treated at pH 5.5 for 5 min at 37°C, and adjusted to neutral pH. An aliquot of each sample was floated on sucrose step gradients, and the top four fractions (T) and bottom three fractions (B) of each gradient were pooled, trichloroacetic acid precipitated, and analyzed by SDS-PAGE. (B) Multiple experiments performed as for panel A were quantitated by phosphorimaging and graphed as the percentage of the total E1* found in the top of the gradient (black bars). The error bars represent the standard deviations of three experiments for the buffer control, MAb E1f, and MAb E1n samples and the range of two experiments for the MAb E1a-1 and nonspecific (NS) control samples. In a separate determination, the amount of HT present in an aliquot of each sample (prior to gradient floatation) was quantitated by SDS-PAGE and is expressed as a percentage of the total E1*. The error bars represent the range of two measurements for each condition. The percent HT was not determined for the nonspecific antibody samples. (C) Immunodepletion with MAb E1f at neutral pH. Radiolabeled ectodomains were incubated at pH 7.0 for 1 h at 4°C with 5 μg of a MAb against E2 (E2-1), MAb E1f, or MAb E1a-1 per ml or in buffer alone. The immune complexes were removed by binding to zysorbin. The immunodepleted samples were then mixed with liposomes (1 mM), treated at pH 5.5 for 10 min at 37°C, and analyzed on sucrose step gradients as described for panel A.
These results suggested that MAb E1f was binding to functional E1* and that binding to the fusion peptide inhibited the protein's membrane binding and trimerization. It was important to confirm that antibody binding occurred at neutral pH and not during the low-pH treatment used to trigger protein-membrane insertion. We therefore pretreated ectodomain samples at neutral pH with MAb E1f, an anti-E2 MAb (E2-1), or the acid-conformation-specific MAb E1a-1. The antigen-antibody complexes were then removed, and the immunodepleted samples were mixed with liposomes, treated at low pH, and tested by gradient floatation as before (Fig. 3C). Pretreatment with E2-1 removed E2* but had no effect on E1* membrane association. As predicted, MAb E1a-1 did not immunodeplete, confirming that under depletion conditions E1* was in the neutral conformation. In contrast, MAb E1f removed almost all of the E1* from the sample at neutral pH, and thus E1f immunodepletion prevented subsequent acid-triggered E1*-liposome interaction. Together these data demonstrate that, unlike the model for class I proteins, the fusion peptide region of the native E1* protein is fully accessible at neutral pH. Blocking the interaction between the fusion peptide and liposomes prevented E1* membrane insertion and HT formation.
Low-pH inactivation studies of E1*.
The class I proteins become inactivated when the fusion peptide is exposed in the absence of target membranes, while our data indicated that the E1* fusion peptide is constitutively exposed at neutral pH without inactivation. We therefore tested whether low-pH treatment of E1* would induce inactivation when performed either in the absence of target membranes or in the presence of cholesterol-deficient target membranes. Controls showed that when cholesterol-containing liposomes were present, low pH caused efficient E1* HT formation and acid-epitope exposure (Fig. 4, lane 2), while no conformational changes were detected at neutral pH (Fig. 4, lane 1). In the absence of membranes, low-pH treatment produced neither HT nor exposure of the acid-specific epitope, even if the sample was assayed at acid pH to detect any reversible conformational changes (Fig. 4, lanes 3 and 4). However, such low-pH treatment in the absence of membranes did not inactivate E1*, since subsequent addition of cholesterol-containing liposomes at acid pH resulted in both E1* HT formation and acid-epitope exposure (lane 5). Importantly, acid treatment of E1* in the presence of membranes lacking cholesterol did not trigger low-pH-dependent conformational changes (lane 6), and the protein was fully active when subsequently treated with cholesterol-containing liposomes at acid pH (lane 7). Thus, a specific interaction of E1* with cholesterol-containing membranes is involved in the irreversible membrane insertion of the fusion peptide, and this insertion and the subsequent conformational changes are blocked by the E1f MAb.
DISCUSSION
We here provide evidence for a multistep mechanism of regulation of viral fusion peptide insertion into a target membrane. On intact SFV particles, the fusion peptide is hidden at neutral pH and becomes exposed when the E1-E2 dimer is dissociated by low-pH treatment. Such low-pH conditions would normally trigger the insertion of the newly exposed fusion peptide into a cholesterol-containing target membrane and lead to membrane fusion. In contrast, the E1* ectodomain is monomeric and thus can be assayed for accessibility of the fusion peptide independent of effects on dimer interactions. Our data with MAb E1f showed that the fusion peptide on E1* was solvent accessible even at pH 8.0. The exposure of this region of the ectodomain did not result in stable association with target membranes or cause the protein to aggregate at the concentrations tested, even though the MAb E1f epitope is sufficiently hydrophobic to become buried in the target membrane at low pH (1). Stable E1* membrane insertion required both low pH and cholesterol-containing target liposomes and was blocked by prebinding MAb E1f to E1* at neutral pH. Even when treated at low pH in the presence of membranes without cholesterol, E1* did not become inactivated at the concentrations tested and remained competent to undergo subsequent low-pH-triggered insertion into cholesterol-containing membranes. The lack of response to low pH was not simply the result of a reversible transition, since there was no acid epitope exposure or HT formation in a sample maintained at acid pH (Fig. 4). Together our data suggest that exposure and insertion of the SFV fusion peptide into target membranes are not coupled events. Instead, low pH first triggers the dissociation of the E2-E1 dimer. The exposed fusion peptide then requires low pH and cholesterol for stable membrane insertion.
In addition to these findings, a number of other features of the membrane interaction of the SFV fusion protein differ from those of the general paradigm for class I fusion proteins. The class I proteins are metastable, with a native conformation that is considerably more labile than the final postfusion conformation. Studies of HA and several other class I proteins showed that fusion peptide exposure, membrane binding, and membrane fusion can be triggered by treatment at elevated temperature or with urea (4, 27, 30, 40). These denaturing conditions allow the protein to switch to its final, most stable conformation, and thus mediate fusion. Similar to the class I fusion proteins, SFV E1 is metastable in that it is significantly more stable in the low-pH-triggered HT conformation than in the native neutral-pH conformation (12). However, treatment with heat or urea does not trigger HT formation or fusion of SFV. Even denaturant treatment of the monomeric E1* protein in the presence of liposomes failed to generate the HT (12). For the class I proteins, exposure of the fusion peptide causes the protein to bind membrane or detergents, similar to an integral membrane protein (29, 32). In contrast, the SFV E1 crystal structure shows that the fusion peptide is solvent accessible in the native ectodomain (23), while biochemical studies show that the SFV fusion peptide does not act generally hydrophobic but displays a more specific membrane interaction (1). In keeping with the exposure and nonspecific aggregation of its hydrophobic fusion peptide, the ectodomain of influenza virus HA is inactivated at low pH in the absence of membranes (8). In contrast, the SFV E1 ectodomain was not inactivated by acid pH in the absence of a target membrane (Fig. 4). Thus, the activation and membrane insertion of the SFV fusion protein appear to require more specific triggering by low pH and cholesterol-containing membranes.
Recent work on the E protein from TBE shows many mechanistic similarities with the SFV E1 protein, in keeping with their shared identity as class II fusion proteins (16). The TBE fusion reaction, although not strictly cholesterol dependent, is promoted by the presence of cholesterol. The TBE fusion peptide is hidden in the native E homodimer and exposed by dimer dissociation at low pH. The native E protein is less stable than the final E HT, but similar to SFV the conversion to the HT is specifically triggered by low pH and not by denaturant treatment (35). Fusion is blocked by mutations in the fusion peptide loop (3), and membrane insertion of the E ectodomain is inhibited by a MAb that maps to the fusion peptide (36). Since E maintains its dimeric interaction in the ectodomain form, the fusion peptide is hidden at neutral pH and, thus, the independence of fusion peptide exposure and membrane insertion is more difficult to test than in the case of SFV. However, it is clear that the E ectodomain is not inactivated by low-pH treatment in the absence of target membranes and that insertion into target membranes and trimerization are promoted by sterol (36, 37). Moreover, while the ectodomain dimer dissociates to a monomer at low pH, in the absence of target membranes this transition is reversible, suggesting that the E fusion peptide is not irreversibly triggered (36). Taken together, the data on SFV and TBE indicate that the class II fusion proteins have a more complex and regulated mechanism of membrane insertion than simple control of fusion peptide accessibility to the target membrane. It is worth noting that the regulation of class I fusion proteins may also be more complex, since recent studies demonstrated that the exposure of the HA fusion peptide was reversible under conditions of low protein concentration in the absence of target membranes (22).
The differences between the behavior of the full-length E1 protein in the virus particle and the soluble E1 ectodomain are important to our understanding of the class II fusion reaction. Unlike E1*, in the absence of target membranes viral E1 forms an HT at low pH and becomes inactivated (14, 39). Full-length E1 might insert into the viral membrane at low pH, thus forming a fusion-incompetent HT. Limited insertion into the virus membrane would explain why the kinetics and yield of the E1 HT are both significantly increased by the presence of a cholesterol-containing target membrane (reviewed in reference 17). Alternatively, it could be that the close interactions and symmetries of E1 in the virus particle lead to HT formation at low pH without any need for a fusion peptide-membrane interaction. Analogously, there might be several reasons for the strict requirement for cholesterol-containing membranes in E1* HT formation. It could be that the interaction of the fusion peptide with cholesterol target membranes specifically changes the E1* conformation to allow trimerization. Alternatively, the membrane association of E1* might serve as a “platform” to orient the monomeric E1* molecules and increase their local concentration, thus permitting them to trimerize. Interestingly, the E1* HT localizes to sterol-rich detergent-resistant regions of the target membrane, suggesting a possible association of the protein with cholesterol and a potential protein concentration mechanism (1). Clearly, the insertion of E1* into the target bilayer specifically requires low pH and cholesterol, and in the absence of either factor the protein remains active but does not convert to the membrane-bound form. This cholesterol requirement is thus similar to that of the full-length molecule, in which cholesterol acts to couple the membrane insertion of E1 to the pH-dependent refolding reaction that drives membrane fusion.
Our data show that simple exposure of the SFV fusion peptide, as normally occurs during low-pH treatment of virus, does not lead to membrane insertion. How might E1's membrane interaction be regulated? While the stable membrane insertion of E1 clearly depends on the initial fusion peptide-membrane interaction and is blocked by MAb E1f, recent data suggest that additional regions of E1 may also be involved in regulating membrane insertion. The cholesterol requirement for SFV fusion and infection can be significantly decreased by single point mutations within domain II of the E1 protein (6, 38). One of these mutations, P226S, is localized to a loop region that is distant from the fusion peptide in the linear sequence of E1 but closely associated with the fusion peptide in the native E1 structure (23, 38). Recent electron microscopy studies of the membrane-bound E1* HT have demonstrated that membrane insertion of E1 is a strikingly cooperative process leading to rings of five to six HT (13). It is also possible that the conformation of the fusion peptide changes upon insertion into a cholesterol-containing membrane, leading to the observed irreversible membrane association of E1*. One model that incorporates these data would be that the exposed fusion peptide and/or the adjacent P226 loop acts as a sensor for cholesterol in the target membrane. An interaction with cholesterol would then allow the protein to respond to low pH by irreversibly inserting into the target membrane, perhaps stabilized by changes in fusion peptide conformation and by the cooperative interactions visualized by electron microscopy. Refolding to the HT would then complete the fusion reaction. Further characterization of the membrane insertion of E1 will allow us to define the roles of specific protein domains and the novel means by which the soluble E1 ectodomain converts to its membrane-bound state.
Acknowledgments
We thank Aimo Salmi for providing MAbs E1f and E1n, Brigid Reilly for expert technical assistance, and the members of the Kielian lab for helpful discussions and critical reading of the manuscript.
This work was supported by a grant to M.K. from the U.S. Public Health Service (R01 GM52929) and by Cancer Center Core Support grant NIH/NCI P30-CA13330. D.L.G. was supported through the Medical Scientist Training Program (NIH T32 GM 07288).
REFERENCES
- 1.Ahn, A., D. L. Gibbons, and M. Kielian. 2002. The fusion peptide of Semliki Forest virus associates with sterol-rich membrane domains. J. Virol. 76:3267-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, A., M. R. Klimjack, P. K. Chatterjee, and M. Kielian. 1999. An epitope of the Semliki Forest virus fusion protein exposed during virus-membrane fusion. J. Virol. 73:10029-10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, and F. X. Heinz. 2001. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol. 75:4268-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carr, C. M., C. Chaudhry, and P. S. Kim. 1997. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc. Natl. Acad. Sci. USA 94:14306-14313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatterjee, P. K., C. H. Eng, and M. Kielian. 2002. Novel mutations that control the sphingolipid and cholesterol dependence of the Semliki Forest virus fusion protein. J. Virol. 76:12712-12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee, P. K., M. Vashishtha, and M. Kielian. 2000. Biochemical consequences of a mutation that controls the cholesterol dependence of Semliki Forest virus fusion. J. Virol. 74:1623-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J., K. H. Lee, D. A. Steinhauer, D. J. Stevens, J. J. Skehel, and D. C. Wiley. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409-417. [DOI] [PubMed] [Google Scholar]
- 8.Doms, R. W., A. Helenius, and J. White. 1985. Membrane fusion activity of the influenza virus hemagglutinin. J. Biol. Chem. 260:2973-2981. [PubMed] [Google Scholar]
- 9.Duffus, W. A., P. Levy-Mintz, M. R. Klimjack, and M. Kielian. 1995. Mutations in the putative fusion peptide of Semliki Forest virus affect spike protein oligomerization and virus assembly. J. Virol. 69:2471-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 11.Garoff, H., A.-M. Frischauf, K. Simons, H. Lehrach, and H. Delius. 1980. Nucleotide sequence of cDNA coding for Semliki Forest virus membrane glycoproteins. Nature 288:236-241. [DOI] [PubMed] [Google Scholar]
- 12.Gibbons, D. L., A. Ahn, P. K. Chatterjee, and M. Kielian. 2000. Formation and characterization of the trimeric form of the fusion protein of Semliki Forest virus. J. Virol. 74:7772-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibbons, D. L., I. Erk, B. Reilly, J. Navaza, M. Kielian, F. A. Rey, and J. Lepault. 2003. Visualization of the target-membrane-inserted fusion protein of Semliki Forest virus by combined electron microscopy and crystallography. Cell 114:573-583. [DOI] [PubMed] [Google Scholar]
- 14.Gibbons, D. L., and M. Kielian. 2002. Molecular dissection of the Semliki Forest virus homotrimer reveals two functionally distinct regions of the fusion protein. J. Virol. 76:1194-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammar, L., S. Markarian, L. Haag, H. Lankinen, A. Salmi, and H. R. Cheng. 2003. Prefusion rearrangements resulting in fusion peptide exposure in Semliki Forest virus. J. Biol. Chem. 278:7189-7198. [DOI] [PubMed] [Google Scholar]
- 16.Heinz, F. X., and S. L. Allison. 2000. Structures and mechanisms in flavivirus fusion. Adv. Virus Res. 55:231-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kielian, M., P. K. Chatterjee, D. L. Gibbons, and Y. E. Lu. 2000. Specific roles for lipids in virus fusion and exit: examples from the alphaviruses, p. 409-455. In H. Hilderson and S. Fuller (ed.), Subcellular biochemistry, vol. 34. Fusion of biological membranes and related problems. Plenum Publishers, New York, N.Y. [DOI] [PubMed]
- 18.Kielian, M., and A. Helenius. 1985. pH-induced alterations in the fusogenic spike protein of Semliki Forest virus. J. Cell Biol. 101:2284-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kielian, M., S. Jungerwirth, K. U. Sayad, and S. DeCandido. 1990. Biosynthesis, maturation, and acid activation of the Semliki Forest virus fusion protein. J. Virol. 64:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kielian, M., M. R. Klimjack, S. Ghosh, and W. A. Duffus. 1996. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J. Cell Biol. 134:863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klimjack, M. R., S. Jeffrey, and M. Kielian. 1994. Membrane and protein interactions of a soluble form of the Semliki Forest virus fusion protein. J. Virol. 68:6940-6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leikina, E., C. Ramos, I. Markovic, J. Zimmerberg, and L. V. Chernomordik. 2002. Reversible stages of the low-pH-triggered conformational change in influenza virus hemagglutinin. EMBO J. 21:5701-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 24.Levy-Mintz, P., and M. Kielian. 1991. Mutagenesis of the putative fusion domain of the Semliki Forest virus spike protein. J. Virol. 65:4292-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marquardt, T., and A. Helenius. 1992. Misfolding and aggregation of newly synthesized proteins in the endoplasmic reticulum. J. Cell Biol. 117:505-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 100:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paterson, R. G., C. J. Russell, and R. A. Lamb. 2000. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology 270:17-30. [DOI] [PubMed] [Google Scholar]
- 28.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2A resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 29.Ruigrok, R. W. H., A. Aitken, L. J. Calder, S. R. Martin, J. J. Skehel, S. A. Wharton, W. Weis, and D. C. Wiley. 1988. Studies on the structure of the influenza virus haemagglutinin at the pH of membrane fusion. J. Gen. Virol. 69:2785-2795. [DOI] [PubMed] [Google Scholar]
- 30.Ruigrok, R. W. H., S. R. Martin, S. A. Wharton, J. J. Skehel, P. M. Bayley, and D. C. Wiley. 1986. Conformational changes in the hemagglutinin of influenza virus which accompany heat-induced fusion of virus with liposomes. Virology 155:484-497. [DOI] [PubMed] [Google Scholar]
- 31.Salminen, A., J. M. Wahlberg, M. Lobigs, P. Liljeström, and H. Garoff. 1992. Membrane fusion process of Semliki Forest virus. II. Cleavage- dependent reorganization of the spike protein complex controls virus entry. J. Cell Biol. 116:349-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skehel, J. J., P. M. Bayley, E. B. Brown, S. R. Martin, M. D. Waterfield, J. M. White, I. A. Wilson, and D. C. Wiley. 1982. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc. Natl. Acad. Sci. USA 79:968-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 34.Stegmann, T., J. M. White, and A. Helenius. 1990. Intermediates in influenza-induced membrane fusion. EMBO J. 9:4231-4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stiasny, K., S. L. Allison, C. W. Mandl, and F. X. Heinz. 2001. Role of metastability and acidic pH in membrane fusion by tick-borne encephalitis virus. J. Virol. 75:7392-7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stiasny, K., S. L. Allison, J. Schalich, and F. X. Heinz. 2002. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J. Virol. 76:3784-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stiasny, K., C. Koessl, and F. X. Heinz. 2003. Involvement of lipids in different steps of the flavivirus fusion mechanism. J. Virol. 77:7856-7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vashishtha, M., T. Phalen, M. T. Marquardt, J. S. Ryu, A. C. Ng, and M. Kielian. 1998. A single point mutation controls the cholesterol dependence of Semliki Forest virus entry and exit. J. Cell Biol. 140:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wahlberg, J. M., R. Bron, J. Wilschut, and H. Garoff. 1992. Membrane fusion of Semliki Forest virus involves homotrimers of the fusion protein. J. Virol. 66:7309-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wharton, S. A., J. J. Skehel, and D. C. Wiley. 2000. Temperature dependence of fusion by Sendai virus. Virology 271:71-78. [DOI] [PubMed] [Google Scholar]
- 41.White, J., J. Kartenbeck, and A. Helenius. 1982. Membrane fusion activity of influenza virus. EMBO J. 1:217-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White, J. M. 1990. Viral and cellular membrane fusion proteins. Annu. Rev. Physiol. 52:675-697. [DOI] [PubMed] [Google Scholar]
- 43.White, J. M., and I. A. Wilson. 1987. Anti-peptide antibodies detect steps in a protein conformational change: low-pH activation of the influenza hemagglutinin. J. Cell Biol. 105:2887-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, W., S. Mukhopadhyay, S. V. Pletnev, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2002. Placement of the structural proteins in Sindbis virus. J. Virol. 76:11645-11658. [DOI] [PMC free article] [PubMed] [Google Scholar]