Abstract
One model for retroviral transduction suggests that template switching between viral RNAs and polyadenylation readthrough sequences is responsible for the generation of acute transforming retroviruses. For this study, we examined reverse transcription products of human immunodeficiency virus (HIV)-based vectors designed to mimic postulated transduction intermediates. For maximization of the discontinuous mode of DNA synthesis proposed to generate transductants, sequences located between the vectors' two long terminal repeats (vector “body” sequences) and polyadenylation readthrough “tail” sequences were made highly homologous. Ten genetic markers were introduced to indicate which products had acquired tail sequences by a process we term transductive recombination. Marker segregation patterns for over 100 individual products were determined, and they revealed that more than half of the progeny proviruses were transductive recombinants. Although most crossovers occurred in regions of homology, about 5% were nonhomologous and some included insertions. Ratios of encapsidated readthrough and polyadenylated transcripts for vectors with wild-type and inactivated polyadenylation signals were compared, and transductive recombination frequencies were found to correlate with the readthrough transcript prevalence. In assays in which either vector body or tail could serve as a recombination donor, recombination between tail and body sequences was at least as frequent as body-body exchange. We propose that transductive recombination may contribute to natural HIV variation by providing a mechanism for the acquisition of nongenomic sequences.
Retroviral transduction of cellular genes is well documented (5, 58) and was best characterized by the discovery of the cellular counterparts of the oncogenes in acute transforming retroviruses (49). These oncogenes were found embedded in viral genomes, replacing part of the viral coding sequences and rendering most acute transforming retroviruses replication defective. Scattered evidence, including a drug-resistant patient isolate and vectors studied in cultured cells, suggests the possibility of transduction by human immunodeficiency virus type 1 (HIV-1) (43, 53), but the ability of HIV to perform the specific steps required for transduction has not been addressed experimentally.
Several models have been proposed for oncogene transduction (4, 15, 16, 19, 29, 38, 41, 44, 50, 54, 56, 61). Most suggest that the fusing of host and viral sequences resulted from a series of rare events that are intrinsic to the viral replication strategy (52). One model for transduction is as follows. First, a retrovirus integrates just upstream of the sequence that will be transduced. Retroviral polyadenylation signals are leaky (18), and readthrough of such signals fuses sequences adjacent to the proviral insertion to the viral genomic RNA's 3′ end. Such readthrough transcripts can be encapsidated and serve as templates for normal reverse transcription products whose synthesis bypasses the RNA readthrough 3′ ends (19, 55). Alternately and on rare occasions, cellular sequences at the 3′ ends of readthrough RNAs become patched into the retroviral genome via nonhomologous recombination between a viral genomic RNA and the readthrough RNA or its alternately spliced derivative, resulting in host gene transduction (29, 54, 62).
For this study, reverse transcription products of HIV-1-based vectors designed to mimic the readthrough RNAs of putative transduction intermediates were analyzed. The vectors used in this study were composed of two parts: a body, defined as the genetic elements between 5′ and 3′ long terminal repeats (LTRs), and a tail, composed of sequences that formed a 3′ extension when 3′ LTR polyadenylation signals were read through (Fig. 1A). Experimental evidence suggests that homologous retroviral recombination is 2 to 3 orders of magnitude more frequent than nonhomologous recombination (1, 62). Thus, to enhance the frequency of transduction-type, or transductive, recombination, these vectors were designed with extended homologies between donor sequences in the vector 3′ tail regions and acceptor sequences in vector bodies.
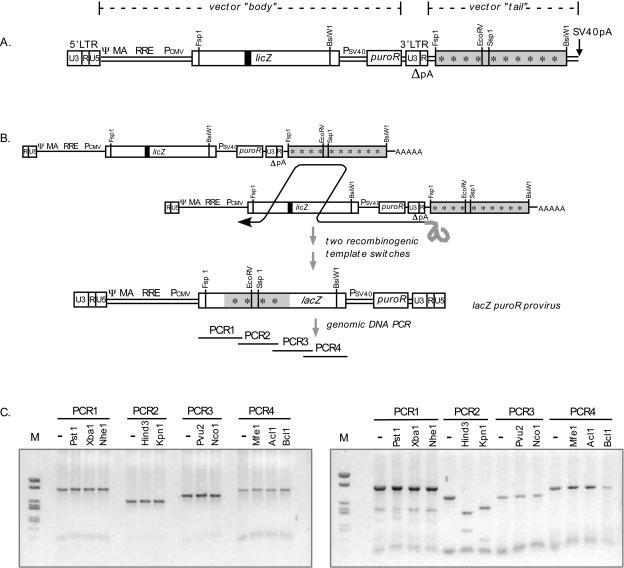
FIG. 1.
HIV-1 homologous transduction vector and protocol for determining template switch junctions. (A) Constitutive readthrough vector. The vector illustrated is LicPuro-äc with a downstream lacZ marked by synonymous point mutations (indicated by 10 asterisks). The 117-bp internal deletion in the body lacZ is indicated by a filled box and designated licZ. Ψ, packaging signal; MA, matrix coding sequence with frameshift mutation; RRE, Rev-responsive elements; Pcmv, cytomegalovirus immediate-early promoter; lacZ, β-galactosidase gene; Psv40, SV40 early promoter; puroR, puromycin resistance gene. (B) Transductive recombination. A predicted template switch pattern for the generation of a blue provirus is shown. PCR1 to PCR4 indicate the products used for the analyses described below. (C) Restriction pattern for a white proviral clone (left) and a blue proviral clone (right). Each of the four PCR products was digested with the indicated restriction enzymes. Undigested DNA was loaded as a size control (lane −) for each sample. Note that the sense primer for PCR1 and the antisense primer for PCR4 were located in 5′- and 3′-terminal regions of lacZ that were absent from the tail. All digestion products of each PCR product in the left panel retained the size of the undigested control, indicating that none of the tail genetic markers was present and that this was a nonrecombinant clone. In the right panel, most PCR products remained undigested except for those in the HindIII and KpnI lanes of PCR2. Digestion with these indicated the presence of tail-derived sequences, demonstrating that the provirus had the structure of the last of the two-crossover blue clones represented in Fig. 2B. M, DNA size marker pUC19/ApaLI+pUC19/HaeIII.
For the studies presented here, vector bodies contained a selectable marker gene and a lacZ expression cassette in which coding sequences were disrupted by a 117-base deletion (this disrupted gene was designated licZ). The 3′ tail contained a 2.6-kb internal portion of lacZ, which included and flanked the 117-base region deleted from the body. If two template switches occurred between licZ in a vector's body (the recombination acceptor region) and the fragment of lacZ at the 3′ end of either the same or the copackaged RNA (the recombination donor), a functional lacZ gene could be generated (Fig. 1B). This would yield drug-resistant colonies that stained blue with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), which would be diagnostic of transductive recombination. Transductive recombination was assessed both by X-Gal staining, which monitored a form of recombination that has been called patch repair (27, 45), and by the segregation of markers engineered to differentiate between vector body and tail sequences. The prevalence of packaged polyadenylation readthrough transcripts was compared to transductive recombination rates, and the effects of donor sequence context were assessed.
MATERIALS AND METHODS
Plasmids.
The HIV-1 helper pCMVΔR8.2 (28) encodes all trans-acting proteins except the envelope protein. For the generation of the vector used in this study, LicPuro-äc, the 3′ LTR polyadenylation sequence in LicPuro-ac, previously called HIV L̂cPuro-ac (31), was deleted. This caused a constitutive readthrough into tail sequences, including a 2.6-kb FspI-BsiWI fragment of lacZ and a simian virus 40 (SV40) polyadenylation signal. The tail lacZ allele was marked with 10 point mutations (represented by “ä” in the vector name), creating sites for PstI, XbaI, NheI, HindIII, KpnI, PvuII, NcoI, MfeI, AclI, and BclI. Intervals between markers ranged from 175 to 287 bp. The native polyadenylation signal vector, LicPuro(pA)äc, was derived from LicPuro-äc by restoring the 3′ LTR polyadenylation signal. In the double pA vector, LicPuro(2pA)äc (2pA stands for double pA), an SV40 polyadenylation signal was placed immediately downstream of the 3′ LTR of LicPuro(pA)äc, before the lacZ-containing tail. LicPuro was identical to HIV LacPuro except for a 117-bp internal lacZ deletion; these have been described previously (31). The trans repair vector HIV ac-äc contained two copies of a 1.8-kb FspI-SacI fragment of lacZ that differed from one another by a synonymous point mutation within the region corresponding to the 117-bp deletion. Construction details are available upon request.
Transfection and virus harvesting.
All cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Gemini). Virions were generated by transiently transfecting ET cells with pCMVΔR8.2 (28) and transducing vector expression plasmids by calcium phosphate precipitation (9). ET cells are a 293T derivative that constitutively express an ecotropic envelope (35). The medium was replaced at 16 to 24 h post transfection, and virion-containing medium was collected at 36 to 48 h posttransfection. Residual cells were removed by centrifugation at 4°C for 5 min at 3,000 × g. Virus stocks were stored at −70°C prior to use. At least two stocks from separate transfections were prepared for each vector.
Infection and lacZ activity assay.
mATRC1/293 cells (293 cells that stably express an ecotropic receptor [25]) were infected for 1 h in the presence of 8 μg of Polybrene (Sigma)/ml. Puromycin (Sigma) selection, cell fixation, and X-Gal (Sigma) staining were performed as described previously (35), except that infected cells in each 60-mm-diameter dish were subcultured into two 100-mm-diameter dishes before selection. Blue and white colonies from all dishes were summed for each infection experiment. Infections with each stock were performed in duplicate or triplicate.
Cell cloning, PCR, and restriction analysis.
For the isolation of cell clones, infections were performed at a very low multiplicity of infection so that well-dispersed colonies formed, and puromycin selection was performed without prior subculturing. Clones were transferred to 24-well plates and subsequently expanded. lacZ expression was assayed by X-Gal staining of cells in duplicate dishes. Each clone's DNA was isolated by use of a Wizard Genomic DNA purification kit (Promega). For the determination of the structures of integrated proviruses, four separate PCRs for each DNA preparation were performed for 30 cycles each with the Expand long template system (Roche). PCR products were digested with appropriate restriction enzymes, resolved in 1% agarose gels, and visualized by ethidium bromide staining.
RNA quantification.
Virion RNAs were purified from filtered media as described previously (47) and were assayed by RNase protection assays to quantify ratios of copackaged RNA segments. Phosphorimager background values were subtracted and molarities were normalized by dividing the values by the number of radiolabeled residues in each product, as previously described (47). For the differentiation of two RNAs with a single probe, the probe spanned and included sequences that were complementary to the licZ deletion (31).
Calculations. (i) lacZ inactivation.
Blue colony frequencies were corrected for lacZ inactivation as described previously (31). Briefly, the proportion of LacPuro single replication cycle products that remained unstained by X-Gal was determined for parallel controls. These values were used to calculate an inactivation baseline to adjust observed staining values for the percentage of white colonies predicted to arise independent of recombination status.
(ii) Normalizing transductive recombination frequencies.
For the generation of Fig. 3A, restriction enzyme cutting frequencies at each position were initially summed separately for blue and white clones. Data from white and blue clones were then weighted to reflect blue clones among the total clone ratios within large populations, and the values for white and blue products were summed. To account for differing interval lengths, we divided the target sequence into 10 regions, with each centered at the restriction site and extending to the midpoints between adjacent sites. Region lengths ranged from 171 to 263 bp (225 bp on average). Thus, frequency data were divided by 225 and multiplied by 100 to derive the relative cleavage frequency at each position per 225 bases (see Fig. 3A). Values ranged from 4.6 to 33.4%, averaging 22.4%.
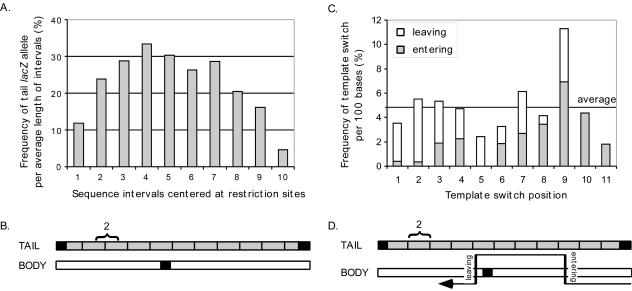
FIG. 3.
Frequency of tail lacZ allele sequences in proviral clones and distribution of template switch sites. (A) Population-wide tail allele frequency at various intervals, based on restriction analysis and calculated as described in Materials and Methods. (B) Schematic representation of tail and body lacZ alleles. The deleted regions of the tail and body alleles are shown as filled (black) boxes. The 10 genetic markers are indicated with short vertical lines. The interval indicated by “2” exemplifies how intervals relative to the body and tail lacZ alleles were defined to span genetic markers. (C) Distribution of crossovers, separated into entering and leaving points and localized to intervals as defined below. The average frequencies of template switching per 100 bases are indicated. (D) Template switch intervals, categorized as either entering or leaving points in relation to which lacZ allele was used as the template switch acceptor (see text for details). The positions of the 11 intervals relative to the body and tail lacZ alleles are exemplified by interval 2.
(iii) Crossover interval distribution.
For Fig. 3C, the numbers of entering and leaving points were tabulated for each region for all blue clones and all white clones, respectively, and were summed after the data were normalized to reflect blue clone/total clone ratios in larger populations. Since the 11 regions varied in length, the values were divided by the interval length to yield the probability of a template switch per 100 bases.
RESULTS
Genetic recombination using a vector resembling a transduction intermediate.
A specialized vector was constructed to allow the comprehensive detection of transductive recombinants (Fig. 1A). This vector was designed to assay the repair of the inactivated lacZ allele (termed licZ) in the vector body by two crossovers between the vector body and homologous sequences engineered into the tail of the same or copackaged RNA (Fig. 1B). The tail lacZ sequences of this vector, LicPuro-äc, were genetically marked with 10 synonymous single base substitutions (indicated by 10 asterisks in Fig. 1A and by “äc” in the vector name). The mutations were relatively evenly spaced and introduced 10 restriction sites separated by an average of 239 bp. Target cells were transduced by HIV virions containing this vector, and ratios of X-Gal-stained to total (stained plus unstained) puromycin-resistant colonies were determined. The rate of blue colony formation for LicPuro-äc was 24.6%, which was slightly lower than values reported for a parental vector without markers (31), possibly reflecting the suppressive effects of genetic heterogeneity on recombination (the body and tail lacZ alleles differed by ∼1%) (1, 37).
These assays were performed with lentiviral vectors that included sequences engineered downstream of proviral structures, and thus expression cassettes for these vectors could not readily be introduced into cells by retroviral transduction, the favored method for establishing vector expression in many retroviral genetic recombination studies. Because the assays for this study relied on transient transfections for vector expression plasmid introduction, DNA-level recombination was a major concern and a potential confounding factor. Thus, the following controls were performed to assess how much LacZ-positive signal could be ascribed to DNA-level recombination during transfection. The first involved the cotransfection of a tail-less lacZ deletion-containing LicPuro vector expression plasmid and the pCMVΔR8.2 helper plasmid with a molar excess of the nonretroviral mammalian lacZ expression plasmid pCH110 and the determination of the blue colony titer of the resulting virions as previously described (31). This control for possible DNA-level recombination between markers on two cotransfected plasmids yielded a very low (<0.1%) level of blue colony formation in the cells that were subsequently infected with the resulting virions. The blue colony formation rate of these control cotransfections was >250-fold lower than the level observed for products of the LicPuro-äc vector. This control suggested that low levels of DNA-level recombination did occur. However, even when the DNA-level recombination donor was present at higher ratios to the acceptor than would occur in the LicPuro-äc transductive recombination experiments, the level of DNA recombination was only about 0.25% of that which resulted from template switching during reverse transcription.
A second control for DNA-level recombination involved a new vector, LicPuro(2pA)äc, which differed from LicPuro-äc only by the insertion of two polyadenylation signals—that of the native HIV LTR and a secondary one from SV40—into the region between the body and tail portions of LicPuro-äc (Fig. 1A). This design was intended to minimize the formation of readthrough tail-containing transcripts while leaving the putative substrates for DNA-level recombination of LicPuro-äc unchanged. LicPuro(2pA)äc was cotransfected with the pCMVΔR8.2 helper, and blue colony titers for products of the resulting virions were determined. Blue colonies were observed in roughly 0.3% of all LicPuro(2pA)äc products, or nearly 100-fold less frequently than for LicPuro-äc. Because blue colonies due to DNA-level recombination during transfection should arise at the same frequency for both LicPuro(2pA)äc and LicPuro-äc, this control suggests that no more than roughly 1% of the recombinants could possibly be ascribed to two crossovers at the DNA level. However, even the 1% figure is likely an overestimate of DNA-level recombination, since among LicPuro(2pA)äc products, blue colonies could result from either DNA-level recombination or from residual polyadenylation signal readthrough and transductive recombination. As further addressed below, low levels of polyadenylation readthrough were detectable for LicPuro(2pA)äc.
Taken together, these controls suggest that although some plasmid DNA-level recombination occurred when the reported transient transfection approaches for vector expression plasmid introduction were taken, ≥99% of the recombinants studied here resulted from viral replication processes.
Transductive template switch frequencies and provirus structures.
The LicPuro-äc assay for transductive recombination was designed with no selective advantage for blue colony formation. Thus, it appeared likely that blue colony frequencies would underestimate transductive recombination, because the staining phenotype would not differentiate any recombinant products that contained the body-derived deleted interval from nonrecombinant white colonies. For use of the introduced genetic markers to more accurately quantify transductive recombinants, 103 individual proviral products were single-cell cloned and mapped with restriction enzymes as follows. Genomic DNAs from each of 32 blue and 71 white clones were amplified by use of four primer pairs to generate four consecutive end-overlapping lacZ PCR products (Fig. 1B, PCR1 through -4). Uninfected cell DNA served as a negative control and generated no PCR products. The vector PCR products were digested with restriction enzymes that were diagnostic of each of the 10 genetic markers and separated in 1% agarose gels to deduce crossover intervals. Sample gels for one blue and one white clone are shown in Fig. 1C. The structure of a clone that remained unstained with X-Gal yet which the genetic markers revealed to be recombinant is used as an example of the resulting product structures (Fig. 2A).
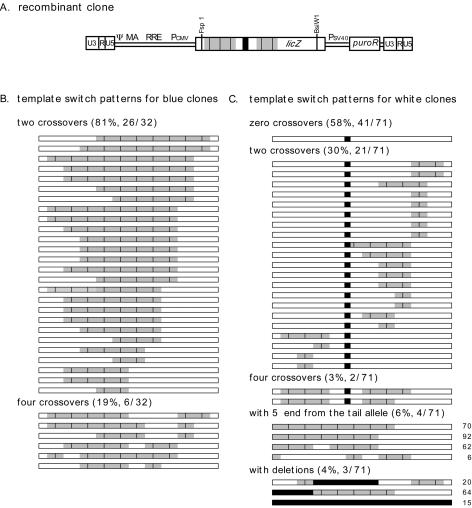
FIG. 2.
Schematic representation of template switch patterns from restriction and sequencing analyses. (A) Schematic drawing of one of the white clone products with four crossovers. Origins of regions are indicated by shading. Uncolored boxes represent regions from the body allele, and shaded regions were derived from the tail lacZ. Short vertical lines mark restriction sites. (B) Profiles for 32 blue clones. (C) Profiles for 71 white clones. Each horizontal bar represents the region of lacZ from an individual proviral clone that was characterized by restriction analysis, and for some, by sequencing. Deletions are shown as filled (black) regions. For the final seven white clones, the clone numbers used in Fig. 4 are indicated to the right.
Marker segregation results for all 32 of the analyzed blue clones are schematically presented in Fig. 2B. All deduced structures were consistent with the prediction that blue colony formation would require at least two template switches between body and tail lacZ alleles in regions flanking the 117-base deletion. Of the 32 blue clones, 81% (26 of 32) had two template switches and 19% (6 of 32) had four crossovers (Fig. 2B).
Structures of the 71 white clones are summarized in Fig. 2C. For 64 of these 71 clones, the lengths of all four PCR products, including the one with the 117-base deletion, were the same as those of the parental vector's body. For 41 of these 64, all restriction patterns were identical to those for the body, suggesting that no crossovers between the body and tail had occurred. For the remaining 23 clones whose PCR product lengths resembled those of the vector body, the 117-base deletion was retained but two or four crossovers were detected in regions flanking the deletion. The remaining seven white clone proviruses had abnormal structures in that they failed to be amplified by at least one of the four PCRs; the structures of these clones were determined separately by sequencing and are further described below. Of the white provirus-containing clones, 58% (41 of 71) displayed no evidence of template switches, 30% (21 of 71) had two crossovers, 3% (2 of 71) had four crossovers, 6% (4 of 71) had an odd number of template switches, and 4% (3 of 71) contained deletions (Fig. 2C).
These marker segregation data were used to calculate transductive recombination frequencies. The ratio of blue colonies to the total number of puromycin-resistant colonies for LicPuro-äc was 24.6%. For the generation of a more accurate figure that included the prevalence of transductive recombinants among white colonies, this phenotypic recombinant rate was integrated with the restriction analysis data. Because the blue and white clones analyzed above were not sampled in the same proportions as they arose in larger populations of transductants, the recombinant data were weighted to reflect the 24.6% blue composition of larger pools (see Materials and Methods). Such calculations suggested that overall, 56% of all puromycin-resistant products of LicPuro-äc were transductive recombinants (Table 1).
TABLE 1.
Frequency of template switching in characterized proviral clones
| Category | No. of clones | No. of crossovers per clone (total no. of crossovers) | Recombinant prevalence (%) (no. of recombinant clones) |
|---|---|---|---|
| Blue | 32 | 2.4 (76) | 100 (32) |
| White | 71 | 0.9 (67) | 42 (30) |
| Totala | 93 | 1.3 (118) | 56 (52) |
Values in the total category were standardized according to the blue/white ratio of clones from large-scale infections (0.305).
The restriction data were also used to compare the frequency of template switching in these assays to reported recombinogenic template switch rates for HIV-1 that were determined by other means. On average, reverse transcriptase (RT) switched templates 1.3 times during the replication of the 2.6-kb lacZ target sequence (Table 1), which is equivalent to at least four crossovers per wild-type length of the HIV-1 genome (9.2 kb). This value is consistent with recombination frequencies reported for two-vector body-body recombination assays (21, 31).
Figure 3A displays the average frequencies at which tail allele sequences were observed in various portions of proviral lacZ for intervals that spanned the genetic markers, as shown in Fig. 3B (calculations are described in Materials and Methods). The allele prevalence data in Fig. 3A fit a fairly normal distribution. However, the additional analysis described below was necessary to address crossover frequencies at specific intervals.
Distribution of template switch sites.
It has been suggested that template switches do not occur at uniform frequencies but are highly context dependent (2, 60). To address whether or not template switch hot spots within the lacZ target were detectable, we classified crossover sites as either entering or leaving points, with entering points defined as sites at which RT switched to the tail and leaving points defined as sites where synthesis switched back to the body (Fig. 3D). Among the 11 regions delimited by the target's 5′ and 3′ ends and by the 10 restriction markers, the probabilities of a template switch ranged from 1.8 to 11.3%, with a 4.8% average value (Fig. 3C). Values for most intervals were close to the average. However, regions 5 (2.4%) and 11 (1.8%) were twofold below average. The low value for region 5 was likely due to the presence of the 117-base deletion in one lacZ allele, and that for region 11 likely reflected its location at the 3′ end of lacZ. The template switch probability for region 9 was 2.4-fold above average, suggesting that it may be a hot spot.
Structures of the seven aberrant white clone proviruses.
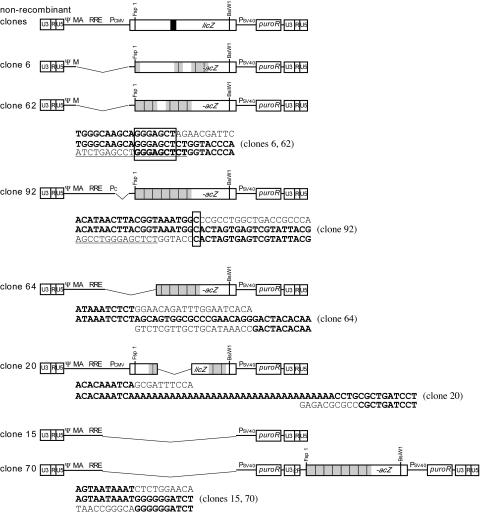
Provirus structures for the seven abnormal clones first detected by the PCR analyses described above are presented in Fig. 4. Sequencing analysis revealed that transductive recombination had occurred for clones 70, 92, 62, and 6 but that DNA synthesis at the tail had proceeded beyond the region where the tail and body were homologous. For three of these, a subsequent nonhomologous switch between extended tail sequences and the vector body contributed to proviral synthesis.
FIG. 4.
Structures of the seven aberrant proviruses. Shaded regions represent intervals from the tail lacZ; vertical lines mark restriction sites. Deletions are represented by disruptions of horizontal lines, with their lengths approximately to scale. Sequence alignments of the donor sequence (bottom sequence), the sequence present in the proviral clone (center), and the acceptor sequence (top) for deletion junctions are shown under each proviral structure. Donor and acceptor sequences that match that of the provirus are shown in bold; any homology between the donor and the acceptor at the transfer site is boxed; 3′ R sequences are underlined. The structure of a nonrecombinant white clone is shown at the top. Clones 6 and 62 differ in lacZ region crossovers but share the features of a reverse-transcribing tail beyond the lacZ portion into the 3′ R and a rejoining body using the same seven bases of donor-acceptor identity. Clone 92 is a nonhomologous recombinant between the tail region upstream of the lacZ sequences and the body, with a single base identity at the junction. Clones 64 and 20 have deletions with insertions. For clone 64, the insertion was 20 bases from the U5-PBS junction; for clone 20, the insertion was a 34-base homopolymeric A region followed by 4 bases of unknown origin. Clones 15 and 70 share a unique deletion that lacks junctional homology but differ in that clone 70 possesses a three-LTR structure.
Clone 70 was unique among those studied here in that the vector's original 3′ LTR was present between the product's two terminal LTRs, thus constituting a third central LTR in the provirus. Its formation likely involved a template switch from the body lacZ of one genomic RNA template to the tail of the copackaged RNA, followed by minus-strand synthesis through the 3′ LTR and the puromycin resistance cassette and into the 3′ end of the body lacZ. We detected only one three-LTR structure among the 103 clones analyzed. Similar three-LTR structures are frequent products in avian retrovirus nonhomologous transduction assays (A. Doshi and J. Coffin, personal communication).
The remaining three white clone proviruses contained lacZ-disrupting deletions. Nonhomologous junctions fell into the following three groups: those with short stretches of sequence identity (seven nucleotides in clones 6 and 62 and one nucleotide in clone 92), those that lacked any homology at switch junctions (clones 15 and 70), and those with sequence insertions (clones 64 and 20). Two separate pairs of identical template switch junctions were observed among the seven independent proviruses containing deletions: one involved microhomology (clones 6 and 62) while the other contained no junctional homology (clones 15 and 70).
Frequencies of native HIV-1 polyadenylation signal readthrough and transductive recombination.
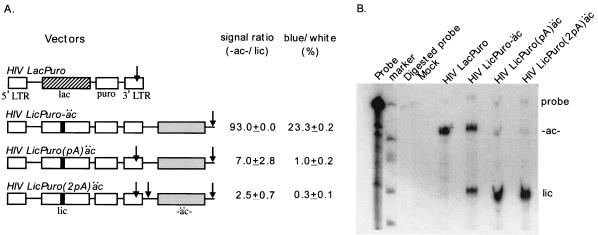
The high frequency of transductive recombination observed with this system allowed us to modify the vectors to test the hypothesis that transductive recombination frequencies correlate with the efficiency of polyadenylation readthrough. The vector used for these experiments, LicPuro(pA)äc, differed from LicPuro-äc by the presence of a wild-type polyadenylation signal (designated pA) in the 3′ LTR (Fig. 5A). The intention was for most transcripts to be cleaved and polyadenylated at the 3′ LTR but for some readthrough transcript formation and polyadenylation at the downstream SV40 polyadenylation signal to occur. Upon infection of fresh cells, recombination between this vector's body and a rare encapsidated readthrough tail should generate blue colonies after puromycin selection and X-Gal staining.
FIG. 5.
HIV-1 polyadenylation signal readthrough and readthrough RNA-related recombination. (A) Proviral structure, lacZ allele ratio from RNase protection assay, and blue/white ratio for vectors used to determine polyadenylation readthrough. Listed from the top downward are the wild-type lacZ vector (LacPuro), the constitutive readthrough vector (LicPuro-äc), the natural readthrough vector [LicPuro(pA)äc], and the double pA control vector [LicPuro(2pA)äc]. Arrows indicate the positions of pA signals within the vector. The RNase protection ratios of tail to body lacZ alleles and recombination rates are listed to the right. (B) RNase protection assay with viral RNAs from vectors shown in panel A. Probe, undigested probe. Signals corresponding to the protected body and tail lacZ alleles are indicated by “lic” and “-ac-,” respectively; the LacPuro body contains uninterrupted lacZ. Mock, medium from cells transfected with helper plasmid only.
The ratio of body-only to readthrough LicPuro(pA)äc RNAs in the virion population was determined by an RNase protection assay (Fig. 5B) and was compared to controls (Fig. 5A). One control vector was LicPuro(2pA)äc (Fig. 5A; also described above), which contained both the wild-type 3′ LTR pA signal and an SV40 polyadenylation signal between the body and tail lacZ alleles. For the LicPuro(pA)äc wild-type 3′ LTR vector, the prevalence of readthrough transcripts among packaged RNAs was about 7% that of the LicPuro-äc constitutive readthrough vector. The readthrough signal of the two-central-pA vector, LicPuro(2pA)äc, was about 2.8-fold lower than that of LicPuro(pA)äc (Fig. 5A).
Transductive recombination with these vectors was compared by using the blue-white colony phenotype assay. The frequency of transductive recombination with native polyadenylation signal vectors, as measured by X-Gal staining, was about 1%, which was roughly 4% as frequent as that with the constitutive readthrough vector. As noted above, the frequency of blue colony formation for the vector with two polyadenylation signals between the body and tail was approximately 0.3%.
Tail sequences are highly accessible to the recombination machinery.
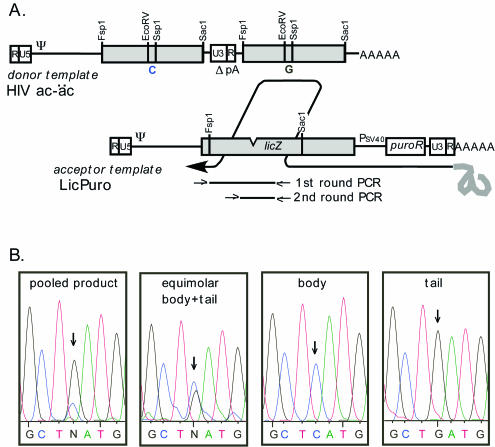
The donor template regions in the extended 3′ tails of the vectors used here were located outside of the RNA regions that ordinarily contribute to provirus formation. Thus, we established an assay to compare how well sequences at this ectopic site could compete with sequences between LTRs and serve as a recombination substrate. For this assay, LicPuro, a vector with the deleted licZ allele in its body but without an appended tail, was coexpressed with a trans repair vector called HIV ac-äc, which contained two potential donor template copies of an extended internal portion of lacZ (Fig. 6A). One donor copy (the body allele) was located between the two LTRs, while the other (the tail allele) was placed downstream of the mutant 3′ LTR. These two lacZ donor segments were identical except for a synonymous point mutation introduced into the region that was removed by the 117-bp deletion. Therefore, in a two-vector recombination assay using LicPuro and the trans repair vector, either the body or the tail HIV ac-äc lacZ allele could serve as the patch repair donor (Fig. 6A).
FIG. 6.
Comparison of body and readthrough tail sequences as participants in patch repair. (A) Schematic representation of how template switching with readthrough RNA during reverse transcription could yield a puromycin-resistant vector with functional lacZ. Note that the donor RNA template contains two copies of the internal portion of the lacZ gene, either of which could serve as a patch repair donor. Patch repair with the tail lacZ allele is shown. The approximate locations of the first- and second-round PCRs for amplifying pooled proviral DNAs are shown beneath the acceptor template. (B) Chromatograms from sequencing of PCR products from pooled genomic DNA (pooled product), from plasmid containing the body lacZ allele only (body), from plasmid containing the tail lacZ allele only (tail), and from HIV ac-äc plasmid DNA (equimolar body + tail). Arrows indicate the nucleotide positions that differed between the body and tail lacZ alleles.
Virions produced by cotransfection of LicPuro, HIV ac-äc, and a helper plasmid generated about 3% blue colonies. For determination of the fraction that had been repaired by each donor allele, genomic DNAs extracted from over 3,000 pooled puromycin-resistant colonies were analyzed. These DNAs were first PCR amplified with primers specific for proviruses generated through two crossover events (Fig. 6A, first-round PCR) and then were reamplified with nested primers identical to those used for PCR2 in Fig. 1B. The recombinant provirus products were gel purified and used directly for automatic sequencing to estimate the relative contribution of the two lacZ alleles (Fig. 6B). A control PCR and sequencing were performed in parallel for samples in which the two alleles were either unique or present in equimolar proportions. Among trans repair products, the signal representing the residue provided by the tail lacZ allele (nucleotide G) was significantly higher than that representing the body lacZ allele (nucleotide C), suggesting that the HIV ac-äc tail served as the patch repair donor at least as well as the body allele (Fig. 6B).
DISCUSSION
We have examined transduction-like events that can occur during single cycles of HIV-1 DNA synthesis. By these approaches, the calculated prevalence of homologous transductive recombinants among products of a constitutive readthrough vector was 56%. These findings demonstrate that the discontinuous mode of minus-strand DNA synthesis proposed to mediate retroviral transduction can occur with remarkable frequency (52).
Considering that sequences unlinked to sites of retroviral integration can be transduced (30, 53) and that several different models for transduction have been proposed, the mechanism described here is clearly not the only way a retrovirus can gain sequences outside its genome. However, our observations here suggest that HIV-1 reverse transcription complexes have a far greater capacity to recruit and utilize encapsidated homologous donor template sequences, regardless of their context, than has previously been recognized. RT complexes that simultaneously include both donor and acceptor templates have been reported as reverse transcription intermediates in studies of template switching at template ends by use of reconstituted reactions in vitro (34). More recent findings confirmed that an acceptor template can be “docked” to an elongating reverse transcription complex prior to the actual template switch from the donor template (40). The findings reported here, which show that the majority of all proviruses display evidence of nonlinear DNA synthesis, suggest that during HIV-1 replication, elongating RT may often, if not continually, exist in a conformation that is capable of simultaneously engaging both acceptor and donor template regions.
When all detectable crossovers were summed, the fraction that were nonhomologous was nearly 5%, which is significantly higher than some previously suggested frequencies but is roughly consistent with other reports of genetic rearrangements (57, 62). The spectrum of nonhomologous products was similar to those reported for oncogene-virus 3′ junctions (11, 14, 17, 23, 61) and other retroviral genetic rearrangements, including the occasional transduction of cellular RNA sequences within deletions (32, 33, 36, 57).
Several interesting features were observed among nonhomologous junctions. Clone 20 had an insertion of over 30 contiguous As within a deletion. Homopolymeric As have been reported for the 3′ junctions of some v-Oncs (20) and for some experimental recombinants (38, 39, 54, 62). These A residues may have been acquired by transductive recombination with the RNAs' 3′ polyadenylate. The source of another insertion with nonhomologous junctions (clone 64) was a 19-base distal portion of vector sequences, likely acquired by nonhomologous patch repair from the copackaged RNA. Such long-distance duplication of genetic material from one portion of the genome to another has been reported for a drug-resistant patient-derived HIV-1 isolate (24).
The use of homologous donor and acceptor regions resulted in high transductive recombination rates, which made it possible to modify the vectors to quantify the effects of polyadenylation readthrough rates on patch repair. The products of native polyadenylation signal readthrough comprised 7% of all virion RNAs in our system, which is within the reported range for HIV-1 polyadenylation signal readthrough (3, 6, 8, 10, 12, 13, 22, 46). The good correlation observed between patch repair frequencies and packaged RNA levels demonstrates that the frequency of transductive recombination was roughly proportionate to the amount of encapsidated readthrough transcript.
The high level of transductive recombination observed here suggests that sequences outside the normal context can contribute readily to provirus formation. To test this notion, we compared the ability of body and tail sequences to serve as patch repair donors in an intermolecular recombination assay. The results suggested that the tail provided repair sequences somewhat more frequently than did the body. A possible explanation for this finding is that sequences between LTRs only transiently exist as minus-strand recombination donor templates. Once they are reverse transcribed, they are converted to minus-strand DNA, which could not serve as a source of patch repair. At a minimum, these findings confirm that packaged sequences that do not reside in the normal genome context can readily contribute to provirus formation.
The data presented here suggest that transductive recombination may patch sequences into HIV genomes fairly often. However, there have been relatively few reports of natural transduction by HIV-1 (24, 43, 53). Reasons for this may include the fact that our system accelerated transduction by introducing donor-acceptor sequence homology. Also, at 9.7 kb, our readthrough vectors were only slightly longer than native HIV genomes, while some natural readthrough RNAs may be too lengthy to be packaged with wild-type efficiencies. Other reasons that there have been so few reports of transduction by HIV likely include HIV's tendency to kill cells, its requirement for central genome portions such as Rev-responsive elements for genome propagation, and the fact that purifying selection in vivo eliminates transductants that lack survival advantages.
Nonetheless, provocative examples of possible transduction can be identified by searching human databases by use of BLAST with certain HIV variants. Speculatively, we hypothesize that some of these may be transductants. An example is AVT-rich env variable region extension variants. The additional glycosylation signals some extensions encode may contribute to immune escape and therefore provide a selective advantage. However, the conservation in these extensions of rare AVT codons (where V = A, C, or G) over more abundant codons for the same amino acids is difficult to explain. Whereas AAT, ACT, and AGT are minor codons for their respective amino acids in other regions of HIV, these codons are highly overrepresented in env extensions (7, 26). AAT is the most common trinucleotide in human microsatellite DNA (51). Thus, it is intriguing that database searching reveals that many AAT-rich HIV-1 env variants (for example, GenBank accession numbers AB02999, AJ404200, and Z34513 [42, 48, 59]) display more similarity to highly abundant human microsatellite sequences than to other portions of HIV. It has previously been postulated that these V2 extensions might somehow be products of a host polymerase (7). The results we report here raise the interesting possibility that some such insertions may indeed be products of host DNA synthesis and that their mechanism of incorporation may be transductive recombination.
Acknowledgments
We thank David Friedman and John Moran for critical reading of the manuscript and Ashita Doshi and John Coffin for sharing results prior to publication.
This work was supported by NIH grant GM 63479.
REFERENCES
- 1.An, W., and A. Telesnitsky. 2002. Effects of varying sequence similarity on the frequency of repeat deletion during reverse transcription of a human immunodeficiency virus type 1 vector. J. Virol. 76:7897-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, J. A., E. H. Bowman, and W. S. Hu. 1998. Retroviral recombination rates do not increase linearly with marker distance and are limited by the size of the recombining subpopulation. J. Virol. 72:1195-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashe, M. P., P. Griffin, W. James, and N. J. Proudfoot. 1995. Poly(A) site selection in the HIV-1 provirus: inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev. 9:3008-3025. [DOI] [PubMed] [Google Scholar]
- 4.Besmer, P., J. E. Murphy, P. C. George, F. H. Qiu, P. J. Bergold, L. Lederman, H. W. Snyder, Jr., D. Brodeur, E. E. Zuckerman, and W. D. Hardy. 1986. A new acute transforming feline retrovirus and relationship of its oncogene v-kit with the protein kinase gene family. Nature 320:415-421. [DOI] [PubMed] [Google Scholar]
- 5.Bishop, J. M. 1983. Cellular oncogenes and retroviruses. Annu. Rev. Biochem. 52:301-354. [DOI] [PubMed] [Google Scholar]
- 6.Bohnlein, S., J. Hauber, and B. R. Cullen. 1989. Identification of a U5-specific sequence required for efficient polyadenylation within the human immunodeficiency virus long terminal repeat. J. Virol. 63:421-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosch, M. L., A. C. Andeweg, R. Schipper, and M. Kenter. 1994. Insertion of N-linked glycosylation sites in the variable regions of the human immunodeficiency virus type 1 surface glycoprotein through AAT triplet reiteration. J. Virol. 68:7566-7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, P. H., L. S. Tiley, and B. R. Cullen. 1991. Efficient polyadenylation within the human immunodeficiency virus type 1 long terminal repeat requires flanking U3-specific sequences. J. Virol. 65:3340-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cepko, C., and W. Pear. 2003. Transduction of genes using retrovirus vectors. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y. [Online.]
- 10.Cherrington, J., and D. Ganem. 1992. Regulation of polyadenylation in human immunodeficiency virus (HIV): contributions of promoter proximity and upstream sequences. EMBO J. 11:1513-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chesters, P. M., K. Howes, J. C. McKay, L. N. Payne, and K. Venugopal. 2001. Acutely transforming avian leukosis virus subgroup J strain 966: defective genome encodes a 72-kilodalton Gag-Myc fusion protein. J. Virol. 75:4219-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeZazzo, J. D., J. E. Kilpatrick, and M. J. Imperiale. 1991. Involvement of long terminal repeat U3 sequences overlapping the transcription control region in human immunodeficiency virus type 1 mRNA 3′ end formation. Mol. Cell. Biol. 11:1624-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeZazzo, J. D., J. M. Scott, and M. J. Imperiale. 1992. Relative roles of signals upstream of AAUAAA and promoter proximity in regulation of human immunodeficiency virus type 1 mRNA 3′ end formation. Mol. Cell. Biol. 12:5555-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girod, A., A. Drynda, F. L. Cosset, G. Verdier, and C. Ronfort. 1996. Homologous and nonhomologous retroviral recombinations are both involved in the transfer by infectious particles of defective avian leukosis virus-derived transcomplementing genomes. J. Virol. 70:5651-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldfarb, M. P., and R. A. Weinberg. 1981. Generation of novel, biologically active Harvey sarcoma viruses via apparent illegitimate recombination. J. Virol. 38:136-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodrich, D. W., and P. H. Duesberg. 1990. Evidence that retroviral transduction is mediated by DNA not by RNA. Proc. Natl. Acad. Sci. USA 87:3604-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hajjar, A. M., and M. L. Linial. 1993. A model system for nonhomologous recombination between retroviral and cellular RNA. J. Virol. 67:3845-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herman, S. A., and J. M. Coffin. 1986. Differential transcription from the long terminal repeats of integrated avian leukosis virus DNA. J. Virol. 60:497-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herman, S. A., and J. M. Coffin. 1987. Efficient packaging of readthrough RNA in ALV: implications for oncogene transduction. Science 236:845-848. [DOI] [PubMed] [Google Scholar]
- 20.Huang, C. C., N. Hay, and J. M. Bishop. 1986. The role of RNA molecules in transduction of the proto-oncogene c-fps. Cell 44:935-940. [DOI] [PubMed] [Google Scholar]
- 21.Jetzt, A. E., H. Yu, G. J. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2000. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 74:1234-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klasens, B. I., A. T. Das, and B. Berkhout. 1998. Inhibition of polyadenylation by stable RNA secondary structure. Nucleic Acids Res. 26:1870-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, S. Y., T. M. Howard, and S. Rasheed. 1998. Genetic analysis of the rat leukemia virus: influence of viral sequences in transduction of the c-ras proto-oncogene and expression of its transforming activity. J. Virol. 72:9906-9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lobato, R. L., E. Y. Kim, R. M. Kagan, and T. C. Merigan. 2002. Genotypic and phenotypic analysis of a novel 15-base insertion occurring between codons 69 and 70 of HIV type 1 reverse transcriptase. AIDS Res. Hum. Retrovir. 18:733-736. [DOI] [PubMed] [Google Scholar]
- 25.Malhotra, S., A. G. Scott, T. Zavorotinskaya, and L. M. Albritton. 1996. Analysis of the murine ecotropic leukemia virus receptor reveals a common biochemical determinant on diverse cell surface receptors that is essential to retrovirus entry. J. Virol. 70:321-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masciotra, S., S. M. Owen, D. Rudolph, C. Yang, B. Wang, N. Saksena, T. Spira, S. Dhawan, and R. B. Lal. 2002. Temporal relationship between V1V2 variation, macrophage replication, and coreceptor adaptation during HIV-1 disease progression. AIDS 16:1887-1898. [DOI] [PubMed] [Google Scholar]
- 27.Mikkelsen, J. G., and F. S. Pedersen. 2000. Genetic reassortment and patch repair by recombination in retroviruses. J. Biomed. Sci. 7:77-99. [DOI] [PubMed] [Google Scholar]
- 28.Naldini, L., U. Blomer, F. H. Gage, D. Trono, and I. M. Verma. 1996. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc. Natl. Acad. Sci. USA 93:11382-11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nilsen, T. W., P. A. Maroney, R. G. Goodwin, F. M. Rottman, L. B. Crittenden, M. A. Raines, and H.-J. Kung. 1985. c-erbB activation in ALV-induced erythroblastosis: novel RNA processing and promoter insertion result in expression of an amino-truncated EGF receptor. Cell 41:719-726. [DOI] [PubMed] [Google Scholar]
- 30.Olsen, J. C., C. Bova-Hill, D. P. Grandgenett, T. P. Quinn, J. P. Manfredi, and R. Swanstrom. 1990. Rearrangements in unintegrated retroviral DNA are complex and are the result of multiple genetic determinants. J. Virol. 64:5475-5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onafuwa, A., W. An, N. D. Robson, and A. Telesnitsky. 2003. Human immunodeficiency virus type 1 genetic recombination is more frequent than that of Moloney murine leukemia virus despite similar template switching rates. J. Virol. 77:4577-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parthasarathi, S., A. Varela-Echavarria, Y. Ron, B. D. Preston, and J. P. Dougherty. 1995. Genetic rearrangements occurring during a single cycle of murine leukemia virus vector replication: characterization and implications. J. Virol. 69:7991-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pathak, V. K., and H. M. Temin. 1990. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc. Natl. Acad. Sci. USA 87:6024-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peliska, J. A., and S. J. Benkovic. 1992. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science 258:1112-1118. [DOI] [PubMed] [Google Scholar]
- 35.Pfeiffer, J. K., R. S. Topping, N. H. Shin, and A. Telesnitsky. 1999. Altering the intracellular environment increases the frequency of tandem repeat deletion during Moloney murine leukemia virus reverse transcription. J. Virol. 73:8441-8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pulsinelli, G. A., and H. M. Temin. 1991. Characterization of large deletions occurring during a single round of retrovirus vector replication: novel deletion mechanism involving errors in strand transfer. J. Virol. 65:4786-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quinones-Mateu, M. E., Y. Gao, S. C. Ball, A. J. Marozsan, A. Abraha, and E. J. Arts. 2002. In vitro intersubtype recombinants of human immunodeficiency virus type 1: comparison to recent and circulating in vivo recombinant forms. J. Virol. 76:9600-9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raines, M. A., N. J. Maihle, C. Moscovici, L. Crittenden, and H. J. Kung. 1988. Mechanism of c-erbB transduction: newly released transducing viruses retain poly(A) tracts of erbB transcripts and encode C-terminally intact erbB proteins. J. Virol. 62:2437-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robson, N. D., and A. Telesnitsky. 1999. Effects of 3′ untranslated region mutations on plus-strand priming during Moloney murine leukemia virus replication. J. Virol. 73:948-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roda, R. H., M. Balakrishnan, J. K. Kim, B. P. Roques, P. J. Fay, and R. A. Bambara. 2002. Strand transfer occurs in retroviruses by a pause-initiated two-step mechanism. J. Biol. Chem. 277:46900-46911. [DOI] [PubMed] [Google Scholar]
- 41.Roebroek, A. J., J. A. Schalken, C. Onnekink, H. P. Bloemers, and W. J. Van de Ven. 1987. Structure of the feline c-fes/fps proto-oncogene: genesis of a retroviral oncogene. J. Virol. 61:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sala, M., G. Zambruno, J. P. Vartanian, A. Marconi, U. Bertazzoni, and S. Wain-Hobson. 1994. Spatial discontinuities in human immunodeficiency virus type 1 quasispecies derived from epidermal Langerhans cells of a patient with AIDS and evidence for double infection. J. Virol. 68:5280-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato, H., Y. Tomita, K. Ebisawa, A. Hachiya, K. Shibamura, T. Shiino, R. Yang, M. Tatsumi, K. Gushi, H. Umeyama, S. Oka, Y. Takebe, and Y. Nagai. 2001. Augmentation of human immunodeficiency virus type 1 subtype E (CRF01_AE) multiple-drug resistance by insertion of a foreign 11-amino-acid fragment into the reverse transcriptase. J. Virol. 75:5604-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwartz, J. R., S. Duesberg, and P. H. Duesberg. 1995. DNA recombination is sufficient for retroviral transduction. Proc. Natl. Acad. Sci. USA 92:2460-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartzberg, P., J. Colicelli, and S. Goff. 1985. Recombination between a defective retrovirus and homologous sequences in host DNA: reversion by patch repair. J. Virol. 53:719-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott, J. M., and M. J. Imperiale. 1996. Reciprocal effects of splicing and polyadenylation on human immunodeficiency virus type 1 pre-mRNA processing. Virology 224:498-509. [DOI] [PubMed] [Google Scholar]
- 47.Shin, N. H., D. Hartigan-O'Connor, J. K. Pfeiffer, and A. Telesnitsky. 2000. Replication of lengthened Moloney murine leukemia virus genomes is impaired at multiple stages. J. Virol. 74:2694-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shioda, T., S. Oka, X. Xin, H. Liu, R. Harukuni, A. Kurotani, M. Fukushima, M. K. Hasan, T. Shiino, Y. Takebe, A. Iwamoto, and Y. Nagai. 1997. In vivo sequence variability of human immunodeficiency virus type 1 envelope gp120: association of V2 extension with slow disease progression. J. Virol. 71:4871-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stehelin, D., H. E. Varmus, J. M. Bishop, and P. K. Vogt. 1976. DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA. Nature 260:170-173. [DOI] [PubMed] [Google Scholar]
- 50.Stuhlmann, H., M. Dieckmann, and P. Berg. 1990. Transduction of cellular neo mRNA by retrovirus-mediated recombination. J. Virol. 64:5783-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subramanian, S., R. K. Mishra, and L. Singh. 2003. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 4:R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugden, B. 1993. How some retroviruses got their oncogenes. Trends Biochem. Sci. 18:233-235. [DOI] [PubMed] [Google Scholar]
- 53.Sun, G., P. K. O'Neil, H. Yu, Y. Ron, B. D. Preston, and J. P. Dougherty. 2001. Transduction of cellular sequence by a human immunodeficiency virus type 1-derived vector. J. Virol. 75:11902-11906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swain, A., and J. M. Coffin. 1992. Mechanism of transduction by retroviruses. Science 255:841-845. [DOI] [PubMed] [Google Scholar]
- 55.Swain, A., and J. M. Coffin. 1989. Polyadenylation at correct sites in genome RNA is not required for retrovirus replication or genome encapsidation. J. Virol. 63:3301-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swanstrom, R., R. C. Parker, H. E. Varmus, and J. M. Bishop. 1983. Transduction of a cellular oncogene: the genesis of Rous sarcoma virus. Proc. Natl. Acad. Sci. USA 80:2519-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varela-Echavarria, A., C. M. Prorock, Y. Ron, and J. P. Dougherty. 1993. High rate of genetic rearrangement during replication of a Moloney murine leukemia virus-based vector. J. Virol. 67:6357-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Varmus, H. E. 1984. The molecular genetics of cellular oncogenes. Annu. Rev. Genet. 18:553-612. [DOI] [PubMed] [Google Scholar]
- 59.Vidal, N., M. Peeters, C. Mulanga-Kabeya, N. Nzilambi, D. Robertson, W. Ilunga, H. Sema, K. Tshimanga, B. Bongo, and E. Delaporte. 2000. Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J. Virol. 74:10498-10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wooley, D. P., L. A. Bircher, and R. A. Smith. 1998. Retroviral recombination is nonrandom and sequence dependent. Virology 243:229-234. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, J., and H. M. Temin. 1993. 3′ junctions of oncogene-virus sequences and the mechanisms for formation of highly oncogenic retroviruses. J. Virol. 67:1747-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, J., and H. M. Temin. 1993. Rate and mechanism of nonhomologous recombination during a single cycle of retroviral replication. Science 259:234-238. [DOI] [PubMed] [Google Scholar]