Abstract
Telomeres are critical for cell survival and functional integrity. Oxidative DNA damage induces telomeric instability and cellular senescence that are associated with normal aging and segmental premature aging disorders such as Werner Syndrome and Rothmund-Thompson Syndrome, caused by mutations in WRN and RECQL4 helicases respectively. Characterizing the metabolic roles of RECQL4 and WRN in telomere maintenance is crucial in understanding the pathogenesis of their associated disorders. We have previously shown that WRN and RECQL4 display a preference in vitro to unwind telomeric DNA substrates containing the oxidative lesion 8-oxoguanine. Here, we show that RECQL4 helicase has a preferential activity in vitro on telomeric substrates containing thymine glycol, a critical lesion that blocks DNA metabolism, and can be modestly stimulated further on a D-Loop structure by TRF2, a telomeric shelterin protein. Unlike that reported for telomeric D-Loops containing 8-oxoguanine, RECQL4 does not cooperate with WRN to unwind telomeric D-Loops with thymine glycol, suggesting RECQL4 helicase is selective for the type of oxidative lesion. RECQL4’s function at the telomere is not yet understood, and our findings suggest a novel role for RECQL4 in the repair of thymine glycol lesions to promote efficient telomeric maintenance.
Keywords: RECQL4, WRN, thymine glycol, telomeres
1. INTRODUCTION
The five human RecQ helicases RECQL1, WRN, BLM, RECQL4 and RECQL5 are functionally significant in DNA maintenance, replication and DNA repair. In particular, WRN and RECQL4 have been shown to be important proteins for telomere maintenance [1, 2]. Telomeres protect human chromosome ends from degradation, and shortened and unstable telomeres are frequently associated with aging [3-5]. In accordance, cells from patients with segmental premature aging syndromes Werner syndrome (WS) and Rothmund-Thompson syndrome (RTS) deficient in WRN and RECQL4, respectively, exhibit telomere instability [2, 6]. While WRN has been extensively studied in regard to its role in DNA and telomere maintenance [6-11], much less is known about the roles of RECQL4.
Telomeres can become unstable due to DNA damage, particularly that resulting from oxidative damage (1) (2-4). Oxidative DNA damage can be caused by external agents, for example by chemical oxidants and ionizing radiation, or endogenously, mainly by reactive oxygen species (ROS) formed by cellular aerobic metabolism (5). Various oxidative DNA lesions exist such as: 6-diamino-4-hydroxy-formamidopyrimidine and 4,6-diamino-5- formamidopyrimidine (commonly known as Fapy G and Fapy A lesions, respectively), hydroxyl-methylamine and 8-oxoguanine which are commonly studied, and thymine glycol which is less well-characterized. In addition to exhibiting unstable telomeres, WS and RTS cells also show hypersensitivity to oxidative agents (6,7), together suggesting a critical role for these RecQ helicases in the repair of oxidative lesions at the telomeres.
Generally, guanine is considered to be the most commonly oxidized base, due to its low oxidation potential. However, thymine glycol (Tg) is acknowledged as the most common oxidation product of the thymine base (8). Because Tg base pairs efficiently with its normal Watson-Crick partner adenine, this lesion is only weakly mutagenic relative to other oxidative lesions (9). Critically, however, Tg has been shown to be lethal to cells by strongly inhibiting DNA replication in vitro and in vivo in both E. coli (10,11) and human cells (12). Although human DNA translation polymerase eta (POLη) is reportedly capable of accurately replicating across Tg lesions in vitro (13,14), it was also shown that traditional replicative and repair polymerases stall just one base beyond the Tg, inhibiting further replication and elongation (10,11,15). Crystallography has demonstrated that replication arrest is actually caused by the local helical distortion created by base-pairing at a Tg lesion (8,15), which also prevents efficient repair of these lesions at sites of double strand breaks (16). Tg damage therefore presents a critical barrier to cell survival.
Supporting these observations is a multi-species comparative study which demonstrates that Tg damage in vivo is correlated to lifespan (17). Interestingly, telomere length is also correlated with longevity in animals, human cells and individuals (18-20). As WRN and RECQL4 are implicated strongly in telomere maintenance, and telomere length and integrity are disrupted by oxidative damage (21-23), one could speculate that their loss prevents efficient telomere maintenance around oxidative lesions such as Tg and leads to telomere instability, telomere loss and ultimately cellular senescence that are observed in WS and RTS patient cells under oxidative stress (7,24,25).
We recently reported that RECQL4 is involved in telomere maintenance in vivo, is functionally stimulated by a telomeric shelterin protein TRF2, and shows a preference to unwind a telomeric D-Loop containing 8-oxoguanine lesions in vitro (26) similar to WRN (27). To explore their possible role in the repair of replication-blocking Tg lesions in telomeres, we developed unique telomeric substrates containing Tg lesions and investigated in vitro the functional capacity of RECQL4 and WRN to unwind telomeric D-Loops and replication forks containing Tg lesions. We show that unlike WRN, RECQL4 shows a clear preference to unwind substrates containing Tg and that this activity is stimulated on Tg D-Loops in the presence of TRF2. We therefore propose that RECQL4 is functionally important for the cellular response to Tg lesions at the telomeres.
2. MATERIALS AND METHODS
2.1 Preparation of oligonucleotides
Synthetic telomeric D-Loops were constructed as described in Figure 1 and Table 1. Unmodified oligonucleotides were manufactured and PAGE-purified by Integrated DNA Technologies (Coralville, IA, USA). All modified oligonucleotides containing thymine glycol were synthesized and PAGE-purified by The Midland Certified Reagent Company (Midland, TX, USA). Oligonucleotides were labeled, annealed and characterized as described previously (27). Non-telomeric D-loop containing thymine glycol was also constructed in a similar manner using the oligonucleotides Tg2 mix, BB mix and BT (Table 1).
FIGURE 1. Construction of telomeric D-Loops with thymine glycols.
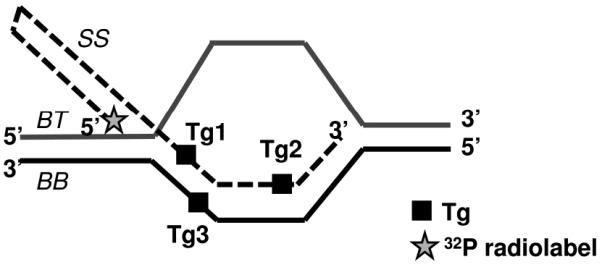
Structures of the telomeric D-Loops used in this study. A black square (■) indicates where thymine bases were modified to thymine glycols in the various substrates, outlined in Table 1. DL does not contain any modified bases, while DL-Tg1, DL-Tg2 or DL-Tg3 contains a modified base at a position Tg1, Tg2 or Tg3. A star (✩) indicates the position of 32P radiolabel.
Table 1. Oligonucleotide names and sequences.
Sequences are given for the synthetic oligodeoxynucleotides with telomeric and complementary sequences used in this study. The thymine base modified to a thymine glycol is indicated by [Tg]. Telomeric D-Loops (Figure 1) consisted of a 33 base pair (bp) duplex and were constructed as follows: DL: SS, BB, BT; DL-Tg1: SS-Tg1, BB, BT; DL-Tg2: SS-Tg2, BB, BT; DL-Tg3: SS, BB-Tg, BT. Non telomeric D-loop was constructed using SS-Tg2 mix, BB mix and BT.
| Oligo name |
Sequence |
|---|---|
| SS | (5′)CACCATCCAGTTCTCTTTTGAGAACTGGATGGTGTTAGGGTTAGGGTTAGGG TTAGGGTTAACGCTC(3′) |
| SS- Tg1 |
(5′)CACCATCCAGTTCTCTTTTGAGAACTGGATGGTG[Tg]TAGGGTTAGGGTTAG GGTTAGGGTTAACGCTC(3′) |
| SS- Tg2 |
(5′)CACCATCCAGTTCTCTTTTGAGAACTGGATGGTGTTAGGGTTAGGGTTAGGG TTAGGG[Tg]TAACGCTC(3′) |
| BB | (5′)TCAAGCTCGGTCTGCAGTCAGGATGATTGTGAGCGTTAACCCTAACCCTAAC CCTAACCCTAATCTGCACTCGAGACTCACGTCCTGGTCACG-3′ |
| BB- Tg |
(5′)TCAAGCTCGGTCTGCAGTCAGGATGATTGTGAGCGTTAACCCTAACCCTAAC CC[Tg]AACCCTAATCTGCACTCGAGACTCACGTCCTGGTCACG(3′) |
| BT | (5′)CGTGACCAGGACGTGAGTCTCGAGTGCAGACCTTTTTTTTTTTTTTTTTTTTT TTTTTTTTTTACAATCATCCTGACTGCAGACCGAGCTTGA(3′) |
| SS- Tg2 mix |
(5′)CACCATCCAGTTCTCTTTTGAGAACTGGATGGTGTATCACATTGCGTTGATGG GACCG[Tg]TAACGCTC(3′) |
| BB mix |
(5′)TCAAGCTCGGTCTGCAGTCAGGATGATTGTGAGCGTTAACGGTCCCATCAACGCAA TGTGATATCTGCACTCGAGACTCACGTCCTGGTCACG(3′) |
2.2 Proteins
Wild-type human RECQL4 and helicase-dead RECQL4 (K508M) with a C-terminal His9 tag in the pGEX6p1 vector (GE Healthcare) were expressed and purified from Escherichia coli Rosetta2 (DE3) (Novagen) in-house as described previously (Rossi et al, 2010). Recombinant histidine-tagged wild-type WRN protein and recombinant histidine-tagged human TRF2 and TRF1 proteins were purified using a baculovirus/insect cell expression as described previously (28,29). Protein concentration was determined by the Bradford assay (Bio-Rad), and purity was determined by SDS-PAGE and Coomassie staining. NEIL1 was commercially sourced from New England Biolabs (Ipswich, MA).
2.3 Helicase and Strand Annealing Assays
To measure helicase activity, RECQL4 protein (at concentrations shown in the figure legends) was incubated with 0.5 nM radio-labeled telomeric D-Loop for 30 minutes at 37°C in a reaction volume of 20 μl reaction buffer containing 30 mM Tris pH 7.4, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 100 μg/ml BSA, 10% glycerol, 5 mM ATP, and 12.5nM unlabeled single stranded primer SS. WRN and BLM protein were incubated with the same substrates for 30 minutes at 37°C in a reaction volume of 20 μl reaction buffer containing (40mM Tris, pH 8.0, 4mM MgCl2, 5 mM DTT, 2mM ATP, and 100 μg/ml BSA, without excess unlabeled SS). As a control for substrate stability, E. coli UvrD helicase activity was measured on the telomeric D-Loops in a 3mM ATP 1X Helicase Buffer M reaction solution (BioHelix, Beverly, MA). Where indicated, TRF2 was incubated with RECQL4 (concentrations given in the figure legends) on ice for 5 minutes before adding radio-labeled substrate to the reaction. To measure strand annealing efficiency, RECQL4 protein was incubated with radio-labeled single stranded primer SS or SS-Tg2 (0.5 nM, Table 1) and its unlabeled complementary primer BB (1 nM, Table 1) for 30 seconds to 30 minutes at 37°C in a volume of 20 μl reaction buffer made of the same constituents as for helicase reactions above excluding ATP and excess single stranded primer. All reactions were stopped by addition of 10 μl of 3× native stop dye (50 mM EDTA, 40% glycerol, 0.9% SDS, 0.05% bromophenol blue, and 0.05% xylene cyanol). The reaction products were separated by electrophoresis on an 8% native polyacrylamide / 20 mM Tris, 10 mM boric acid, and 0.5 mM EDTA (0.5×TBE) gel in 0.5× TBE running buffer at 150V for 2 h, exposed to a PhosphorImager screen (GE Healthcare, Piscataway, NJ), and imaged with a Typhoon scanner (GE Healthcare, Piscataway, NJ). ImageQuant version 5.2 (GE Healthcare, Piscataway, NJ) was employed to analyze the phosphoimages and calculate the percentage of unwound and annealed substrate in each reaction, normalizing against background, as described previously (27). Assays were done at least in triplicate, and representative gels are shown.
2.4 ATPase assays
RECQL4 (80 nM) was incubated with 90 nM of different structures of DNA and 1.25 μCi [γ-32P] ATP (PerkinElmer Life Sciences) in 10 μl helicase assay buffer with 50 μM cold ATP for 1 h at 37 °C. SS, Tg-containing ssDNA, SS-Tg1, SS-Tg2, DL and Tg-containing D-loops, DL-Tg1, DL-Tg2 are used as a DNA cofactor as indicated in Figure and Figure legends. Reactions were stopped by addition of 5 μl of 0.5 M EDTA and placed on ice. 2.5 μl from each reaction was spotted on a cellulose polyethyleneimine thin-layer chromatography (TLC) sheet (JTBaker), and resolved in 0.8 M LiCl/1 M formic acid solution. The chromatography sheet was exposed to a PhosphorImager plate (GE Healthcare) and analyzed as described above. Assays were done at least in triplicate, and representative TLC image are shown.
2.5 Exonuclease Assays
WRN and E. coli EXOIII (at concentrations shown in the figure legends) was incubated with either 0.5 nM undamaged telomeric D-Loop (DL) or 0.5 nM thymine glycol telomeric D-Loops (DL-Tg1, DL-Tg2, and DL-Tg3) in a 20 μl reaction buffer containing 40 mm Tris, pH 8.0, 4 mm MgCl2, 5 mm DTT, 2 mm ATP, and 1 mg/ml BSA for 30 min at 37°C, and stopped with 10 μl of a 80% formamide, 0.01% SDS, 0.05% bromophenol blue/Orange G buffer. Reactions were denatured at 95°C for 5 min and run on a pre-heated 15% denaturing (7M urea) PAGE at 15 watts for 55 min. Gels were imaged and analyzed for exonuclease-degraded substrate as described above for enzymatic assays. Assays were performed at least in triplicate.
2.6 EMSA Binding Assays
RECQL4 (at concentrations shown in the figure legends) was incubated with either 0.5 nM undamaged telomeric D-Loop (DL) or 0.5 nM thymine glycol telomeric D-Loop (DL-Tg2) in a 10 μl EMSA buffer containing 30 mM Tris-HCl, pH 7.4, 1 mM DTT, 100 μg/ml BSA, and 50 mM KCl for 15 min on ice. Reactions were stopped with 5 μl non-reducing stop dye (50% glycerol and 0.05% bromphenol blue) and kept on ice prior to loading onto a 5% polyacrylamide / 20 mM Tris, 10 mM acetic acid, and 0.5 mM EDTA (0.5×TAE) gel and run in 0.5×TAE at 200V for 1.5 h at 4°C. Gels were imaged and analyzed for the percentage of bound substrate as described above for enzymatic assays. Assays were performed in triplicate.
2.7 NEIL1 incision Assays
To measure the effect of RECQL4 or WRN activity on NEIL1 incision capacity, 1:1-1:8 and 1:1-1:20 molar ratios of RECQL4 and WRN respectively were incubated with 2.5 nM NEIL1 for 10 min at 30°C, then incubated with 0.5 nM radio-labeled telomeric D-Loop or forked duplex substrates containing Tg for another 30 min at 37°C. Reactions were terminated by adding a stop buffer containing 30 mM NaOH, 80% formamide and a bromo-phenol blue/xylene cyanol dye mixture. The contents of the reaction were denatured at 90°C for 5 min and separated by electrophoresis on a 15% denaturing polyacrylamide gel. Assays were performed in triplicate, and gels were imaged and analyzed for the percentage of incised substrate as described above for helicase and annealing assays.
3. RESULTS
3.1 RECQL4 shows a preference to unwind telomeric D-Loops containing Tg
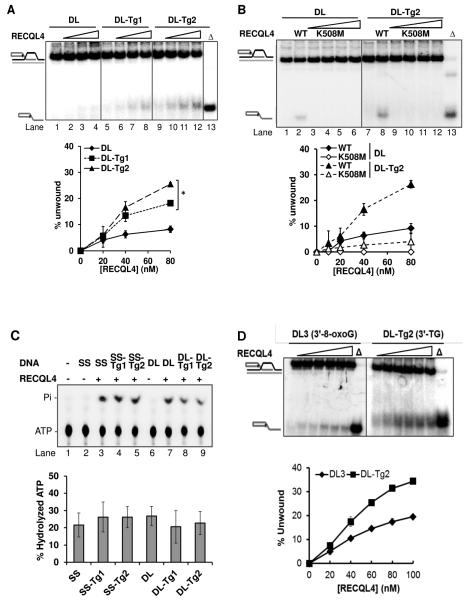
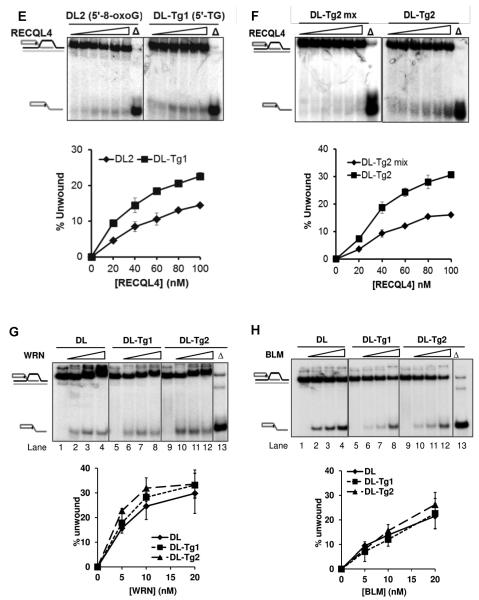
We and others have shown previously that BLM, WRN and RECQL4 have functional roles in telomere maintenance and can potentially unwind telomeric substrates in vitro. They also exhibit a preference for a telomeric structure containing an oxidized guanine base (8-oxoguanine) over an undamaged structure. While 8-oxoguanine is a mutational lesion, thymine glycol (Tg) is a replication-blocking lesion and therefore a highly critical lesion to repair for cellular survival. We investigated whether these RecQ helicases showed altered activity in vitro for telomeric substrates containing a Tg lesion. It has already been reported that the RECQL4 helicase shows preference for a telomeric sequence in the D-Loop structure, as it fails to unwind a non-telomeric D-Loop (26). As reported previously (30,31), RECQL4 required 25× excess single-stranded DNA (ssDNA) in all helicase reactions in order to visualize the helicase activity on the D-Loops and telomeric forks used here. Even then, RECQL4 showed a very weak helicase activity of just 3% at 20 nM on the undamaged D-Loop, DL (Figure 2A). Even at a higher concentration, RECQL4 was able to unwind only 6% of this D-loop. However, on D-Loops containing Tg (DL-Tg1 and DL-Tg2), RECQL4 exhibited a 3-4 fold increase in activity compared with the undamaged D-Loop (DL) in the presence of excess ssDNA (Figure 2A). A similar level of activity was also observed for RECQL4 on an additional D-Loop, DL-Tg3, which contained a Tg lesion in the bottom strand (data not shown). To confirm whether the increase in unwound products of Tg containing D-Loops was due to RECQL4’s helicase activity, we used RECQL4 K508M, a helicase-dead mutant, in the helicase assay. The results showed that the K508M failed to exhibit helicase activity on both DL and DL-Tg2, while wild type RECQL4 could unwind both these substrates. (Figure 2B). We also tested if a thymine glycol lesion affected the ATPase activity of RECQL4. We found that there was no significant difference in ATPase activity with or without a Tg lesion in either ssDNA or D-loops (Figure 2C). To investigate the specificity of RECQL4 towards Tg containing D-loops, we compared its helicase activity on Tg and 8-oxoguanine (8-oxoG) containing D-loops. We compared the efficiency of helicase activity when placing the modifications at similar positions towards either 3′- or 5′- end of the invading strand, thus eliminating any differences in activity that might arise from differences in sequence context of the lesions. On comparing the D-loops containing either Tg (DL-Tg2) or 8-oxoG (DL3) located towards 3′-end of the invading strand, the activity on DL-Tg2 was greater by almost 2-fold than that observed on the DL3 (Figure 2D). Similar results were obtained by comparing D-loops containing 8-oxoG (DL2) and Tg (DL-Tg1) in which the lesions are located towards 5′-end of the invading strand (Figure 2E). We further tested if RECQL4 could unwind non-telomeric D-loops containing Tg. RECQL4 had higher activity on the telomeric D-loop containing Tg (DL-Tg2) than on the non-telomeric D-loop (DL-Tg2 mix) (Figure 2F). As an additional control for the substrate stability, we tested E. coli UvrD helicase on DL and DL-Tg2 (Figure S1). UvrD showed no enhanced unwinding activity on the Tg D-Loop. Altogether, our findings indicate that RECQL4 has a unique preference to unwind telomeric D-Loops that contain a Tg lesion. This preference by RECQL4 was in contrast to WRN and BLM which showed no altered unwinding activity when a Tg lesion was present (Figures 2G and 2H).
FIGURE 2. RECQL4 shows unique preference towards telomeric D-Loops containing thymine glycol.
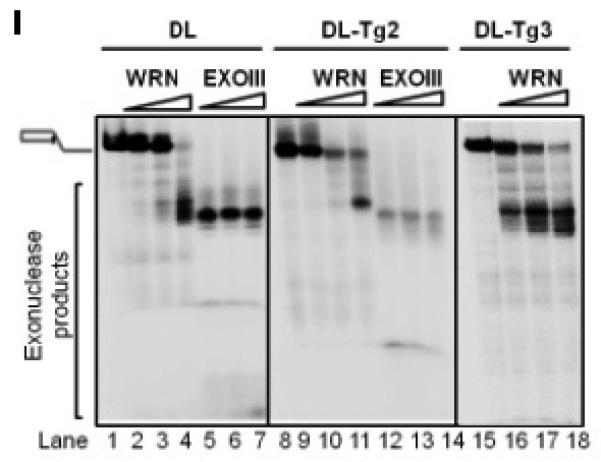
(A) Gel and quantitative analysis showing the unwinding capacity of 20, 40 and 80 nM RECQL4 on D-Loops with and without thymine glycol (Tg) lesions: DL (an undamaged D-Loop; lanes 1-4), DL-Tg1 (containing a Tg lesion toward the 5′ end of the invading strand; lanes 5-8) and DL-Tg2 (containing a Tg lesion toward the 3′ end of the invading strand; lanes 9-12), each at 0.5 nM concentration in the reaction. “Δ” (lane 13) indicates heat-denatured substrate. (B) Gel and quantitative analysis showing the unwinding capacities of RECQL4 WT and K508M on D-Loops with and without Tg. Unwinding activity of 10, 20, 40 and 80 nM RECQL4 K508M on DL (lanes 3-6) or DL-Tg2 (lanes 9-12) are compared with those of wild type RECQL4 (lane 2 or 8). (C) A representative phosphor image of thin-layer chromatography and quantitative analysis showing the effect of DNA cofactors with and without Tg lesions on ATP hydrolysis. (lane 2-3,SS; lane 4, SS-Tg1; lane 5, SS-Tg2; lane 6-7, DL; lane 8, DL-Tg1, lane 9, DL-Tg2) (D) Gel and quantitative analysis with increasing concentrations of RECQL4 (20-100 nM) on telomeric D-loops containing either 8-oxoguanine (DL3) or Tg (DL-Tg2) located towards the 3′- end of the invading strand. (E) Gel and quantitative analysis with increasing concentrations of RECQL4 (20-100 nM) on telomeric D-loops containing either 8-oxoguanine (DL2) or Tg (DL-Tg1) located towards 5′- end of the invading strand. (F) Gel and quantitative analysis with increasing concentrations of RECQL4 (20-100 nM) on telomeric D-loop containing Tg (DL-Tg2) and non-telomeric D-loop containing Tg (DL-Tg2 mix). (G and H) Gel and quantitative analysis showing the comparable unwinding capacity of 5, 10 and 20 nM WRN (G) and BLM (H) on the same substrates as in A, as indicated. (I) Representative gel showing the effect of Tg lesions on the activity of 5, 10 and 20 nM WRN exonuclease (lanes 1-4 DL; lanes 8-11 DL-Tg2; and additionally lanes 15-18 DL-Tg3) compared with E. coli EXOIII (lanes 5-7 DL; lanes 12-14 DL-Tg2). *P<0.05 by a t-test.
Previous reports show that WRN exonuclease activity is blocked by Tg and other oxidative lesions in fork and overhang substrates (32,33). We therefore tested the exonuclease activity of WRN on the D-Loop substrates containing the Tg lesion, DL-Tg2 and DL-Tg3. As a marker we used the activity of E. coli exonuclease EXOIII, a duplex-specific 3′-5′ exonuclease which cleaves at the duplex in the SS oligonucleotide (34) and is not blocked by a Tg lesion. Consistent with the literature, WRN exonuclease was blocked at Tg lesions located on the invading strand of DL-Tg2 (Figure 2I), as shown by decreased mobility of longer exonuclease products for WRN (lanes 9-11) compared with EXOIII (lanes 12-14) and compared with its exonuclease products in the undamaged D-Loop DL (lanes 2-4). WRN exonuclease was not blocked when the Tg lesion was in the bottom BB-Tg strand of DL-Tg3 (far right panel, Figure 2I). This strand-specific effect by other types of oxidative lesions on WRN exonuclease has been shown for forked and recessed substrates (35), but it is a novel observation that WRN exonuclease is inhibited by Tg on a telomeric D-Loop structure.
The cause of RECQL4’s enhanced unwinding activity is not clear. To clarify, we first tested the binding ability of RECQL4 on undamaged or damaged D-loops. Binding capacity was no greater on the Tg-containing D-Loop (Figure 3A), thus the enzyme is not more effectively bound or recruited by this lesion. However, RECQL4 is known to have a strong helicase-independent annealing activity (30), and it has also been reported that substrates containing Tg are less efficiently annealed in vitro (11). We therefore investigated RECQL4’s annealing capacity on D-Loops containing Tg. Over a time course of 30 seconds to 30 minutes, we observed similar annealing kinetics using a low concentration of RECQL4 on the Tg-containing D-Loop as on the undamaged D-Loop despite a lower saturation point on the former (Figure 3B). This small difference may not account for the 4-fold increase of helicase activity observed, particularly as no difference in its annealing capacity was measurable between the substrates under conditions identical to those used to measure its helicase activity (Figure 3C).
FIGURE 3. Comparative binding and annealing efficiency of RECQL4 on telomeric D-Loops with and without thymine glycol.
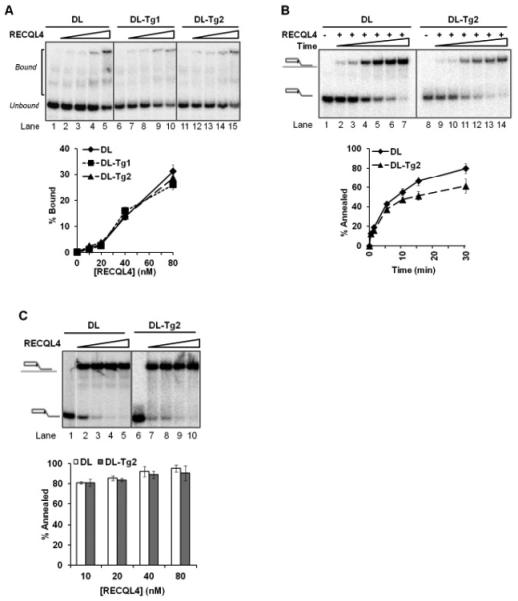
(A) Gel and quantitative analysis showing the binding capacity of 10, 20, 40 and 80 nM RECQL4 on an undamaged D-Loop, DL (lanes 1-5), compared with two Tg-containing D-Loops, DL-Tg1 and DL-Tg2 (lanes 6-10 and 11-15, respectively). (B) Gel and quantitative analysis showing the annealing efficiency of 1 nM RECQL4 on DL (lanes 1-7) compared with DL-Tg2 (lanes 8-14) over a time period of 30 seconds to 30 minutes. (C) Gel and quantitative analysis showing the annealing efficiency of RECQL4 on the same substrates (DL lanes 1-5, DL-Tg2 lanes 6-10) done in conditions used in the helicase assays (minus ssDNA), at 10-80 nM RECQL4 over 30 minutes. Error bars show standard deviation over at least three independent assays.
3.2 RECQL4 does not cooperate with WRN to unwind D-Loops containing Tg
We reported previously that RECQL4 and WRN cooperate on D-Loops containing 8-oxoguanine (26). Here, we investigated whether the two proteins may also cooperate to unwind D-Loops containing Tg. As in the previous study, the assay was performed under conditions optimal for WRN helicase without excess ssDNA, such that no helicase activity was detectable from RECQL4, thereby restricting visualization of helicase activity to that of WRN only. Our data shows that RECQL4 and WRN do not cooperate to unwind D-Loops containing Tg, as the amount of unwound substrate detected from WRN helicase was decreased in the presence of RECQL4 (Figure 4A). This may be caused either by RECQL4’s annealing activity or a non-catalytic effect in these reaction conditions, as a helicase-dead mutant K508M which exhibits similar annealing to the wild-type (30) also decreased the amount of substrate unwound compared with WRN helicase alone on both substrates (Figure 4B). No helicase activity was detectable from either wild-type or mutant RECQL4 at this concentration. Quantitative analysis of WRN’s unwinding in their presence (Figure 4C) indicates that the decrease observed was significant by a T-test (P<0.05) for both wild-type RECQL4 and the helicase-dead mutant K508M.
FIGURE 4. Functional interaction of RECQL4 with WRN to unwind D-Loops containing a thymine glycol lesion.
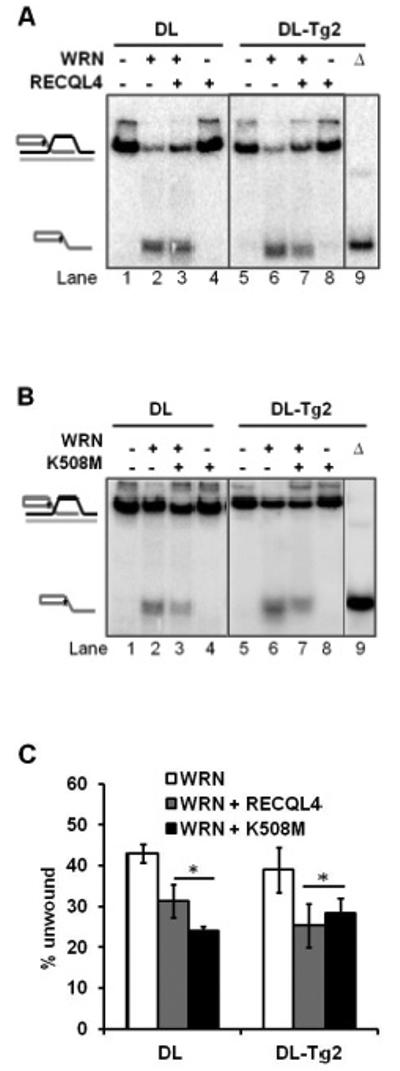
(A) Gel showing the unwinding capacity of 10 nM WRN on an undamaged D-Loop, DL (lanes 1-4), and a Tg-lesion containing D-Loop, DL-Tg2 (lanes 5-8), with (+) or without (−) 10 nM wild type RECQL4 in the reaction mixture. Δ (lane 9) indicates heat-denatured substrate. (B) Gel showing a comparable effect of 10 nM helicase-dead mutant RECQL4 (K508M) on 10 nM WRN on the same substrates as in A. (C) Quantitative analysis showing the effect of either wild-type or mutant (K508M) RECQL4 on WRN helicase. Neither wild-type nor mutant RECQL4 exhibited any detectable helicase activity on the substrate alone. Error bars show standard deviation over at least three independent assays. *P<0.05 by a t-test.
3.3 TRF2 directly stimulates RECQL4 helicase on D-Loops containing Tg
The shelterin proteins, which include double-stranded binding proteins TRF1 and TRF2, bind to and stabilize telomeric D-Loop structures (36). The shelterins are known to regulate WRN on telomeric substrates with oxidative damage (28,37). We therefore investigated whether TRF2 could functionally interact with RECQL4 on telomeric D-Loops containing Tg. Using the helicase assay with excess ssDNA, we incubated RECQL4 with increasing concentrations of TRF1 and TRF2 prior to addition of the substrate. While TRF1 did not affect RECQL4 helicase activity in this study (not shown), TRF2 did stimulate RECQL4 on the Tg-containing D-Loop, DL-Tg2 (Figure 5A). RECQL4 helicase activity on DL-Tg2 was enhanced roughly 2-fold at 1:1 molar ratio concentration (P<0.05). This functional interaction on an oxidatively damaged telomeric substrate is a novel observation. TRF2 and RECQL4 have been shown previously to physically interact by co-immunoprecipitation and cellular colocalization studies (26). On the other hand, TRF2 does not stimulate WRN on the D-loop substrate containing Tg (DL-Tg2) (Figure S2). Instead, inhibition was observed at higher concentrations of TRF2, possibly due to the competition in substrate binding. TRF2 is also known to bind to telomeric D-Loops (38), but shows decreased binding affinity for D-Loops containing 8-oxoguanine (39). To explore whether TRF2 stimulated RECQL4 helicase by binding to the Tg D-Loops and recruiting RECQL4, we used electrophoretic mobility shift assays to look for cooperative binding of TRF2 and RECQL4 on an undamaged and a Tg-containing D-Loop. Both TRF2 and TRF1 showed 50% decreased binding capacity to D-Loop substrates with Tg lesions (Figure 5B; P<0.05 by a t-test). At the concentration at which TRF2 stimulated RECQL4 helicase, RECQL4 showed no greater capacity to bind either D-Loop but instead competed with TRF2 for the substrate, together binding less substrate than RECQL4 was able to bind on its own (P<0.05; Figure 5C). These observations indicate that TRF2 stimulation of RECQL4 helicase was not via enhancement of its binding capacity to the Tg D-Loop.
FIGURE 5. Functional interaction of TRF2 with RECQL4 on telomeric D-Loops containing thymine glycol.
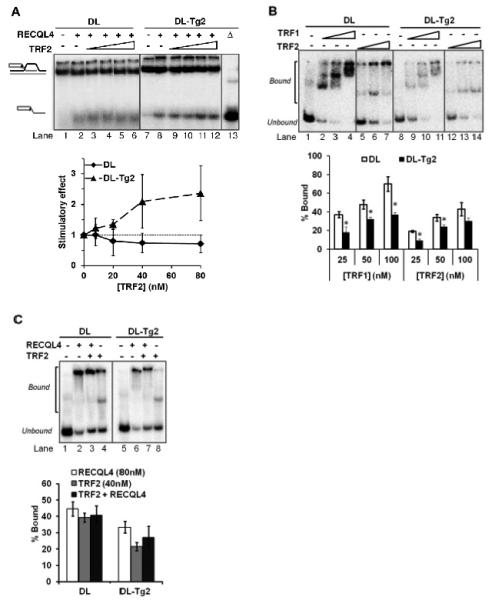
(A) Gel and quantitative analysis showing the effect of 8, 20, 40 and 80 nM TRF2 on 80 nM RECQL4 on an undamaged D-Loop, DL (lanes 1-6), and a Tg-containing D-Loop, DL-Tg2 (lanes 7-12). Δ (lane 13) indicates heat-denatured substrate. The graph represents the stimulatory effect on RECQL4’s helicase activity by TRF2. Stimulatory effect is calculated relative to the helicase activity without TRF2 (Lane 2 for DL, lane 8 for DL-Tg2). (B) Gel showing the effect of Tg lesions on shelterin protein binding to D-Loops. 25, 50 and 100 nM TRF1 or TRF2 was incubated with 0.5 nM DL (lanes 1-7) or DL-Tg2 (lanes 8-14) as described in the methods. The percent of substrate DL (white bars) and DL-Tg2 (black bars) bound by each protein was quantified over three repeats. (C) Gel and quantitative analysis showing the effect of TRF2 on RECQL4 binding to D-Loops, where 80 nM RECQL4 binding alone (lanes 2 and 6) are compared with the binding of 80 nM RECQL4 in the presence of 40 nM TRF2 (lanes 3 and 7) on DL and DL-Tg2 as indicated. The binding capacity of 40 nM TRF2 alone on these substrates is also shown (lanes 4 and 8). Error bars show standard deviation over at least three independent assays. *P<0.05 by a t-test.
3.4 WRN but not RECQL4 interacts with NEIL1 to remove Tg lesions on telomeric DNA
While the nucleotide excision repair pathway is also implicated in Tg repair (40), the main pathway responsible for the removal of Tg lesions in human cells is base excision repair (BER) (41). BER is initiated by removal of one (in short-patch BER) to ten (in long-patch) nucleotides 3′ of the lesion by one of several human DNA glycosylases; of these, NEIL1 and NTH1 specifically remove Tg (42,43). Among the human RecQ helicases, exclusively WRN has been shown to enhance the incision activity of NEIL1 on a variety of oxidized synthetic DNA substrates (44,45). WRN’s functional interaction with NEIL1 on Tg-containing substrates however, particularly on telomeric substrates, is not currently reported. Additionally, while RECQL4 is shown to modulate the activity of key BER proteins (46,47), its role in the response to Tg lesions either via NEIL1 or independently is also not yet known. We therefore investigated whether RECQL4 and WRN were able to stimulate NEIL1 incision of Tg on telomeric substrates. As predicted, WRN was able to stimulate NEIL1 incision of Tg on the D-Loop structure (Figure 6, right panel). To accommodate for the lower helicase activity, greater concentrations of RECQL4 was introduced to the NEIL1 reaction, but this range of RECQL4 had no effect on NEIL1 incision activity (Figure 6, left panel). In fact, at a 1:20 molar ratio, RECQL4 inhibited NEIL1, possibly due to binding competition. These findings further demonstrate the exclusivity of WRN’s interaction with the NEIL1 glycosylase and are a novel observation on the D-Loop structure.
FIGURE 6. Effect of RECQL4 and WRN on NEIL1 incision of thymine glycol in telomeric D-Loops and forks.
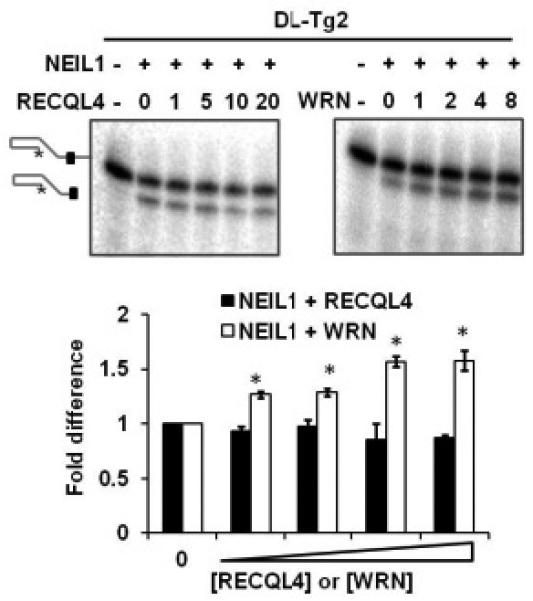
Representative gels and quantitative analysis showing the effect of RECQL4 (black bars) or WRN (white bars) on NEIL1 incision activity of a thymine glycol lesion on a telomeric D-Loop DL-Tg2. 2.5 nM NEIL1 was incubated with 0.5 nM 5′-labeled substrate and increasing concentrations of RECQL4 (at molar concentrations of 1, 5, 10 and 20 against NEIL1) or WRN (at molar concentrations of 1, 2, 4 and 8 against NEIL1). Error bars show standard deviation over at least three independent assays. *P<0.05 by a t-test.
4. DISCUSSION
Characterizing the metabolic roles of RECQL4 and WRN in telomere maintenance is crucial to understanding the pathogenesis of their associated disorders. The D-Loop is a generally accepted telomeric structure that is formed to protect chromosome ends from degradation and is unwound during telomeric repair, replication and transcription (48,49). WRN, BLM and RECQL4 are all recognized to operate in telomeric maintenance and function in D-Loop processing (26,50-56). The other RecQ helicases, RECQL1 and RECQL5, are not yet reported to have a similar function. Indeed, RECQL5 showed no ability to unwind the telomeric D-Loops (Figure S3). RECQL4, WRN and BLM have been shown to have preferential helicase activity on telomeric D-Loops containing oxidative damage in the form of 8-oxoguanine (26,27). Thymine glycol (Tg), however, the most common type of oxidative damage on thymine bases (and in one study more common than 8-oxoguanine (57)), is a particularly critical lesion that blocks DNA metabolism (10-12,15,58). We have developed novel synthetic substrates containing Tg base modifications in the telomeric sequence. Our in vitro findings indicate that while RECQL4 has a much weaker helicase activity and more limited substrate specificity than WRN and BLM, it has considerable partiality to unwind telomeric substrates containing Tg lesions, suggesting a novel function for RECQL4 in the cellular response to replication-blocking Tg lesions.
The RecQ helicases have overlapping substrate specificity, and therefore cooperative roles in DNA maintenance are predicted. RECQL4, for example, stimulates BLM helicase on a long DNA fork substrate in vitro, but does not on more complex telomeric D-Loops (26,59). RECQL4 does stimulate WRN, however, on a D-Loop containing 8-oxoguanine (26). Conversely, in this study we did not observe cooperation between RECQL4 and WRN on a D-Loop containing thymine glycol. Rather, RECQL4 may have inhibited WRN’s helicase activity through DNA binding, or the strong annealing activity of RECQL4 in the absence of ssDNA in the reaction re-annealed a fraction of the unwound substrate. Our observations indicate that the cooperative nature of the RecQ helicases is likely to be highly complex, particularly on non-B form DNA structures like D-Loops. The cooperative nature of the RecQ helicases needs further exploration and is of great interest in understanding mechanisms of genome stability.
The study of RECQL4 is in its infancy relative to the other RecQ family members, since its helicase activity was only recently uncovered (30,60), therefore, specifically how RECQL4 might promote telomeric maintenance at sites of Tg damage is not known. Tg lesions can impair DNA replication and transcription (12,17,61). Dysregulated transcription in the absence of proper telomere maintenance is shown to cause telomeric loss and cellular senescence (62). Single strand break intermediates persisting from inefficient BER can inhibit both replication and transcription (63). BER is the major pathway for removing Tg lesions; both RECQL4 and WRN are implicated in BER (46,47,64), and RECQL4-deficient cells exhibit inefficient DNA break repair (46,65).
The specific role of RECQL4 in BER is not yet known though it has been shown to interact with APE1, FEN1, POLβ and PARP1 (46,47). Its role specifically at sites of Tg has never before been reported. Biochemical data reported in the literature consistently and uniquely implicates WRN in a cooperative mechanism with the BER glycosylase NEIL1 by stimulating its incision activity in vitro on a variety of substrates containing oxidative lesions (45), and cellular studies place WRN and NEIL1 in the same functional pathway (44). Cooperative analysis using Tg lesions or D-Loop structures have not yet been reported. Consistent with previously reported trends, our findings in this study indicate that WRN can stimulate NEIL1’s incision of Tg on a telomeric D-Loop, but RECQL4 cannot. Differences in helicase activities between WRN and RECQL4 should not exclusively account for this distinction, since NEIL1 prefers to incise a double-stranded substrate (45,66). Therefore our data supports previous findings that among the RecQ helicases WRN exclusively functions with NEIL1 at oxidative lesions. It is possible that RECQL4 may be beneficial for the repair mechanism by unwinding around the site and creating access to the Tg lesion. For example, as Tg damage is induced in a dose-dependent manner by H2O2 (67), for example, and persistence of repair intermediates contributes to H2O2 toxicity (which is observed in RTS cells (7)), it is feasible that RECQL4 may have a key role in promoting efficient BER specifically at Tg lesions. Furthermore, it is likely that this function is critical prior to telomeric replication, as RECQL4 could not unwind a telomeric fork substrate with a Tg lesion in the translocating strand (data not shown), consistent with the literature reporting strand-specific helicase obstruction by Tg (33).
4.1 Conclusions
In the future, crystallographic analysis may further characterize the physical interactions with the Tg lesion and with TRF2 that are shown to stimulate RECQL4’s helicase activity. Perhaps the helical distortion induced by Tg lesions is a more optimal conformation on which the RECQL4 helicase can specifically act; certainly this distortion is not optimal for TRF2 or TRF1 binding, which may result in unprotected and unstable telomeres. Structurally, RECQL4 is unique compared with WRN and the other RecQ helicases in that it lacks the RQC domain; this structural difference may affect the functional interactions we have observed. Of particular interest will be to investigate whether Tg lesions or Tg-derived repair intermediates accumulate in the telomeres of RTS cells, and whether these cells are hypersensitive to Tg-inducing agents. Currently these investigations prove challenging, as no compound is known to exclusively induce Tg lesions in human cells, and while the detection of other oxidative damage like 8-oxoguanine is common, the immunocytological detection of Tg at the telomeres in live human cells using currently available antibodies is not well developed. These will be interesting hypotheses to explore as technology and telomere research advances.
Supplementary Material
HIGHLIGHTS.
RECQL4 preferentially unwinds telomeric substrates containing thymine glycol (Tg)
TRF2, a telomeric shelterin protein, stimulates RECQL4 helicase in presence of Tg
RECQL4 binding is unchanged but TRF2 binds Tg-containing D-Loops with less affinity
WRN, but not RECQL4, stimulates NEIL1 glycosylase incision of the Tg lesion
ACKNOWLEDGEMENTS
We thank Huiming Lu and Christopher Dunn, NIA, for critically reading the manuscript. We also thank Marie Rossi and Lale Dawut, NIA, for biochemical methods instruction and technical help, Christopher Dunn and Tomasz Kulikowicz, NIA, for purification of the RecQ helicase proteins, and Patricia Opresko, University of Pittsburgh, for purified shelterin proteins. This work was supported entirely by the Intramural Research Program of the NIH, National Institute on Aging.
Abbreviations
- Tg
thymine glycol
- ROS
reactive oxygen species
- WS
Werner Syndrome
- RTS
Rothmund-Thompson syndrome
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Liu L, Trimarchi JR, Smith PJS, Keefe DL. Aging Cell. 2002;1:40–46. doi: 10.1046/j.1474-9728.2002.00004.x. [DOI] [PubMed] [Google Scholar]
- 2.Richter T, Zglinicki T. v. Experimental Gerontology. 2007;42:1039–1042. doi: 10.1016/j.exger.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Rubio MA, Davalos AR, Campisi J. Experimental Cell Research. 2004;298:17–27. doi: 10.1016/j.yexcr.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Rhee DB, Lu J, Bohr CT, Zhou F, Vallabhaneni H, de Souza-Pinto NC, Liu Y. PLoS Genet. 2010;6:e1000951. doi: 10.1371/journal.pgen.1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hasty P, Campisi J, Hoeijmakers J, van Steeg H, Vijg J. Science. 2003;299:1355–1359. doi: 10.1126/science.1079161. [DOI] [PubMed] [Google Scholar]
- 6.Harrigan JA, Wilson DM, Prasad R, Opresko PL, Beck G, May A, Wilson SH, Bohr VA. Nucleic Acids Research. 2006;34:745–754. doi: 10.1093/nar/gkj475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Werner SR, Prahalad AK, Yang J, Hock JM. Biochemical and Biophysical Research Communications. 2006;345:403–409. doi: 10.1016/j.bbrc.2006.04.093. [DOI] [PubMed] [Google Scholar]
- 8.Aller P, Duclos S, Wallace SS, Doublié S. Journal of Molecular Biology. 2011;412:22–34. doi: 10.1016/j.jmb.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayes RC, Petrullo LA, Huang H, Wallace SS, LeClerc JE. Journal of Molecular Biology. 1988;201:239–246. doi: 10.1016/0022-2836(88)90135-0. [DOI] [PubMed] [Google Scholar]
- 10.Ide H, Kow YW, Wallace SS. Nucleic Acids Research. 1985;13:8035–8052. doi: 10.1093/nar/13.22.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark JM, Beardsley GP. Biochemistry. 1987;26:5398–5403. doi: 10.1021/bi00391a027. [DOI] [PubMed] [Google Scholar]
- 12.Tornaletti S, Maeda LS, Lloyd DR, Reines D, Hanawalt PC. Journal of Biological Chemistry. 2001;276:45367–45371. doi: 10.1074/jbc.M105282200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kusumoto R, Masutani C, Iwai S, Hanaoka F. Biochemistry. 2002;41:6090–6099. doi: 10.1021/bi025549k. [DOI] [PubMed] [Google Scholar]
- 14.Takata K.-i., Shimizu T, Iwai S, Wood RD. Journal of Biological Chemistry. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 15.Aller P, Rould MA, Hogg M, Wallace SS, Doublié S. Proceedings of the National Academy of Sciences. 2007;104:814–818. doi: 10.1073/pnas.0606648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou R-Z, Blanco L, Garcia-Diaz M, Bebenek K, Kunkel TA, Povirk LF. Nucleic Acids Research. 2008;36:2895–2905. doi: 10.1093/nar/gkn126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adelman R, Saul RL, Ames BN. Proceedings of the National Academy of Sciences. 1988;85:2706–2708. doi: 10.1073/pnas.85.8.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allsopp RC, Vaziri H, Patterson C, Goldstein S, Younglai EV, Futcher AB, Greider CW, Harley CB. Proceedings of the National Academy of Sciences. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Njajou OT, Cawthon RM, Damcott CM, Wu SH, Ott S, Garant MJ, Blackburn EH, Mitchell BD, Shuldiner AR, Hsueh WC. Proceedings of the National Academy of Sciences. 2007;104:12135–12139. doi: 10.1073/pnas.0702703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heidinger BJ, Blount JD, Boner W, Griffiths K, Metcalfe NB, Monaghan P. Proceedings of the National Academy of Sciences. 2012;109:1743–1748. doi: 10.1073/pnas.1113306109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Zglinicki T, Saretzki G, Döcke W, Lotze C. Experimental Cell Research. 1995;220:186–193. doi: 10.1006/excr.1995.1305. [DOI] [PubMed] [Google Scholar]
- 22.von Zglinicki T. Trends in Biochemical Sciences. 2002;27:339–344. doi: 10.1016/s0968-0004(02)02110-2. [DOI] [PubMed] [Google Scholar]
- 23.Morin GB. Experimental Gerontology. 1997;32:375–382. doi: 10.1016/s0531-5565(96)00164-7. [DOI] [PubMed] [Google Scholar]
- 24.Chang S, Multani AS, Cabrera NG, Naylor ML, Laud P, Lombard D, Pathak S, Guarente L, DePinho RA. Nat Genet. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- 25.Szekely AM, Bleichert F, Nümann A, Van Komen S, Manasanch E, Ben Nasr A, Canaan A, Weissman SM. Molecular and Cellular Biology. 2005;25:10492–10506. doi: 10.1128/MCB.25.23.10492-10506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh AK, Rossi ML, Singh DK, Dunn C, Ramamoorthy M, Croteau DL, Liu Y, Bohr VA. Journal of Biological Chemistry. 2012;287:196–209. doi: 10.1074/jbc.M111.295063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh A, Rossi ML, Aulds J, Croteau D, Bohr VA. Journal of Biological Chemistry. 2009;284:31074–31084. doi: 10.1074/jbc.M109.027532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Opresko PL, Otterlei M, Graakj+|r J, Bruheim P, Dawut L, K++lvraa S, May A, Seidman MM, Bohr VA. Molecular Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 29.Orren DK, Brosh RM, Nehlin JO, Machwe A, Gray MD, Bohr VA. Nucleic Acids Research. 1999;27:3557–3566. doi: 10.1093/nar/27.17.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossi ML, Ghosh AK, Kulikowicz T, Croteau DL, Bohr VA. DNA Repair. 2010;9:796–804. doi: 10.1016/j.dnarep.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu X, Liu Y. EMBO J. 2009;28:568–577. doi: 10.1038/emboj.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Machwe A, Ganunis R, Bohr VA, Orren DK. Nucleic Acids Research. 2000;28:2762–2770. doi: 10.1093/nar/28.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suhasini AN, Sommers JA, Mason AC, Voloshin ON, Camerini-Otero RD, Wold MS, Brosh RM. Journal of Biological Chemistry. 2009;284:18458–18470. doi: 10.1074/jbc.M109.012229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demple B, Harrison L. Annual Review of Biochemistry. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 35.Bukowy Z, Harrigan JA, Ramsden DA, Tudek B, Bohr VA, Stevnsner T. Nucleic Acids Research. 2008;36:4975–4987. doi: 10.1093/nar/gkn468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Lange T. Genes & Development. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 37.Nora GJ, Buncher NA, Opresko PL. Nucleic Acids Research. 2010;38:3984–3998. doi: 10.1093/nar/gkq144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stansel RM, de Lange T, Griffith JD. EMBO J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Opresko PL, Fan J, Danzy S, Wilson DM, Bohr VA. Nucleic Acids Research. 2005;33:1230–1239. doi: 10.1093/nar/gki273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bessho T. Nucleic Acids Research. 1999;27:979–983. doi: 10.1093/nar/27.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dianov GL, Thybo T, Dianova II, Lipinski LJ, Bohr VA. Journal of Biological Chemistry. 2000;275:11809–11813. doi: 10.1074/jbc.275.16.11809. [DOI] [PubMed] [Google Scholar]
- 42.Bandaru V, Sunkara S, Wallace SS, Bond JP. DNA Repair. 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 43.Dizdaroglu M, Karahalil B, Sentürker S, Buckley TJ, Roldán-Arjona T. Biochemistry. 1998;38:243–246. doi: 10.1021/bi9819071. [DOI] [PubMed] [Google Scholar]
- 44.Das A, Boldogh I, Lee JW, Harrigan JA, Hegde ML, Piotrowski J, de Souza Pinto N, Ramos W, Greenberg MM, Hazra TK, Mitra S, Bohr VA. Journal of Biological Chemistry. 2007;282:26591–26602. doi: 10.1074/jbc.M703343200. [DOI] [PubMed] [Google Scholar]
- 45.Popuri V, Croteau DL, Bohr VA. DNA Repair. 2010;9:636–642. doi: 10.1016/j.dnarep.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schurman SH, Hedayati M, Wang Z, Singh DK, Speina E, Zhang Y, Becker K, Macris M, Sung P, Wilson DM, Croteau DL, Bohr VA. Human Molecular Genetics. 2009;18:3470–3483. doi: 10.1093/hmg/ddp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woo LL, Futami K, Shimamoto A, Furuichi Y, Frank KM. Experimental Cell Research. 2006;312:3443–3457. doi: 10.1016/j.yexcr.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 48.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 49.Neidle S, Parkinson GN. Current Opinion in Structural Biology. 2003;13:275–283. doi: 10.1016/s0959-440x(03)00072-1. [DOI] [PubMed] [Google Scholar]
- 50.Bachrati CZ, Borts RH, Hickson ID. Nucleic Acids Research. 34:2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 52.Du X, Shen J, Kugan N, Furth EE, Lombard DB, Cheung C, Pak S, Luo G, Pignolo RJ, DePinho RA, Guarente L, Johnson FB. Molecular and Cellular Biology. 2004;24:8437–8446. doi: 10.1128/MCB.24.19.8437-8446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghosh A, Liu Y, Bohr VA. In: Role of RecQ Helicases in Nuclear DNA Repair and Telomere Maintenance Cellular Senescence and Tumor Suppression. Adams PD, Sedivy JM, editors. Springer; New York: 2010. pp. 45–62. [Google Scholar]
- 54.Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. Journal of Biological Chemistry. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 55.Orren DK, Theodore S, Machwe A. Biochemistry. 2002;41:13483–13488. doi: 10.1021/bi0266986. [DOI] [PubMed] [Google Scholar]
- 56.Verdun RE, Karlseder J. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 57.Loft S, Poulson HE. Acta Biochimica Polonica. 1998;45:133–144. [PubMed] [Google Scholar]
- 58.McNulty JM, Jerkovic B, Bolton PH, Basu AK. Chemical Research in Toxicology. 1998;11:666–673. doi: 10.1021/tx970225w. [DOI] [PubMed] [Google Scholar]
- 59.Kumar Singh D, Popuri V, Kulikowicz T, Shevelev I, Ghosh AK, Ramamoorthy M, Rossi ML, Janscak P, Croteau DL, Bohr VA. Nucleic Acids Research. 2012 doi: 10.1093/nar/gks349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suzuki T, Kohno T, Ishimi Y. Journal of Biochemistry. 2009;146:327–335. doi: 10.1093/jb/mvp074. [DOI] [PubMed] [Google Scholar]
- 61.Kao JY, Goljer I, Phan TA, Bolton PH. Journal of Biological Chemistry. 1993;268:17787–17793. [PubMed] [Google Scholar]
- 62.Maicher A, Kastner L, Dees M, Luke B. Nucleic Acids Research. 2012;40:6649–6659. doi: 10.1093/nar/gks358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kathe SD, Shen G-P, Wallace SS. Journal of Biological Chemistry. 2004;279:18511–18520. doi: 10.1074/jbc.M313598200. [DOI] [PubMed] [Google Scholar]
- 64.Ahn B, Harrigan JA, Indig FE, Wilson DM, Bohr VA. Journal of Biological Chemistry. 2004;279:53465–53474. doi: 10.1074/jbc.M409624200. [DOI] [PubMed] [Google Scholar]
- 65.Singh DK, Karmakar P, Aamann M, Schurman SH, May A, Croteau DL, Burks L, Plon SE, Bohr VA. Aging Cell. 2010;9:358–371. doi: 10.1111/j.1474-9726.2010.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parsons JL, Zharkov DO, Dianov GL. Nucleic Acids Research. 2005;33:4849–4856. doi: 10.1093/nar/gki816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alanazi M, Leadon SA, Mellon I. Nucleic Acids Research. 2002;30:4583–4591. doi: 10.1093/nar/gkf588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.