Abstract
Egr3 is a nerve growth factor (NGF)-induced transcriptional regulator that is essential for normal sympathetic nervous system development. Mice lacking Egr3 in the germline have sympathetic target tissue innervation abnormalities and physiologic sympathetic dysfunction similar to humans with dysautonomia. However, since Egr3 is widely expressed and has pleiotropic function, it has not been clear whether it has a role within sympathetic neurons and if so, what target genes it regulates to facilitate target tissue innervation. Here, we show that Egr3 expression within sympathetic neurons is required for their normal innervation since isolated sympathetic neurons lacking Egr3 have neurite outgrowth abnormalities when treated with NGF and mice with sympathetic neuron-restricted Egr3 ablation have target tissue innervation abnormalities similar to mice lacking Egr3 in all tissues. Microarray analysis performed on sympathetic neurons identified many target genes deregulated in the absence of Egr3, with some of the most significantly deregulated genes having roles in axonogenesis, dendritogenesis, and axon guidance. Using a novel genetic technique to visualize axons and dendrites in a subpopulation of randomly labeled sympathetic neurons, we found that Egr3 has an essential role in regulating sympathetic neuron dendrite morphology and terminal axon branching, but not in regulating sympathetic axon guidance to their targets. Together, these results indicate that Egr3 has a sympathetic neuron autonomous role in sympathetic nervous system development that involves modulating downstream target genes affecting the outgrowth and branching of sympathetic neuron dendrites and axons.
Introduction
The sympathetic nervous system (SNS) is a subdivision of the autonomic nervous system that regulates organ homeostasis and it is the target of many developmental and degenerative diseases in vertebrates, including humans. SNS development is a complex process mediated by multiple transcriptional regulators, morphogens, and target tissue-derived growth factors that direct precursor migration, noradrenergic specification, differentiation, and target tissue innervation (Goridis and Rohrer, 2002; Rohrer, 2003; Howard, 2005). Once precursors migrate to form discrete sympathetic ganglia, axons extend along blood vessels, which produce diffusible factors such as Artemin (Honma et al., 2002) and neurotrophin 3 (Francis et al., 1999; Kuruvilla et al., 2004) that are required for normal axon growth along blood vessels. Additional target tissue-derived trophic factors such as nerve growth factor (NGF) (Crowley et al., 1994; Glebova and Ginty, 2004) and NGF-induced morphogens such as Wnt5a (Bodmer et al., 2009; Ho et al., 2012) are required for normal patterning of sympathetic innervation within target tissues.
NGF has long been known as an essential target tissue-derived survival factor for sympathetic neurons (Levi-Montalcini and Booker, 1960; Levi-Montalcini and Cohen, 1960; Crowley et al., 1994); however, more recent studies have demonstrated its additional role in target tissue innervation (Albers et al., 1994; Hassankhani et al., 1995; Patel et al., 2000; Glebova and Ginty, 2004). NGF function is mediated by tyrosine kinase receptor TrkA signaling, which activates Ras/mitogen activated protein kinase (MAPK), Src/protein kinase C (PKC), and phosphotidylinositol triphosphate (PI3) kinase signaling pathways (Skaper, 2008). Although there is considerable cross-talk among the different signaling pathways, PI3 kinase signaling has a major role in neuron survival, whereas Ras/MAPK signaling is primarily involved in neuronal differentiation and neurite outgrowth (Klesse and Parada, 1999; Atwal et al., 2000; Huang and Reichardt, 2001). However, very little is known about how NGF signaling pathways regulate gene expression within sympathetic neurons to facilitate its major functions in axon and dendrite outgrowth during target tissue innervation.
Egr3 is an example of a transcriptional regulator that is induced by NGF and coupled to MAPK signaling in sympathetic neurons. Mice lacking Egr3 in all tissues have sympathetic innervation abnormalities and dysautonomia, but whether its expression in sympathetic neurons is required for normal target tissue innervation and/or what target genes it regulates is not known (Eldredge et al., 2008; Li et al., 2011). Here, we show that loss of Egr3 expression specifically in sympathetic neurons leads to abnormal neurite outgrowth in response to NGF signaling in vitro and sympathetic neuron dendrite and axon abnormalities in vivo. We identified several Egr3-regulated target genes with known roles in axonogenesis and dendritogenesis, and using a genetic labeling strategy to visualize a random subset of sympathetic neurons, we found that Egr3 is important for dendrite development and axon innervation, but not for axon guidance to target tissues. Thus, Egr3 has a sympathetic neuron autonomous role in gene regulation involved in the outgrowth and branching of dendrites and axons, which likely explains the diminished sympathetic innervation and physiologic dysautonomia that occurs in its absence.
Materials and Methods
Animals
Mice with Bax and Egr3 germline loss-of-function mutations were generated as previously described (Knudson et al., 1995; Tourtellotte and Milbrandt, 1998). CaMKII-iCre BAC transgenic mice were obtained from G. Schutz (Casanova et al., 2001). DβH-Cre BAC transgenic mice were recovered from the GENSAT sperm cryorepository (MMRRC Stock# 032081-UCD) using in vitro fertilization and oocytes derived from C57BL/6 female mice. Isogenic C57BL/6J CAG-FlpE mice were obtained from the RIKEN animal resource (#RBRC01834) (Kanki et al., 2006), and Rosa-eGFP (JAX# 005670), Ai14 (JAX# 007908), and StLa (JAX# 010633) Cre-recombinase reporter mice were obtained from The Jackson Laboratory (Soriano, 1999; Belteki et al., 2005; Nam and Benezra, 2009; Madisen et al., 2010). All mice were back-crossed at least four generations to the C57BL/6 genetic background.
Egr3-Cre knockin mice.
An 8.3 kb HindIII genomic DNA fragment (spanning 50605023–50613323, GenBank NT_039606.8) isolated from 129Sv/J mouse genomic DNA that contained both coding exons, the intron, and the 3′UTR of Egr3 was used to generate the targeting vector (Tourtellotte and Milbrandt, 1998). The endogenous translation start codon was converted to an NcoI site and the coding sequence for Cre-recombinase was exchanged with the endogenous exon 1, intron, and exon 2 coding sequence of Egr3 using homologous recombination in Escherichia coli (recombineering) (Liu et al., 2003). A PGK-Neo (Neo) positive-selection cassette containing a novel BglII restriction site and flanked by forward-oriented Frt (F) sites was inserted downstream of the nlsCre coding sequence. The targeting construct was electroporated into 129/ola-derived HM-1 embryonic stem (ES) cells and G418-resistant clones were screened for homologous recombination. ES cell clones were screened using long-range PCR flanking the 3′ recombination arm of the targeting construct with primers OT1182 (TTGCATCGCATTGTCTGA) and OT1114 (AAGCACCCCTGTCAAGTTTC). PCR-positive clones were additionally screened by Southern blotting with a 700 nt probe located 5′ of the 5′ recombination arm (spanning 50604041–50604741, GenBank, NT_039606.8), which was hybridized to genomic DNA restricted with BglII endonuclease to yield a 7.1 kb wild-type band and a 4.3 kb targeted band. Three appropriately targeted clones were injected into isogenic C57BL/6J blastocysts using standard methods (Magin et al., 1992); chimeric mice were mated to C57BL/6J female mice and germline heterozygote mice (Egr3+/Neo) were mated to CAG-FlpE mice (Kanki et al., 2006) to remove the frt-flanked Neo positive-selection cassette from the germline and to generate the final Egr3-Cre allele (designated Egr3+/Cre).
Egr3-flx mice.
The Egr3-flx mice were generated using the same 8.3 kb HindIII genomic DNA fragment used to generate the Egr3-Cre knockin targeting construct. A forward-oriented loxP site containing a novel BglII restriction site was targeted into the intron using homologous recombination in E. coli (insertion site at position 778, GenBank, NT_039606). A PGK-Neo (Neo) positive selection cassette flanked by forward-oriented Frt (F) sites and a single forward-oriented loxP site positioned 3′ to the F-Neo-F cassette was targeted immediately downstream of the TGA translation stop codon in exon 2 using homologous recombination in E. coli (insertion site at 2463 nt, GenBank, NT_039606). The targeting construct was electroporated into 129/ola-derived HM-1 ES cells and G418-resistant clones were subjected to screening by long-range PCR and Southern blotting. Targeted clones were identified by 3′ long-range PCR using primers OT1111 (TACGAAGTTATTAGGTGGATCC) and OT1114 that spanned the 3′ recombination arm. Correctly targeted clones were screened by Southern blotting using the same probe that was used for generating Egr3-Cre mice, which yielded a 7.2 kb fragment for the wild-type allele and a 4.5 kb fragment for the Neo allele on BglII restricted genomic DNA. Three targeted clones were injected into isogenic C57BL/6J blastocysts using standard methods (Magin et al., 1992). Germline heterozygous Egr3+/Neo mice were mated to CAG-FlpE mice to excise the frt-flanked Neo positive-selection cassette (Kanki et al., 2006) to generate the final Egr3+/f allele (designated Egr3+/f).
DβH-CreIRESτlacZ (DCτlZ) transgenic mice:
The transgenic mice were generated using B6SJL embryos and are similar to sympathetic reporter mice that were previously generated by us (Eldredge et al., 2008). However, these new sympathetic reporter mice express Cre-recombinase and the axon localized τlacZ reporter (Callahan and Thomas, 1994) transcribed as a single bicistronic mRNA under control of the human dopamine β-hydroxylase (DβH) promoter (Mercer et al., 1991).
All mice were genotyped by PCR with genomic DNA isolated from tail biopsy tissue. The sequences of the genotyping primers and PCR conditions are available upon request. All experimental procedures involving mice complied with the Public Health Service Policy on Humane Care and Use of Laboratory Animals and all animal-related protocols were approved by the Northwestern University Institutional Animal Care and Use Committee.
Tissue preparation
Anesthetized mice were perfused through the heart with 0.1 m phosphate buffered, pH 7.2, 4% paraformaldehyde (PFA) and tissues were postfixed at 4°C for 1–4 h. For frozen sections, tissues were cryoprotected overnight at 4°C in graded (15–30%) sucrose, embedded in OCT and sectioned at 12 μm thickness. For quantitative studies involving stereologic neuron counting, PFA fixed tissues were embedded in paraffin and serial sectioned at 16 μm. For some experiments, tissues were isolated fresh for RNA extraction.
Ganglion neuron counts
SCG neuron numbers from adult (∼8 weeks) DCτlZ+; Egr3+/− and DCτlZ+; Egr3f/− mice were determined using unbiased stereology and optical dissector methods (StereoInvestigator; Microbrightfield) on every fifth serial section as previously described (Albert et al., 2005; Eldredge et al., 2008).
Immunohistochemistry
Immunofluorescence staining for tyrosine hydroxylase (TH; rabbit anti-TH, 1:5000; Millipore Bioscience Research Reagents), Renin (goat anti-renin, 1:150; Santa Cruz Biotechnology), and PECAM (hamster anti-PECAM (2H8), 1:250; a gift from William A. Muller, Northwestern University, Chicago, IL) were performed on frozen tissue sections. Immunohistochemistry staining for Egr3 (rabbit anti-Egr3, 1:1000; Santa Cruz Biotechnology) was performed on paraffin tissue sections. βIII tubulin staining (mouse anti-tubulin, TU-20, 1:1000; Millipore Bioscience Research Reagents) was used to label neurons grown in culture. Species-appropriate secondary antibodies conjugated to Cy3 (Jackson ImmunoResearch) or Alexa488 (Invitrogen) were used to localize primary antibody binding.
Semiquantitative sympathetic axon terminal innervation
Quantification of target tissue innervation on adult DCτlZ+; Egr3+/− and DCτlZ+; Egr3f/− mice was performed using fluorescence densitometry. Fluorescent images were captured with a Zeiss LS510 confocal microscope using identical aperture and photomultiplier tube voltage settings to ensure accurate comparison between tissues from both conditions. The density of TH-positive terminals was calculated using MetaMorph software (Molecular Devices) as the ratio of total immunofluorescence in a specified area and averaged for six separate confocal areas per section. Representative sympathetic target tissues, such as spleen, pineal gland, heart, and kidney, were analyzed in areas with the highest density of axons identified in control tissues. The “relative innervation” in DCτlz+; Egr3f/− mice was expressed as a mean percentage of control (DCτlZ+; Egr3+/−) from 3 to 4 animals of each genotype as indicated.
LacZ enzyme histochemistry
Whole mount lacZ enzyme histochemistry was performed on adolescent (∼4 weeks) DβH-Cre+; Egr3+/+; StLa+/f and DβH-Cre+; Egr3−/−; StLa+/f mice. Adolescent mice were examined since adult tissues had high nonspecific lacZ background staining in whole-mounted tissues and SNS development is complete by this age. Tissues were dissected and postfixed in 2% PFA, 0.2% glutaraldehyde, 5 mm EGTA, and 0.01% NP-40 in PBS-Mg at 4°C and reacted overnight at 37°C in reaction buffer (1 mg/ml X-gal, 5 mm potassium ferrocyanide, 5 mm potassium ferricyanide). Whole tissues were cleared by stepping them through increasing concentrations of glycerol (20, 50, 80, and 100%; 4 d each) mixed in 1% potassium hydroxide (KOH). The tissues were imaged with a Nikon SMZ1500 stereomicroscope and a Nikon DS-Ri1 digital camera. Focal image planes for superior cervical ganglion (SCG) neurons were projected to flat-field using the Zerene Stacker software program (Zerene Systems). Neuron tracings were drawn using the DrawingSlate II (GTCO CalComp Peripherals) drawing pad with TabletWorks software. Quantification of dendrite morphology was done using MetaMorph software on calibrated images. The axon was identified as the only neurite that extended out of the ganglia while all other neurites were classified as dendrites. Only dendrites longer than the diameter of the soma were counted as primary dendrites. The maximum extent of the dendritic arbor was calculated as the diameter of the largest concentric circle that could encompass the entire dendritic arbor. Calculation of divergent axons emanating from the external carotid artery was measured by counting axons that crossed a defined border that was parallel to and three times the width of the artery. Quantification of sympathetic axon branching in the submandibular gland was calculated by counting the branch points of axons within the target tissue once they breached the tissue edge.
Primary sympathetic neuron cultures
SCG neurons from newborn (P0) Egr3+/+ and Egr3−/− mice were used for neuron culture. SCG neurons were dissociated with type IV collagenase (1 mg/ml, Sigma), followed by 0.25% trypsin-EDTA and plated on poly-l-lysine- and laminin-coated coverslips with media containing DMEM, 10% fetal bovine serum, 1% penicillin/streptomycin, and 2 ng/ml NGF. Neurons were differentiated for 2 days as previously described (Eldredge et al., 2008). Neurite outgrowth parameters were measured using MetaMorph software on calibrated digital images of βIII tubulin-positive neurons.
Illumina BeadChip gene expression profiling
Microarray studies were performed on total cellular RNA isolated from P0 SCG from Egr3+/+; Bax−/− and Egr3−/−; Bax−/− mice. As neurons do not innervate target tissues properly in Egr3−/− mice and hence some neurons die, the Bax-deficient genetic background (Bax−/−) was used to inhibit neuron apoptosis and therefore avoid detecting apoptosis-related genes that are a secondary consequence of Egr3 loss. Total RNA was isolated from SCG using Trizol extraction (Invitrogen) and the integrity of the RNA samples was verified using an Agilent 2100 Bioanalyzer. Microarray analysis was performed at the National Institutes of Health Neuroscience Microarray Consortium using the Illumina BeadChip platform and MouseWG-6 v.2.0 BeadChip microarrays. Briefly, cRNA for array hybridization was generated using 500 ng of total cellular RNA from each experimental condition using the TotalPrep RNA Amplification kit (Applied Biosystems). Then 1500 ng of amplified heat denatured biotin-labeled cRNA was mixed with hybridization buffer (Illumina), the mixture was hybridized to the arrays at 58°C for 20 h, the arrays were washed and stained according to the manufacturer's specifications, and they were scanned using the Illumina iScan. RNA isolated from three individual mice of each genotype was used for hybridization to a MouseWG-6 v.2.0 slide with six independent arrays each containing 45,281 probe targets. Using the BeadStudio software, the raw dataset was processed by subtracting background noise and normalizing using an average normalization parameter. The error model used to determine significantly differentially expressed genes was Illumina custom. The dataset was filtered using the cutoff of difference p value < 0.01. The list of differentially expressed genes was analyzed using the Gene Ontology (GO) software program Database for Annotation, Visualization and Integrated Discovery (DAVID). The background reference list used was MouseWG-6_V1_1_R4_11234304_A. The microarray data have been deposited in NCBI's Gene Expression Omnibus and are accessible through NCBI GEO Series accession number GSE41290 (Edgar et al., 2002).
Quantitative real-time PCR
Quantitative real-time PCR (qPCR) was performed as previously described (Albert et al., 2005). Briefly, total cellular RNA was isolated from P0 SCG from control Egr3+/+; Bax−/− mice and experimental Egr3−/−; Bax−/− mice. cDNA was synthesized using Superscript III reverse transcriptase (Invitrogen) and random octamer/oligo-dT priming according to recommendations by the manufacturer. Relative gene expression was determined using Sybr Green (Invitrogen) fluorescence with non-intron spanning primers. The primers spanned and amplified the following coding regions of target genes: apbb1 (amplified 2184–2304 nt, GenBank, NM_001253885), b3galt6 (amplified 466–712 nt, GenBank, NM_080445), efnb1 (amplified 1467–1663 nt, GenBank, NM_010110), esd (amplified 352–731 nt, GenBank, NM_016903), gins2 (amplified 506–814 nt, GenBank, NM_178856), hs3st2 (amplified 996–1300 nt, GenBank, NM_001081327), insc (amplified 410–670 nt, GenBank, NM_173767), irak1 (amplified 1234–1383 nt, GenBank, NM_001177973), lmo7 (amplified 2754–2971 nt, GenBank, NM_201529), mxra8 (amplified 300–511 nt, GenBank, NM_024263), nf1 (amplified 5041–5316 nt, GenBank, NM_010897), ngfr (amplified 723–904 nt, GenBank, NM_033217), notch1 (amplified 6532–6836 nt, GenBank, NM_008714), pabpc1l (amplified 1029–1188 nt, GenBank, NM_001114079), pde3a (amplified 728–993 nt, GenBank, NM_018779), plxnd1 (amplified 3584–3800 nt, GenBank, NM_026376), ptpn11 (amplified 564–679 nt, GenBank, NM_001109992), rccd1 (amplified 1152–1439 nt, GenBank, NM_173445), ret (amplified 965–1206 nt, GenBank, NM_001080780), slc15a2 (amplified 2343–2649 nt, GenBank, NM_021301), slit2 (amplified 5249–5407 nt, GenBank, NM_178804), sox9 (amplified 807–1016 nt, GenBank, NM_011448), and zfp292 (amplified 2973–3235 nt, GenBank, NM_013889). Standard curves were generated for each primer pair using mouse genomic DNA and the target gene expression results were normalized to GAPDH expression for each sample analyzed. Results are represented as fold-change expression relative to the control condition and represent 3–4 replicates from different animals.
Statistical analysis
Values are expressed as mean ± SEM and p values are determined by Student's t test unless otherwise designated. For the χ2 test the expected random distribution was calculated by dividing the total N evenly among the number of given conditions.
Results
Egr3 has a sympathetic neuron autonomous role in target tissue innervation
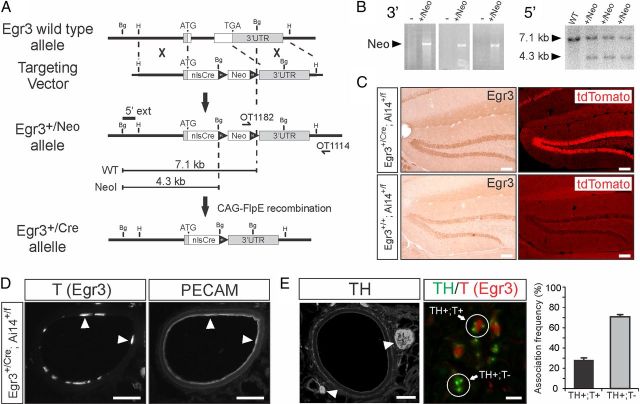
Egr3 expression is regulated by NGF signaling in sympathetic neurons and in the absence of Egr3 mice have physiologic dysautonomia caused by a failure of sympathetic neurons to properly innervate target tissues (Eldredge et al., 2008). In addition, mice lacking both Egr3 and Bax (the latter, which prevents apoptotic neuron death) have innervation abnormalities that resemble those occurring in mice lacking both NGF and Bax (Glebova and Ginty, 2004; Li et al., 2011), further suggesting that Egr3 regulates, at least in part, gene expression induced by NGF signaling in sympathetic neurons to influence target tissue innervation. However, previous studies indicate that Egr3 is also expressed in blood vessels along which sympathetic axons travel (Shneider et al., 2009) and in Schwann cells that myelinate some axons in nerves through which unmyelinated sympathetic axons pass during development (Gao et al., 2007). Moreover, Egr3 may be expressed in other tissues that sympathetic axons encounter during development, and its expression in those tissues may be transient, raising the possibility that Egr3 has a complex role in target tissue innervation that may not solely depend on its expression in sympathetic neurons. To identify cells that express Egr3 at any time during their development, we generated Egr3-Cre knockin mice using gene targeting to replace the endogenous Egr3 coding sequence with the coding sequence for nuclear localized Cre-recombinase (nlsCre) (Fig. 1A,B). Egr3+/Cre mice were mated with Ai14+/f reporter mice, which constitutively express the red fluorescent protein tdTomato (T) in cells that express Cre-recombinase (Madisen et al., 2010). In the hippocampal dentate gyrus, where Egr3 is robustly and transiently expressed by neuronal activity (Li et al., 2007), T fluorescence was constitutively coexpressed with Egr3 in Egr3+/Cre; Ai14+/f double heterozygous dentate gyrus granule neurons as expected (Fig. 1C). We examined T fluorescence in peripheral tissues known to interact with sympathetic axons from newborn and adult Egr3+/Cre; Ai14+/flx double heterozygous mice and found strong labeling in arterial vascular endothelial cells (identified by colocalization with the endothelial cell marker, PECAM; Fig. 1D), which are in close proximity with sympathetic axons that course along the surface of arterial vessels (identified by the sympathetic marker, TH; Fig. 1E, left). Within perivascular nerves, the Egr3 reporter was present in 100% of Schwann cells (identified by colocalization with the Schwann cell marker, S100; data not shown and Gao et al., 2007). Although sympathetic axons are unmyelinated, they are bundled with myelinated axons in mixed motor and sensory nerves, where they often come in close contact with Schwann cells. For example, within the sciatic nerve we found that 28% of TH+ axons were closely associated with Schwann cells that expressed the Egr3 tracer (Fig. 1E). The association frequency is very likely higher since the analysis only assessed association in single tissue sections and not along entire individual axons. Together, these results indicate that Egr3 is expressed in cells that could influence SNS development apart from its presumed autonomous function within sympathetic neurons.
Figure 1.
Design and validation of Egr3-Cre knockin mice to trace historical Egr3 expression in tissues. A, Gene targeting in ES cells was used to replace both exons and the intron of Egr3 with the open reading frame encoding nuclear localized Cre-recombinase (nlsCre). The Egr3-Cre (Egr3+/Cre) allele was generated by removing the frt-site flanked Neo-positive selection cassette from the germline using CAG-FlpE mice. B, Left, Electroporated G418 selected ES cell clones were screened using long-range PCR to amplify across the 3′ recombination arm (primers: OT1182 and OT1114). Right, PCR confirmed ES cell clones were screened by Southern blotting using a 713 nt probe located upstream of the 5′ recombination arm (5′ ext), which was hybridized to BglII restricted genomic DNA. C, To validate the appropriate expression and function of Cre-recombinase, Egr3+/Cre mice were mated to Ai14 Cre-recombinase reporter mice that irreversibly express fluorescent tdTomato protein after Cre-recombinase mediated loxP-site recombination (Madisen et al., 2010). Dentate gyrus granule neurons expressed Egr3 protein in (left) Egr3+/Cre; Ai14+/f and Egr3+/+; Ai14+/f mice, but only in (right) Egr3+/Cre; Ai14+/f mice did they also express the tdTomato reporter. D, In Egr3+/Cre; Ai14+/f double heterozygous mice, Egr3 reporter (T) was observed in arteries along which sympathetic nerves travel (splenic artery shown here), which colocalized in endothelial cells with the vascular endothelial marker PECAM (arrowheads). The tdTomato reporter is localized primarily in the endothelial nucleus whereas the PECAM protein is expressed on the endothelial membrane surface. E, Left, Sympathetic nerves labeled with TH and T (data not shown) were observed coursing along the adventitia of arteries (arrowheads). Right, Within nerves (sciatic nerve shown here) T was colocalized in S100+ Schwann cells (data not shown) and a fraction (28%) of TH+ unmyelinated axons was closely associated with T+ Schwann cells (TH+; T+) in sectioned nerves. Scale bars: C, 200 μm; D, E, left, 50 μm; E, right, 5 μm.
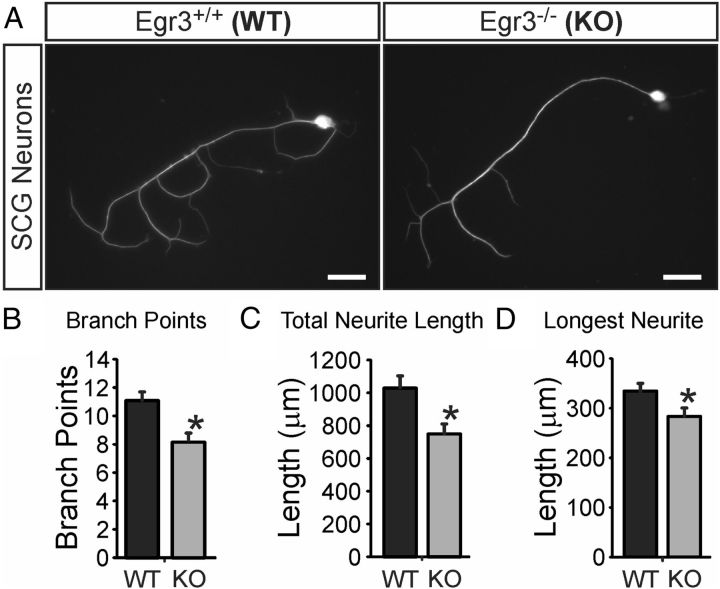
Unlike NGF, which is required for sympathetic neuron survival and neurite outgrowth, Egr3 is not required for neuron survival in isolated sympathetic neurons treated with NGF (Eldredge et al., 2008). Thus, if Egr3 has a role in mediating gene regulation induced by NGF signaling, it may have a role in neurite outgrowth. To test whether Egr3 has an autonomous role in sympathetic neurite outgrowth in response to NGF, sympathetic neurons were isolated from the SCG of newborn Egr3+/+ (wild-type) and Egr3−/− (Egr3 knock-out; KO) mice and grown in dissociated cultures treated with NGF (Fig. 2A). Compared with wild-type SCG neurons, those lacking Egr3 had significantly fewer neurite branch points (Fig. 2B), decreased total neurite length (Fig. 2C), and a reduction in the length of the longest neurite (Fig. 2D). These data indicate that Egr3 has a moderate, but significant autonomous role in neurite extension and branching in response to NGF, which may be correlated with the abnormal sympathetic target tissue innervation abnormalities observed in germline Egr3 KO mice.
Figure 2.
Egr3 is required for normal neurite outgrowth and branching in NGF-treated sympathetic neurons. A, Dissociated Egr3−/− (KO) sympathetic neurons have impaired neurite outgrowth in response to NGF treatment. Neurites from Egr3 KO neurons have significantly fewer branch points (B), decreased total neurite length (C), and decreased longest neurites compared with Egr3+/+ wild-type (WT) neurons (D). *p < 0.05, N > 60 neurons of each genotype analyzed. Scale bar: A, 50 μm.
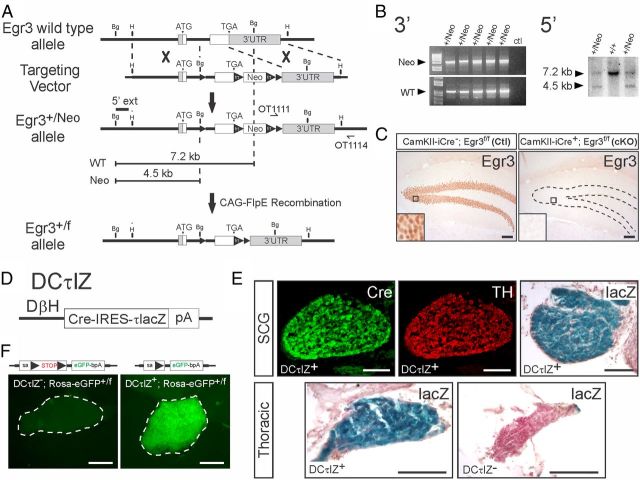
To further examine whether Egr3 has a sympathetic neuron autonomous role in target tissue innervation, we generated two novel mouse strains. We generated mice with a conditional Egr3 allele by inserting loxP sites flanking exon 2 (flx allele), which contains the zinc-finger DNA binding domain required for transcriptional activation by the Egr3 protein. When loxP sites are recombined, the resulting mutation is similar to the germline loss-of-function mutation we generated in Egr3 KO mice (Tourtellotte and Milbrandt, 1998) (Fig. 3A,B). Homozygous Egr3f/f mice were indistinguishable from wild-type mice, they expressed normal levels of Egr3, and removal of exon2 by loxP-site recombination led to complete loss of Egr3 protein in Cre-recombinase-expressing cells as expected (Fig. 3C). In addition, we generated transgenic mice (DCτlZ) using the human DβH promoter (Mercer et al., 1991) to drive the expression of a bicistronic transcript in noradrenergic neurons that encodes Cre-recombinase and axon localized lacZ (τlacZ) (Fig. 3D). DCτlZ transgenic mice expressed high levels of Cre-recombinase and lacZ in sympathetic neurons (Fig. 3E), and functional tests demonstrated efficient loxP-site recombination by activation of eGFP expression in sympathetic neurons from reporter mice (Fig. 3F).
Figure 3.
Design and validation of Egr3-flx conditional knock-out and sympathetic neuron Cre-driver mice. A, The mouse Egr3 genomic locus was modified to contain forward-oriented loxP sites in intron 1 and downstream of the stop codon using homologous recombination in E. coli. A frt-flanked Neo selection cassette was inserted upstream of the 3′ loxP site and the Egr3-flx (Egr3+/f) allele was generated by removing the Neo selection cassette from the germline using CAG-FlpE mice. B, Left, Electroporated G418 selected ES cell clones were screened using long-range PCR with primers that flanked the 3′ recombination arm (primers: OT1111 and OT1114). Right, PCR confirmed ES cell clones were screened by Southern blotting analysis using a 713 nt probe (5′ ext) located upstream of the 5′ recombination arm that was hybridized to BglII restricted genomic DNA. C, To validate the conditional knock-out allele, CaMKII-iCre transgenic mice (Casanova et al., 2001) were mated to Egr3+/f mice to ablate Egr3 in postnatal forebrain and hippocampal dentate gyrus neurons. As expected, Egr3 protein was robustly expressed in CaMKII-iCre−; Egr3f/f (left; Ctl) dentate gyrus neurons but was completely absent in CaMKII-iCre+; Egr3f/f (right; cKO) neurons. D, The human DβH promoter was used to generate transgenic mice that express a Cre-recombinase and axon localized β-galactosidase (τlacZ) bicistronic message in noradrenergic neurons (designated DCτlZ transgenic mice). E, Cre-recombinase and lacZ expression were detectable in all TH+ sympathetic neurons. F, To validate the efficiency of DCτlZ mice to recombine loxP sites in sympathetic neurons, they were mated to Rosa-eGFP reporter mice (Belteki et al., 2005). DCτlZ−; Rosa-eGFP+/f mice showed no expression of eGFP, whereas DCτlZ+; Rosa-eGFP+/f mice showed expression of eGFP in all sympathetic neurons (SCG shown here). Scale bars: C, 200 μm; E, F, 100 μm.
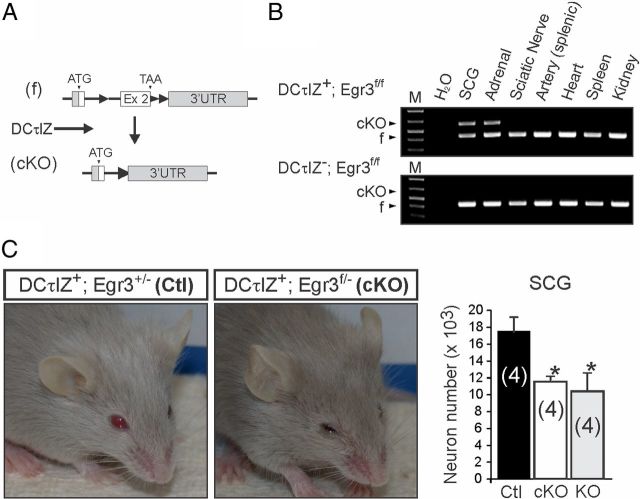
Using DCτlZ transgenic mice and Egr3-flx mice, we ablated Egr3 expression in sympathetic neurons to examine its neuron-specific role in target tissue innervation (Fig. 4A). Mice with the genotype DCτlZ+; Egr3f/f showed loxP-site recombination specifically in sympathetic neurons and adrenal chromaffin cells, but not in blood vessels, nerves, or other tissues examined (Fig. 4B). Since Egr3 heterozygous mice have no identifiable abnormalities, and to minimize mosaicism in recombined sympathetic neurons, we mated Egr3+/f mice to Egr3+/− mice to generate Egr3f/− mice. Mice with specific ablation of Egr3 in sympathetic neurons (DCτlZ+; Egr3f/−, subsequently referred to as cKO mice) lacked the characteristic scoliosis and gait ataxia seen in germline Egr3 KO mice, the latter which is caused by loss of Egr3 in muscle spindle stretch receptors (Tourtellotte and Milbrandt, 1998; Tourtellotte et al., 2001; Chen et al., 2002b). However, the cKO mice had prominent SNS abnormalities including blepharoptosis and significant neuron loss in the SCG compared with DCτlZ+; Egr3+/− (referred to as Ctl) mice, and similar to the blepharoptosis and neuron loss identified in germline Egr3 KO mice (Fig. 4C). In addition, Egr3 cKO mice had peripheral sympathetic target tissue innervation abnormalities that closely recapitulated the innervation abnormalities observed in germline Egr3 KO mice (Li et al., 2011). For example, sympathetic innervation (demonstrated by TH immunofluorescence) to heart, spleen, kidney, and pineal gland was significantly reduced in Egr3 cKO mice (Fig. 5). These results clearly demonstrate that Egr3 expression by sympathetic neurons is essential for their normal target tissue innervation and that loss of Egr3 expression within sympathetic neurons recapitulates the SNS abnormalities observed in mice lacking Egr3 expression in all tissues.
Figure 4.
Egr3 ablation specifically in noradrenergic neurons leads to abnormal SNS development. A, DCτlZ+ transgenic mice were mated to Egr3+/f mice to ablate Egr3 in sympathetic neurons, but not in peripheral cells such as Schwann cells and arterial endothelial cells with which sympathetic axons interact during development. B, PCR amplification of the flx (f) and cKO allele using genomic DNA isolated from DCτlZ+; Egr3f/f tissues showed loxP recombination to generate the cKO allele specifically in tissues expressing DβH (SCG neurons and adrenal chromaffin cells) but not in other tissues tested. The flx allele PCR product remains detectable in the SCG and adrenal glands from DCτlZ+; Egr3f/f mice since these tissues contain several cell types (glial cells, endothelial cells, fibroblasts, adrenal cortical cells, etc.) that do not express DβH and hence, do not express Cre-recombinase. As expected, no cKO PCR product was detected in any of the tissues tested from DCτlZ; Egr3f/f mice. C, Left, Compared with DCτlZ+; Egr3+/− mice (Ctl), DCτlZ+; Egr3f/− (cKO) mice had bilateral ptosis similar to the ptosis that is observed in germline Egr3 KO mice, which is caused by disruption of sympathetic innervation from the SCG to the levator muscles of the eyelids. Right, Loss of Egr3 specifically in sympathetic neurons (cKO) resulted in ∼30% loss of SCG neurons, similar to the neuron loss observed in germline Egr3 KO mice, which lacked Egr3 in all tissues. *p < 0.05, N = 4 animals of each genotype analyzed.
Figure 5.
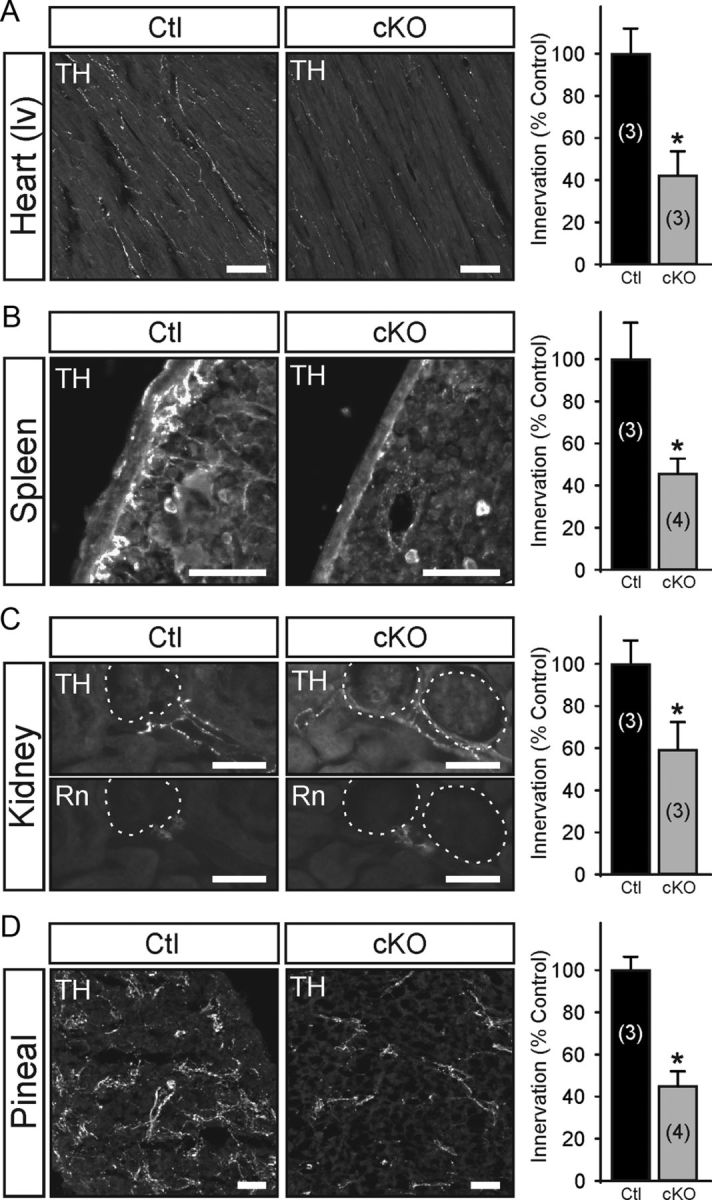
Egr3 ablation in sympathetic neurons impairs their target tissue innervation. Tissues known to have abnormal target tissue innervation in germline Egr3 KO mice, where Egr3 is absent in all tissues (Eldredge et al., 2008), were examined in DCτlZ+; Egr3f/− (cKO) mice using semiquantitative TH immunofluorescence. Compared with DCτlz+; Egr3+/− (Ctl) mice, sympathetic innervation to the heart (A, left ventricle, lv) was reduced by 58%, in the spleen by 57% (B), in the kidney by 41% (C), and in the pineal gland (D) by 56% in DCτlZ+; Egr3f/− (cKO) mice. Unlike the heart and pineal gland where TH+ fibers innervated the tissues diffusely, in the spleen the majority of the innervation was confined to the subcapsular space and in the kidney it was localized primarily to the juxtaglomerular organ highlighted by immunofluorescent staining for renin (Rn). *p < 0.02, N = 3–4 animals of each genotype analyzed, as indicated in parentheses. Scale bars: 50 μm.
Egr3-mediated gene regulation in sympathetic neurons
In vitro studies from Egr3 KO sympathetic neurons and in vivo studies with Egr3 cKO mice indicate that it has a sympathetic neuron autonomous role in target tissue innervation, but the target genes it regulates within them are not known. To characterize Egr3-mediated gene expression in sympathetic neurons we used Illumina BeadChip microarray analysis. SCGs from three newborn (P0) Egr3+/+; Bax−/− and Egr3−/−; Bax−/− mice were isolated to obtain total cellular RNA to perform the array analysis in triplicate. Since Egr3 does not have a direct role in neuron survival (Eldredge et al., 2008), the Bax-deficient genetic background was used to inhibit neuron apoptosis-associated gene induction resulting from abnormal target tissue innervation (Li et al., 2011). The analysis yielded 197 transcripts that were significantly downregulated (p < 0.01) and 240 transcripts that were significantly upregulated (p < 0.01) in the absence of Egr3 from a total of 45,281 target sequences represented on the array (<1% of sequences on the array were differentially expressed). The integrity of the microarray dataset was determined by examining 30 upregulated and downregulated genes with highly significant difference scores (small difference p values) using qPCR. Using qPCR analysis on independently isolated RNA, 77% (23/30) of the genes tested were confirmed to be deregulated (Table 1).
Table 1.
qPCR-confirmed deregulated genes sampled from 30 genes with the smallest difference p value in the microarray dataset of differentially expressed genes in Egr3-deficient sympathetic neurons. (23 confirmed genes shown)
| Official gene symbol | NCBI gene ID (mouse) | Microarray |
qPCR fold change (± SEM) | |
|---|---|---|---|---|
| Difference p value | Fold change | |||
| Downregulated | ||||
| Sox9 | 20682 | 2.74 × 10−6 | −2.25 | −2.32 ± 0.08 |
| Plxnd1 | 67784 | 5.07 × 10−6 | −1.82 | −2.28 ± 0.05 |
| Esd | 13885 | 5.99 × 10−6 | −3.80 | −1.64 ± 0.12 |
| Gins2 | 272551 | 2.22 × 10−5 | −1.88 | −1.51 ± 0.07 |
| Ret | 19713 | 5.28 × 10−5 | −1.63 | −1.94 ± 0.06 |
| Insc | 233752 | 2.94 × 10−4 | −2.32 | −2.77 ± 0.48 |
| Notch1 | 18128 | 3.92 × 10−4 | −1.63 | −2.07 ± 0.30 |
| Efnb1 | 13641 | 2.75 × 10−3 | −1.40 | −2.24 ± 0.34 |
| Lmo7 | 380928 | 3.00 × 10−3 | −2.90 | −2.59 ± 0.53 |
| Hs3st2 | 195646 | 5.42 × 10−3 | −2.68 | −1.60 ± 0.25 |
| Ngfr | 18053 | 8.50 × 10−3 | −1.48 | −2.86 ± 0.19 |
| Upregulated | ||||
| B3galt6 | 117592 | 3.68 × 10−8 | 7.47 | 2.58 ± 0.11 |
| Mxra8 | 74761 | 6.73 × 10−7 | 2.20 | 1.60 ± 0.17 |
| Nf1 | 18015 | 2.05 × 10−5 | 2.87 | 2.26 ± 0.37 |
| Apbb1 | 11785 | 7.44 × 10−5 | 1.54 | 1.78 ± 0.16 |
| Pabpc1l | 381404 | 2.90 × 10−4 | 4.77 | 3.67 ± 0.23 |
| Slit2 | 20563 | 5.32 × 10−4 | 1.70 | 1.63 ± 0.20 |
| Slc15a2 | 57738 | 8.59 × 10−4 | 3.72 | 2.24 ± 0.55 |
| Irak1 | 16179 | 1.57 × 10−3 | 2.80 | 2.00 ± 0.17 |
| Zfp292 | 30046 | 1.95 × 10−3 | 2.79 | 1.50 ± 0.08 |
| Pde3a | 54611 | 6.81 × 10−3 | 3.91 | 1.97 ± 0.20 |
| Rccd1 | 269955 | 7.72 × 10−3 | 3.59 | 4.43 ± 0.45 |
| Ptpn11 | 19247 | 8.65 × 10−3 | 1.55 | 1.73 ± 0.27 |
Results represent qPCR-confirmed deregulated genes from three replicate cDNA samples (p < 0.05).
Using GO analysis to categorize the differentially expressed genes based on their known function and then applying a candidate target gene approach on qPCR-confirmed deregulated genes, we identified several functional categories of deregulated genes that were significantly over represented (p < 0.001) in the array dataset using DAVID functional annotation analysis (Huang da et al., 2009). We focused on genes in significantly enriched categories with ascribed functions related to SNS development (GO: 0048485), axonogenesis (GO: 00074904), dendrite morphogenesis (GO: 0048813), and axon guidance (GO: 0007411) since abnormal sympathetic target tissue innervation, not related to neuron survival, appears to be a dominant function of Egr3 in sympathetic neurons (Eldredge et al., 2008; Li et al., 2011). Since sympathetic neurons require Egr3 for neurite outgrowth (in vitro) and target tissue innervation (in vivo), it was not surprising to identify significantly deregulated genes with GO functions related to SNS development and axonogenesis (Table 2). Unexpectedly, however, some of the most significantly deregulated genes have ascribed functions that relate to dendritogenesis and axon guidance (Table 2). These results raise the possibility that target tissue innervation abnormalities in Egr3 KO mice could be related to abnormal sympathetic axon guidance as growing axons traverse blood vessels and navigate intermediate peripheral targets during early development. Similarly, as previous studies have shown that NGF is important for dendritogenesis in vivo (Snider, 1988; Ruit et al., 1990), Egr3 may regulate gene expression involved in sympathetic neuron dendritogenesis, which could further contribute to the profound physiologic dysautonomia observed in Egr3 KO mice. For example, qPCR confirmed genes deregulated in Egr3-deficient sympathetic neurons with previously documented roles in dendritogenesis and axon guidance (Table 2), such as Ngfr (Hartmann et al., 2004; Gascon et al., 2005; Salama-Cohen et al., 2005; Zagrebelsky et al., 2005), Notch1 (Redmond et al., 2000; Salama-Cohen et al., 2006; Breunig et al., 2007), Efnb1 (Moreno-Flores et al., 2002), Ptpn11 (Chacón et al., 2010), and Slit2 (Furrer et al., 2007; Dimitrova et al., 2008; Hocking et al., 2010), could contribute to abnormal SNS development in the absence of Egr3, but whether abnormalities in dendritogenesis and/or axon guidance exist in Egr3 KO mice has not been examined.
Table 2.
Selected qPCR-confirmed deregulated genes in Egr3-deficient sympathetic neurons and their GO determined functional associations
| Official gene symbol | NCBI gene ID (mouse) | Microarray |
qPCR fold change (±SEM) | SNS development | Axonogenesis | Dendritogenesis | Axon guidance | |
|---|---|---|---|---|---|---|---|---|
| Difference p value | Fold change | |||||||
| Downregulated | ||||||||
| Plxnd1 | 67784 | 5.07 × 10−6 | −1.82 | −2.28 ± 0.05 | X | |||
| Ret | 19713 | 5.28 × 10−5 | −1.63 | −1.94 ± 0.06 | X | X | X | |
| Notch1 | 18128 | 3.92 × 10−4 | −1.63 | −2.0 ± 0.30 | X | X | X | |
| Efnb1 | 13641 | 2.75 × 10−3 | −1.40 | −2.24 ± 0.34 | X | X | X | |
| Ngfr | 18053 | 8.50 × 10−3 | −1.48 | −2.86 ± 0.19 | X | X | X | X |
| Upregulated | ||||||||
| Nf1 | 18015 | 2.05 × 10−5 | 2.87 | 2.26 ± 0.37 | X | X | ||
| Apbb1 | 11785 | 7.44 × 10−5 | 1.54 | 1.78 ± 0.16 | X | |||
| Slit2 | 20563 | 5.32 × 10−4 | 1.70 | 1.63 ± 0.20 | X | X | X | |
| Ptpn11 | 19247 | 8.65 × 10−3 | 1.55 | 1.73 ± 0.27 | X | X | X | |
Fold change compared to control with negative fold-change values representing genes downregulated in the absence of Egr3 and positive fold-change values representing genes upregulated in the absence of Egr3. All qPCR fold changes are significant, p < 0.05, three replicate cDNA samples.
In vivo genetic tracing of sympathetic neuron dendrite morphology and axon guidance
Previous studies have used either retrograde tracers (Rubin, 1985; Smolen and Beaston-Wimmer, 1986) or direct intracellular sympathetic neuron injections with fluorescent dyes (Yawo, 1987; Voyvodic, 1989) to label dendrites and axons in vivo, but the yield of analyzable neurons is often low. As some of the identified target genes may have a role in dendritogenesis, dendrite morphology was examined in vivo using a genetic tracing technique that generates mosaic loxP recombination to sparsely label neurons and their processes in Cre-recombinase reporter mice. DβH-Cre BAC transgenic mice (recovered from the GENSAT repository; MMRRC Stock# 032081-UCD and previously uncharacterized) were found to inefficiently recombine loxP sites in sympathetic neurons. For example, when they were mated to StLa reporter mice, which express axon localized lacZ (τlacZ) after Cre-mediated loxP recombination (Nam and Benezra, 2009), <1% of sympathetic neurons were labeled in sympathetic ganglia (Fig. 6A; SCG shown). The labeling pattern was similar to Golgi-stained neurons or neurons labeled by a Thy1 driven transgene reporter (Feng et al., 2000) in that the entire neuron and its processes were labeled. This sparse labeling pattern made it possible to compare in detail individual sympathetic neuron dendrite and axon morphology between wild-type and Egr3 KO mice.
Figure 6.
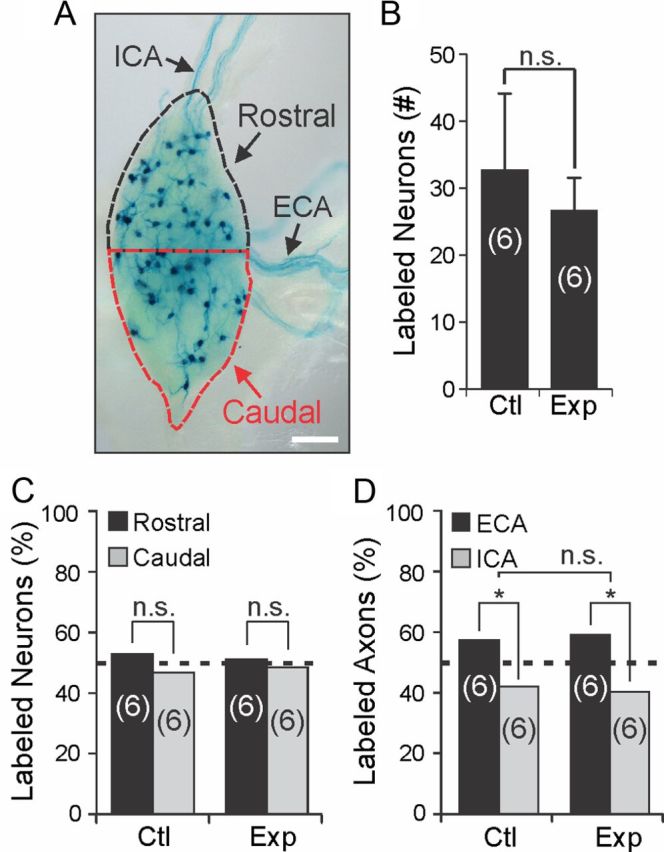
In vivo genetic tracing highlights randomly labeled sympathetic neurons and their dendrite morphology and axon trajectories. A, Sympathetic neurons are labeled in a mosaic pattern in DβH-Cre+; StLa+/f SCG where <1% of the neurons are labeled by the τlacZ reporter (neurons and their processes labeled by lacZ histochemistry shown). iCA axons arise primarily from neurons located in the rostral 50% of the ganglion (black dashed line) and eCA axons arise primarily from neurons located in the caudal 50% of the ganglion (red dashed line). B, No significant difference in the number of lacZ-labeled SCG neurons was detected between DβH-Cre+; Egr3+/+; StLa+/f (Ctl) and DβH-Cre+; Egr3−/−; StLa+/f (Exp) SCG (N = 6 mice of each genotype analyzed). C, To examine whether lacZ-labeled neurons were randomly distributed, the distribution frequency of neurons located in rostral and caudal regions was determined. No difference in their distribution frequency relative to an expected random 50% distribution was detected between rostral and caudal regions of the SCG in either Ctl or Exp SCG. (N = 6 mice of each genotype analyzed; n.s., not significant, χ2 test). D, Since the SCG provides greater innervation to target tissues innervated by axons that travel along the external carotid artery, the frequency distribution of lacZ-stained axons was significantly different from a random (50%) distribution, as expected in both Ctl (χ2, *p < 0.03) and Exp (χ2, *p < 0.02) mice. However, no significant difference was seen in the ECA–ICA frequency distributions between Ctl and Exp mice (n.s., not significant; χ2 test; frequency distributions are shown as percentage of total lacZ-stained axons for each genotype, N = 6 animals of each genotype analyzed). Scale bar: A, 200 μm.
Although there was never a visually similar pattern of labeled neurons in sympathetic ganglia in any of the mice examined, to determine whether sympathetic neurons were randomly labeled, we first quantified sympathetic neurons that were lacZ labeled in DβH-Cre+; Egr3+/+; StLa+/f (referred to as Ctl) and DβH-Cre+; Egr3−/−; StLa+/f (referred to as Exp) mice and we found no significant difference between them (Fig. 6B). To determine whether the loxP-site recombination in sympathetic neurons was stochastic, and hence whether labeling of SCG neurons was random, we examined the percentage of neurons labeled in different regions of the SCG. SCG neurons that give rise to internal carotid axons (ICA) generally reside in the rostral half of the ganglion, whereas those giving rise to the external carotid axons (ECA) generally reside in the caudal half of the ganglion (Bowers and Zigmond, 1979). The SCGs were divided into rostral and caudal portions (Fig. 6A) and the percentage of lacZ-stained neurons in both halves was determined. No significant difference from the expected random distribution of neurons (50% in rostral and 50% in caudal) was identified between rostral and caudal portions of the SCG in Ctl (χ2, p = 0.39) or Exp mice (χ2, p = 0.75) (Fig. 6C). Similarly, since SCG neurons give rise to more eCA axons that innervate target tissues compared with iCA axons, the distribution frequency of labeled eCA axons was greater than in iCA axons in both Ctl (χ2, p < 0.03) and Exp (χ2, p < 0.02) mice as expected (Fig. 6D). However, there was no significant difference in the frequency distribution of axons between Ctl and Exp mice (χ2, p = 0.66). These results indicate that sympathetic neurons are randomly labeled in these reporter mice and that the random labeling pattern is retained in Egr3 KO mice.
Abnormal sympathetic neuron dendritogenesis in Egr3 KO mice
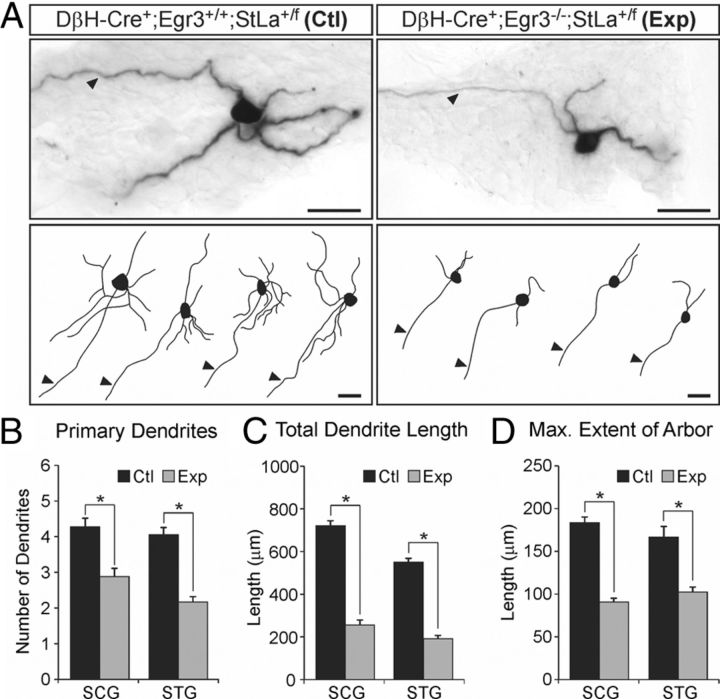
Chemical clearing of whole lacZ-stained sympathetic ganglia made it possible to visualize dendrites in detail and to compare them between Ctl and Exp mice (Fig. 7A). Neuron tracings from flat-field projected images of labeled sympathetic neurons demonstrated that neurons from Exp mice had significantly fewer primary dendrites compared with neurons from Ctl mice in both the SCG (p < 0.00001) and stellate ganglion (STG) (p < 0.00001) (Fig. 7B). Similarly, labeled neurons had decreased total dendrite length in Exp mice in the SCG (p < 0.00001) and STG (p < 0.00001) (Fig. 7C) as well as a reduction in the maximum extent of the dendritic arbor in SCG (p < 0.00001) and STG (p < 0.00001) compared with neurons from Ctl mice (Fig. 7D). Finally, in agreement with previously published results indicating that sympathetic neurons are atrophic in the absence of Egr3 (Eldredge et al., 2008; Li et al., 2011), we observed that labeled neurons in Exp mice were also significantly atrophic compared with Ctl mice (mean soma area in Ctl = 769.9 ± 45.6 μm2 and in Exp = 560.5 ± 32.7 μm2, p < 0.0004). Thus, in addition to target tissue innervation abnormalities, Egr3 has a role in dendrite morphogenesis that may further contribute to physiological sympathetic dysfunction in Egr3 KO mice. Since germline Egr3 KO mice have ∼30% neuron loss that occurs during late embryogenesis and early postnatal life (Eldredge et al., 2008), these data do not rule out the possibility that neuron loss may influence the dendritogenesis of remaining neurons. Similarly, since these studies were performed using germline KO mice, the data do not definitively demonstrate that dendritogenesis abnormalities are due to loss of Egr3 expression within sympathetic neurons.
Figure 7.
Abnormal dendrite morphogenesis in sympathetic neurons lacking Egr3. A, Top, lacZ-stained neurons in Ctl (DβH-Cre+; Egr3+/+; StLa+/f) and Exp (DβH-Cre+; Egr3−/−; StLa+/f) sympathetic ganglia were labeled in a sparse mosaic pattern with their axons and dendrites clearly visible. In labeled neurons, processes that projected out of the ganglia were defined as axons (arrowhead), while the remainder of the processes confined to the ganglia were defined as dendrites. Bottom, Representative tracings from neuron profiles of flat-field (arrowheads) projected images demonstrated attenuated dendritic processes in Exp sympathetic neurons compared with those from Ctl mice. B–D, Dendrite morphology was quantified in Exp and Ctl neurons from SCG and STG. Compared with Ctl neurons those from Exp mice showed significantly decreased total number of dendrites per neuron (B) significantly decreased total dendrite length (C) and a reduction in maximum extent of the dendritic arbor in sympathetic neurons from both SCG and STG (D). *p < 0.00001, N = 3–4 animals analyzed of each genotype. Scale bars: A, 50 μm.
Role of Egr3 in axon guidance and target tissue innervation
Gene expression analysis indicated that several genes well established to have a role in axon guidance such as Plxnd1 (Chauvet et al., 2007), Efnb1 (Orioli and Klein, 1997; Flanagan and Vanderhaeghen, 1998; Bush and Soriano, 2009), and Slit2 (Nguyen Ba-Charvet et al., 1999; Brose and Tessier-Lavigne, 2000; Plump et al., 2002) and some genes less well recognized to be involved in axon guidance including Ngfr (Wong et al., 2002; Lim et al., 2008) and Ret (Enomoto et al., 2001; Kramer et al., 2006; Honma et al., 2010) are deregulated in sympathetic neurons that lack Egr3 (Table 2). This raises the possibility that target tissue innervation abnormalities may result from abnormal axon guidance as they traverse the periphery to innervate target tissues during development. However, as sympathetic target tissue innervation is diminished, but not completely absent in Egr3 KO mice (Eldredge et al., 2008; Li et al., 2011), axon guidance abnormalities would likely exist in only a subset of axons, if at all. Previous analysis using sympathetic axon reporter mice to label all sympathetic axons in the absence of Egr3 was not informative because it was impossible to visualize a small number of potentially misguided axons among the entire labeled axon population (Eldredge et al., 2008). However, using mosaic neuron/axon labeling in Ctl and Exp mice, where only a small subset of sympathetic axons are randomly labeled, it was possible to carefully examine individual axon trajectories in Exp mice.
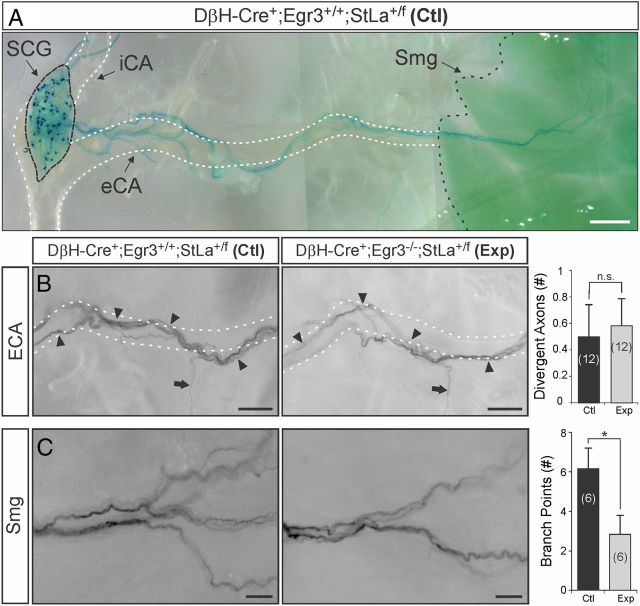
To determine whether sympathetic axon guidance was disrupted in Egr3 KO mice, a whole-mount preparation was developed to examine axons projecting along their entire trajectory, from the SCG along the external carotid artery to the submandibular gland (Smg) (Fig. 8A). The Smg was chosen as a representative target tissue to study since it receives robust sympathetic innervation from the SCG and innervation to it is highly abnormal in Egr3 KO mice (Eldredge et al., 2008). In Ctl and Exp mice, labeled sympathetic ECAs followed the course of the external carotid artery (Fig. 8B, arrowheads) and only occasional axons were observed to diverge from the artery en route to the Smg (Fig. 8B, arrow). There was no statistical difference between the number of divergent axons arising from the SCG to innervate the Smg between Ctl and Exp mice (p = 0.39, N = 12 Ctl and N = 12 Exp mice analyzed) (Fig. 8B). These results indicate that loss of Egr3 does not result in axon misguidance to the Smg and therefore does not explain the diminished sympathetic target tissue innervation observed in Egr3 KO mice.
Figure 8.
Egr3 has a role in target tissue innervation that is independent of axon guidance. A, Chemical clearing and whole-mount preparations using genetic axon tracing with DβH-Cre driver and StLa reporter mice made it possible to visualize a random subset of SCG neurons and their entire axon trajectories as they innervate the Smg. The Smg has profound innervation abnormalities in Egr3 KO mice (Eldredge et al., 2008) making this an informative preparation for testing whether innervation abnormalities are due to axon guidance defects or branching within the target tissue. B, Sparsely labeled axons were traced along the external carotid artery (eCA; white dashed lines) in Ctl (DβH-Cre+; Egr3+/+; StLa+/f) and Exp (DβH-Cre+; Egr3−/−; StLa+/f) mice. In both Ctl and Exp mice sympathetic axons coursed along the artery (arrowheads) to innervate the Smg, but occasional axons diverged from the artery to innervate tissues other than the Smg (arrow). There was no significant difference in the number of sympathetic axons traveling from the SCG to the Smg that diverged from the eCA between Ctl and Exp mice. C, In the Smg from Ctl and Exp mice lacZ-stained sympathetic axons entered the target tissue normally. However, the extent of axon branching within the target tissue was decreased in Exp relative to Ctl mice. *p < 0.015; N = 6–12 animals studied of each genotype as indicated. Scale bars: A, 500 μm; B, 250 μm; C, 100 μm. Dashed lines highlight the common carotid artery, internal carotid artery (iCA) and external carotid artery (eCA).
Egr3 has a clear neuron autonomous role in target tissue innervation that is unrelated to neuron survival or axon guidance, but how sympathetic target tissue innervation is impaired in Egr3 KO mice remains unclear. Based on in vitro results demonstrating that Egr3 has a role in neurite extension and branching, target tissue innervation could be impaired if axons from Egr3 KO neurons fail to extend and branch properly within tissues. Interestingly, mice lacking both NGF and Bax have innervation abnormalities that are characterized by ineffective axon invasion and branching within most tissues (Glebova and Ginty, 2004) and similar results were observed in mice lacking both Egr3 and Bax (Li et al., 2011). To determine whether Egr3 has a role in axon branching within target tissues, SCG-Smg whole-mount preparations were used to examine axon innervation to the gland. A significant decrease in axon branching within the gland in Exp relative to Ctl mice was observed (Fig. 8C). Thus, terminal axon branching within target tissues, and not neuron loss or axon guidance, appears to explain the relatively diminished target tissue innervation that occurs in sympathetic neurons lacking Egr3.
Discussion
The SNS is an essential part of the autonomic nervous system, which is impaired in many human neurodegenerative and metabolic diseases such as diabetes. Since the underlying cause of sympathetic neuron loss in these diseases is largely unknown and they lead to substantial suffering and premature death (Vinik et al., 2011), a greater understanding of critical signaling pathways involved in establishing and maintaining sympathetic target tissue innervation homeostasis is of considerable interest. NGF and signaling through its high-affinity TrkA receptor is essential for sympathetic neuron survival, target tissue innervation, and postnatal viability in mice (Crowley et al., 1994; Smeyne et al., 1994; Glebova and Ginty, 2004) so it is not surprising that rare mutations in NGFβ (HSAN type V, OMIM #608654) and TrkA (HSAN type IV, OMIM #256800) also lead to severe developmental abnormalities of the SNS in humans (Rotthier et al., 2012). How these essential mediators regulate downstream signaling in sympathetic neurons or whether any downstream effectors are involved in human disease is not known.
In previous studies we found that Egr3 is regulated by NGF signaling in sympathetic neurons and that it has an essential role in SNS development in mice (Eldredge et al., 2008; Li et al., 2011). However, the studies were performed in germline Egr3 knock-out mice, where Egr3 was absent in all tissues. In addition to sympathetic neurons, Egr3 is expressed in a variety of cell types where it is induced by signaling molecules that mediate its substantial pleiotropism. For example, Egr3 expression is regulated by neuregulin-1 (Nrg1) in developing myoblasts, where it has an important role in regulating skeletal muscle stretch receptor morphogenesis (Tourtellotte and Milbrandt, 1998; Hippenmeyer et al., 2002), it is regulated by factors such as brain-derived neurotrophic factor in cortical neurons, where it has a role in regulating NMDA receptor and the plasticity associated Arc gene expression involved in learning and memory (Li et al., 2005, 2007; Kim et al., 2012), and it is regulated by T-cell receptor signaling, where it has a role in immune function (Carter et al., 2007; Li et al., 2012). To determine how Egr3 expression may specifically influence sympathetic neuron development, we examined its expression pattern in tissues known to interact with developing sympathetic axons. Using Egr3-Cre knockin mice (Egr3+/Cre) and Cre-dependent reporter mice (Ai14) it was possible to trace the historical expression of Egr3, even if expression is transient and/or present in lineage precursors during development. In newborn and adult mice, we found reporter expression localized in Schwann cells and arterial vascular endothelial cells, but not in other tissues that may be encountered by developing sympathetic axons during their outgrowth to target tissues. Thus, Egr3 may have a functional role in Schwann cells or endothelial cells, which could influence sympathetic target tissue innervation. We confirmed a cell autonomous function for Egr3 within sympathetic neurons using isolated Egr3-deficient sympathetic neurons, which showed significant neurite outgrowth and branching abnormalities compared with wild-type neurons in response to NGF treatment. Similarly, using Egr3 conditional knock-out and DβH-CreIRESτLacZ (DCτlZ) cre-driver transgenic mice to ablate Egr3 function in sympathetic neurons in vivo, but not in other cells such as Schwann cells or vascular endothelial cells, we found that Egr3 cKO mice had sympathetic target tissue innervation abnormalities that were indistinguishable from Egr3 KO mice. Our studies did not address the role of Egr3 expression in Schwann cells or vascular endothelial cells. Nevertheless, previous studies indicate that Egr3 modulates the expression of the low-affinity NGF receptor (Ngfr) in Schwann cells where it has a role in peripheral axon myelination (Cosgaya et al., 2002; Gao et al., 2007) and it appears to mediate vascular endothelial growth factor signaling in vascular endothelial cells (Suehiro et al., 2010). Thus, a sympathetic neuron autonomous phenotype that is similar to the phenotype observed in germline KO mice indicates that Egr3 function in Schwann and vascular endothelial cells is not likely related to SNS development.
To identify target genes that are regulated by Egr3 in sympathetic neurons we used microarray analysis on SCG isolated from newborn wild-type and Egr3 knock-out mice. We identified several hundred genes that were significantly upregulated and downregulated in the absence of Egr3 (p < 0.01 and 1.2-fold cutoff). The list of deregulated genes appeared to be relatively reliable since >75% of the 30 most significantly deregulated genes were confirmed by qPCR analysis and all of them were found to be deregulated in the same direction (upregulated or downregulated) as indicated by the microarray data (Table 1). Our current results indicate that Egr3 has a role in neurite outgrowth that appears to correlate with abnormal dendrite development and terminal axon branching in vivo. Thus, the large list of deregulated genes was refined using GO analysis on qPCR confirmed deregulated genes to organize them by relevant function (Table 2). Some of the genes that were significantly deregulated in the absence of Egr3 have been previously shown to be involved in SNS development including Ngfr (Lee et al., 1994; Bamji et al., 1998; Brennan et al., 1999), Notch1 (Tsarovina et al., 2008), Ret (Enomoto et al., 2001; Tsui-Pierchala et al., 2002), Nf1 (Vogel et al., 1995; Vogel and Parada, 1998), and Ptpn11 (shp-2) (Chen et al., 2002a). More specifically, many of the genes regulated by Egr3 in sympathetic neurons have roles in neurite outgrowth such as Ngfr (Yamashita et al., 1999; Bentley and Lee, 2000; Domeniconi et al., 2005), Ephrin-B1 (Tanaka et al., 2004), Notch1 (Sestan et al., 1999; Huang et al., 2005), Ret (Enomoto et al., 2001; Zhang et al., 2006), Nf1 (Romero et al., 2007), Apbb1 (Ikin et al., 2007), Ptpn11 (shp-2) (Chen et al., 2002a; Perrinjaquet et al., 2010), and Slit2 (Wang et al., 1999; Ozdinler and Erzurumlu, 2002). For example, Ret is a receptor for the glial cell line-derived neurotrophic factor (GDNF) family of ligands, which include GDNF, neurturin, artemin (ARTN), and persephin (Airaksinen et al., 1999; Baloh et al., 2000; Airaksinen and Saarma, 2002). Ret knock-out mice have sympathetic neuron migration abnormalities (Enomoto et al., 2001) and one of its ligands, ARTN has an essential role in mediating sympathetic axon growth along blood vessels (Honma et al., 2002). Since Egr3 is first expressed in postmigratory sympathetic neurons, it is not surprising that loss of Egr3 does not result in neuron migration abnormalities (Eldredge et al., 2008). However, decreased Ret expression in postmigratory Egr3-deficient neurons could result in abnormalities in GDNF and ARTN signaling. GDNF and ARTN knock-out mice have partial loss of sympathetic neurons and target tissue innervation (Moore et al., 1996; Honma et al., 2002), most likely caused by their well established role in sympathetic neurite outgrowth and branching (Ebendal et al., 1995; Honma et al., 2002; Niwa et al., 2002; Yan et al., 2003). In addition, Ngfr is also required for normal sympathetic target tissue innervation (Lee et al., 1994). Thus, Egr3-mediated regulation of Ngfr expression in sympathetic neurons could modulate some aspects of NGF signaling that may be essential for modulating a feedforward signaling loop that is dependent on NGF-induced Egr3 expression. Negative regulators of sympathetic neurite outgrowth, which were upregulated in the absence of Egr3, such as Ptpn11 (shp-2), Apbb1, and Nf1 may further contribute to decreased sympathetic axon and dendrite outgrowth in vivo (Chen et al., 2002a; Ikin et al., 2007; Romero et al., 2007). NGF has a clear role in sympathetic neuron dendrite development in vivo (Snider, 1988; Ruit et al., 1990) although our studies did not determine which particular target genes may predominate.
The microarray analysis also identified several genes with a role in axon guidance such as Plxnd1 and Efnb1 (downregulated) and Slit2 (upregulated), suggesting that Egr3 could modulate axon guidance. While we cannot rule out that Egr3 has a role in early sympathetic axon guidance, our analysis showed only axons with abnormal branching and no evidence of axon guidance abnormalities along blood vessels. Many molecules with axon guidance functions have been shown to have other functions in neuronal processes including synapse formation and dendritic spine formation as well as axon and dendritic growth (Miller and Kaplan, 2003). For example, Slit2, which was upregulated in Egr3-deficient sympathetic neurons, can increase axon branching of sensory neurons (Wang et al., 1999; Ozdinler and Erzurumlu, 2002). Slit proteins along with their Robo receptors are also involved in dendrite development in cortical and retinal ganglion neurons (Whitford et al., 2002; Dimitrova et al., 2008). Therefore, this molecule could influence axon and dendrite development in sympathetic neurons apart from any specific role in axon guidance.
In summary, Egr3 is an NGF-induced transcriptional regulator with a sympathetic neuron autonomous function in dendrite morphogenesis and axon branching. Mice lacking Egr3 have physiologic sympathetic dysautonomia that appears to be caused by disrupted expression of a relatively small network of genes, some of which have functions related to dendrite and axon development. These results raise the possibility that similar regulatory networks may be impaired in humans with SNS disease, the majority of which are not understood and poorly treated.
Footnotes
This work was supported by National Institutes of Health Grants (NIH) R01-NS040748, K02-NS046468, and K26-OD026099 (W.G.T.). D.H.Q. was supported by NIH Grant F31-NS066721. We thank X. Gao for assistance in generating the DCτlZ transgenic mice and L. Eldredge, A. Hosanoge, and Q. Chen for technical assistance. G. Schutz generously provided the CaMKII-iCre transgenic mice.
References
- Airaksinen MS, Saarma M. The GDNF family: signalling, biological functions and therapeutic value. Nat Rev Neurosci. 2002;3:383–394. doi: 10.1038/nrn812. [DOI] [PubMed] [Google Scholar]
- Airaksinen MS, Titievsky A, Saarma M. GDNF family neurotrophic factor signaling: four masters, one servant? Mol Cell Neurosci. 1999;13:313–325. doi: 10.1006/mcne.1999.0754. [DOI] [PubMed] [Google Scholar]
- Albers KM, Wright DE, Davis BM. Overexpression of nerve growth factor in epidermis of transgenic mice causes hypertrophy of the peripheral nervous system. J Neurosci. 1994;14:1422–1432. doi: 10.1523/JNEUROSCI.14-03-01422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert Y, Whitehead J, Eldredge L, Carter J, Gao X, Tourtellotte WG. Transcriptional regulation of myotube fate specification and intrafusal muscle fiber morphogenesis. J Cell Biol. 2005;169:257–268. doi: 10.1083/jcb.200501156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwal JK, Massie B, Miller FD, Kaplan DR. The TrkB-Shc site signals neuronal survival and local axon growth via MEK and P13-kinase. Neuron. 2000;27:265–277. doi: 10.1016/s0896-6273(00)00035-0. [DOI] [PubMed] [Google Scholar]
- Baloh RH, Enomoto H, Johnson EM, Jr, Milbrandt J. The GDNF family ligands and receptors-implications for neural development. Curr Opin Neurobiol. 2000;10:103–110. doi: 10.1016/s0959-4388(99)00048-3. [DOI] [PubMed] [Google Scholar]
- Bamji SX, Majdan M, Pozniak CD, Belliveau DJ, Aloyz R, Kohn J, Causing CG, Miller FD. The p75 neurotrophin receptor mediates neuronal apoptosis and is essential for naturally occurring sympathetic neuron death. J Cell Biol. 1998;140:911–923. doi: 10.1083/jcb.140.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belteki G, Haigh J, Kabacs N, Haigh K, Sison K, Costantini F, Whitsett J, Quaggin SE, Nagy A. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 2005;33:e51. doi: 10.1093/nar/gni051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley CA, Lee KF. p75 is important for axon growth and Schwann cell migration during development. J Neurosci. 2000;20:7706–7715. doi: 10.1523/JNEUROSCI.20-20-07706.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer D, Levine-Wilkinson S, Richmond A, Hirsh S, Kuruvilla R. Wnt5a mediates nerve growth factor-dependent axonal branching and growth in developing sympathetic neurons. J Neurosci. 2009;29:7569–7581. doi: 10.1523/JNEUROSCI.1445-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers CW, Zigmond RE. Localization of neurons in the rat superior cervical ganglion that project into different postganglionic trunks. J Comp Neurol. 1979;185:381–391. doi: 10.1002/cne.901850211. [DOI] [PubMed] [Google Scholar]
- Brennan C, Rivas-Plata K, Landis SC. The p75 neurotrophin receptor influences NT-3 responsiveness of sympathetic neurons in vivo. Nat Neurosci. 1999;2:699–705. doi: 10.1038/11158. [DOI] [PubMed] [Google Scholar]
- Breunig JJ, Silbereis J, Vaccarino FM, Sestan N, Rakic P. Notch regulates cell fate and dendrite morphology of newborn neurons in the postnatal dentate gyrus. Proc Natl Acad Sci U S A. 2007;104:20558–20563. doi: 10.1073/pnas.0710156104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brose K, Tessier-Lavigne M. Slit proteins: key regulators of axon guidance, axonal branching, and cell migration. Curr Opin Neurobiol. 2000;10:95–102. doi: 10.1016/s0959-4388(99)00066-5. [DOI] [PubMed] [Google Scholar]
- Bush JO, Soriano P. Ephrin-B1 regulates axon guidance by reverse signaling through a PDZ-dependent mechanism. Genes and development. 2009;23:1586–1599. doi: 10.1101/gad.1807209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan CA, Thomas JB. Tau-beta-galactosidase, an axon-targeted fusion protein. Proc Natl Acad Sci U S A. 1994;91:5972–5976. doi: 10.1073/pnas.91.13.5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter JH, Lefebvre JM, Wiest DL, Tourtellotte WG. Redundant role for early growth response transcriptional regulators in thymocyte differentiation and survival. J Immunol. 2007;178:6796–6805. doi: 10.4049/jimmunol.178.11.6796. [DOI] [PubMed] [Google Scholar]
- Casanova E, Fehsenfeld S, Mantamadiotis T, Lemberger T, Greiner E, Stewart AF, Schütz G. A CaMKIIalpha iCre BAC allows brain-specific gene inactivation. Genesis. 2001;31:37–42. doi: 10.1002/gene.1078. [DOI] [PubMed] [Google Scholar]
- Chacón PJ, Arevalo MA, Tébar AR. NGF-activated protein tyrosine phosphatase 1B mediates the phosphorylation and degradation of I-kappa-Balpha coupled to NF-kappa-B activation, thereby controlling dendrite morphology. Mol Cell Neurosci. 2010;43:384–393. doi: 10.1016/j.mcn.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Chauvet S, Cohen S, Yoshida Y, Fekrane L, Livet J, Gayet O, Segu L, Buhot MC, Jessell TM, Henderson CE, Mann F. Gating of Sema3E/PlexinD1 signaling by neuropilin-1 switches axonal repulsion to attraction during brain development. Neuron. 2007;56:807–822. doi: 10.1016/j.neuron.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Hammonds-Odie L, Perron J, Masters BA, Bixby JL. SHP-2 mediates target-regulated axonal termination and NGF-dependent neurite growth in sympathetic neurons. Dev Biol. 2002a;252:170–187. doi: 10.1006/dbio.2002.0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HH, Tourtellotte WG, Frank E. Muscle spindle-derived neurotrophin 3 regulates synaptic connectivity between muscle sensory and motor neurons. J Neurosci. 2002b;22:3512–3519. doi: 10.1523/JNEUROSCI.22-09-03512.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgaya JM, Chan JR, Shooter EM. The neurotrophin receptor p75NTR as a positive modulator of myelination. Science. 2002;298:1245–1248. doi: 10.1126/science.1076595. [DOI] [PubMed] [Google Scholar]
- Crowley C, Spencer SD, Nishimura MC, Chen KS, Pitts-Meek S, Armanini MP, Ling LH, McMahon SB, Shelton DL, Levinson AD, et al. Mice lacking nerve growth factor display perinatal loss of sensory and sympathetic neurons yet develop basal forebrain cholinergic neurons. Cell. 1994;76:1001–1011. doi: 10.1016/0092-8674(94)90378-6. [DOI] [PubMed] [Google Scholar]
- Dimitrova S, Reissaus A, Tavosanis G. Slit and Robo regulate dendrite branching and elongation of space-filling neurons in Drosophila. Dev Biol. 2008;324:18–30. doi: 10.1016/j.ydbio.2008.08.028. [DOI] [PubMed] [Google Scholar]
- Domeniconi M, Zampieri N, Spencer T, Hilaire M, Mellado W, Chao MV, Filbin MT. MAG induces regulated intramembrane proteolysis of the p75 neurotrophin receptor to inhibit neurite outgrowth. Neuron. 2005;46:849–855. doi: 10.1016/j.neuron.2005.05.029. [DOI] [PubMed] [Google Scholar]
- Ebendal T, Tomac A, Hoffer BJ, Olson L. Glial cell line-derived neurotrophic factor stimulates fiber formation and survival in cultured neurons from peripheral autonomic ganglia. J Neurosci Res. 1995;40:276–284. doi: 10.1002/jnr.490400217. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldredge LC, Gao XM, Quach DH, Li L, Han X, Lomasney J, Tourtellotte WG. Abnormal sympathetic nervous system development and physiological dysautonomia in Egr3-deficient mice. Development. 2008;135:2949–2957. doi: 10.1242/dev.023960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto H, Crawford PA, Gorodinsky A, Heuckeroth RO, Johnson EM, Jr, Milbrandt J. RET signaling is essential for migration, axonal growth and axon guidance of developing sympathetic neurons. Development. 2001;128:3963–3974. doi: 10.1242/dev.128.20.3963. [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Flanagan JG, Vanderhaeghen P. The ephrins and Eph receptors in neural development. Annu Rev Neurosci. 1998;21:309–345. doi: 10.1146/annurev.neuro.21.1.309. [DOI] [PubMed] [Google Scholar]
- Francis N, Farinas I, Brennan C, Rivas-Plata K, Backus C, Reichardt L, Landis S. NT-3, like NGF, is required for survival of sympathetic neurons, but not their precursors. Dev Biol. 1999;210:411–427. doi: 10.1006/dbio.1999.9269. [DOI] [PubMed] [Google Scholar]
- Furrer MP, Vasenkova I, Kamiyama D, Rosado Y, Chiba A. Slit and Robo control the development of dendrites in Drosophila CNS. Development. 2007;134:3795–3804. doi: 10.1242/dev.02882. [DOI] [PubMed] [Google Scholar]
- Gao X, Daugherty RL, Tourtellotte WG. Regulation of low affinity neurotrophin receptor (p75(NTR)) by early growth response (Egr) transcriptional regulators. Mol Cell Neurosci. 2007;36:501–514. doi: 10.1016/j.mcn.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascon E, Vutskits L, Zhang H, Barral-Moran MJ, Kiss PJ, Mas C, Kiss JZ. Sequential activation of p75 and TrkB is involved in dendritic development of subventricular zone-derived neuronal progenitors in vitro. Eur J Neurosci. 2005;21:69–80. doi: 10.1111/j.1460-9568.2004.03849.x. [DOI] [PubMed] [Google Scholar]
- Glebova NO, Ginty DD. Heterogeneous requirement of NGF for sympathetic target innervation in vivo. J Neurosci. 2004;24:743–751. doi: 10.1523/JNEUROSCI.4523-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goridis C, Rohrer H. Specification of catecholaminergic and serotonergic neurons. Nat Rev Neurosci. 2002;3:531–541. doi: 10.1038/nrn871. [DOI] [PubMed] [Google Scholar]
- Hartmann M, Brigadski T, Erdmann KS, Holtmann B, Sendtner M, Narz F, Lessmann V. Truncated TrkB receptor-induced outgrowth of dendritic filopodia involves the p75 neurotrophin receptor. J Cell Sci. 2004;117:5803–5814. doi: 10.1242/jcs.01511. [DOI] [PubMed] [Google Scholar]
- Hassankhani A, Steinhelper ME, Soonpaa MH, Katz EB, Taylor DA, Andrade-Rozental A, Factor SM, Steinberg JJ, Field LJ, Federoff HJ. Overexpression of NGF within the heart of transgenic mice causes hyperinnervation, cardiac enlargement, and hyperplasia of ectopic cells. Dev Biol. 1995;169:309–321. doi: 10.1006/dbio.1995.1146. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Shneider NA, Birchmeier C, Burden SJ, Jessell TM, Arber S. A role for neuregulin1 signaling in muscle spindle differentiation. Neuron. 2002;36:1035–1049. doi: 10.1016/s0896-6273(02)01101-7. [DOI] [PubMed] [Google Scholar]
- Ho HY, Susman MW, Bikoff JB, Ryu YK, Jonas AM, Hu L, Kuruvilla R, Greenberg ME. Wnt5a-Ror-Dishevelled signaling constitutes a core developmental pathway that controls tissue morphogenesis. Proc Natl Acad Sci U S A. 2012;109:4044–4051. doi: 10.1073/pnas.1200421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking JC, Hehr CL, Bertolesi GE, Wu JY, McFarlane S. Distinct roles for Robo2 in the regulation of axon and dendrite growth by retinal ganglion cells. Mech Dev. 2010;127:36–48. doi: 10.1016/j.mod.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Honma Y, Araki T, Gianino S, Bruce A, Heuckeroth R, Johnson E, Milbrandt J. Artemin is a vascular-derived neurotropic factor for developing sympathetic neurons. Neuron. 2002;35:267–282. doi: 10.1016/s0896-6273(02)00774-2. [DOI] [PubMed] [Google Scholar]
- Honma Y, Kawano M, Kohsaka S, Ogawa M. Axonal projections of mechanoreceptive dorsal root ganglion neurons depend on Ret. Development. 2010;137:2319–2328. doi: 10.1242/dev.046995. [DOI] [PubMed] [Google Scholar]
- Howard MJ. Mechanisms and perspectives on differentiation of autonomic neurons. Dev Biol. 2005;277:271–286. doi: 10.1016/j.ydbio.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EJ, Li H, Tang AA, Wiggins AK, Neve RL, Zhong W, Jan LY, Jan YN. Targeted deletion of numb and numblike in sensory neurons reveals their essential functions in axon arborization. Genes Dev. 2005;19:138–151. doi: 10.1101/gad.1246005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikin AF, Sabo SL, Lanier LM, Buxbaum JD. A macromolecular complex involving the amyloid precursor protein (APP) and the cytosolic adapter FE65 is a negative regulator of axon branching. Mol Cell Neurosci. 2007;35:57–63. doi: 10.1016/j.mcn.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki H, Suzuki H, Itohara S. High-efficiency CAG-FLPe deleter mice in C57BL/6J background. Exp Anim. 2006;55:137–141. doi: 10.1538/expanim.55.137. [DOI] [PubMed] [Google Scholar]
- Kim JH, Roberts DS, Hu Y, Lau GC, Brooks-Kayal AR, Farb DH, Russek SJ. Brain-derived neurotrophic factor uses CREB and Egr3 to regulate NMDA receptor levels in cortical neurons. J Neurochem. 2012;120:210–219. doi: 10.1111/j.1471-4159.2011.07555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klesse LJ, Parada LF. Trks: signal transduction and intracellular pathways. Microsc Res Tech. 1999;45:210–216. doi: 10.1002/(SICI)1097-0029(19990515/01)45:4/5<210::AID-JEMT4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Knudson CM, Tung KS, Tourtellotte WG, Brown GA, Korsmeyer SJ. Bax-deficient mice with lymphoid hyperplasia and male germ cell death. Science. 1995;270:96–99. doi: 10.1126/science.270.5233.96. [DOI] [PubMed] [Google Scholar]
- Kramer ER, Knott L, Su F, Dessaud E, Krull CE, Helmbacher F, Klein R. Cooperation between GDNF/Ret and ephrinA/EphA4 signals for motor-axon pathway selection in the limb. Neuron. 2006;50:35–47. doi: 10.1016/j.neuron.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Kuruvilla R, Zweifel LS, Glebova NO, Lonze BE, Valdez G, Ye H, Ginty DD. A neurotrophin signaling cascade coordinates sympathetic neuron development through differential control of TrkA trafficking and retrograde signaling. Cell. 2004;118:243–255. doi: 10.1016/j.cell.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Lee KF, Bachman K, Landis S, Jaenisch R. Dependence on p75 for innervation of some sympathetic targets. Science. 1994;263:1447–1449. doi: 10.1126/science.8128229. [DOI] [PubMed] [Google Scholar]
- Levi-Montalcini R, Booker B. Destruction of the sympathetic ganglia in mammals by an antiserum to a nerve-growth protein. Proc Natl Acad Sci U S A. 1960;46:384–391. doi: 10.1073/pnas.46.3.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi-Montalcini R, Cohen S. Effects of the extract of the mouse submaxillary salivary glands on the sympathetic system of mammals. Ann N Y Acad Sci. 1960;85:324–341. doi: 10.1111/j.1749-6632.1960.tb49963.x. [DOI] [PubMed] [Google Scholar]
- Li L, Carter J, Gao X, Whitehead J, Tourtellotte WG. The neuroplasticity-associated arc gene is a direct transcriptional target of early growth response (Egr) transcription factors. Mol Cell Biol. 2005;25:10286–10300. doi: 10.1128/MCB.25.23.10286-10300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Yun SH, Keblesh J, Trommer BL, Xiong H, Radulovic J, Tourtellotte WG. Egr3, a synaptic activity regulated transcription factor that is essential for learning and memory. Mol Cell Neurosci. 2007;35:76–88. doi: 10.1016/j.mcn.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Eldredge LC, Quach DH, Honasoge A, Gruner K, Tourtellotte WG. Egr3 dependent sympathetic target tissue innervation in the absence of neuron death. PLoS One. 2011;6:e25696. doi: 10.1371/journal.pone.0025696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Miao T, Sebastian M, Bhullar P, Ghaffari E, Liu M, Symonds AL, Wang P. The transcription factors egr2 and egr3 are essential for the control of inflammation and antigen-induced proliferation of B and T cells. Immunity. 2012;37:685–696. doi: 10.1016/j.immuni.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YS, McLaughlin T, Sung TC, Santiago A, Lee KF, O'Leary DD. p75(NTR) mediates ephrin-A reverse signaling required for axon repulsion and mapping. Neuron. 2008;59:746–758. doi: 10.1016/j.neuron.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magin TM, McWhir J, Melton DW. A new mouse embryonic stem cell line with good germ line contribution and gene targeting frequency. Nucleic Acids Res. 1992;20:3795–3796. doi: 10.1093/nar/20.14.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer EH, Hoyle GW, Kapur RP, Brinster RL, Palmiter RD. The dopamine beta-hydroxylase gene promoter directs expression of E. coli lacZ to sympathetic and other neurons in adult transgenic mice. Neuron. 1991;7:703–716. doi: 10.1016/0896-6273(91)90274-4. [DOI] [PubMed] [Google Scholar]
- Miller FD, Kaplan DR. Signaling mechanisms underlying dendrite formation. Curr Opin Neurobiol. 2003;13:391–398. doi: 10.1016/s0959-4388(03)00072-2. [DOI] [PubMed] [Google Scholar]
- Moore MW, Klein RD, Fariñas I, Sauer H, Armanini M, Phillips H, Reichardt LF, Ryan AM, Carver-Moore K, Rosenthal A. Renal and neuronal abnormalities in mice lacking GDNF. Nature. 1996;382:76–79. doi: 10.1038/382076a0. [DOI] [PubMed] [Google Scholar]
- Moreno-Flores MT, Martín-Aparicio E, Avila J, Díaz-Nido J, Wandosell F. Ephrin-B1 promotes dendrite outgrowth on cerebellar granule neurons. Mol Cell Neurosci. 2002;20:429–446. doi: 10.1006/mcne.2002.1128. [DOI] [PubMed] [Google Scholar]
- Nam HS, Benezra R. High levels of Id1 expression define B1 type adult neural stem cells. Cell Stem Cell. 2009;5:515–526. doi: 10.1016/j.stem.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Ba-Charvet KT, Brose K, Marillat V, Kidd T, Goodman CS, Tessier-Lavigne M, Sotelo C, Chédotal A. Slit2-Mediated chemorepulsion and collapse of developing forebrain axons. Neuron. 1999;22:463–473. doi: 10.1016/s0896-6273(00)80702-3. [DOI] [PubMed] [Google Scholar]
- Niwa H, Hayakawa K, Yamamoto M, Itoh T, Mitsuma T, Sobue G. Differential age-dependent trophic responses of nodose, sensory, and sympathetic neurons to neurotrophins and GDNF: potencies for neurite extension in explant culture. Neurochem Res. 2002;27:485–496. doi: 10.1023/a:1019896502774. [DOI] [PubMed] [Google Scholar]
- Orioli D, Klein R. The Eph receptor family: axonal guidance by contact repulsion. Trends Genet. 1997;13:354–359. doi: 10.1016/s0168-9525(97)01220-1. [DOI] [PubMed] [Google Scholar]
- Ozdinler PH, Erzurumlu RS. Slit2, a branching-arborization factor for sensory axons in the mammalian CNS. J Neurosci. 2002;22:4540–4549. doi: 10.1523/JNEUROSCI.22-11-04540.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel TD, Jackman A, Rice FL, Kucera J, Snider WD. Development of sensory neurons in the absence of NGF/TrkA signaling in vivo. Neuron. 2000;25:345–357. doi: 10.1016/s0896-6273(00)80899-5. [DOI] [PubMed] [Google Scholar]
- Perrinjaquet M, Vilar M, Ibáñez CF. Protein-tyrosine phosphatase SHP2 contributes to GDNF neurotrophic activity through direct binding to phospho-Tyr687 in the RET receptor tyrosine kinase. J Biol Chem. 2010;285:31867–31875. doi: 10.1074/jbc.M110.144923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plump AS, Erskine L, Sabatier C, Brose K, Epstein CJ, Goodman CS, Mason CA, Tessier-Lavigne M. Slit1 and Slit2 cooperate to prevent premature midline crossing of retinal axons in the mouse visual system. Neuron. 2002;33:219–232. doi: 10.1016/s0896-6273(01)00586-4. [DOI] [PubMed] [Google Scholar]
- Redmond L, Oh SR, Hicks C, Weinmaster G, Ghosh A. Nuclear Notch1 signaling and the regulation of dendritic development. Nat Neurosci. 2000;3:30–40. doi: 10.1038/71104. [DOI] [PubMed] [Google Scholar]
- Rohrer H. The role of bone morphogenetic proteins in sympathetic neuron development. Drug News Perspect. 2003;16:589–596. doi: 10.1358/dnp.2003.16.9.829341. [DOI] [PubMed] [Google Scholar]
- Romero MI, Lin L, Lush ME, Lei L, Parada LF, Zhu Y. Deletion of Nf1 in neurons induces increased axon collateral branching after dorsal root injury. J Neurosci. 2007;27:2124–2134. doi: 10.1523/JNEUROSCI.4363-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotthier A, Baets J, Timmerman V, Janssens K. Mechanisms of disease in hereditary sensory and autonomic neuropathies. Nat Rev Neurol. 2012;8:73–85. doi: 10.1038/nrneurol.2011.227. [DOI] [PubMed] [Google Scholar]
- Rubin E. Development of the rat superior cervical ganglion: ganglion cell maturation. J Neurosci. 1985;5:673–684. doi: 10.1523/JNEUROSCI.05-03-00673.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruit KG, Osborne PA, Schmidt RE, Johnson EM, Jr, Snider WD. Nerve growth factor regulates sympathetic ganglion cell morphology and survival in the adult mouse. J Neurosci. 1990;10:2412–2419. doi: 10.1523/JNEUROSCI.10-07-02412.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama-Cohen P, Arévalo MA, Meier J, Grantyn R, Rodríguez-Tébar A. NGF controls dendrite development in hippocampal neurons by binding to p75NTR and modulating the cellular targets of Notch. Mol Biol Cell. 2005;16:339–347. doi: 10.1091/mbc.E04-05-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama-Cohen P, Arévalo MA, Grantyn R, Rodríguez-Tébar A. Notch and NGF/p75NTR control dendrite morphology and the balance of excitatory/inhibitory synaptic input to hippocampal neurones through Neurogenin 3. J Neurochem. 2006;97:1269–1278. doi: 10.1111/j.1471-4159.2006.03783.x. [DOI] [PubMed] [Google Scholar]
- Sestan N, Artavanis-Tsakonas S, Rakic P. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- Shneider NA, Mentis GZ, Schustak J, O'Donovan MJ. Functionally reduced sensorimotor connections form with normal specificity despite abnormal muscle spindle development: the role of spindle-derived neurotrophin 3. J Neurosci. 2009;29:4719–4735. doi: 10.1523/JNEUROSCI.5790-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaper SD. The biology of neurotrophins, signalling pathways, and functional peptide mimetics of neurotrophins and their receptors. CNS Neurol Disord Drug Targets. 2008;7:46–62. doi: 10.2174/187152708783885174. [DOI] [PubMed] [Google Scholar]
- Smeyne RJ, Klein R, Schnapp A, Long LK, Bryant S, Lewin A, Lira SA, Barbacid M. Severe sensory and sympathetic neuropathies in mice carrying a disrupted Trk/NGF receptor gene. Nature. 1994;368:246–249. doi: 10.1038/368246a0. [DOI] [PubMed] [Google Scholar]
- Smolen AJ, Beaston-Wimmer P. Dendritic development in the rat superior cervical ganglion. Brain Res. 1986;394:245–252. doi: 10.1016/0165-3806(86)90100-8. [DOI] [PubMed] [Google Scholar]
- Snider WD. Nerve growth factor enhances dendritic arborization of sympathetic ganglion cells in developing mammals. J Neurosci. 1988;8:2628–2634. doi: 10.1523/JNEUROSCI.08-07-02628.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Suehiro J, Hamakubo T, Kodama T, Aird WC, Minami T. Vascular endothelial growth factor activation of endothelial cells is mediated by early growth response-3. Blood. 2010;115:2520–2532. doi: 10.1182/blood-2009-07-233478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Ohashi R, Nakamura R, Shinmura K, Kamo T, Sakai R, Sugimura H. Tiam1 mediates neurite outgrowth induced by ephrin-B1 and EphA2. EMBO J. 2004;23:1075–1088. doi: 10.1038/sj.emboj.7600128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourtellotte WG, Milbrandt J. Sensory ataxia and muscle spindle agenesis in mice lacking the transcription factor Egr3. Nat Genet. 1998;20:87–91. doi: 10.1038/1757. [DOI] [PubMed] [Google Scholar]
- Tourtellotte WG, Keller-Peck C, Milbrandt J, Kucera J. The transcription factor Egr3 modulates sensory axon-myotube interactions during muscle spindle morphogenesis. Dev Biol. 2001;232:388–399. doi: 10.1006/dbio.2001.0202. [DOI] [PubMed] [Google Scholar]
- Tsarovina K, Schellenberger J, Schneider C, Rohrer H. Progenitor cell maintenance and neurogenesis in sympathetic ganglia involves Notch signaling. Mol Cell Neurosci. 2008;37:20–31. doi: 10.1016/j.mcn.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Tsui-Pierchala BA, Milbrandt J, Johnson EM., Jr NGF utilizes c-Ret via a novel GFL-independent, inter-RTK signaling mechanism to maintain the trophic status of mature sympathetic neurons. Neuron. 2002;33:261–273. doi: 10.1016/s0896-6273(01)00585-2. [DOI] [PubMed] [Google Scholar]
- Vinik AI, Maser RE, Ziegler D. Autonomic imbalance: prophet of doom or scope for hope? Diabet Med. 2011;28:643–651. doi: 10.1111/j.1464-5491.2010.03184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel KS, Parada LF. Sympathetic neuron survival and proliferation are prolonged by loss of p53 and neurofibromin. Mol Cell Neurosci. 1998;11:19–28. doi: 10.1006/mcne.1998.0670. [DOI] [PubMed] [Google Scholar]
- Vogel KS, Brannan CI, Jenkins NA, Copeland NG, Parada LF. Loss of neurofibromin results in neurotrophin-independent survival of embryonic sensory and sympathetic neurons. Cell. 1995;82:733–742. doi: 10.1016/0092-8674(95)90470-0. [DOI] [PubMed] [Google Scholar]
- Voyvodic JT. Peripheral target regulation of dendritic geometry in the rat superior cervical ganglion. J Neurosci. 1989;9:1997–2010. doi: 10.1523/JNEUROSCI.09-06-01997.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Brose K, Arnott D, Kidd T, Goodman CS, Henzel W, Tessier-Lavigne M. Biochemical purification of a mammalian slit protein as a positive regulator of sensory axon elongation and branching. Cell. 1999;96:771–784. doi: 10.1016/s0092-8674(00)80588-7. [DOI] [PubMed] [Google Scholar]
- Whitford KL, Marillat V, Stein E, Goodman CS, Tessier-Lavigne M, Chédotal A, Ghosh A. Regulation of cortical dendrite development by Slit-Robo interactions. Neuron. 2002;33:47–61. doi: 10.1016/s0896-6273(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Wong ST, Henley JR, Kanning KC, Huang KH, Bothwell M, Poo MM. A p75(NTR) and Nogo receptor complex mediates repulsive signaling by myelin-associated glycoprotein. Nat Neurosci. 2002;5:1302–1308. doi: 10.1038/nn975. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Tucker KL, Barde YA. Neurotrophin binding to the p75 receptor modulates Rho activity and axonal outgrowth. Neuron. 1999;24:585–593. doi: 10.1016/s0896-6273(00)81114-9. [DOI] [PubMed] [Google Scholar]
- Yan H, Newgreen DF, Young HM. Developmental changes in neurite outgrowth responses of dorsal root and sympathetic ganglia to GDNF, neurturin, and artemin. Dev Dyn. 2003;227:395–401. doi: 10.1002/dvdy.10294. [DOI] [PubMed] [Google Scholar]
- Yawo H. Changes in the dendritic geometry of mouse superior cervical ganglion cells following postganglionic axotomy. J Neurosci. 1987;7:3703–3711. doi: 10.1523/JNEUROSCI.07-11-03703.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagrebelsky M, Holz A, Dechant G, Barde YA, Bonhoeffer T, Korte M. The p75 neurotrophin receptor negatively modulates dendrite complexity and spine density in hippocampal neurons. J Neurosci. 2005;25:9989–9999. doi: 10.1523/JNEUROSCI.2492-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhu W, Wang YG, Liu XJ, Jiao L, Liu X, Zhang ZH, Lu CL, He C. Interaction of SH2-Bbeta with RET is involved in signaling of GDNF-induced neurite outgrowth. J cell science. 2006;119:1666–1676. doi: 10.1242/jcs.02845. [DOI] [PubMed] [Google Scholar]