Abstract
The sirtuins are a protein family named after the first identified member, S. cerevisiae Sir2p. Sirtuins are protein deacetylases whose activity is dependent on NAD+ as a cosubstrate. They are structurally defined by two central domains that together form a highly conserved catalytic center, which catalyzes the transfer of an acetyl moiety from acetyllysine to NAD+, yielding nicotinamide, the unique metabolite O-acetyl-ADP-ribose and deacetylated lysine. One or more sirtuins are present in virtually all species from bacteria to mammals. Here we describe a phylogenetic analysis of sirtuins. Based on their phylogenetic relationship, sirtuins can be grouped into over a dozen classes and subclasses. Humans, like most vertebrates, have seven sirtuins: SIRT1-SIRT7. These function in diverse cellular pathways, regulating transcriptional repression, aging, metabolism, DNA damage responses and apoptosis. We show that these seven sirtuins arose early during animal evolution. Conserved residues cluster around the catalytic center of known sirtuin family members.
Keywords: deacetylase, evolution, molecular phylogeny, SIR2, sirtuin
INTRODUCTION
The sirtuins are a family of NAD+-dependent deacetylases. They are named after their founding member budding yeast Sir2p, a histone deacetylase (Braunstein et al., 1993; 1996) first discovered in a genetic screen for genes required for transcriptional silencing of the budding yeast mating type loci (Ivy et al., 1985; 1986; Klar et al., 1979). In animals, sirtuins have also been implicated in transcriptional silencing (Astrom et al., 2003; Newman et al., 2002; Pruitt et al., 2006; Rosenberg and Parkhurst, 2002; Tissenbaum and Guarente, 2001; Vaquero et al., 2004; 2007), aging (Rogina and Helfand, 2004; Tissenbaum and Guarente, 2001; Wood et al., 2004) and metabolic regulation (Schwer and Verdin, 2008). The sirtuin family appears to be virtually ubiquitous throughout all kingdoms of life and the number of distinct sirtuins within an organism ranges from as little as one in bacteria to seven in vertebrates.
The biological functions of sirtuins have been summarized in several recent reviews (Dali-Youcef et al., 2007; Haigis and Guarente, 2006; Longo and Kennedy, 2006; North and Verdin, 2004; Schwer and Verdin, 2008; Yamamoto et al., 2007). The founding member, budding yeast Sir2p was initially shown to be required for the transcriptional silencing of the mating type loci, and subsequently also implicated in transcriptional silencing at telomere proximal sites (Aparicio et al., 1991) and at ribosomal repeats (Bryk et al., 1997; Fritze et al., 1997; Gottlieb and Esposito, 1989). Sir2p forms complexes with different protein cofactors depending on the target site. At telomeres and the mating type loci, Sir2p forms a complex with Sir3p and Sir4p (Aparicio et al., 1991), while at rDNA sites Sir2p associates with Net1p and Cdc14p to form the regulator of nucleolar silencing and telophase exit (RENT) complex (Shou et al., 1999; Straight et al., 1999). In budding yeast, Sir2p action at ribosomal repeats is required to prevent illegitimate recombination leading to extrachomsomal ribosomal DNA circles, the accumulation of which is associated with aging (Gottlieb and Esposito, 1989; Kaeberlein et al., 1999; Sinclair and Guarente, 1997).
In animals, sirtuins have been implicated in a wide variety of processes, including transcriptional silencing (Astrom et al., 2003; Newman et al., 2002; Pruitt et al., 2006; Rosenberg and Parkhurst, 2002; Tissenbaum and Guarente, 2001; Vaquero et al., 2004; 2007), aging (Rogina and Helfand, 2004; Tissenbaum and Guarente, 2001; Wood et al., 2004), metabolic regulation (Schwer and Verdin, 2008), and apoptosis (Cohen et al., 2004; Dai et al., 2007; Greiss et al., 2008; Luo et al., 2001; Vaziri et al., 2001; Wang et al., 2006). Many of these functions are not related to histone deacetylation, and multiple non-histone substrates have been identified. Mammalian sirtuins (SIRT1-7) share the conserved sirtuin domain but vary in subcellular localization and function. SIRT1, SIRT6 and SIRT7 localize to the nucleus, SIRT3, SIRT4 and SIRT5 are mitochondrial, while SIRT2 is predominantly cytoplasmic (Michishita et al., 2005). SIRT1, the best characterized mammalian sirtuin, is predominantly nuclear but also has roles in the cytoplasm (Jin et al., 2007; Tanno et al., 2007). SIRT1 interacts with and regulates a number of histone and non-histone protein substrates including p53 (Luo et al., 2001; Vaziri et al., 2001), NF-κB (Yeung et al., 2004), PPARγ (Picard et al., 2004), PGC-1α (Rodgers et al., 2005) and Foxo transcription factors (Brunet et al., 2004; Motta et al., 2004; van der Horst et al., 2004). It has roles in developmental and aging regulation (Yamamoto et al., 2007), and recent evidence also points towards a role in ensuring efficient DNA double-strand break repair (Oberdoerffer et al., 2008). SIRT6 has been found to deacetylate histone H3 and is linked to transcriptional regulation and the maintenance of genomic stability (Kawahara et al., 2009; Lombard et al., 2008; Michishita et al., 2008). SIRT7, which is present in the nucleolus, regulates RNA-PolI mediated expression of ribosomal RNA genes and plays a role in angiogenesis (Ford et al., 2006; Potente et al., 2007). SIRT2 may have a role in cell cycle regulation, is able to deacetylate histone H4, and appears to act as a tumor suppressor in certain gliomas (Dryden et al., 2003; Hiratsuka et al., 2003; Inoue et al., 2006; Vaquero et al., 2006). Little is known about the mitochondrial sirtuins SIRT3, SIRT4 and SIRT5, although specific substrates that have been identified so far include acetyl coenzyme A synthetase 2, glutamate dehydrogenase and cytochrome c respectively (Haigis et al., 2006; Hallows et al., 2006; Schlicker et al., 2008; Schwer et al., 2006).
Sirtuin classification
Sirtuins were first grouped into five major classes (I, II, III, IV and U) (Frye, 2000): classes I-IV each include at least one of the seven sirtuins present in humans; class U includes sirtuins from archaea and bacteria. The last classification of sirtuins included human, D. melanogaster, C. elegans, three yeast species, rice, plasmodium, leishmania, trypanosoma, and several bacteria and archaea (Frye, 2000). Since then a large number of genome sequences from all kingdoms of life have become available, allowing for a much more comprehensive analysis of sirtuin phylogeny. For instance, it is now possible to analyze sirtuins from all major classes of animals, and thus determine the evolutionary origins of the seven sirtuins encoded in the human genome.
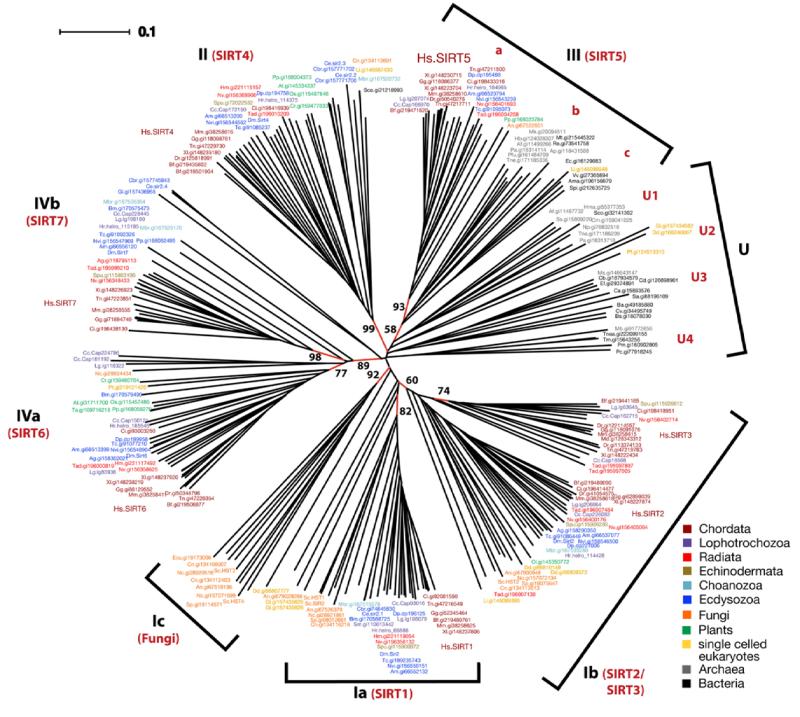
To analyze the evolutionary relationships of sirtuins we searched for all sirtuins present in 77 representative species of animals, plants, bacteria and archaea. We identified sirtuin members in all species we examined with the exception of two red algae and several archaea (Supplementary File. 1). At present we cannot rule out that the lack of sirtuin homologs in these species may be due to incomplete sequencing or mistakes in gene predictions, although this seems unlikely in the case of archaea due to the small size of those genomes. Using sequences representing the catalytic core of crystallized human SIRT2, Archaeoglobus fulgidus Af1 and S. cerevisae Hst2 sirtuins as templates (Finnin et al., 2001; Min et al., 2001; Zhao et al., 2003b) we aligned 240 sirtuins and used Neighbor Joining methods to construct a phylogenetic tree (Table 1, Fig. 1, Supplementary File 2). The results from this analysis were largely confirmed by a similar analysis using full-length protein sequences (Supplementary File 3). Our analysis mainly confirms the classification of Frye; however, we suggest splitting class Ib sirtuins into two subgroups defined by vertebrate SIRT2 and SIRT3 family members, respectively (Table 1, Fig. 1, Supplementary File 2). It appears likely that the SIRT3 family form a separate subgroup within class Ib sirtuins that originated rather recently, as the SIRT3 group contains only animal species (Fig. 1, Supplementary File 2). Furthermore, the group of “undifferentiated” sirtuins (U), which is not closely related to any vertebrate sirtuin, is comprised of a number of unrelated but clearly defined groups (U1 to U4). U1 is mostly comprised of archaeal sirtuins (grey), while U2 sirtuins are encoded by phylogenetically unrelated single-celled eukaryotes (yellow), such as Dictyostelium discoideum (Dd, amoebozoa), Giardia lamblia (Gl, excavata) and Plasmodium falciparum (Pf, alveolata). U3 and U4 sirtuins are predominantly bacterial (black). Our analysis also establishes that group III is more diverse than previously found, and can be split into three subgroups which we term IIIa, IIIb and IIIc: group IIIa is almost exclusively comprised of animal sequences, which are predominantly mitochondrially localized, and include human SIRT5; group IIIb sirtuins are predominantly archaeal; and IIIc sirtuins are mostly bacterial. Group IIIa/SIRT5 mammalian sirtuins are related to the IIIc bacterial group, suggesting that IIIa/SIRT5 group sirtuins might be evolutionarily ancient. The majority of sirtuins can be clustered around the seven human sirtuin family members The only other clusters besides the U1 to U4 group are a group of sirtuins exclusive to fungi (class Ic), which include the S. cerevisiae Hst3 and Hst4 sirtuin family members. The Ic sirtuin family likely arose early during fungal evolution, as such sirtuins were found in all fungal species we examined, including members of ascomycota, basidiomycota and microsporidia (Table 1, Fig. 1, Supplementary File 2).
Table 1.
Sirtuin complement of analyzed eukaryotes. Numbers indicate one or more SIRT1-SIRT7 related sirtuins, question marks indicate sirtuins that could not be assigned to groups defined by mammalian sirtuins. The number of question marks indicates the number of unassigned sirtuins. Question marks between the SIRT6 and SIRT7 column signify class IV sirtuins that could not clearly be placed into the SIRT6 or SIRT7 groups. Colors correspond to the classification described in Fig. 1. “F” signifies class Ic sirtuins that are specific to Fungi. 1) to 5) indicates various phylogenetic groups: 1) urochordata; 2) crustacea; 3) basidiomycota; 4) microsporidia; 5) bryophyta
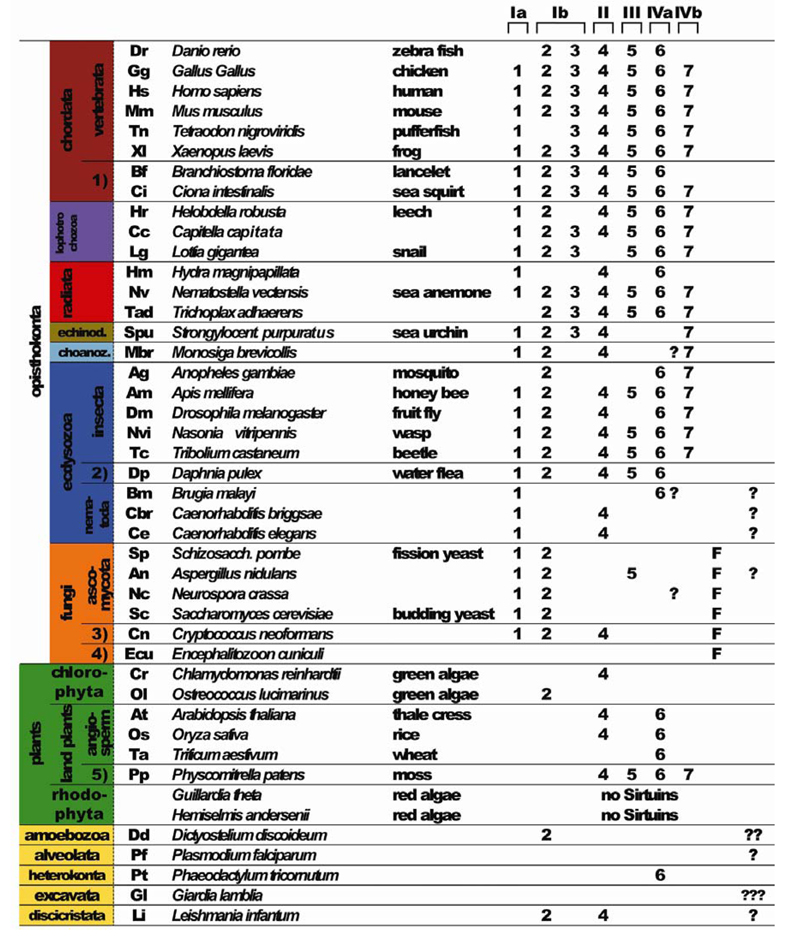
Fig. 1.
Unrooted phylogenetic tree of all aligned sirtuin sequences. The tree was constructed using SplitsTree 4 (Huson and Bryant, 2006). Classes defined by Frye (2000) are shown in black, classes added in this review are shown in red. Branches for which bootstrap values are shown are also in red. The SplitsTree file with all bootstrap values can be down-loaded as Supplementary File 2.
Evolutionary considerations
According to Baldauf (2003) eukaryotes can be categorized into 8 major phylogenetic groups: opisthokonts includes animals, fungi and choanoflagellates; plants make up another major group; and the remaining 6 groups are mostly single celled organisms, including amoebozoa (containing Dictyostelium discoideum) and discicristates (containing Leishmania). To investigate the evolutionary origins of the seven groups defined by human sirtuins we commenced our analysis by looking for sirtuins within the group of the opisthokonts. This group includes animals, fungi and choanoflagellates such as the recently sequenced Monosiga brevicollis (Mbr, light blue) (King et al., 2008). Monosiga sirtuins can be clearly found in the SIRT1, SIRT2 and SIRT4 and SIRT7 group, although a second class IV Monosiga sirtuin cannot be clearly assigned to the SIRT6 or the SIRT7 group (Fig. 1, Table 1). With very few exceptions fungal sirtuins (orange) cluster within class I sirtuins. Besides the class Ic group, which is fungus-specific, fungal sirtuins are also found within the SIRT1 and the SIRT2 groups. Interestingly, we also found a single fungal representative in class IV sirtuins that cannot clearly be grouped with SIRT6 or SIRT7 like sequences. Furthermore, there is a single fungal representative each within the SIRT4 and the SIRT5 group. Most fungi analyzed contain 5 sirtuins, with the exception of S. pombe (encoding 3 sirtuins), and Encephalitozoon cuniculi (Katinka et al., 2001) that encodes only one sirtuin of the fungus-specific class. Thus sirtuins encoded by fungi and Choanoflagellates are represented in all groups except for SIRT3, arguing for an early radiation of sirtuins in the evolution of Opisthokonts (Fig. 1, Table 1).
We next looked at the plants, a further major group of eukaryotes that includes vascular plants, mosses, and red and green algae. Surprisingly, we found that the moss Physcomitrella patens (Rensing et al., 2008) encodes sirtuin SIRT4- to SIRT7-like sequences, while only SIRT4 and SIRT6 were found in the angiosperms we analyzed (Fig. 1, Table 1). Interestingly, one green alga, Ostreococcus lucimarinus, encodes a SIRT2 homolog, which appears to be lost in higher plants. Surprisingly, we were not able to find any sirtuins in red algae, although at present we cannot be sure whether this is due to incomplete sequence information. Members of five further groups of eukaryotes, namely amoebozoa [Dictyostelium discoideum (Eichinger et al., 2005; Gardner et al., 2002)], alveolata [Plasmodium falciparum (Gardner et al., 2002)], heterokonta [Phaeodactylum tricornutum (Bowler et al., 2008)], excavata (Giardia lamblia) and discicristata, (Leishmania infantum) contain divergent sirtuins that cannot be firmly assigned phylogenetically, as well as SIRT2, SIRT4 and SIRT6 family members (Fig. 1, Table 1).
We next searched for sirtuins encoded in animals with radial symmetry (radiata), which include examples of cnidaria (corals, sea anemones and jellyfish) such as the starlet sea anemone Nematostella vectensis and Hydra magnipapilata. Amongst this group of animals, the placozoan Trichoplax adhaerens is a representative of a basal eumetazoan lineage (all animal clades except sponges) that diverged before the separation of cnidarians and bilaterians (Srivastava et al., 2008). Trichoplax is comprised of a flat disc of cells with two epithelial layers sandwiching a layer of multinucleate fiber cells. Within this species we could find homologs of all human sirtuin groups except for SIRT1. Thus relatives of all seven sirtuin groups are already present in animals with radial symmetry, and indeed all of them can be found in Nematostella. The absence of a SIRT1 homolog in Trichoplax and of several sirtuins in Hydra could also be due to incomplete sequence information. Nevertheless, our analysis clearly indicates that representatives of all seven sirtuin groups were present in the common ancestor of all animals.
The two most important non-vertebrate animal models are the nematode C. elegans and the fruit fly D. melanogaster. Insects, forming part of the larger group of arthropods, and nematodes are classified as members of the ecdysozoa (Der Ou et al., 2007). Consistent with this grouping, we find that all ecdysozoan species we analyzed failed to encode a sirtuin of the SIRT3 group, which we deduce must have been lost early in the evolution of ecdysozoa. Further, the focused analysis of nematode sequences provides evidence for extensive loss of sirtuins at an early evolutionary stage. Fully sequenced C. elegans and C. briggsae genomes (Stein et al., 2003) contain clear SIRT1 and SIRT4 homologs, while an additional divergent sirtuin in both Caenorhabditis species does not cluster with any other sirtuin groups. Interestingly, another nematode Brugia malayi (Ghedin et al., 2007) does not contain a SIRT4 homolog, although clear SIRT1, SIRT6 and SRIT7 homologs are encoded. Within arthropods, several insects including the honey bee Apis mellifera (Solignac et al., 2007), the parasitoid wasp Nasonia vitripennis, and the red flour beetle Tribolium castaneum (Richards et al., 2008) contain homologs of all sirtuins except SIRT3, while the nearly completely sequenced Drosophila melanogaster genome additionally lacks a SIRT5 homolog, and the mosquito Anopheles gambiae encodes only three sirtuins. Thus, the loss of specific sirtuin groups is characteristic of nematodes and arthropod lineages.
Structural features and biochemical function
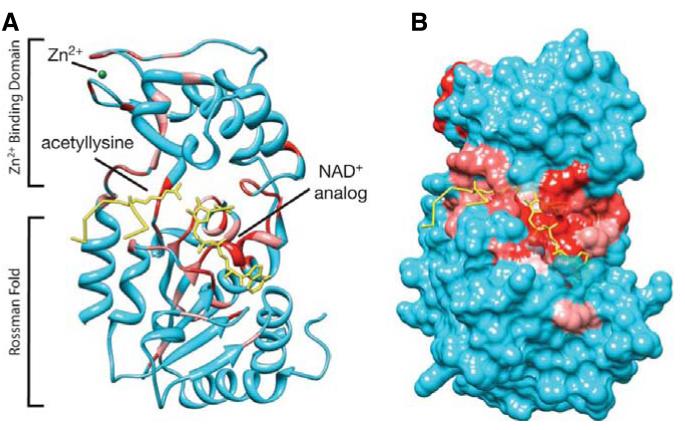
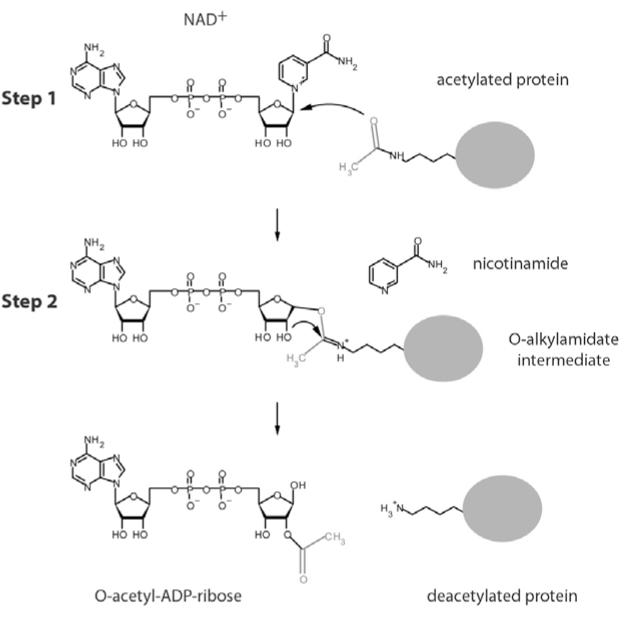
A number of sirtuins from different organisms have been crystallized (Avalos et al., 2002; Chang et al., 2002; Finnin et al., 2001; Min et al., 2001; Zhao et al., 2003a; 2003b). They all consist of a highly conserved catalytic core of approximately 250 amino acids, composed of a NAD+-binding Rossman fold domain, and a Zn2+-binding domain containing four highly conserved cysteine residues (Fig. 2). The catalytic site is situated inside a hydrophobic channel formed at the interface of the two domains, wherein the end of the acetyllysine side chain is arranged in close proximity to the nicotinamide ribose of NAD+ (Fig. 2A, Supplementary File 5). In contrast to class I, II and IV histone deacetylases where zinc participates in catalysis to produce free acetate and deacetylated lysine (Holbert and Marmorstein, 2005), sirtuins do not use zinc in their catalytic center, but it rather has a structural role. Sirtuins transfer the acetyl moiety from the ε-amino group of lysine to the nicotinamide ribose of NAD+ (Fig. 3). The sirtuin reaction is thought to proceed in two stages: firstly the acetyl oxygen replaces nicotinamide either by an SN1 or more likely an SN2 mechanism (Hu et al., 2008) to produce an O-alkyamidate intermediate and nicotinamide product; in the second step the acetyl group is transferred to ADP-ribose to form O-acetyl-ADP-ribose and deacetylated lysine (Fig. 3). O-acetyl-ADP-ribose is a metabolite uniquely generated by sirtuin deacetlyases (Imai et al., 2000; Landry et al., 2000; Sauve et al., 2001; Smith et al., 2000; Tanner et al., 2000; Tanny and Moazed, 2001).
Fig. 2.
Structural context of conserved residues. Structure of budding yeast Hst2p catalytic domain in complex with acetyllysine histone H4 peptide (yellow) and a non-hydrolyzable NAD+ analogue (yellow) (Zhao et al., 2004). Amino acids 4 to 204 and 213 to 287 of the structure were used. Amino acids 205 to 212 (WLREKITT) were excluded because they are unique to Hst2p and not conserved in any other sirtuin. Conserved residues were visualized using Chimera software (Meng et al., 2006; Pettersen et al., 2004); highly conserved residues are shown in red. A rotational 360° view of the structure is shown in Supplementary File 5 (ribbon view, Fig. 1A) and Supplementary File 6 (surface view, Fig. 1B).
Fig. 3.
Deacetylation mechanism catalyzed by sirtuins. Sirtuin-mediated deacetylation proceeds in two steps. In the first step nicotinamide is cleaved, yielding an O-alkylamidate intermediate. In the second step the nicotinamide ribose 2′OH group attacks the intermediate, yielding deacetylated lysine and O-acetyl-ADP-ribose.
Numerous studies have found that some sirtuins also possess ADP-ribosyl transferase activity in addition to their protein deacetylase activity, amongst them are yeast Sir2p, trypanosoma Sir2, and mammalian SIRT4 and SIRT6 (Haigis et al., 2006; Kowieski et al., 2008; Liszt et al., 2005; Tanny et al., 1999; Tsang and Escalante-Semerena, 1998). For SIRT4, only ADP-ribosylation but no deacetylation activity has so far been reported (Ahuja et al., 2007; Haigis et al., 2006). However, a recent study using yeast Sir2p and Hst1p, as well as several mammalian sirtuins including SIRT4, has questioned the physiological significance of sirtuincatalyzed ADP-ribosylation. Du et al. found that the ADP-ribosyl transferase activity of sirtuins is far weaker than their protein deacetylation activity and several orders of magnitude below that of the bacterial ADP-ribosyl transferase, diphtheria toxin. ADP-ribosylation may therefore be an insignificant side reaction (Du et al., 2009).
Generating a multiple sequence alignment (Supplementary File 4) allowed us to visualize the level of conservation of amino acids on the published budding yeast Hst2 structure. It is apparent that almost all highly conserved residues (red) are involved in the formation of the catalytic channel, the binding of NAD+, and in the coordination of the acetyllysine (Fig. 2B, Supplementary File 6). Apart from the amino acids forming the catalytic channel, the most highly conserved residues are four structurally important cysteines that coordinate Zn2+- binding. Although no clear target consensus sequences have yet been identified in sirtuin substrates, it is possible that sirtuins recognize their targets through interactions that lie outside the catalytic domain (Blander et al., 2005; Khan and Lewis, 2005; Mead et al., 2007). While the majority of sirtuin proteins consist solely of a catalytic domain, the class Ia subgroup that includes the human SIRT1 homolog of budding yeast Sir2p, also contain extensive N- and C-terminal domains (Frye, 2000). It is interesting to speculate that these domains are needed to interact with binding partners, or for conferring specificity for substrate recognition.
CONCLUSION
Despite the recent explosion in the number of reports on sirtuins there are still substantial gaps in our understanding of sirtuin function. Our evolutionary analysis provides clear evidence that all seven sirtuin families are ancient in animal evolution. Thus, a clearer understanding of the function of sirtuins may benefit from the analysis of simple organisms. For instance, the use of RNAi technology in the cnidarian Nematostella vectensis, may aid in elucidating the key functions related to individual sirtuins (Pankow and Bamberger, 2007). Given that sirtuins have been selectively and extensively lost during evolution, especially in insects, nematodes and plants, it appears likely that the loss of individual sirtuins might be compensated for redundant functions conferred by remaining sirtuin family members. This might indeed explain the relatively weak phenotypes associated with the reported murine single sirtuin knockouts, that all permit development into adult mice. Furthermore, functional redundancy between sirtuins may account for the failure to confirm the physiological importance of sirtuin-mediated deacetylation events of known in vitro substrates. Thus, to understand the core functions of sirtuins it will be necessary to analyze strains with mutations in multiple sirtuins using genetically tractable model organisms containing few sirtuin homologs, such as fruit flies, nematodes or Arabidopsis. Additionally, very little is known about the mechanism(s) of substrate recognition by sirtuins, and whether this requires additional proteins. Budding yeast Sir2p is known to be part of different complexes with distinct biological functions (Aparicio et al., 1991; Shou et al., 1999; Straight et al., 1999). It will therefore be interesting to assess whether this is a common theme in sirtuin regulation. Again, a phylogenetic approach might provide helpful insights. For instance, while sirtuin conservation is highest around the catalytic center, more extensive modeling of individual sirtuin classes combined with structural studies might identify conserved surface residues likely to be important for mediating interactions to provide either substrate specificity or binding specificity for essential protein cofactors. Given the increasing evidence of sirtuin-mediated protection from aging and aging-related neurodegenerative disease, the development of compounds that specifically activate or inhibit individual sirtuin members could be aided by comparative phylogenetic and structural analyses.
ACKNOWLEDGMENTS
We are grateful to Ashley Craig for helpful comments and to Jim Procter and Monika Rella for help with sequence analysis and visualization. Funding was provided by a CRUK CDA fellowship and a WT project grant to Anton Gartner.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, Maechler P, Verdin E. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J. Biol. Chem. 2007;282:33583–33592. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- Astrom SU, Cline TW, Rine J. The Drosophila melanogaster sir2+ gene is nonessential and has only minor effects on position-effect variegation. Genetics. 2003;163:931–937. doi: 10.1093/genetics/163.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avalos JL, Celic I, Muhammad S, Cosgrove MS, Boeke JD, Wolberger C. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol. Cell. 2002;10:523–535. doi: 10.1016/s1097-2765(02)00628-7. [DOI] [PubMed] [Google Scholar]
- Baldauf SL. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- Blander G, Olejnik J, Krzymanska-Olejnik E, McDonagh T, Haigis M, Yaffe MB, Guarente L. SIRT1 shows no substrate specificity in vitro. J. Biol. Chem. 2005;280:9780–9785. doi: 10.1074/jbc.M414080200. [DOI] [PubMed] [Google Scholar]
- Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- Braunstein M, Sobel RE, Allis CD, Turner BM, Broach JR. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- Chang JH, Kim HC, Hwang KY, Lee JW, Jackson SP, Bell SD, Cho Y. Structural basis for the NAD-dependent deacetylase mechanism of Sir2. J. Biol. Chem. 2002;277:34489–34498. doi: 10.1074/jbc.M205460200. [DOI] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- Dai JM, Wang ZY, Sun DC, Lin RX, Wang SQ. SIRT1 interacts with p73 and suppresses p73-dependent transcriptional activity. J. Cell Physiol. 2007;210:161–166. doi: 10.1002/jcp.20831. [DOI] [PubMed] [Google Scholar]
- Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J. Sirtuins: the ‘magnificent seven’, function, metabolism and longevity. Ann. Med. 2007;39:335–345. doi: 10.1080/07853890701408194. [DOI] [PubMed] [Google Scholar]
- Der Ou, H D, Lohr F, Vogel V, Mantele W, Dotsch V. Structural evolution of C-terminal domains in the p53 family. EMBO J. 2007;26:3463–3473. doi: 10.1038/sj.emboj.7601764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Jiang H, Lin H. Investigating the ADP-ribosyltransferase activity of sirtuins with NAD analogs and 32P-NAD. Biochemistry. 2009;48:2878–2890. doi: 10.1021/bi802093g. [DOI] [PubMed] [Google Scholar]
- Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol. Cell Biol. 2003;23:3173–3185. doi: 10.1128/MCB.23.9.3173-3185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnin MS, Donigian JR, Pavletich NP. Structure of the histone deacetylase SIRT2. Nat. Struct. Biol. 2001;8:621–625. doi: 10.1038/89668. [DOI] [PubMed] [Google Scholar]
- Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritze CE, Verschueren K, Strich R, Easton ER. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, Crabtree J, Allen JE, Delcher AL, Guiliano DB, Miranda-Saavedra D, et al. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- Greiss S, Hall J, Ahmed S, Gartner A. C. elegans SIR-2.1 translocation is linked to a proapoptotic pathway parallel to cep-1/p53 during DNA damage-induced apoptosis. Genes Dev. 2008;22:2831–2842. doi: 10.1101/gad.482608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Guarente LP. Mammalian sirtuins - emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. USA. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka M, Inoue T, Toda T, Kimura N, Shirayoshi Y, Kamitani H, Watanabe T, Ohama E, Tahimic CG, Kurimasa A, et al. Proteomics-based identification of differentially expressed genes in human gliomas: down-regulation of SIRT2 gene. Biochem. Biophys. Res. Commun. 2003;309:558–566. doi: 10.1016/j.bbrc.2003.08.029. [DOI] [PubMed] [Google Scholar]
- Holbert MA, Marmorstein R. Structure and activity of enzymes that remove histone modifications. Curr. Opin. Struct. Biol. 2005;15:673–680. doi: 10.1016/j.sbi.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Hu P, Wang S, Zhang Y. Highly dissociative and concerted mechanism for the nicotinamide cleavage reaction in Sir2Tm enzyme suggested by Ab Initio QM/MM molecular dynamics simulations. J. Am. Chem. Soc. 2008;130:16721–16728. doi: 10.1021/ja807269j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Inoue T, Hiratsuka M, Osaki M, Yamada H, Kishimoto I, Yamaguchi S, Nakano S, Katoh M, Ito H, Oshimura M. SIRT2, a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress. Oncogene. 2006;26:945–957. doi: 10.1038/sj.onc.1209857. [DOI] [PubMed] [Google Scholar]
- Ivy JM, Hicks JB, Klar AJ. Map positions of yeast genes SIR1, SIR3 and SIR4. Genetics. 1985;111:735–744. doi: 10.1093/genetics/111.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivy JM, Klar AJ, Hicks JB. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol. Cell. Biol. 1986;6:688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q, Yan T, Ge X, Sun C, Shi X, Zhai Q. Cytoplasm-localized SIRT1 enhances apoptosis. J. Cell Physiol. 2007;213:88–97. doi: 10.1002/jcp.21091. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AN, Lewis PN. Unstructured conformations are a substrate requirement for the Sir2 family of NAD-dependent protein deacetylases. J. Biol. Chem. 2005;280:36073–36078. doi: 10.1074/jbc.M508247200. [DOI] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar AJ, Fogel S, Macleod K. MAR1 - a Regulator of the HMa and HMα Loci in Saccharomyces cerevisiae. Genetics. 1979;93:37–50. doi: 10.1093/genetics/93.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowieski TM, Lee S, Denu JM. Acetylation-dependent ADP-ribosylation by Trypanosoma brucei Sir2. J. Biol. Chem. 2008;283:5317–5326. doi: 10.1074/jbc.M707613200. [DOI] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Schwer B, Alt FW, Mostoslavsky R. SIRT6 in DNA repair, metabolism and ageing. J. Intern. Med. 2008;263:128–141. doi: 10.1111/j.1365-2796.2007.01902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006;126:257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Mead J, McCord R, Youngster L, Sharma M, Gartenberg MR, Vershon AK. Swapping the gene-specific and regional silencing specificities of the Hst1 and Sir2 histone deacetylases. Mol. Cell. Biol. 2007;27:2466–2475. doi: 10.1128/MCB.01641-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng EC, Pettersen EF, Couch GS, Huang CC, Ferrin TE. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics. 2006;7:339. doi: 10.1186/1471-2105-7-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–279. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Newman BL, Lundblad JR, Chen Y, Smolik SM. A Drosophila homologue of Sir2 modifies position-effect variegation but does not affect life span. Genetics. 2002;162:1675–1685. doi: 10.1093/genetics/162.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 2004;5:224. doi: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankow S, Bamberger C. The p53 tumor suppressor-like protein nvp63 mediates selective germ cell death in the sea anemone Nematostella vectensis. PLoS ONE. 2007;2:e782. doi: 10.1371/journal.pone.0000782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado DO, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potente M, Ghaeni L, Baldessari D, Mostoslavsky R, Rossig L, Dequiedt F, Haendeler J, Mione M, Dejana E, Alt FW, et al. SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev. 2007;21:2644–2658. doi: 10.1101/gad.435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SH, Watkins DN, Herman JG, Baylin SB. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, Gibbs R, Bucher G, Friedrich M, Grimmelikhuijzen CJ, et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MI, Parkhurst SM. Drosophila Sir2 is required for heterochromatic silencing and by euchromatic Hairy/E(Spl). bHLH repressors in segmentation and sex determination. Cell. 2002;109:447–458. doi: 10.1016/s0092-8674(02)00732-8. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Celic I, Avalos J, Deng H, Boeke JD, Schramm VL. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry. 2001;40:15456–15463. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J. Mol. Biol. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. USA. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Shevchenko A, Charbonneau H, Deshaies RJ. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles - a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solignac M, Zhang L, Mougel F, Li B, Vautrin D, Monnerot M, Cornuet JM, Worley KC, Weinstock GM, Gibbs RA. The genome of Apis mellifera: dialog between linkage mapping and sequence assembly. Genome Biol. 2007;8:403. doi: 10.1186/gb-2007-8-3-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, et al. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J. Biol. Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- Tanny JC, Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. USA. 2001;98:415–420. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Tsang AW, Escalante-Semerena JC. CobB, a new member of the SIR2 family of eucaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2. J. Biol. Chem. 1998;273:31788–31794. doi: 10.1074/jbc.273.48.31788. [DOI] [PubMed] [Google Scholar]
- van der Horst A, Tertoolen LG, Vries-Smits LM, Frye RA, Medema RH, Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2SIRT1. J. Biol. Chem. 2004;279:28873–28879. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng EE, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1). functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, Nemoto S, Finkel T, Gu W, Cress WD, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat. Cell Biol. 2006;8:1025–1031. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2 - a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Schoonjans K, Auwerx J. Sirtuin functions in health and disease. Mol. Endocrinol. 2007;21:1745–1755. doi: 10.1210/me.2007-0079. [DOI] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Chai X, Clements A, Marmorstein R. Structure and autoregulation of the yeast Hst2 homolog of Sir2. Nat. Struct. Biol. 2003a;10:864–871. doi: 10.1038/nsb978. [DOI] [PubMed] [Google Scholar]
- Zhao K, Chai X, Marmorstein R. Structure of the yeast Hst2 protein deacetylase in ternary complex with 2′-O-acetyl ADP ribose and histone peptide. Structure. 2003b;11:1403–1411. doi: 10.1016/j.str.2003.09.016. [DOI] [PubMed] [Google Scholar]
- Zhao K, Harshaw R, Chai X, Marmorstein R. Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD+-dependent Sir2 histone/protein deacetylases. Proc. Natl. Acad. Sci. USA. 2004;101:8563–8568. doi: 10.1073/pnas.0401057101. [DOI] [PMC free article] [PubMed] [Google Scholar]