SUMMARY
DNA interstrand crosslinks (ICLs) are highly toxic because they block the progression of replisomes. The Fanconi Anemia (FA) proteins, encoded by genes that are mutated in FA, are important for repair of ICLs. The FA core complex catalyzes the monoubiquitination of FANCD2, and this event is essential for several steps of ICL repair. However, how monoubiquitination of FANCD2 promotes ICL repair at the molecular level is unknown. Here, we describe a highly conserved protein, KIAA1018/MTMR15/FAN1, that interacts with, and is recruited to sites of DNA damage by, the monoubiquitinated form of FANCD2. FAN1 exhibits endonuclease activity toward 5′ flaps and has 5′ exonuclease activity, and these activities are mediated by an ancient VRR_nuc domain. Depletion of FAN1 from human cells causes hypersensitivity to ICLs, defects in ICL repair, and genome instability. These data at least partly explain how ubiquitination of FANCD2 promotes DNA repair.
INTRODUCTION
DNA interstrand crosslinks (ICLs) are formed when bifunctional agents covalently link the two strands in a double helix. ICLs are toxic lesions that prevent strand separation necessary for transcription and DNA replication. ICLs can be induced by drugs and also by endogenous metabolites. Crosslinking agents such as mitomycin-C (MMC) and cisplatin generate a mixture of monoadducts and ICLs in cells but cellular toxicity correlates with the number of ICLs. Although ICLs can be repaired in G1, the major route for ICL repair appears to occur in S phase (Akkari et al., 2000; Rothfuss and Grompe, 2004; Taniguchi et al., 2002). Various models for the repair of ICLs have been suggested (McCabe et al., 2009; Moldovan and D’Andrea, 2009), and recent studies proposed that ICL repair requires two forks to converge on the ICL (Räschle et al., 2008) (Figure S1 available online).
Forks that stall at ICLs recruit signaling complexes including the Fanconi Anemia (FA) proteins and FA-associated proteins (Moldovan and D’Andrea, 2009) (Figure S1). Fanconi Anemia is an inherited recessive condition characterized by developmental defects, skeletal abnormalities, bone marrow failure, and cancer predisposition (Wang, 2007). FA falls into 13 complementation groups, and the relevant FA genes have been cloned (Patel and Joenje, 2007; Wang, 2007). Nevertheless, FA patients exist where mutations in known FA genes could not be found. The central components of the FA pathway are FANCD2 and its paralogue FANCI, which together form the “ID” complex (Garcia-Higuera et al., 2001; Smogorzewska et al., 2007). These two proteins are monoubiquitinated at Lys561 and Lys523, respectively, in S phase and in response to ICLs (Figure S1) (Garcia-Higuera et al., 2001; Taniguchi et al., 2002). This reaction is catalyzed by the E3 ubiquitin ligase FANCL subunit of the FA core complex, which comprises FANCA, B, C, E, F, G, L, and M, and also requires the FA-associated proteins FAAP100 and FAAP24 (Ciccia et al., 2007; Collis et al., 2008; Ling et al., 2007). Furthermore, loss of FANCD2 monoubiquitination is observed in many FA patients (Moldovan and D’Andrea, 2009).
Monoubiquitination of FANCD2 is necessary for ICL repair but the underlying molecular mechanisms are unclear. The monoubiquitinated form of the ID complex may recruit ICL repair proteins, but as yet no ligands for ubiquitinated FANCD2 have been reported. It was reported that monoubiquitination of FANCD2 is required for the “unhooking” of the ICL in a cell-free repair system (Knipscheer et al., 2009) (Figure S1). Unhooking involves incisions on either side of the ICL, one of which is catalyzed by the structure-specific nuclease MUS81-EME1 (Figure S1) (Hanada et al., 2007; Hanada et al., 2006). MUS81-EME1 creates a one-ended double-strand break (DSB) that can be used later to initiate homologous recombination (HR). The identity of the nuclease that catalyzes the second incision to enable unhooking of the ICL is unclear. XPF-ERCC1 has been implicated, but this is controversial (Bergstralh and Sekelsky, 2008; Bhagwat et al., 2009). After unhooking, the resulting gap is filled in by translesion synthesis, which also appears to require FANCD2 ubiquitination (Knipscheer et al., 2009), and the unhooked lesion is removed by excision repair. The DSBs generated by unhooking are resected and one of them initiates HR to complete ICL repair (Figure S1). Successful HR-mediated repair of the MUS81-generated DSB depends on processing of DNA repair intermediates by the SLX4 complex. SLX4 acts as a scaffold for XPF-ERCC1, MUS81-EME1, and SLX1. Cells lacking, or depleted of, SLX4 (Fekairi et al., 2009; Muñoz et al., 2009; Svendsen et al., 2009) or XPF-ERCC1 (Niedernhofer et al., 2004) cannot efficiently repair the DSBs created by MUS81 after ICL induction and exhibit defects in HR-mediated repair of DSBs. In this study, we report the identification of FAN1, a nuclease recruited to sites of DNA damage by monoubiquitinated FANCD2 that is important for repair of ICLs.
RESULTS
Domain Organization of KIAA1018/MTMR15/FAN1
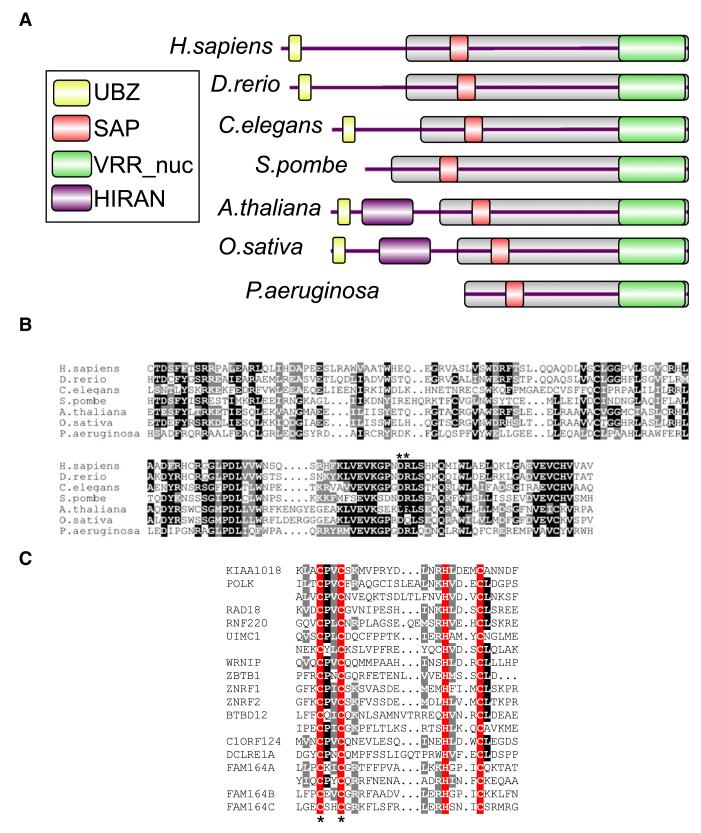
We noticed an uncharacterized human protein, KIAA1018/MTMR15, in the human sequence databases, that has a UBZ-type ubiquitin-binding domain domain, a SAP-type DNA binding domain, and a putative nuclease domain termed the “VRR_nuc” domain (Figure 1A), initially referred to as “domain of unknown function 994” (DUF994) (Iyer et al., 2006). Orthologs of KIAA1018 are found in prokaryotes and most eukaryotes with the notable exception of budding yeast (Figure 1A).
Figure 1. The KIAA1018/MTMR15/FAN1 Family of Proteins.
(A) Schematic representation of the domain architecture of KIAA1018/MTMR15/FAN1 orthologs from different species. The relevant protein identification codes are as follows: Homo sapiens Q9Y2M0, Danio rerio Q1LWH4, Caenorhabditis elegans P90740, Schizosaccharomyces pombe Q9Y804, Arabidopsis thaliana Q9SX69, Oryza sativa B9FRR6, and Pseudomonas aeruginosa Q9I2N0.
(B) Alignment of the VRR_nuc domain of FAN1. Identical residues are shaded in black, and similar residues are shaded in gray. The asterisks denote conserved residues Asp981 and Arg982 mutated in the FAN1-DR mutant.
(C) Alignment of the UBZ domain of FAN1. Identical residues are shaded in black, and similar residues are shaded in gray. The conserved Cys and His residues that define the two dyads of the UBZ domain are shaded in red. Asterisks denote the conserved Cys44 and Cys47 residues in the first dyad.
We suspected that KIAA1018 is involved in DNA damage responses for a number of reasons. KIAA1018 is the only VRR-nuc domain-containing protein in eukaryotes but many bacteria and bacteriophages have genes that encode solely VRR_nuc domains. Although the functions of these genes are unknown, most of them are located in operons that include known DNA repair enzymes, hence the name VRR_nuc (virus-type replication-repair nuclease) (Iyer et al., 2006). The VRR_nuc domains contain a PD-(D/E)XK motif found in the active site of many restriction nucleases (Kosinski et al., 2005) (Figure 1B). We thus suspected that KIAA1018 might act as a repair endonuclease. A putative role for KIAA1018 in DNA repair is also implied by the presence of a UBZ4-type ubiquitin-binding domain that belongs to the RAD18 family of zinc fingers, a domain commonly found in DNA damage response proteins such as DNA polymerase κ (POL κ), RAD18, and WRNIP (Figure 1C) (Hofmann, 2009). Furthermore, KIAA1018 was also found to interact with the MLH1 DNA mismatch repair protein in a genome wide screen (Cannavo et al., 2007). We therefore decided to test whether KIAA1018, which we refer to hereafter as FAN1 for reasons that will become clear later, has nuclease activity and whether it is involved in DNA repair.
FAN1 Has Structure-Specific Endonuclease Activity
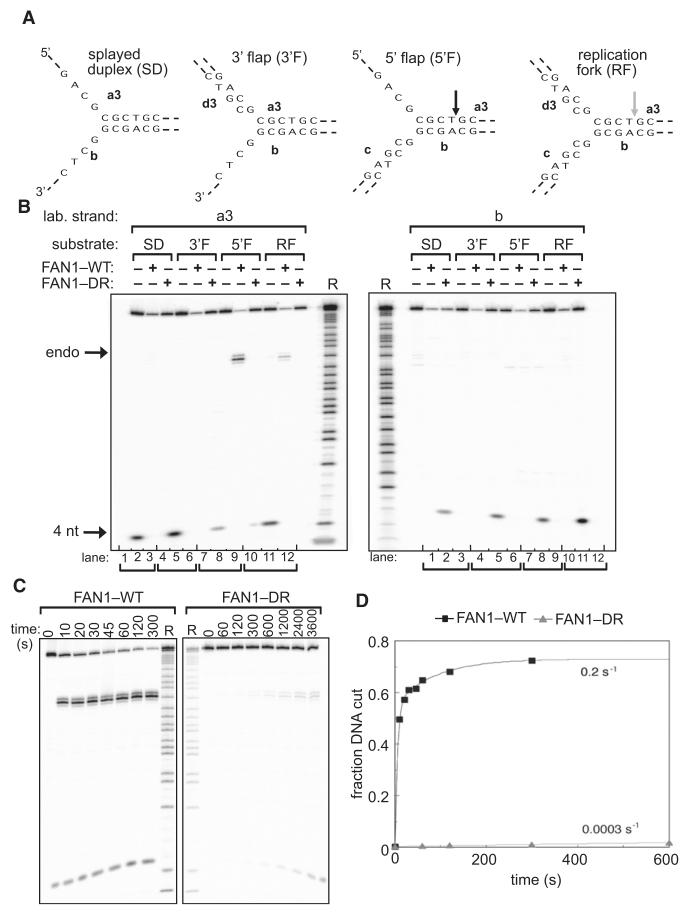
To test for nuclease activity, we expressed recombinant FAN1, fused to an N-terminal NUS-His6 tag, in bacteria and purified it through three steps of ion exchange chromatography (Figure S2A). In parallel, we purified a mutant version of FAN1 where the conserved Asp981 and Arg982 residues (indicated by asterisks in Figure 1B) found in the VRR_nuc domain were mutated to alanine (“DR” mutant). We next tested the ability of FAN1 to cleave a range of branched DNA substrates that resemble DNA repair and replication intermediates. These included a splayed duplex, a 3′ flap, a 5′ flap, and a nicked three-way junction that mimics a DNA replication fork (Figure 2A). All substrates were 32P labeled at the 5′ end of the a3 strand or the b strand as indicated in Figure 2A. After incubation with wild-type or mutant FAN1, reaction products were separated by gel electrophoresis under denaturing conditions.
Figure 2. FAN1 Has Structure-Specific Endonuclease Activity.
Recombinant human FAN1 was incubated with synthetic DNA structures: splayed duplex (SD; oligos a3, b), 3′ flap (3′F; oligonucleotides a3, b, d3), 5′ flap (5′F; oligos a3, b, c), or a replication fork (RF)-like structure (oligos a3, b, c, d3), each radioactively 5′ 32P-labeled on the strands indicated. WT refers to wild-type FAN1, and DR refers to the Asp981Ala-Arg982Ala FAN1 mutant.
(A) Schematic diagram of the DNA substrates used in (B). Sites of DNA cleavage are indicated by arrows.
(B) Reaction products (10 min incubation) were subjected to denaturing PAGE. Purine-specific chemical sequencing ladders (R) were derived from oligonucleotides a3 or b.
(C) FAN1 was incubated with the 5′ flap shown in (A) for the time indicated (s, seconds), and reaction products were subjected to denaturing PAGE.
(D) Progress curves of cleavage of the 5′ flap construct incubated with wild-type (black squares) and DR (gray triangles) mutant FAN1. The data have been fitted to a single (DR) or double (wild-type) exponential functions (lines). From these data, we have calculated observed rates of cleavage of >0.2 s−1 and 0.0003 s−1 for wild-type and DR enzymes, respectively.
See also Figure S2.
As shown in Figure 2B, FAN1 displayed strong endonuclease activity toward the 5′ flap structure and weaker activity toward the replication fork model. Cleavage affected only one strand of these structures and occurred in the double-stranded region on the same strand as the flap, 4 nucleotides (nt) 3′ to the branch point (Figure 2A). Selectivity of FAN1 for these DNA structures, as opposed to specificity for DNA sequence, was confirmed by analysis of the cleavage of an analogous set of branched DNA structures composed of strands with alternative sequence (Figures 2B and 2C). The endonuclease activity of the FAN1 DR mutant was severely reduced compared with wild-type protein (Figure 2B;Figures S2B and S2C), resulting in cleavage rates approximately 1000-fold lower than those for wild-type protein (Figures 2C and 2D). FAN1 did not exhibit endonuclease activity toward four-way junctions (data not shown).
FAN1 Has 5′ Exonuclease Activity
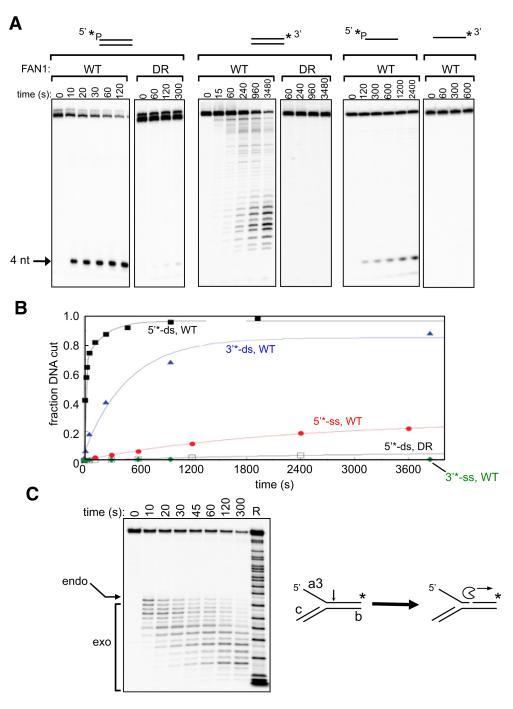
Incubation of wild-type FAN1 with 5′ 32P-labeled branched substrates also produced a short 4 nt radioactive fragment (Figures 2B and 2C), suggesting that FAN1 might possess an additional endo- or exonuclease activity. To further investigate this, we incubated FAN1 with linear double-stranded (dsDNA) or single-stranded (ssDNA) DNA in which one of the oligonucleotides was radioactively 5′ or 3′ 32P labeled. We observed a clear 5′ to 3′ exonuclease activity that initiates 4 nt from the 5′ end and cleaves every phosphate bond thereafter but with varying intensity (Figure 3A). The exonuclease activity of FAN1 toward ssDNA required that the 5′ end be phosphorylated (Figure 3A). FAN1 exhibited potent 5′ exonuclease activity toward DNA substrates with a recessed 5′ end, indicating that a blunt dsDNA end is not required for exonuclease activity (Figure S3). The exonuclease activity of FAN1 was severely reduced by mutation of Asp981 and Arg982 in the VRR_nuc domain (Figures 3A and 3B). Quantitation of these data showed that the rate of initiation of the exonuclease activity of FAN1 toward dsDNA (0.09 s−1; Figure 3B) was approximately half that of the endonuclease toward a 5′ flap (>0.2 s−1; Figure 2D). The calculated rates of initiation using wild-type enzyme (Figure 3B) were 0.09 s−1 for dsDNA (5′ 32P), 0.002 s−1 for dsDNA (3′ 32P), 0.0005 s−1 for ssDNA (5′ 32P), and too low to measure for ssDNA (3′ 32P). The rate of cleavage of dsDNA (5′ 32P) with the DR mutant was 0.0003 s−1, around 300-fold lower than that of wild-type FAN1 (Figure 3B).
Figure 3. FAN1 has 5′ Exonuclease Activity.
Recombinant human FAN1 was incubated for the time indicated (s, seconds) with dsDNA (oligonucleotide a3, a3-cp), ssDNA (oligonucleotide a3), or a 5′ flap (5′F; oligonucleotides a3, b, c) radioactively 5′ or 3′ 32P-labeled on the a3 strand as shown (asterisks). WT refers to wild-type FAN1, and DR refers to the Asp981Ala-Arg982Ala FAN1 mutant. Reaction products were subjected to denaturing PAGE.
(A) Cleavage of linear DNA substrates.
(B) The cleavage products were quantitated. “Fraction DNA cut” is the ratio of the relevant cleavage product to total DNA (cleaved plus uncleaved DNA). The data are plotted as a function of time and are fitted to single or double exponential functions.
(C) Activity of WT FAN1 on radioactively 3′ 32P-labeled 5′ flap. R refers to a purine-specific chemical sequencing ladder derived from the labeled strand. See also Figure S3.
These results raised the possibility that the endonuclease activity of FAN1 on branched substrates might be coupled with a 5′-3′ exonuclease activity. We therefore examined FAN1-mediated cleavage of a 5′ flap in which the a3 strand containing the flap was radioactively labeled at the 3′ end (Figure 3C). This experiment clearly revealed that the endonucleolytic incision described above (Figures 2B and 2C) was followed by a 5′-3′ exonuclease activity that with time generated ever-shorter products (Figure 3C). Cleavage was observed at each phosphate bond but with varying intensity. Taken together, these data show that FAN1 has a 5′ to 3′ exonuclease activity that initiates 4 nt from the 5′ end on single- and double-stranded DNA and 4 nt from the branchpoint on 5′ flaps.
FAN1 Interacts with FANCD2 and FANCI
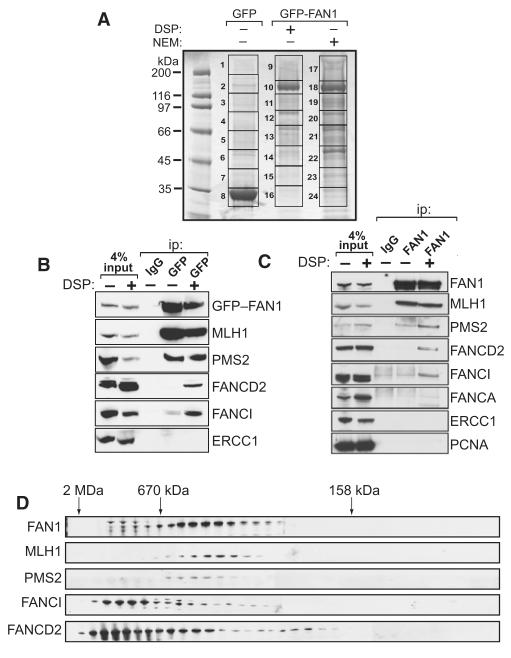
In an attempt to link the endonuclease activity of FAN1 toward branched DNA structures with known DNA repair pathways, we aimed to find FAN1-interacting proteins. Plasmids expressing GFP-FAN1 or GFP alone, both under the control of a tetracycline-inducible promoter, were stably integrated in HEK293 cells. Cells were lysed after induction either in the presence of the reversible protein crosslinker dithiobis (succinimidyl propionate) (DSP) or the deubiquitinase inhibitor N-ethyl maleimide (NEM). Extracts were subjected to immunoprecipitation with GFP-Trap beads, and protein-protein crosslinking was reversed with dithiothreitol. After SDS-PAGE, strong bands at the expected molecular weights of GFP-FAN1 and GFP were observed in the respective lanes (Figure 4A). In addition, a range of other proteins was found in GFP-FAN1 but not GFP precipitates. Mass fingerprinting revealed that most of these proteins are involved in DNA repair. Both components of the MLH1-PMS2 complex involved in mismatch repair were found in GFP-FAN1 precipitates when cells were lysed in NEM or DSP (Table S1) (Cannavo et al., 2007). We also found FANCD2 and FANCI, but only when DSP was included in the lysis buffer. The presence of ubiquitin in the FANCD2-containing band indicated that the ubiquitinated form of FANCD2 might coprecipitate with FAN1 (Table S1). The specificity of the FAN1 protein interactions was independently confirmed by analysis of the immunoprecipitates of FLAG-FAN1 under similar conditions (data not shown), and only the proteins identified in both experiments are shown in Table S1.
Figure 4. FAN1 Interacts with DNA Repair Proteins.
(A) HEK293 Flp-In cells that stably express GFP-FAN1 were lysed in the presence of dithiobis (succinimidyl propionate) (DSP) or N-ethyl maleimide (NEM). These extracts together with extracts of cells that express GFP only were subjected to immunoprecipitation with GFP-Trap beads, and after extensive washing precipitates were subjected to SDS-PAGE. The gel was fixed and stained with Colloidal Blue. The gel lanes were cut into slices, as indicated, and the proteins were digested with trypsin before mass spectrometric fingerprinting.
(B) HEK293 Flp-In cells that stably express GFP-FAN1 were lysed in the presence or absence of DSP and extracts were subjected to immunoprecipitation with control anti-HA (IgG) or GFP-Trap beads. Precipitates were analyzed by western blotting with the antibodies indicated. Input represents 4% of the extract used for immunoprecipitation.
(C) HEK293 cells were lysed in the presence or absence of DSP, and extracts were subjected to immunoprecipitation with anti-HA (IgG) or with anti-FAN1 antibodies. Precipitates were analyzed by western blotting with the antibodies indicated. Input represents 4% of the extract used for immunoprecipitation.
(D) Extracts of HEK293 cells were analyzed by size-exclusion chromatography on a HiLoad 26/60 Superdex 200 column in buffer containing 0.2 M NaCl, and every third fraction was denatured and analyzed by western blotting with the indicated antibodies. The elution positions of Dextran blue (2 MDa), thyroglobulin (670 kDa), and bovine γ-globulin (158 kDa) are shown.
See also Table S1.
FAN1 interactors were confirmed by a number of experiments. First, western blotting detected MLH1, FANCD2, and FANCI in GFP-FAN1 but not GFP precipitates (Figure 4B). FANCD2 and FANCI were only found in GFP-FAN1 precipitates when DSP was present in the lysis buffer. To examine endogenous complexes, we raised antibodies in sheep against human FAN1. These antibodies recognized a protein of the expected molecular mass (114 kDa) in extracts of HEK293 cells that was not detected when cells were transfected with FAN1-specific small interfering RNA (siRNA) duplexes (as described later). These antibodies were used to immunoprecipitate FAN1 from HEK293 cell extracts. Endogenous MLH1, FANCD2, and FANCI were detected in anti-FAN1 immunoprecipitates (Figure 4C) but not in precipitates with an antibody against an unrelated epitope (HA). Again, FANCD2 and FANCI were only found in GFP-FAN1 precipitates when DSP was present in the lysis buffer. These interactions were not affected by ethidium bromide or by treatment of immunoprecipitates with DNase I or benzonase (data not shown), excluding the possibility that these interactions are DNA dependent. Abundant DNA repair proteins such as ERCC1 or PCNA, and other FA proteins such as FANCA were not detected in anti-FAN1 immunoprecipitates (Figures 4B and 4C).
Size-exclusion chromatography of HEK293 cell extracts showed that FAN1 elutes in two subcomplexes; one of these overlaps with MLH1 and elutes slower than the 670 kDa marker, while the other subcomplex elutes faster than the 670 kDa marker and overlaps with FANCD2 and FANCI (Figure 4D). It is interesting that FAN1 in the latter subcomplex migrates more slowly on SDS-PAGE than the form of FAN1 that coelutes with MLH1, and this may represent a posttranslationally modified form of FAN1. Taken together, these data show that FAN1 binds to MLH1, FANCD2, and FANCI. The acronym FAN1 stands for “FANCD2/FANCI-associated nuclease 1.”
The UBZ Domain of FAN1 Interacts with FANCD2
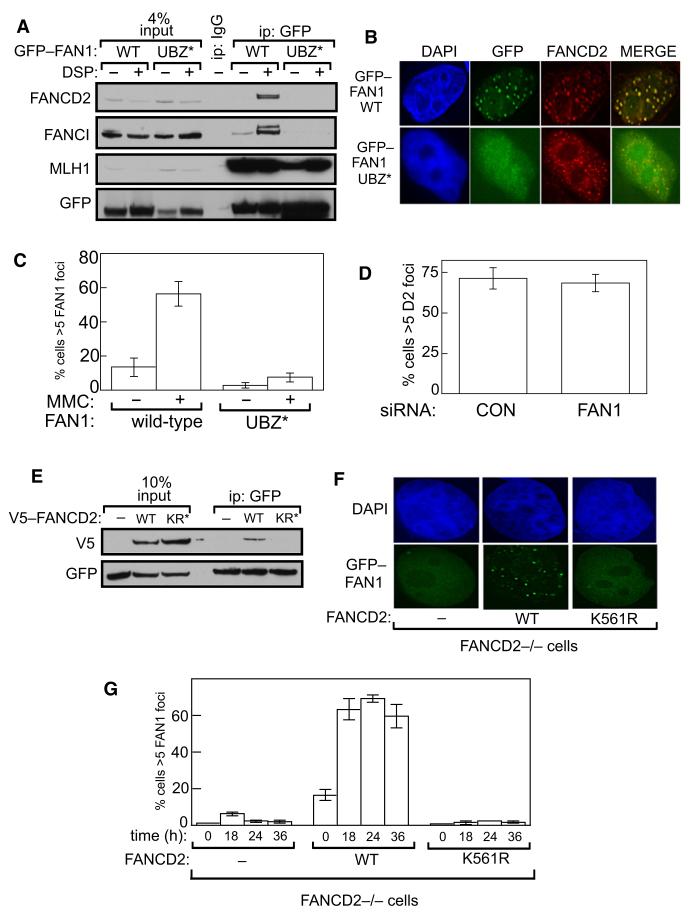
FAN1 has a UBZ domain of the RAD18 type that is found in DNA repair proteins such as WRNIP, POL κ, and RAD18 (Figure 1C). The POL κ UBZ domain binds to monoubiquitinated PCNA (Bienko et al., 2005), but we could not detect PCNA in FAN1 precipitates (Figure 4C). We hypothesized that instead, the UBZ domain of FAN1 binds to the monoubiquitinated form of FANCD2, since we detected FANCD2 and ubiquitin in FAN1 precipitates. To test this, we mutated both of the two conserved cysteine residues in the first dyad of the FAN1 UBZ domain (Cys44 and Cys47, indicated by asterixes in Figure 1C) to alanine residues (UBZ* mutant). Whereas wild-type GFP-FAN1 transiently expressed in cells coimmunoprecipitated with endogenous FANCD2 and FANCI, the FAN1 UBZ* mutant did not, even though this mutant retained the ability to bind MLH1 (Figure 5A).
Figure 5. The UBZ Domain of FAN1 Binds Monoubiquitinated FANCD2.
(A) HEK293 cells were transiently transfected with pcDNA5.1-GFP-FAN1 wild-type (WT) or pcDNA5.1-GFP-FAN1-UBZ* (Cys44A/Cys47A). After 48 hr, cells were lysed in the presence or absence of DSP, and anti-GFP precipitates were analyzed by western blotting with the antibodies indicated. “Input” represents cell extracts.
(B) U2OS cells, grown on glass coverslips, were transiently transfected with pcDNA5.1-GFP, pcDNA5.1-GFP-FAN1 wild-type (WT), or pcDNA5.1-GFP-FAN1-UBZ* (Cys44A/Cys47A). Cells were treated, or not, with MMC, and after 16 hr GFP-FAN1 foci and FANCD2 foci were detected.
(C) Quantitation of data from (B). The number of cells with more than five GFP-FAN1 foci in a sample of 500 cells was counted.
(D) U2OS cells were transfected with control siRNA or siRNA targeting FAN1, and the number of cells with more than five FANCD2 foci was quantitated after exposure of cells to MMC.
(E) HEK293 cells stably expressing GFP-FAN1 were transiently transfected with pDEST40-lacZ (“–”), pDEST40-V5-FANCD2 wild-type (WT), or pDEST40-V5-FANCD2 K561R. After 48 hr, cells were lysed in the presence of DSP, and anti-GFP precipitates were analyzed by western blotting with the antibodies indicated.
(F) FANCD2−/− (PD20) cells stably transfected with empty vector (–), FANCD2 wild-type (WT), or FANCD2 K561R were transiently transfected with GFP-FAN1. Cells were treated with MMC for 18 hr and then fixed, and GFP-FAN1 foci were visualized.
(G) Same as (F), except that cells were treated with MMC for the times indicated, and the number of cells with more than five FAN1 foci were quantitated after exposure of cells to MMC.
Data in (C), (D), and (G) are represented as mean ± SEM. See also Figure S4.
FANCD2 forms subnuclear “foci” at sites of DNA damage in cells after DNA damage. Endogenous FAN1 formed foci that colocalized with FANCD2 in response to MMC (Figure S4A). GFP-FAN1 transiently transfected into U2OS cells also formed subnuclear foci in MMC-treated cells, and these colocalized with FANCD2 (Figure 5B). The GFP-FAN1 UBZ* mutant, however, did not form subnuclear foci in MMC-treated cells (Figures 5B and 5C). Depletion of FAN1 from cells had no detectable effect on MMC-induced FANCD2 focus formation (Figure 5D). GFP-FAN1 formed foci not just in response to MMC but also in response to HU or IR (Figure S4B). These data show that colocalization of FAN1 at sites of DNA damage with FANCD2 requires the FAN1 UBZ domain.
FAN1 Is Recruited to DNA Damage by Monoubiquitinated FANCD2
We next tested the possibility that it is specifically the monoubiquitinated form of FANCD2 that interacts with FAN1. Although wild-type FANCD2 transiently transfected into cells stably expressing GFP-FAN1 was detected in GFP-FAN1 precipitates, the FANCD2 K561R mutant that cannot be ubiquitinated was not (Figure 5E). These data indicate that FAN1 interacts with the monoubiquitinated form of FANCD2. To test the possibility that the monoubiquitination of FANCD2 might be required to recruit FAN1 to foci, we studied FANCD2−/− (PD20) human cells stably transfected with wild-type FANCD2 or with a FANCD2 K561R mutant (Garcia-Higuera et al., 2001). We found that GFP-FAN1 did not form MMC-induced foci in FANCD2−/− cells, but formation of foci was restored when these cells stably express wild-type FANCD2 (Figures 5F and 5G). Only back-ground levels of GFP-FAN1 foci occurred when the FANCD2 K561R mutant was expressed in these cells, at all time points examined (Figures 5F and 5G).
FAN1 Is Required for Cellular Resistance to Agents that Induce ICLs
Monoubiquitination of FANCD2 is required for ICL repair, but the underlying mechanism is unclear. We hypothesized that the association of FAN1, a nuclease with specificity for branched structures, with the monoubiquitinated form of FANCD2 may provide an explanation. Defective ICL repair causes hypersensitivity to agents that induce ICLs. Depletion of FAN1 with two siRNAs targeting different FAN1 exons (Figure S5A) caused cells to become hypersensitive to cisplatin and MMC compared with control siRNA (Figure 6A). The hypersensitivity to ICL-inducing agents associated with depletion of FAN1 was similar to that observed when FANCA was depleted (Figure 6A). Cells depleted of FAN1 did not show hypersensitivity to DNA-damaging agents such as campthothecin, hydroxyurea, UV light, or ionizing radiation (Figure S5B). In contrast, depletion of the ATR kinase caused hypersensitivity to all of these agents (Figure S5B). Cells defective in mismatch repair are resistant to killing by 6-thioguanine (6-TG) (Swann et al., 1996), and, consistent with this, depletion of MLH1 from HEK293 cells caused cells to become more resistant to 6-TG (Figure S5B). However, depletion of FAN1 did not, so it is unlikely that FAN1 is involved in mismatch repair.
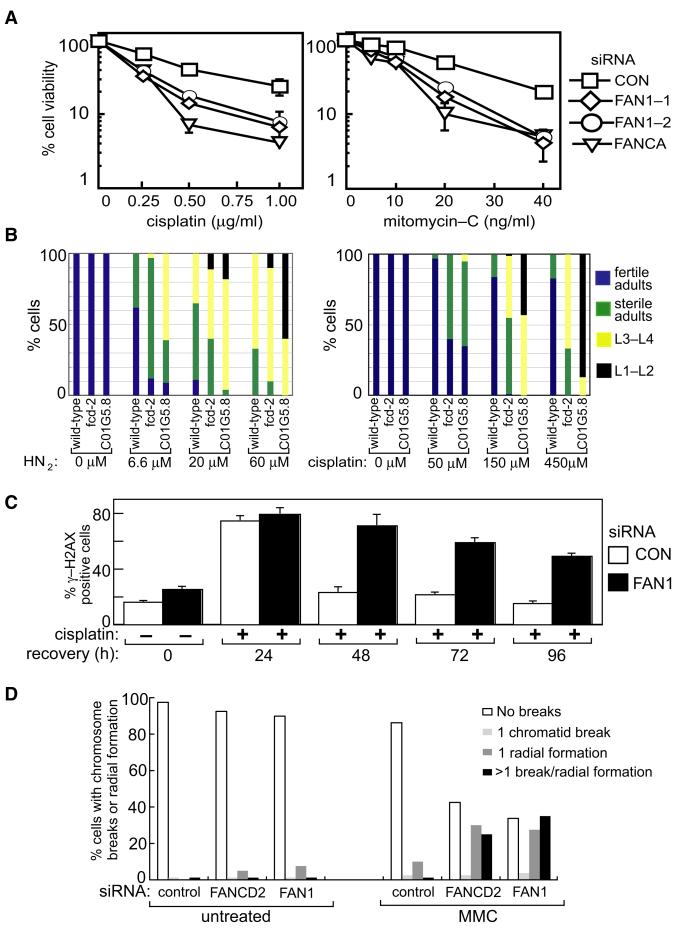
Figure 6. FAN1 Is Required for ICL Repair.
(A) HEK293 cells were transfected with the siRNAs indicated. Clonogenic survival assays were carried out with cisplatin or mitomycin-C (see the Experimental Procedures). For each siRNA, cell viability of untreated cells is defined as 100%.
(B) Synchronized L1 larva stage animals of the relevant genotype were incubated with the indicated concentrations of nitrogen mustard (HN2) or cisplatin. After 48 hr, the extent of developmental progression of the worms was scored, by counting the number of worms in the adult and various larval stages (L1–L2, L3–L4). Adult worms were scored as fertile if they contained fertilized eggs and as sterile if they did not.
(C) HEK293 cells transfected with control siRNA or FAN1 siRNA (FAN1-1) were treated with cisplatin (1 μg/ml) for 2 hr and then allowed to recover for the times indicated. The proportion of cells in each population with more than two γ-H2AX foci at each time point (“γ-H2AX positive”) was determined. The experiment was done three times, and a representative experiment is shown.
(D) The frequency of chromosome breaks and radial chromosomes in metaphase spreads of HEK293 cells transfected with control siRNA or FAN1-1 siRNA was measured before and after exposure to MMC (25 ng/ml; 18 hr) was measured as described previously (Deans and West, 2009).
Data in (A) and (C) are represented as mean ± SEM. See also Figure S5.
We also tested a Caenorhabditis elegans strain harboring a deletion in the C. elegans ortholog of FAN1 (Figures 1A and 1B), encoded by the C01G5.8 locus, for sensitivity to ICL-inducing agents. This deletion does not lead to any overt developmental defects (data not shown). L1-stage worm larvae were exposed to ICL-inducing agents, and the effects on the progression to subsequent larval stages L2, L3, L4 and to adult stages were observed. As shown in Figure 6B worms defective in Ce-fan-1 were hypersensitive to nitrogen mustard (HN2) and cisplatin, even more so than a deletion of the C. elegans fcd-2 Fancd-2 ortholog. These data were confirmed by depletion of Ce-fan-1 by RNA interference (data not shown). Furthermore, the Ce-rend-1 (tm423) deletion resulted in reduced progeny survival when L4 stage animals were exposed to nitrogen mustard (data not shown). Thus, the ICL hypersensitivity associated with defects in FAN1 is evolutionarily conserved.
FAN1 Is Required for Efficient Repair of ICL-Induced DNA Breaks
We next sought to determine at what stage of ICL repair FAN1 acts. ICLs cause monoubiquitination of FANCD2 and FANCI that promotes ICL repair. Monoubiquitination of FANCD2 and FANCI causes reduced electrophoretic mobility (Garcia-Higuera et al., 2001). Exposure of HEK293 cells transfected with control siRNA to cisplatin or MMC resulted in damage-induced monoubiquitination of FANCD2 and FANCI (Figure S5C). Consistent with previous reports, depletion of FANCA abolished FANCD2 and FANCI monoubiquitination. However, depletion of FAN1 had no detectable effect (Figure S5C). Therefore, FAN1 is not required for monoubiquitination of FANCD2 or FANCI.
Exposure of cells to ICL-inducing agents causes DSBs, judged by γ-H2AX foci or pulsed field gels. These DSBs, formed as a result of replication fork cleavage by MUS81 during ICL unhooking (Hanada et al., 2007; Hanada et al., 2006) (Figure S1), initiate the HR step of ICL repair. We next tested whether depletion of FAN1 from cells affected the induction of, or disappearance of γ-H2AX foci induced by ICLs. HEK293 cells were transfected with control siRNA or FAN1 siRNA and were either left untreated or exposed to cisplatin for 2 hr. Cells were washed free of cisplatin and incubated in fresh medium, and γ-H2AX foci were counted at various times during recovery. Around 80% of cells transfected with control siRNA had between 2 and 40 foci 24 hr after transient exposure to cisplatin; cells with more than two γ-H2AX foci were scored as “γ-H2AX positive.” Although the percentage of control siRNA-transfected cells that were γ-H2AX positive declined to almost basal levels by 48 hr (Figure 6C), almost no decrease in the percentage of γ-H2AX-positive cells depleted of FAN1 was observed at this time (Figure 6C). By 96 hr, over 50% of cells depleted of FAN1 were still γ-H2AX positive, although γ-H2AX foci had returned to basal levels in cells treated with control siRNA (Figure 6C). These data show that depletion of FAN1 does not affect DSB induction at replisomes blocked by ICLs but instead causes a defect in DNA repair. Consistent with the persistence of cisplatin-induced γ-H2AX foci, we observed an increase in chromosomal abnormalities in metaphase spreads of FAN1-depleted cells exposed to MMC. FAN1-depleted cells showed a substantial increase in the frequency of cells with more than one chromosome break or radial chromosome, similar to cells depleted of FANCD2 (Figure 6D). These data are consistent with FAN1 acting in the FA pathway.
FAN1 Is Required for Efficient HR but Not for DSB Resection or RAD51 Loading
Human cells solely expressing FANCD2 K561R show reduced HR efficiency (Nakanishi et al., 2005). It is possible that the defect in the resolution of ICL-induced DSBs in FAN1-depleted cells could reflect a defect in HR. We used a reporter system in U2OS cells to measure HR frequency. In this system, an 18 bp sequence recognized by the I-SceI meganuclease is placed between tandem mutant copies of GFP (Nakanishi et al., 2005). HR between these two copies generates a wild-type GFP open reading frame, and functional GFP expression can be detected by FACS analysis. As shown in Figure 7A, depletion of FAN1 with two separate siRNAs reduced the efficiency of I-SceI-induced HR by 50%–60%, similar to the reduction reported for depletion of FA proteins (Nakanishi et al., 2005; Smogorzewska et al., 2007). These data indicate that FAN1 promotes HR in response to DSBs. Depletion of FAN1—or FANCD2—from HEK293 cells did not appear to affect the frequency of MMC-induced sister chromatid exchanges (Figure S6).
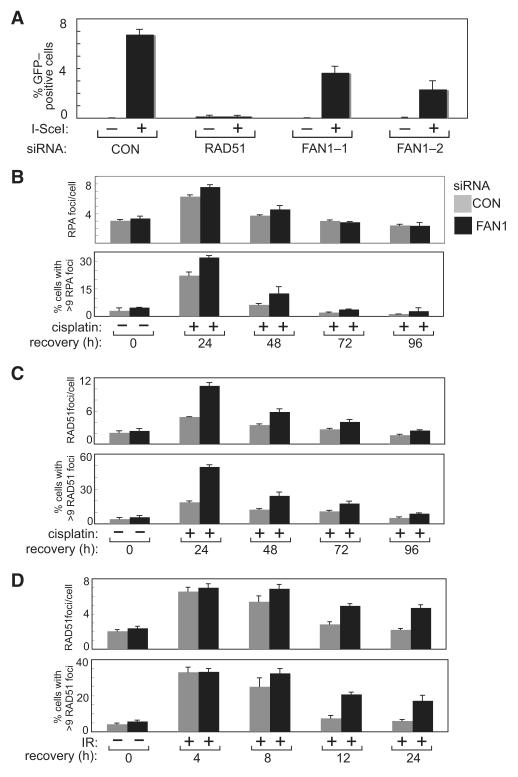
Figure 7. Effect of FAN1 Depletion on HR and Focus Formation by RAD51 and RPA.
(A) U2OS cells, in which an 18 bp sequence recognized by I-SceI was placed between tandem mutant copies of the gene encoding GFP, were transfected with control siRNA (luciferase) and/or siRNAs specifically targeting FAN1 (FAN1-1 or FAN1-2) or RAD51. After 48 hr, cells were transfected with a plasmid expressing I-SceI or with an empty vector, and 24 hr later cells were tested for GFP expression by FACS analysis. The frequency of HR in cells transfected with the various siRNAs was calculated relative to cells transfected with control siRNA.
(B) U2OS cells transfected with control siRNA or FAN1 siRNA (FAN1-1) were treated with cisplatin (1 μg/ml) for 2 hr and then allowed to recover for the times indicated. Cells were then fixed, permeabilized, washed, and blocked before incubation with anti-RPA antibodies. Coverslips were incubated with secondary antibodies, mounted on glass slides, and visualized. The average number of RPA foci per cell was analyzed. The experiment was done three times, and a representative experiment is shown.
(C) Same as (B), except that cells were stained with anti-RAD51 antibodies.
(D) Same as (C), except that cells were exposed to IR (3 Gy) and then allowed to recover for the times indicated.
Data represented as mean ± SEM. See also Figure S6.
We postulated that the exonuclease activity of FAN1 could affect HR by controlling the resection of DSBs generated during ICL repair. Resection of DSBs leads to the generation of ssDNA, and the coating of ssDNA by RPA leads to RPA foci. We analyzed RPA foci after exposure of cells to a pulse of cisplatin to assess DSB resection and found that depletion of FAN1 did not prevent cisplatin-induced RPA focus formation. In fact, depletion of FAN1 caused a slight increase in the average number of RPA foci per cell and in the average number of cells with more than nine RPA foci (Figure 7B). This suggests that FAN1 is not required for resection of DSBs. RPA foci gradually disappeared during the recovery of cells treated with control siRNA or FAN1 siRNA from cisplatin (Figure 7B).
DSB resection is followed by formation of the RAD51 nucleoprotein filament on the resected DSB, so we examined formation of RAD51 foci at various times during recovery of cells from cisplatin. After 24 hr recovery, cells depleted of FAN1 showed an approximately 2.5,fold increase in the number of cells with RAD51 foci, and a similar increase in the number of RAD51 foci per cell, compared with control siRNA (Figure 7C). At subsequent times during recovery, FAN1-depleted cells continued to have around twice as many RAD51 foci as control cells and these data are consistent with a defect in HR. We also tested the effects of depleting FAN1 on RAD51 loading after IR. Four hours after exposure to IR, the number of cells with greater than nine RAD51 foci, and the average number of RAD51 foci per cell, was similar in FAN1-depleted and cells treated with control siRNA (Figure 7D). However, whereas RAD51 foci declined to basal levels by 12-24 hr after IR in control cells, FAN1-depleted cells showed a delay in the disappearance of RAD51 foci. These data suggest that FAN1 depletion leads to failure of a late stage of HR.
DISCUSSION
It has been known for almost a decade that mono–ubiquitination of FANCD2 is required for ICL repair (Garcia-Higuera et al., 2001). However, the molecular role of this ubiquitination event has remained elusive. Here, we report that the FAN1 nuclease is recruited to sites of DNA damage by monoubiquitinated FANCD2 and thus might act as an effector molecule carrying out one or more nucleolytic steps required for ICL repair. The phenotypic consequences of depleting FAN1 from human cells—sensitivity to ICL-inducing agents, chromosome instability in MMC-treated cells and defects in HR—are consistent with a role in ICL repair and are similar to those seen in cells solely expressing FANCD2 K561R (Moldovan and D’Andrea, 2009). These findings might at least in part explain how ubiquitination of FANCD2 promotes ICL repair.
According to our data, FAN1 is recruited to sites of ICLs by monoubiquitinated FANCD2. This is supported by the requirement of the FAN1 UBZ domain (Figure 5B) and the monoubiquitinated form of FANCD2 (Figure 5F) for FAN1 localization. Furthermore, FAN1 and FANCD2 proteins coprecipitate in a manner that depends on FANCD2 K561 and on the FAN1 UBZ domain. Both ubiquitinated and nonubiquitinated FANCD2 were detected in FAN1 immunoprecipitates, even though when FANCD2 K561 is mutated, no FANCD2 is detected in FAN1 immunoprecipitates. This discrepancy may be explained by deubiquitination of a proportion of FANCD2 after cell lysis or by the association of modified FANCD2 with the unmodified form of the protein. Even though DNA damage stimulates FANCD2 ubiquitination, FAN1 interacts with FANCD2 even without exposure of cells to genotoxins. This is probably a reflection of basal FANCD2 monoubiquitination that occurs in the absence of DNA damage in S phase cells (Taniguchi et al., 2002).
Building upon existing models for ICL repair, and what is already known about the role of FANCD2 monoubiquitination in this pathway, it is possible to speculate on where on the ICL repair pathway FAN1 might act. Experiments on the replication of plasmids bearing single ICLs in Xenopus egg extracts showed that FANCD2 monoubiquitination is required for ICL unhooking (Knipscheer et al., 2009), which suggests that FAN1 might act at this point. In this system, it was proposed that initiation of ICL repair requires the convergence of two replication forks at an ICL (Knipscheer et al., 2009). Consequently, an ICL would be located at the intersection of an X structure shown in Figure S7A. In this scenario, cleavage of the leading strand template of one of the forks by MUS81 in concert with cleavage of the same strand on the opposite side of the ICL (which resembles a 5′ flap) by a second nuclease would unhook the ICL (Figure S7A). This nuclease could be FAN1 based on our in vitro data showing that this nuclease preferentially cleaves the double-stranded portion of a synthetic 5′ flap structure (Figure 2A). However, there are conceptual problems with FAN1 acting at this point of the ICL repair pathway. First, this hypothesis predicts that MUS81 and FAN1 are each responsible for 50% of the one-ended DSBs in cells treated with ICL-inducing agents. Assuming that γ-H2AX foci are representative of DSBs, this is not what we observe—depletion of FAN1 does not affect formation of cisplatin-induced DSBs (Figure 6C), whereas deletion of MUS81 abolished all ICL-induced DSBs (Hanada et al., 2006). Second, this hypothesis requires that MUS81 acts on the leading strand template of one of the two stalled replication forks (Figure S7A), but it is difficult to see why MUS81 would not cleave the leading strand template of both forks (Figure S7B). Cleavage of both forks by MUS81 would result in two one-ended DSBs and a linear duplex containing the ICL, and it is unlikely that unhooking of the ICL from this linear duplex would require FAN1. Third, it is not yet clear whether the two-fork model for ICL repair is relevant in vivo, and it is possible that the collision of a single replication fork with an ICL is sufficient to initiate ICL repair (Figure S7C). In this scenario, it is difficult to see how FAN1 could be involved in unhooking since it would have to cleave linear duplex DNA on the 5′ side of the ICL, a structure that is not flap-like in nature. More experiments are required to test whether the one-fork or two-fork models for ICL repair, or both, operate in vivo and to test if the defect in ICL unhooking in the context of the FANCD2 K561R mutant is due to a defect specifically in recruitment of FAN1.
After unhooking, excision repair removes the crosslink adduct and translesion synthesis fills in the gap (Figure S7A). Although FANCD2 ubiquitination appears to be involved in translesion synthesis in the Xenopus cell-free system, it is difficult to see how FAN1 could be involved at this stage, and it is likely that other FANCD2-binding proteins are required. We found that FAN1 interacts with MLH1 that is involved in mismatch repair, consistent with a previous report (Cannavo et al., 2007). At present, the significance of this interaction is not clear, but it may be that the interaction of FAN1 with MLH1 promotes the correction of translesion synthesis-induced mismatches during ICL repair.
Regardless of how exactly MUS81—alone or in conjunction with FAN1 or an as yet unidentified nuclease—unhooks the ICL, at least one and possibly two (one-ended) DSBs are generated (Figure S7). These DSBs are resected and this is a function that could be fulfilled by FAN1 based on our finding that FAN1 has 5′-3′ exonuclease activity that is capable of generating 3′ overhangs (Figure 3A). However, cytological data showing that RPA loading is normal in FAN1-depleted cells (Figure 7B) argue against this role, although potential redundancy between FAN1 and other 5′-3′ exonucleases would need to be investigated.
One of the overhangs generated by DSB resection invades the complementary duplex to initiate D loop formation and HR (Figures 7A and 7B). After extension by DNA synthesis, the invading strand reanneals to the complementary strand in the duplex it came from originally. It is possible that continued DNA synthesis on the parent strand displaces the DNA in front of it. This would generate a 5′ flap that could be cleaved by FAN1. In this scenario, FAN1 is required at a late step in HR, and several observations are consistent with this hypothesis. First, there is a delay in the disappearance of γ-H2AX foci induced by cisplatin or IR in cells treated with FAN1 siRNA compared with control cells. Similar results were reported recently in FA cells (Leskovac et al., 2010). At all time points during recovery from cisplatin and IR, there are more RAD51 foci in FAN1-depleted cells than in control cells (Figure 7C). It is possible that disappearance of foci in FAN1-depleted cells reflects inappropriate repair perhaps by nonhomologous end joining, and this may account for the increase in radial chromosomes seen in MMC-treated cells depleted of FAN1 (Figure 6D). Second, depletion of FAN1 from human cells results in reduced efficiency in I-SceI-induced HR (Figure 7A), and similar results were reported in human cells expressing FANCD2 K561R (Nakanishi et al., 2005). Depletion of FAN1 does not affect RAD51 loading in cisplatin-treated cells (Figure 7C), suggesting that if FAN1 functions at the HR step of ICL repair, then it acts independently of RAD51 or after RAD51 focus formation. Similar to FANCD2 null cells, depletion of FAN1 does not affect the frequency of MMC-induced SCEs (Figure S6). Therefore, if FAN1 does act at the HR step of ICL repair, then its role may be restricted to a subpathway of HR such as synthesis-dependent strand annealing that avoids crossing over.
The SLX4 complex of structure-specific nucleases is also required for the HR step of ICL repair (Fekairi et al., 2009; Muñoz et al., 2009; Svendsen et al., 2009). SLX4-XPF-SLX1-MUS81 can cleave three-way DNA junctions, 3′ flaps, and 5′ flaps in vitro, and so FAN1 specificity overlaps with the SLX4 complex in 5′ flap cleavage. We could find no evidence for an interaction of FAN1 with the SLX4 scaffold (unpublished data). It is not yet clear why two 5′ endonucleases are required during ICL repair, and it will be important to test redundancy between FAN1 and the SLX4 complex.
FAN1 is the only VRR_nuc domain-containing protein in eukaryotic cells. These domains are found in all kingdoms of life, but the functions of most of them are unknown (Iyer et al., 2006). Many bacteria and phages have VRR_nuc domain proteins, and so it appears that the FA repair pathway which appeared relatively late during evolution was built on a more ancient VRR_nuc domain nuclease. It will be interesting to follow up on this hypothesis. Many cytotoxic anticancer agents act by inducing ICLs, and it is possible that nucleases such as FAN1 are good targets for sensitizing cancer cells to killing by ICLs. Finally, although the majority of FA patients have mutations in the known FA genes, FA patients exist where mutations in known FA genes could not be found. In this light, it is likely that FAN1 mutations will be found in some of these patients.
EXPERIMENTAL PROCEDURES
General Methods
Gel filtration and analysis of the resolution of cisplatin-induced DSBs were carried out as described previously (Muñoz et al., 2009). Dithiobis (succinimidyl propionate) (DSP; Pierce) is a homobifunctional and thiol-cleavable crosslinker that was used according to the manufacturer’s instructions. DSP was included in lysis buffer at 2.5 mg/ml, and lysates were incubated for 30 min on ice. Excess DSP was quenched by addition of 1 M Tris-HCl (pH 7.4) to 0.2 M followed by an additional 30 min incubation. Crosslinks were reversed by the inclusion of dithiothreitol in SDS-PAGE sample buffer added to cell extracts or immunoprecipitates before electrophoresis. Details of immunofluorescence are given in the Extended Experimental Procedures.
Antibodies, Cell Lysis, and Immunoprecipiation
The primary antibodies used in this study were the following: FAN1 (this study; sheep S420C, fourth bleed), MLH1 (BD Pharminigen, 554073), PMS2 (Santa Cruz, sc-617), PCNA (Santa Cruz, PC10), FANCI (Bethyl, A301-354), FANCD2 (Abcam, ab2187-50), FANCD2 (Novus, NB100-182; immunofluorescence), FANCA (Cascade Biosciences, abm6202), FANCC (Cascade Biosciences, abp6305), FANCE (a kind gift from K.J. Patel), FANCF, FANCG (kind gifts from Johan De Winter), FLAG (Sigma, M2), Ku80 (Cell Signaling, 2753), RAD51 (Santa Cruz, H-92), RPA70 (Cell Signaling, 2267), and anti-γ-H2AX (Bethyl, A300-081A). The FAN1 antibody, raised in sheep against full-length FAN1 fused to GST at the Scottish Antibody Production Unit (Carluke, Lanarkshire), was affinity purified with immobilized antigen. GFP-Trap beads were from Chromotek. Protein G Sepharose was from GE Healthcare. Cells were lysed in ice-cold buffer: 40 mM HEPES (pH 7.4), 120 mM NaCl, 1% (v/v) Triton X-100, and 1 mM EDTA with protease inhibitors (Roche). For visualization of monoubiquitinated forms of FANCI and FANCD2, 0.5 U/ml of benzonase (Sigma) was included in the lysis buffer, and lysates were incubated on ice for 30 min. All immunoprecipitations were carried out in lysis buffer for 1 hr at 4°C. Endogenous immunoprecipitations were carried out with 2 μg FAN1 antibody coupled to 10 μl protein G Sepharose per 4 mg of whole-cell extract.
Purification of GFP–FAN1 from HEK293 Cells
FlpIn T-Rex cells (Invitrogen) cells stably expressing GFP-FAN1 in a tetracycline-inducible manner were made according to the manufacturer’s instructions with FAN1 in plasmid pcDNA5-FRT-TO-GFP-FAN1. FAN1 was induced and purified according to a previously described protocol (Muñoz et al., 2009).
siRNA
Cells were transfected with the relevant siRNA duplex (100 nM) via the calcium phosphate precipitation method. In Figure 7, U2OS cells were transfected in 96-well plates with siRNAs at a concentration of 20 nM and DharmaFECT 1 (Dharmacon) at a 1:1000 concentration. Cells were incubated at 37°C for 48 hr. The messenger RNA target sequences used for siRNAs were as follows: FANCA (GGGUCAAGAGGGAAAAAUA), FAN1-1 (GUAAGGCUCUUUCAAC GUA; exon 3), FAN1-2 (GCAGGAAGGCAGAGUGGCU; exon 12), MLH1 (GCAUGUGGCUCAUGUUAC), ATR (GGGAGCCUGUUGAGACAAGAU), and FANCD2 (siGenome SMARTPool from Dharmacon).
Oligonucleotides
The following oligonucleotides were used:
a3: 5′-CCTCGATCCTACCAACCAGATGACGCGCTGCTACGTGCTACCGGAAGTCG
b: 5′-CGACTTCCGGTAGCACGTAGCAGCGGCTCGCCACGAACTGCACTCTAGGC
c: 5′-GCCTAGAGTGCAGTTCGTGGCGAGC
d3: 5′-CGTCATCTGGTTGGTAGGATCGAGG
a3-cp: 5′-CGACTTCCGGTAGCACGTAGCAGCGCGTCAACTGGTTGGTAGGATCGAGG
Preparation of DNA Substrates
All substrates and standards were annealed by slow cooling of one radioactively 5′ 32P-labeled oligonucleotide with the relevant unlabeled one(s). In Figure 2, these were splayed duplex (SD), a3, b; 3′ flap (3′F) a3, b, d3; 5′ flap (5′F) a3, b, c; and replication fork analog (RF), a3, b, c, and d3. In Figure 3, these were a3 and a3-cp (dsDNA), a3 (ssDNA). Synthetic structures were then purified by electrophoresis on a native 8% polyacrylamide gel and recovered by the crush and soak method followed by ethanol precipitation.
Nuclease Assays
Purified recombinant FAN1 (35 nM) was preincubated for at least 10 min with radiolabeled DNA substrates (5 nM) at 37°C in 25 mM Tris-HCl (pH 7.5), 10 mM NaCl,15 mM KCl, and 0.1 mg/ml BSA to allow binding to occur. The reaction was started by the addition of 1 mM MnCl2 and stopped by the addition of 2 mM EDTA. The samples were then boiled at 95°C for 10 min and analyzed by denaturing PAGE (15% polyacrylamide and 8 M urea). Gels were dried, exposed to storage Phosphor screens, and analyzed with the ImageGauge software (Fujifilm). For kinetics experiments, the data were plotted as the fraction of DNA in the relevant product bands as a function of time and fitted to either one or two exponential functions. When two exponential functions were used, the rate given is the faster of the two.
Clonogenic Survival Assays
HEK293 cells were seeded in 10 cm2 dishes at 25% confluence and allowed to adhere overnight. Cells were transfected with the relevant siRNA for 48 hr and cells were split and seeded in 10 cm2 dishes (5000 cells/dish). Cells were allowed to adhere for a minimum of 8 hr before cisplatin or mitomycin-C were added at the indicated concentrations for 24 hr. Cells were then washed free of drugs and incubated in fresh medium for 10–14 days before the number of colonies of more than 50 cells in each dish were counted.
C. elegans Genotoxin Sensitivity Assays
Worms were maintained at 20°C on NGM (Nematode Growth Media) agar plates according to standard protocols (Brenner, 1974). Alleles used were fcd-2 (tm1298) and C01G5.8 (tm423). The C01G5.8 mutant was generated and kindly provided by Shoehi Mitani of the National Bioresource Project for the Nematode, Japan. This strain, which has a 411 bp deletion in C01G5.8 that removes exons 6–8, was outcrossed five times with N2 Bristol strain (wild-type) to eliminate secondary mutations. So that ICL sensitivity could be accessed, synchronized L1 larval stage animals of the relevant genotype were incubated at 20°C for 16 hr in 1 ml S-basal buffer (0.1 m NaCl, 0.05M KH2PO4 [pH 6.0], 5 mg/ml cholesterol) containing E. coli OP50 and the indicated concentration of nitrogen mustard (HN2) or cisplatin. After incubation worms were transferred to OP50-seeded NGM plates. After 48 hr, the extent of developmental progression was scored. In each experiment, a minimum of 60 worms was scored, and the results shown are the average of three independent experiments.
GFP HR Assay
Cells were transfected with siRNA in 96-well dishes, and after 48 hr cells were transfected with 0.25 μg I-Sce-I vector and 0.2 μg PEI in 150 μl OptiMEM/well. GFP-positive cells were analyzed with FACS 48 hr after I-Sce-I transfection as previously described (Pierce et al., 1999).
ACKNOWLEDGMENTS
We are grateful to Katja Kratz and Joe Jiricny for experimental advice, for helping us with the purification of FAN1, and for communicating results prior to publication. We thank K.J. Patel, Johan de Winter, and Maureen Hoatlin for kind gifts of antibodies. pDEST40-V5-FANCD2 wild-type and pDEST40-V5-FANCD2 K561R plasmids were kind gifts from Niall Howlett and Thomas Glover. We thank Alan d’Andrea for the kind gifts of FANCD2−/− fibroblasts (PD20 cells) corrected with vector, FANCD2 wild-type, or FANCD2 K561R. We are grateful to James Hastie, Hilary MacLauchlan, and the Antibody Production Team at Division of Signal Transduction Therapy, University of Dundee, to Bob Gourlay, Sanjay Kothiya, and Nick Morrice for help with mass spectrometry, and to the DNA Sequencing Service at the College of Life Sciences (CLS), University of Dundee. We are grateful to the microscopy facility, CLS, University of Dundee for assistance with microscopy. We thank Jim Haber and the J.R. lab for critical reading of the manuscript. This work was funded by the UK Medical Research Council (C.M., T.J.H., and J.R.), Cancer Research UK (A.C.D, A.G., and D.M.J.L.), and the Fanconi Anemia Research Fund (A.J.D. and S.C.W.).
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes Extended Experimental Procedures, seven figures, and one table and can be found with this article online at doi:10.1016/j.cell.2010.06.021.
REFERENCES
- Akkari YMN, Bateman RL, Reifsteck CA, Olson SB, Grompe M. DNA replication is required To elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol. Cell. Biol. 2000;20:8283–8289. doi: 10.1128/mcb.20.21.8283-8289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstralh DT, Sekelsky J. Interstrand crosslink repair: can XPF-ERCC1 be let off the hook? Trends Genet. 2008;24:70–76. doi: 10.1016/j.tig.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Bhagwat N, Olsen AL, Wang AT, Hanada K, Stuckert P, Kanaar R, D’Andrea A, Niedernhofer LJ, McHugh PJ. XPF-ERCC1 participates in the Fanconi anemia pathway of cross-link repair. Mol. Cell. Biol. 2009;29:6427–6437. doi: 10.1128/MCB.00086-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E, Gerrits B, Marra G, Schlapbach R, Jiricny J. Characterization of the interactome of the human MutL homologues MLH1, PMS1, and PMS2. J. Biol. Chem. 2007;282:2976–2986. doi: 10.1074/jbc.M609989200. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani H, Joenje H, McDonald N, de Winter JP, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Collis SJ, Ciccia A, Deans AJ, Horejsí Z, Martin JS, Maslen SL, Skehel JM, Elledge SJ, West SC, Boulton SJ. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol. Cell. 2008;32:313–324. doi: 10.1016/j.molcel.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Deans AJ, West SC. FANCM connects the genome instability disorders Bloom’s Syndrome and Fanconi Anemia. Mol. Cell. 2009;36:943–953. doi: 10.1016/j.molcel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, 3rd, Russell P, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D’Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006;25:4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 2007;14:1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- Hofmann K. Ubiquitin-binding domains and their role in the DNA damage response. DNA Repair (Amst.) 2009;8:544–556. doi: 10.1016/j.dnarep.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Babu MM, Aravind L. The HIRAN domain and recruitment of chromatin remodeling and repair activities to damaged DNA. Cell Cycle. 2006;5:775–782. doi: 10.4161/cc.5.7.2629. [DOI] [PubMed] [Google Scholar]
- Knipscheer P, Räschle M, Smogorzewska A, Enoiu M, Ho TV, Schärer OD, Elledge SJ, Walter JC. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski J, Feder M, Bujnicki JM. The PD-(D/E)XK superfamily revisited: identification of new members among proteins involved in DNA metabolism and functional predictions for domains of (hitherto) unknown function. BMC Bioinformatics. 2005;6:172. doi: 10.1186/1471-2105-6-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leskovac A, Vujic D, Guc-Scekic M, Petrovic S, Joksic I, Slijepcevic P, Joksic G. Fanconi anemia is characterized by delayed repair kinetics of DNA double-strand breaks. Tohoku J. Exp. Med. 2010;221:69–76. doi: 10.1620/tjem.221.69. [DOI] [PubMed] [Google Scholar]
- Ling C, Ishiai M, Ali AM, Medhurst AL, Neveling K, Kalb R, Yan Z, Xue Y, Oostra AB, Auerbach AD, et al. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J. 2007;26:2104–2114. doi: 10.1038/sj.emboj.7601666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe KM, Olson SB, Moses RE. DNA interstrand crosslink repair in mammalian cells. J. Cell. Physiol. 2009;220:569–573. doi: 10.1002/jcp.21811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, D’Andrea AD. How the fanconi anemia pathway guards the genome. Annu. Rev. Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz IM, Hain K, Déclais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol. Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D’Andrea AD, Wang ZQ, Jasin M. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc. Natl. Acad. Sci. USA. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NG, Beverloo HB, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell. Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KJ, Joenje H. Fanconi anemia and DNA replication repair. DNA Repair (Amst.) 2007;6:885–890. doi: 10.1016/j.dnarep.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promoteshomology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Räschle M, Knipscheer P, Knipsheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Schärer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfuss A, Grompe M. Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol. Cell. Biol. 2004;24:123–134. doi: 10.1128/MCB.24.1.123-134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D’Andrea AD, Elledge SJ. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swann PF, Waters TR, Moulton DC, Xu YZ, Zheng QG, Edwards M, Mace R. Role of postreplicative DNA mismatch repair in the cytotoxic action of thioguanine. Science. 1996;273:1109–1111. doi: 10.1126/science.273.5278.1109. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D’Andrea AD. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]