Abstract
Cellular proteins are exposed to oxidative modification and other forms of damage through oxidative stress, disease and as a consequence of aging. This oxidative damage results in loss and or modification of protein function, which in turn compromises cell function and may even cause cell death. Therefore, the removal of damaged proteins is extremely important for the maintenance of normal cell function. The 20S Proteasome functions primarily as a system for removal of such damaged proteins. Unlike the 26S Proteasome, the 20S Proteasome exhibits a high degree of selectivity in degrading the oxidized, or otherwise damaged, forms of cell proteins. The 20S Proteasome is broadly distributed throughout the cell and has a range of specific functions in different organelles which are controlled through a number of Proteasome regulators. It is also activated, and its synthesis is induced, under conditions of enhanced oxidative stress, thus permitting permit greater removal of damaged proteins.
Keywords: 26S Proteasome, 20S Proteasome, oxidative stress, free radicals, hydrogen peroxide, 11S, Pa200, Pa28αβ, PARP, HSP70
I. Introduction
Protein damage is a natural and common aspect of life from multi-cellular endothermic mammals to single celled prokaryotes. It can be caused in many different ways through a wide range of mechanisms. Such protein damage can occur as a result of external stimuli, including exposure to oxidants present in air pollution [1, 2], and pesticides [3], by ozone [4, 5] and by various other chemical agents. Protein damage can also be caused through exposure to radiation such as UV [6] or various forms of ionizing radiation [7]. In addition to external factors, protein damage can also be caused through internally generated oxidants and reactive species which are produced during metabolism [8] or immune responses [9]. Finally, damaged proteins can be formed through transcriptional or translational errors. Such synthesis errors can occur naturally but become significantly more prevalent with age due to a positive feedback loop from accumulating damage to transcriptional and translational machinery [10].
Regardless of the mechanism through which it occurs, protein damage has many severe effects on cell function [11-15] and organismal viability [16]. Due to these severe outcomes, the rapid and efficient removal of damaged proteins is extremely important. In this chapter we discuss the role of various forms of the Proteasome in the removal of damaged proteins. This chapter will have a particular focus on the 20S Proteasome, whose main function is thought to be the removal of damaged proteins. This chapter will also briefly discuss the roles of 26S Proteasome and Immunoproteasome in the removal of damaged proteins.
II. Formation of Oxygen Radicals and Toxic Oxidants
Oxygen is one of the greatest blessings and perhaps curses to complex life, this is what is often referred to as the ‘Oxygen Paradox’[13,17]. Oxygen possesses a vital role as the final acceptor in the electron transport chain. In the electron transport chain, a series of alternating hydrogen and electron carriers ensure that electrons are vectorially transported across the inner mitochondrial membrane, such that protons are transported outwads. The resulting electrochemical gradient is then used for the formation of ATP from ADP + P. In order to maintain electron flow in the inner mitochondrial membrane, electrons must also be removed from the electron transport chain by a terminal electron acceptor. This role is played by the cytochrome c oxidase complex (mitochondrial complex IV) which oxidizes electrons with oxygen to generate water, thereby ‘bleeding’ electrons from the chain, and allowing continual ATP production. The presence of oxygen is, thus, vital for mitochondrial function and overall energy balance in eucaryotes. While there are alternative processes to generate ATP that are independent of oxygen (such as fermentation), these processes are considerably less efficient than oxidative phosphorylation. As a result of the sizable difference in ATP production rates between oxygen-dependent and oxygen-independent, pathways, it has been suggested by some groups that the increased efficiency of oxygen-dependent respiration over fermentation was one of the key factors that permitted the formation of complex multi-cellular life forms [18].
While the role of oxygen as a final electron acceptor permits a greatly enhanced energy production rate, the reaction has the potential to generate a number of highly toxic byproducts. In the electron transport chain electrons pass through a series of electron acceptors, eventually terminating with cytochrome c oxidase, which catalyzes the formation of water. The termination of the electron transport chain seems to occur, safely, with little or no detectable leakage of electrons, however some electrons do leak out at earlier points in the chain [19]. Such leaked electrons can directly react with oxygen to form the superoxide anion radical (O2•-), commonly referred to simply as superoxide. Free radicals such as O2•- possess an unpaired electron in their outer orbital (represented by the dot). These radicals have a stronger tendency to react with other molecules to either donate the exra electron or gain another, in either case restoring their electron pairing. In such oxidation/reduction (or redox) reactions, the electron donor is termed the reductant, while the electron acceptor is the oxidant. Naturally, both the oxidant and the reductant are modified, sometimes reversibly but often irreversibly, and both may go on to react further with surrounding molecules. In many cases such free radical driven, redox chain reactions produce species that cause serious damage to cellular constituents [13].
Superoxide radicals are capable of reacting with iron or copper ions in the cell so converting them from Fe3+ to Fe2+ or Cu2+ to Cu+. This is harmful in itself as it results in the modification of metal ions which form an important catalytic centers of a wide range of cellular enzymes. In addition, the Fe2+ or Cu+ generated by this reaction is then capable of reacting (by the Fenton reaction [20]) with the mild oxidant hydrogen peroxide (H2O2) to form highly toxic hydroxyl radicals (•OH) This is termed the Haber-Weis reaction [21].
Superoxide can instead react with two protons and another electron to form hydrogen peroxide. Hydrogen peroxide is a relatively mild oxidant, but it can cause modification of DNA, lipids, and proteins through reactions catalyzed by transition metals. Hydrogen peroxide also appears to be capable of modifications to some amino acids chains in proteins (e.g. amino acids containing thiol groups [22] or keto-acids such as pyruvate [23]) . More importantly hydrogen peroxide is capable of reacting with additional electrons to form hydroxyl radicals which are highly toxic and capable of a reacting with a wide range of proteins as well as lipids and DNA [24, 25]. Superoxide can also react with nitric oxide radicals (NO•), which are produced constantly as vasodilating agents and signaling molecules [26] to form peroxynitrite (ONOO-). This is detrimental both through the reduction in nitric oxide signaling [27] and through the formation of peroxynitrite which is a powerful oxidant. (common free radical reactions are summarized in Figure 1).
Figure 1.
Formation of Oxygen Radicals and Reactive Oxidants
In addition to oxidative stress occurring through the formation of free radicals and oxidizing agents in metabolism, oxidative stress can occur through contact with a range of environmental factors. One of these environmental factors is air pollution. Air pollution contains a range of metals which can catalyze the formation of reactive oxidizing agents, such as iron, copper, chromium and vanadium, through the Haber-Weis reaction [28-30]. Many particles in air pollution can also produce an inflammatory response, which in part involves the formation of oxygen radicals [31]. UV radiation is another source of oxidative stress, the main source of which is solar irradiation, but there are also a range of lifestyle related sources of exposure. The degree of solar exposure to UV radiation is dependent on a range of environmental factors such as cloud coverage and ozone level. UV radiation can cause the formation of both superoxide (O2•-) and hydroxyl radicals (•OH). In addition it can cause direct modification of proteins within the cell. Similar effects are also seen with infra-red and ionizing radiation [32].
III. Removal of Oxygen Radicals and Toxic Oxidants by the Cell
Various oxygen and nitrogen radicals, as well as other reactive oxygen or nitrogen species, can (directly or indirectly) modify amino acids within proteins, damage vital prosthetic groups, or oxidize key transition metal centers. Although direct modification of amino acids or protein prosthetic groups typically increases local hydrophilicity (often changing or introducing charges), the net effect of protein oxidative modification is typically partial unfolding, which exposes the side chains of many hydrophobic amino acids that are normally buried in the interior of a properly folded protein. Thus, when free radicals, or related reactive species, modify a protein they usually cause an increase in surface hydrophobicity and an increase in accessibility of protein hydrophobic groups.
While some mild modifications might be relatively harmless, other changes will reduce, inhibit or even modify the function of the protein [13, 33, 34]. If severely oxidatively modified proteins are allowed to accumulate, cell function will be progressively compromised [13]. Thus, it is extremely important for any aerobic cell to have a system for removing oxidatively damaged proteins. It is worth noting that both DNA and lipid oxidation also present a serious challenge to maintenance of cell function and aerobic cells have well developed systems for combating these types of damage as well. For the purpose of this chapter however we will focus solely on protein oxidation.
The cells’ first line of defense is the removal of oxygen radicals and the detoxification of reactive oxidants, to stop proteins becoming modified in the first place. Hydrogen peroxide is a relatively mild oxidant, although it can be reduced to generate hydroxyl radicals which are considerably more reactive (figure 1). Because of this it is important for hydrogen peroxide to be removed. This is done primarily through glutathione peroxidase [35], which couples the reduction of hydrogen peroxide into water with the oxidation of glutathione (GSH). The oxidized glutathione will then react with another oxidized glutathione to form glutathione disulfide (GSSG). The glutathione disulfide is later reduced back to two glutathione molecules in a reaction catalyzed by glutathione reductase [36]. Catalase is another powerful enzyme that can remove hydrogen peroxide, but its actions seem limited to peroxisomes in eucaruotic cells.
In addition to hydrogen peroxide, superoxide is also capable of reacting with copper, iron or nitric oxide radicals. As a result of this, even with efficient removal of hydrogen peroxide through glutathione peroxidase, there are other pathways by which protein damage can occur. In response to this cells have a evolved superoxide dismutase (SOD) [37]. SOD catalyzes the formation of hydrogen peroxide from oxygen radicals so decreasing the chance of the superoxide reacting with copper, iron, or nitric oxide. Although it may seem counter-intuitive for cells to produce SOD that generates hydrogen peroxide, it seems that the combination of SOD and glutathione peroxidase ensures that most of the superoxide produced is converted harmlessly to water without going through the hydroxyl radical step in the univalent pathway for oxygen reduction. Minimizing the production of hydroxyl radical, the most powerful oxygen radical of biological significance, seems to be the strategy that our cells employ.
In addition, there is a range of other anti-oxidant mechanisms, largely based on antioxidant compounds derived from fruits and vegetables in the diet, that is utilized by the cell. Ascorbic acid (vitamin C), is one of these, it is involved in a similar reaction to glutathione in which it function as an electron acceptor for oxygen radicals resulting in its conversion to dehydroascorbic acid (DHA). Dehydroascorbic acid may subsequently be reduced back to ascorbic acid by a reaction with glutathione in the endoplasmic reticulum [38]. Another example is vitamin E [19] which functions as a scavenger of peroxyl radicals. Many other phenolic or polyphenolic dietary components have also been suggested as important biological antioxidants.
IV. The Role of 20S Proteasome vs 26S Proteasome in Degrading Oxidized Proteins
Although the first-line defenses of anti-oxidant enzymes and compounds described above are quite effective, they are not perfect, and some oxygen radicals and other reactive oxygen species do get through and cause protein damage. To combat this problem, the cell has mechanisms in place to remove damaged proteins and enable normal cell function to continue. The Proteasome plays an important role in the removal of these damaged proteins. It has been shown that when a cellular protein is damaged (up to a point) it will be preferentially degraded, compared to a non-damaged protein [39, 40]. When Proteasome is removed from the cell, using immunoprecipitation, or depleted using anti-sense RNA or siRNA, most of the cells’ ability to remove damaged proteins is lost, suggesting that Proteasome is important in the removal of damaged cellular proteins [39, 41].
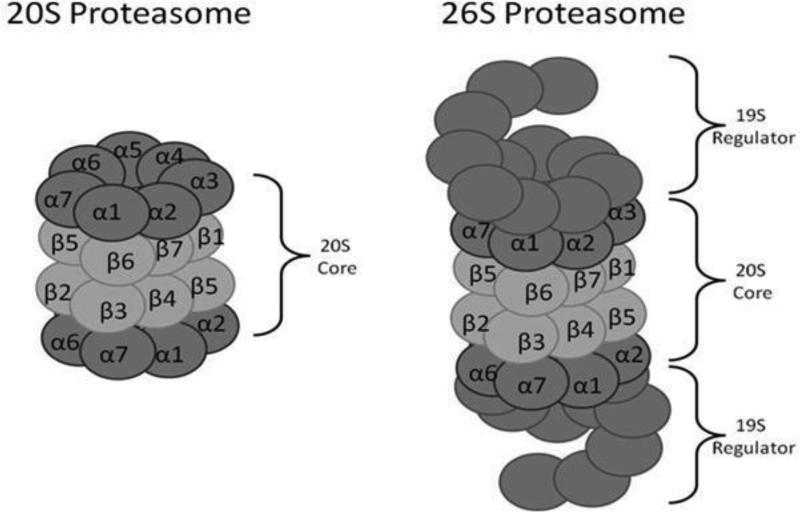
Proteasome comes in different forms, with many different potential activators or regulators. A strong candidate for the removal of damaged proteins is the 20S Proteasome. The four-ringed 20S Proteasome is the basic or core form of the Proteasome, to which two 19S regulators can be added (one at either end of the four-ringed 20S ‘barrel or cylinder’) to form the 26S Proteasome (Figure 2). The 20S Proteasome is a seven hundred kilodalton barrel shaped protein composed of four rings, each made up of seven subunits ranging from twenty to thirty five kilodaltons. The first ring contains alpha subunits1,2,3,4,5,6, and 7 while the second ring contains beta subunits 1,2,3,4,5,6, and 7. The third ring again contains the beta 1-7 subunits, and the fourth ring again has the alpha 1-7 subunits. Thus, the 20S Proteasome barrel or cylinder is symmetrical along its long axis, with the structure, α ring, β ring, β ring, α ring. All the proteolytic activities of the Proteasome are found in the two β rings, where the β1, β2 and β5 subunits each possess’ specific proteolytic activities. In the 26S Proteasome, 19S regulators are bound at either end of the 20S Proteasome barrel (Figure 2). The 19S regulator serves to feed proteins into the Proteasome core and to restrict the proteins fed through to the core, to only those which posses a poly-Ubiquitin tag. Degradation polyubiquinated proteins by the 26S Proteasome occurs in an ATP/Mg2+ dependent manner. In the 20S Proteasome, which is free of 19S regulatory caps, proteins are still capable of binding to the Proteasome and being fed into the core however degradation occurs in a manner that is ATP/Ubiquitin independent [42, 43].
Figure 2.
Structures of the 20S and 26S Proteasomes, based on diagrams published in McNaught et al 2001 [94]
Not only is the degradation of proteins by the 20S Proteasome ATP/Ubiquitin independent, but studies using purified Proteasome have shown that 20S Proteasome will selectively degrade damaged or oxidized proteins over native proteins. The 20S Proteasome shows a selective preference for degradation of oxidized over non-oxidized proteins for a wide range of proteins [39, 41, 44]. The 26S Proteasome by comparison does not appear in mammalian cells, at least, to have any selective preference for degradation of oxidized proteins [39]. In addition to this, while depletion of 20S Proteasome results in an approximately eighty percent loss of capacity to degrade oxidized proteins, there is only a twenty percent reduction in capacity to degrade oxidized proteins, with depletion of 26S Proteasome from a cell [41].
The 20S Proteasome appears to be quite resistant to oxidative stress, in comparison with other proteins. In fact, in studies of its capacity to degrade oxidized proteins. under conditions of oxidative stress, its I50 appears to be four times higher than the level of oxidation to a target protein that would result in optimum selective degradation [44]. This shows that 20S Proteasome can easily function under the non-toxic degrees of oxidative stress to which cells are normally exposed. In comparison, the I50 of purified 26S Proteasome under oxidative stress appears to be four to sixteen times lower than that of the 20S Proteasome [44]. Similarly, in cell culture a level of oxidation that completely blocks ATP-stimulated 26S Proteasome activity does not appear to significantly affect 20S Proteasome activity [45]. As a result, the 26S Proteasome is actually inactivated under very mild degree's of oxidation compared to the 20S Proteasome [44, 45].
In conclusion both 20S and the 26S Proteasome are capable of degrading damaged proteins, however 26S Proteasome does not appear to have any selectivity for damaged over native proteins. Under conditions where protein damage occurs, the 26S Proteasome is easily inactivated compared to the much hardier 20S Proteasome which is able to function at most relevant levels of cellular stress. Finally, if 20S Proteasome is depleted from the cell, the majority of the capacity to remove oxidized proteins is lost, compared to a very minor loss following the removal of the 26S Proteasome. From these results it appears that the 20S Proteasome is the major system for removal of oxidized proteins, compared to a relatively minor role played by the 26S Proteasome.
V. Mechanism of Selective Degradation of Damaged Proteins by the 20S Proteasome
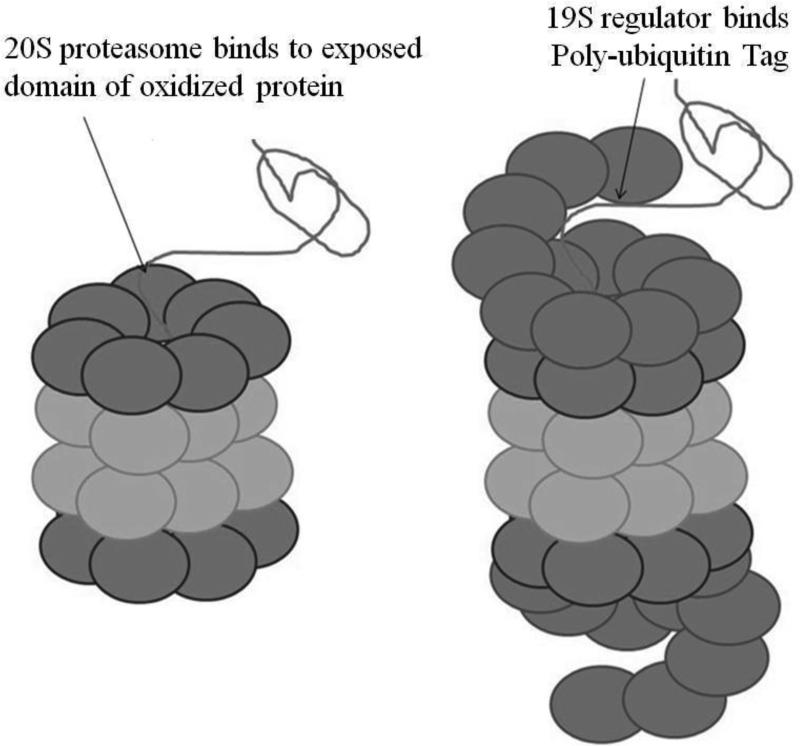
The 20S Proteasome core is composed of an inner pair of rings of β subunits containing the proteoltytic subunits which are positioned within the lumen of the Proteasome core (Fig. 3). Above and below the β rings are rings of α subunits which are not proteolytically active [46]. The α rings provide a scaffold for the proteolytic β rings, and are the binding sites for a variety of activators and regulators (including the 19S regulators). The α rings also function as gates to the Proteasome, these gates prevent correctly folded proteins from entering and becoming degraded [47]. In the case of the 26S Proteasome there are caps on each of the alpha rings known as 19S regulators. The 19S regulators act like magnets for ubiquitin-tagged proteins, whose polyubiquitin tails bind and initiate degradation of the tagged protein substrates. The 19S regulators remove the polyubiquitin tails, force the alpha rings open, and feed the substrate proteins into the Proteasome core β rings where they are degraded [48]. The 26S Proteasome is responsible for most of the turnover of normal proteins within cells. The key factor is the structure of each protein, which determines its susceptibility to polyubiquitinylation by so called ‘E3’ enzymes. Basically, proteins which are highly susceptible to ubiquitinylation are rapidly degraded by the 26S Proteasome and, thus, have short half-lives. In contrast, proteins which are more resistant to ubiquitinylation will have longer cellular half-lives. As the 19S regulator is absent in the 20S Proteasome, substrate proteins must be fed through in a different way.
Figure 3.
Degradation of Proteins by the 20S and 26S Proteasomes
As discussed above, a most proteolysis of damaged or oxidized proteins is dependent on the 20S Proteasome but independent of the 26S Proteasome. While 20S Proteasome is not capable of degrading undamaged ubiquitin-tagged proteins as does the 26S Proteasome, if the proteins are already unfolded then the 20S Proteasome appears to be just is as capable as the 26S Proteasome of degrading such substrates [47]. This indicates proteins are fed into 20S Proteasome in the same way that they are into 26S Proteasome however 20S Proteasome does not have the structures possessed by the 26S Proteasome to recognize and unfold poly-ubiquitinated proteins. The 20S Proteasome does appear to have its own system for recognition and unfolding of target proteins; specifically it will recognize and unfold damaged or oxidized proteins. As discussed earlier, protein damage results in the exposure of hydrophobic domains which would not normally interact with the aqueous environment. These exposed residues are thought to act as sites of recognition for the 20S Proteasome in the same way as polyubiqutin tags act as sites of recognition for the 26S Proteasome (Figure 3). These domains permit binding of the damaged protein to the 20S Proteasome. The protein is then unfolded and fed into the core where it may be degraded [47]. However, in the 26S Proteasome this selectivity for damaged proteins is lost because the outer Proteasome α rings are covered by the 19S regulators which bind polyubiquitin.
VI. The 20S Proteasome in Aging and the Removal of Age-related Protein Damage
Over the course of aging there is a progressive decline in protein synthesis [49]. Despite this decline in synthesis there is actually an increase in protein content with age. This discrepancy is the product of a buildup of damaged non-functioning proteins within cells [49]. This buildup of damaged proteins appears to derive from protein oxidation as well as a range of other modifications including; glycation, methylation, ADP-ribosylation, deamidation. etc. [49]. The increase in damaged proteins in cells is extremely detrimental and decreases the functionality of all pathways in the cell. This forms part of the widely popular “Free Radical Theory of Aging.” This theory suggests that many of the effects of aging are at least in part the product of accumulating oxidative damage within a cell which results in deterioration of cell function and eventually death [50].
Fortunately much of the accumulated damage can be removed and the damaged proteins can be degraded and replaced by non-damaged ones. In fact, a mild degree of modification or damage to a protein makes it a better candidate for degradation by the 20S Proteasome or other proteases [33, 39, 41, 51]. However, if a protein becomes too heavily modified it becomes a very poor candidate for degradation [33, 39, 40]. Thus, while many mildly oxidized proteins are readily degradable, at least some of the age-associated (or time-associated) accumulation of damaged proteins is due to proteins which are so highly modified that they are difficult or impossible to degrade. It has been argued that it is the buildup of these non-degradable damaged proteins that causes age-related effects, however, the accumulation of oxidized proteins in cells is exponential rather than linear over time, indicating that the rise in protein oxidation is not just a product of a buildup of indigestible proteins, but a potentially reversible change in cell function [52].
The accumulation of oxidized or otherwise damaged proteins in cells during aging could be a product either of a rise in damaging conditions or a fall in the rate of removal of damaged proteins. it has been observed that over age there is a rise in mitochondrial generation of oxidants [53]. In addition, it has been shown that there is a decline in protein turnover. This decline in protein turnover is, at least in part, the product of a sharp decrease in 20S Proteasome function over age (which has been shown in a range of different tissues) [54-59]. In addition to a decline in 20S Proteasome function there is also a drop in 20S Proteasome levels over in aging which also reduces protein turnover [60-62]. The decline in 20S Proteasome function is partly the product of an increase in modification or damage to the 20S Proteasome over the course of age [54, 59-61]. For instance, it has been seen that 20S Proteasome isolated from old rats is 50% less proteolytically active than 20S Proteasome isolated from young rats [63]. As a result, not only is there a decrease in the amount of 20S Proteasome present during the aging process but there is also a decrease in the ability of the remaining 20S Proteasome to degrade the accumulating damaged proteins, thus resulting in an overall accumulation of damaged cellular proteins with age. As a result, older rats are less able to remove damaged proteins from their cells and tissues than are younger rats, which goes some way to explain the difference in the levels of protein damage found in the two animals.
In studies of long lived animals, such as two long live bat species [64], it has been argued that a large factor of their longer lifespan, compared to similar shorter lived animals, is a product of higher 20S Proteasome levels [64, 65]. The higher levels of expression of 20S Proteasome results in lower levels of accumulation of oxidized proteins and greater resistance to oxidative stress which has been argued as sufficient to account for the differences in lifespan [64, 65]. Similarly in studies with naked mole rats (which are the longest living rodent species) there was also a very high expression of 20S Proteasome, resulting in greater oxidative stress resistance compared to similar rodents; this was suggested to potentially account for at least part of their extremely long lifespan [66]. Similarly, while mice show a progressive rise in accumulation of protein damage over age, there is almost no change in levels of protein damage over the course of age in the naked mole rat, which is thought to be a product of their enhanced 20S Proteasome levels and activity [66]. These results provide at least correlative and speculative evidence for an important role of the 20S Proteasome as a determinant of lifespan through its ability to minimize the accumulation of damaged proteins in an organism as it ages.
VII. The 20S Proteasome in Adaptation to Protein Damage
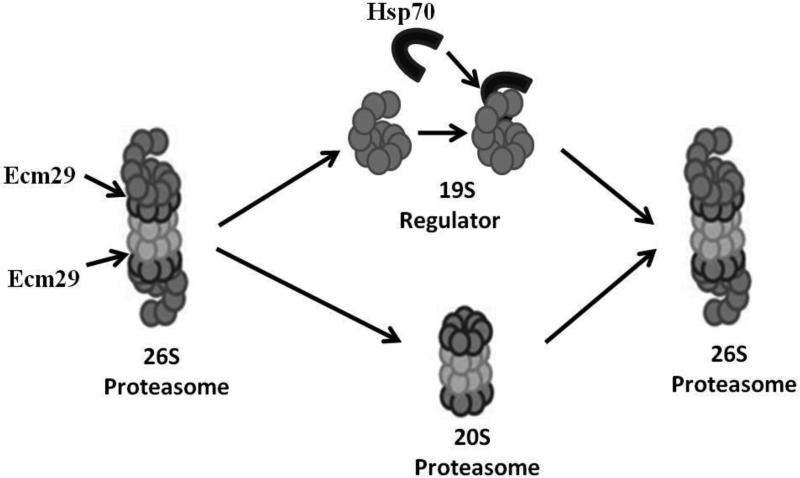
Cells or organisms are frequently exposed to a low level of oxidative stress or protein damage though, occasionally, they will experience much greater stress. Even for a short time, a large stress will result in greater accumulation of damaged proteins, which is highly damaging to the cell. To combat this accumulation, cells appear to have a transient response in which capacity for degrading oxidized proteins is increased initially in a protein synthesis independent manner (0.5 h -5 h after oxidative stress exposure). This is followed by protein synthesis dependent increase in proteolytic capacity (5h - 48h after oxidative stress exposure) [41, 67]. These responses make the cell much more resistant to normally toxic conditions of protein damage. The initial protein synthesis independent (unaffected by cycloheximide) response is mostly blocked when Proteasome inhibitors are used, indicating that Proteasome plays an important role [67]. This increase in capacity to degrade oxidized proteins appears to coincide with a marked reduction in ATP-stimulated proteolytic capacity which is subsequently restored three to five hours later. It appears that this response is the product of a disassociation or disassembly of the 26S Proteasome into free 19S regulators and 20S Proteasomes. After the stress has abated and/or the protein synthesis dependent response has begun, it is important for 26S Proteasome to be reformed so that normal cell function can return. Indeed. three to five hours after the initial stress, the 26S Proteasome appear to re-form from the free 19S regulator and 20S Proteasome (Figure 4). A number of chaperones appear to be involved in this process, and in the mammalian system hsp70 has been shown to be important in stabilizing the 19S regulator after dissociation from 20S Proteasomes, and has been shown to be important for the re-assembly of functional 26S Proteasomes [67]. In yeast, the Ecm29 protein has been shown to bind to the 19S regulator following dissociation (it may actually cause dissociation) and is required for the increases in survival and ability to degrade damaged proteins [68]. We suggest that Ecm29 may cause 26S disassembly, and hsp70 may protect 19S regulators and allow 26S reassembly in both yeast and mammals (Figure 4), and we are currently testing this hypothesis.
Figure 4.
Disassembly and Reassembly of the 26S Proteasome in Response to Oxidative Stress
In addition to the transient response of 26S Proteasome dissociation/reassembly there is also a slower adaptive response to a stress. Over the 5 to 48 hours following oxidative stress, cellular capacity to remove damaged proteins and cellular tolerance to damaging conditions, increases significantly [41, 67, 69]. This response, like the initial 0.5 to 5 hour response, described above, is largely blocked using Proteasome inhibitors [41, 67]. However, unlike the earlier response, this transient adaptive increase in stress-resistance is accompanied by de novo synthesis of 20S Proteasome, and is blocked by siRNA against Proteasome subunit genes, and by general protein synthesis inhibitors such as cycloheximide. Thus, it appears that the adaptive response is at least in part the product of an increase in de novo synthesis of 20S Proteasome. Such transient, and reversible, adaptive responses have also been called hormesis. Importantly, if new 20S Proteasome synthesis is blocked using siRNA then the increase in capacity to degrade oxidized proteins, as well as the enhanced resistance to oxidative stress, are both blocked, indicating that the increase in 20S Proteasome is important for the adaptive response. In addition to 20S Proteasome these is also an increase in synthesis both of the Immunoproteasome and the Pa28αβ Proteasome regulator which have also both been shown to be important for adaptation.
VIII. Removal of Damaged Proteins by the 20S Proteasome in the Nucleus
In addition to its presence in the cytoplasm, there is also a large amount of 20S Proteasome in the nucleus which plays a very important role in stress resistance. As in the cytoplasm, 20S Proteasome in the nucleus is very important in the removal of oxidized or otherwise damaged proteins such that normal cell function can continue. In addition to these roles, because of the specialist nature of many proteins in the nucleus, nuclear 20S Proteasome has additional specific functions. These nuclear-specific functions are regulated through Proteasome regulators that are only found in the nucleus.
The Nuclear 20S Proteasome Regulator: PARP
As in the cytoplasm, the nucleus appears to have a rapid response system to oxidative stress. In fact, in just fifteen minutes following an oxidative stress there is an eighteen fold increase in Proteasome dependent proteolytic activity in the nucleus [70]. It has been shown that this is not due to de novo protein synthesis but due to modifications to the 20S Proteasome to make it not only more proteolytically active but also more capable of specifically degrading oxidized histones [70]. The selective and very specific degradation of oxidized histones is very important for DNA repair during oxidative stress and, therefore, very important for cell survival and minimizing mutations. The histones around which DNA is wrapped play an important role in control of DNA expression [71], and so it is very important that any damage to the histone complexes in the chromatin be repaired as well as the DNA itself. In fact, damaged histones must actually be removed from the DNA before DNA repair can effectively begin, and Proteasome is the enzyme which catalyzes this selective proteolytic removal of damaged histone proteins.
As noted above, following an oxidative stress there is a very large and rapid increase in Proteasome dependent proteolytic activity in the nucleus [70], and this change in Proteasome activity is regulated by another protein called poly-ADP ribose polymerase (PARP). Following stress, PARP functions by transferring ADP moieties off NAD+ onto other proteins [72]. PARP is believed to use this mechanism heavily as a means of signaling DNA damage and inducing its repair [73]. It appears that the 20S Proteasome can also act as a substrate for modification by PARP. It was shown in studies using purified 20S Proteasome that ADP ribosylation of 20S Proteasome by PARP resulted in an increase in selectivity for degradation of oxidized histones, which accounted for the changes in proteolytic selectivity and increased activity observed under oxidative stress of the nucleus. Furthermore, the increase in proteolytic activity in the nucleus under oxidative stress is lost with inhibition of PARP activity indicating that PARP is a key regulator of Proteasome in the nucleus [70].
The Nuclear 20S/26S Proteasome Regulator: Pa200
Another regulator of nuclear 20S Proteasome is the Pa200 (also known as PSME4) Proteasome regulator. This is a 200 kilodalton activator of Proteasome activity (hence Pa200) that is highly conserved throughout the eukaryotic domain [74, 75]. Pa200 is a nuclear-localized regulator of Proteasome. Like PARP, Pa200 appears to be involved in repair of DNA damage. Unlike PARP, however, which appears to assist DNA damage repair through activating the 20S Proteasome to increase preferential degradation of damaged histones, the role which Pa200 plays is not entirely clear [70, 74]. It has been seen in studies using the yeast homologue of Pa200 (Blm10), that mutation of Blm10 results in cells becoming much less resistant to DNA damage [76]. Similarly in mammalian culture, the depletion of Pa200 results in greater DNA damage susceptibility to ionizing radiation [77]. Interestingly, although Pa200 is referred to as a Proteasome activator, because of its ability to enhance the 20S Proteasome's ability to degrade short peptides, it does not appear to enhance the ability of 20S Proteasome to degrade whole proteins [74]. In at least a partial elucidation of its role, it appears that under cell damage through ionizing radiation or hydrogen peroxide treatment, there is a rapid formation of foci of Pa200 on the DNA chromatin. The formation of these DNA foci correlates with an increase in Pa200 binding to either 20S Proteasome or so called hybrid Proteasome. This hybrid Proteasome is Proteasome on which at one end there is a 19S regulator and at the other end the Pa200 regulator [74, 77]. It is unclear whether Pa200 functions mainly as a regulator of 20S Proteasome or as a regulator of 26S Proteasome through the formation of hybrid Proteasomes. It is also unclear whether Pa200 functions primarily though degradation of damaged proteins in the chromatin or through triggering some form of DNA repair mechanism. It is clear, however, that whatever its actual role, it is a highly conserved, regulator of Proteasome in response to oxidation or other damage in the nucleus.
The Nuclear 20S Proteasome Regulator: Pa28γ
The final regulator of 20S Proteasome in the nucleus is the Proteasome regulator Pa28γ (also known as REGγ, 11sγ, or PSME3). There is no evidence at present of a role for Pa28γ in removal of damaged proteins. The Pa28γ regulator forms a homoheptameric ring on either end of the 20S Proteasome. It is nuclear-localized and is a genetic ortholog of the better studied Pa28αβ regulator [78]. It weakly enhances the 20S Proteasomes ability to degrade short peptides, Its main function however is largely unknown. Pa28γ has been shown to be important in cell cycle progression as well as apoptosis [79]. It has also been shown to be a critical regulator of p53 expression during a stress response [80] however its full role is largely unknown.
IX. Alternative Forms of the 20S Proteasome in the Cytoplasm and Other Non-nuclear Compartments and their Role(s) in Removal of Damaged Proteins
Immunoproteasome and Oxidative Stress
Another form of the Proteasome which has been suggested to be involved in proteolytic removal of oxidized proteins is the Immunoproteasome (Figure 5), described in more detail in other chapters. This form of Proteasome differs from 20S Proteasome by the presence of three alternative subunits (β1i, β2i and β5i) sometimes known as (lmp2, mecl-1 and lmp7). These alternative subunits result in differences in the sites of cleavage of proteins degraded by the Immunoproteasome compared to the 20S Proteasome, which will give a different range of peptides [81, 82]. The peptides generated by the Immunoproteasome, because of their composition and structure, are favored as peptides for MHC class 1 antigen presentation [83, 84]. Expression of the Immunoproteasome is strongly induced by the regulator interferon-γ. Interferon-γ is a cell signaling molecule that is important in both innate and adaptive immune response [85]. This along with the localization of the Immunoproteasome to the endoplasmic reticulum has led to the belief that it functions primarily as a means of generating MHC class 1 peptides for cell surface presentation [83, 84]. This theory of the Immunoproteasome's role being primarily that of immune response has however come into question recently with the observation that interferon-γ itself produces an oxidative stress response. It is entirely possible than that interferon-γ induced synthesis of Immunoproteasome might actually be an oxidative stress response rather than an immune response [86].
Figure 5.
Subunit Composition of the 20S Proteasome and the Immunoproteasome
In addition to a hypothesized role for the Immunoproteasome in immune response it appears that it possesses a role in degradation of oxidized proteins. The Immunoproteasome has been shown to be as capable as the 20S Proteasome of selectively degrading oxidized proteins [41] and its expression does increase under conditions of protein damage [41, 55, 87, 88]. It has been hypothesized that protein oxidation might function as a universal marker for selection of proteins for MHC antigen presentation This ‘PrOxI hypothesis’ [83] suggests a potential explanation of, and reason for Immunoproteasome's capacity to selectively degrade oxidized proteins. It also seems that Immunoproteasome can, like the 20S Proteasome, play a role in the removal of damaged proteins as part of cellular damage repair. In support of this it has been shown that the Immunoproteasome, like the 20S Proteasome, is inducible during oxidative stress and in aging. Furthermore, if Immunoproteasome is removed or depleted in certain cells then those cells are more susceptible and less adaptable to oxidative stress [41, 89, 90]. As a result, while the role of the Immunoproteasome remains somewhat controversial, several studies have provided highly suggestive evidence that the Immunoproteasome plays a role in the selective removal of oxidized proteins from the cell.
The cytoplasmic Proteasome Activator Pa28αβ
The Proteasome regulator Pa28αβ, also known as REGαβ, 11Sαβ or PSME1 and PSME2, is an activator of 20S Proteasome activity. Pa28αβ is localized to the cytoplasm and forms a heteroheptameric ring composed of three alpha subunits and four beta subunits [91]. This ring can fit on either end of the core of the 20S Proteasome. When associated with the 20S Proteasome it induces a large increase in the capacity to degrade short peptides [78]. The regulator Pa28αβ has been shown to be important in the process of oxidative stress adaptation. Like 20S Proteasome there is a strong increase in production of Pa28αβ when cells are exposed to an adaptive dose of an oxidant [41]. It has also been shown, using mutants of Pa28αβ and using siRNA to block Pa28αβ expression, that the adaptive response to oxidative stress is notably weaker and there is much less of an increase in the capacity of cells to degrade oxidized proteins without expression of functional Pa28αβ. This indicates that Pa28αβ plays an important role both in oxidative stress adaptation and in stimulating the degradation of oxidized proteins [41]. There is also a strong suggestion for a potential role for Pa28αβ in MHC class I antigen presentation. Like the Immunoproteasome Pa28αβ is regulated by interferon-γ [92]. This has lead to some suggestion that the two might act together with Pa28αβ forming a cap on Immunoproteasome to enable or assist MHC class I antigen presentation of self and non-self antigens [83]. Additionally, it has been shown that increases in Pa28αβ expression result in enhanced recognition of viral antigens by cytotoxic T-cells [93].
X. Summary
Cells accumulate protein damage through exposure to environmental toxins, physiological stresses, protein synthesis errors, and as a product of age. These damaged proteins reduce cell function and, in some cases, can even threaten cell viability. To allow normal cell function to return, and prevent toxicity, the 20S Proteasome functions to remove such damaged proteins and minimize their tendancy to aggregate, cross-link, and accumulate. Unlike the 26S Proteasome, the 20S Proteasome is capable of selectively degrading the oxidatively damaged forms of cellular proteins. It is also more resilient to the conditions where damage would occur (oxidative stress) and with the removal or inhibition of 20S Proteasome cells appear poorly equipped to remove damaged proteins. As a result, while the 26S Proteasome is highly important in regulation of normal cell function, it appears not to play a major role in the removal of damaged proteins.
The removal of damaged proteins by the 20S Proteasome, as well as being important for maintenance of normal cell function, appears to have an important role in aging. It seems that part of the decline in cell and organismal function as a product of age is through an increase in protein damage due to diminished 20S Proteasome levels and activity. As well as aging and normal cell maintanence, the 20S Proteasome appears to be important in transient and reversible adaptation to stress. Thus, 20S Proteasome activity is not static but is directly activated by oxidants, and its synthesis is inducible by (oxidant) signaling molecules such as hydrogen peroxide, resulting in an enhanced resistance to protein damage and enhanced proteolytic removal of damaged proteins.
There are also several regulators of the 20S Proteasome which are important for specific functions in different compartments. PARP binds to 20S Proteasome in response to oxidative stress and enhances removal of damaged histones. Pa200 which also binds to Proteasome in the nucleus appears to have some role in response to DNA damage but its function is unclear. Pa28αβ appears to enhance the Proteasome's proteolytic capacity in the cytoplasm and is important in oxidative stress responses. Similar to the 20S Proteasome, the Immunoproteasome has also been found to selectively degrade oxidized proteins. Immunoproteasome is also transiently and reversibly induced by oxidative stress, and we suggest that it is also important in the overall degradation of damaged proteins.
References
- 1.Halliwell B, Hu ML, Louie S, Duvall TR, Tarkington BK, Motchnik P, Cross CE. Interaction of nitrogen dioxide with human plasma. Antioxidant depletion and oxidative damage. FEBS Lett. 1992;313:62–66. doi: 10.1016/0014-5793(92)81185-o. [DOI] [PubMed] [Google Scholar]
- 2.Menzel DB. The toxicity of air pollution in experimental animals and humans: the role of oxidative stress. Toxicol Lett. 1994;72:269–277. doi: 10.1016/0378-4274(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 3.Abdollahi M, Ranjbar A, Shadnia S, Nikfar S, Rezaie A. Pesticides and oxidative stress: a review. Med Sci Monit. 2004;10:RA141–147. [PubMed] [Google Scholar]
- 4.Cross CE, Motchnik PA, Bruener BA, Jones DA, Kaur H, Ames BN, Halliwell B. Oxidative damage to plasma constituents by ozone. FEBS Lett. 1992;298:269–272. doi: 10.1016/0014-5793(92)80074-q. [DOI] [PubMed] [Google Scholar]
- 5.Cross CE, Reznick AZ, Packer L, Davis PA, Suzuki YJ, Halliwell B. Oxidative damage to human plasma proteins by ozone. Free Radic Res Commun. 1992;15:347–352. doi: 10.3109/10715769209049150. [DOI] [PubMed] [Google Scholar]
- 6.Hu ML, Tappel AL. Potentiation of oxidative damage to proteins by ultraviolet-A and protection by antioxidants. Photochem Photobiol. 1992;56:357–363. doi: 10.1111/j.1751-1097.1992.tb02171.x. [DOI] [PubMed] [Google Scholar]
- 7.Leach JK, Van Tuyle G, Lin PS, Schmidt-Ullrich R, Mikkelsen RB. Ionizing radiation-induced, mitochondria-dependent generation of reactive oxygen/nitrogen. Cancer Res. 2001;61:3894–3901. [PubMed] [Google Scholar]
- 8.Kappus H. Oxidative stress in chemical toxicity. Arch Toxicol. 1987;60:144–149. doi: 10.1007/BF00296968. [DOI] [PubMed] [Google Scholar]
- 9.Baeuerle PA, Rupec RA, Pahl HL. Reactive oxygen intermediates as second messengers of a general pathogen response. Pathol Biol (Paris) 1996;44:29–35. [PubMed] [Google Scholar]
- 10.Orgel LE. The maintenance of the accuracy of protein synthesis and its relevance to ageing. Proc Natl Acad Sci U S A. 1963;49:517–521. doi: 10.1073/pnas.49.4.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies KJA, Delsignore ME, Lin SW. Protein damage and degradation by oxygen radicals. II. Modification of amino acids. J Biol Chem. 1987;262:9902–9907. [PubMed] [Google Scholar]
- 12.Davies KJA, Delsignore ME. Protein damage and degradation by oxygen radicals. III. Modification of secondary and tertiary structure. J Biol Chem. 1987;262:9908–9913. [PubMed] [Google Scholar]
- 13.Davies KJA. Oxidative stress: the paradox of aerobic life. Biochem Soc Symp. 1995;61:1–31. doi: 10.1042/bss0610001. [DOI] [PubMed] [Google Scholar]
- 14.Davies KJA. Protein damage and degradation by oxygen radicals. I. general aspects. J Biol Chem. 1987;262:9895–9901. [PubMed] [Google Scholar]
- 15.Davies KJA, Lin SW, Pacifici RE. Protein damage and degradation by oxygen radicals. IV. Degradation of denatured protein. J Biol Chem. 1987;262:9914–9920. [PubMed] [Google Scholar]
- 16.Copeland JM, Cho J, Lo T, Jr., Hur JH, Bahadorani S, Arabyan T, Rabie J, Soh J, Walker DW. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr Biol. 2009;19:1591–1598. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 17.Davies KJA, Ursini F. The oxygen paradox. Cleup University Press; 1995. [Google Scholar]
- 18.Raymond J, Segre D. The effect of oxygen on biochemical networks and the evolution of complex life. Science. 2006;311:1764–1767. doi: 10.1126/science.1118439. [DOI] [PubMed] [Google Scholar]
- 19.Halliwell B, Gutteridge MC. Free Radicals in Biology and Medicine. 3rd Edition Oxford University Press; 2000. [Google Scholar]
- 20.Fenton HJH. Oxidation of tartaric acid in presence of iron. J. Chem. Soc., Trans. 1894;65:899–910. [Google Scholar]
- 21.Haber F, Weiss J. Über die Katalyse des Hydroperoxydes. Die Naturwissenschaften. 1932;20:948–950. [Google Scholar]
- 22.Brodie AE, Reed DJ. Reversible oxidation of glyceraldehyde 3-phosphate dehydrogenase thiols in human lung carcinoma cells by hydrogen peroxide. Biochem Biophys Res Commun. 1987;148:120–125. doi: 10.1016/0006-291x(87)91084-9. [DOI] [PubMed] [Google Scholar]
- 23.Holleman MAF. Notice sur l'action de l'eau oxygénée sur les acides a-cétoniques et sur les dicétones. ecl Trav Chim Pays Bas Belg. 1904;23:169–171. [Google Scholar]
- 24.Cadet J, Delatour T, Douki T, Gasparutto D, Pouget JP, Ravanat JL, Sauvaigo S. Hydroxyl radicals and DNA base damage. Mutat Res. 1999;424:9–21. doi: 10.1016/s0027-5107(99)00004-4. [DOI] [PubMed] [Google Scholar]
- 25.Wolff SP, Garner A, Dean RT. Free radicals, lipids and protein degradation. Trends in Biochemical Sciences. 1986;11:27–31. [Google Scholar]
- 26.Garthwaite J, Boulton CL. Nitric oxide signaling in the central nervous system. Annu Rev Physiol. 1995;57:683–706. doi: 10.1146/annurev.ph.57.030195.003343. [DOI] [PubMed] [Google Scholar]
- 27.Moncada S, Higgs EA. Molecular mechanisms and therapeutic strategies related to nitric oxide. Faseb J. 1995;9:1319–1330. [PubMed] [Google Scholar]
- 28.Lloyd DR, Carmichael PL, Phillips DH. Comparison of the formation of 8-hydroxy-2′-deoxyguanosine and single- and double-strand breaks in DNA mediated by fenton reactions. Chemical research in toxicology. 1998;11:420–427. doi: 10.1021/tx970156l. [DOI] [PubMed] [Google Scholar]
- 29.Ghio AJ, Pritchard RJ, Lehmann JR, Winsett DW, Hatch GE. Lung inflammation after exposure to nonfibrous silicates increases with chelatable [Fe3+]. Journal of toxicology and environmental health. 1996;49:11–28. doi: 10.1080/009841096160961. [DOI] [PubMed] [Google Scholar]
- 30.Ghio AJ, Stonehuerner J, Dailey LA, Carter JD. Metals associated with both the water-soluble and insoluble fractions of an ambient air pollution particle catalyze an oxidative stress. Inhalation toxicology. 1999;11:37–49. doi: 10.1080/089583799197258. [DOI] [PubMed] [Google Scholar]
- 31.Becker S, Dailey LA, Soukup JM, Grambow SC, Devlin RB, Huang YC. Seasonal variations in air pollution particle-induced inflammatory mediator release and oxidative stress. Environmental health perspectives. 2005;113:1032–1038. doi: 10.1289/ehp.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schröder P, Krutmann J. The Handbook of Environmental Chemistry, Volume 2. Springer-Verlag; Berlin Heidelberg: 2005. Environmental Oxidative Stress – Environmental Sources of ROS. pp. 19–31. [Google Scholar]
- 33.Davies KJA. Protein modification by oxidants and the role of proteolytic enzymes. Biochem Soc Trans. 1993;21:346–353. doi: 10.1042/bst0210346. [DOI] [PubMed] [Google Scholar]
- 34.Davies KJA. An overview of oxidative stress. IUBMB Life. 2000;50:241–244. doi: 10.1080/713803723. [DOI] [PubMed] [Google Scholar]
- 35.Mills GC. The purification and properties of glutathione peroxidase of erythrocytes. J Biol Chem. 1959;234:502–506. [PubMed] [Google Scholar]
- 36.Carlberg I, Mannervik B. Glutathione reductase. Methods Enzymol. 1985;113:484–490. doi: 10.1016/s0076-6879(85)13062-4. [DOI] [PubMed] [Google Scholar]
- 37.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 38.Nishikimi M, Yagi K. Biochemistry and molecular biology of ascorbic acid biosynthesis. Subcell Biochem. 1996;25:17–39. doi: 10.1007/978-1-4613-0325-1_2. [DOI] [PubMed] [Google Scholar]
- 39.Davies KJA. Degradation of oxidized proteins by the 20S Proteasome. Biochimie. 2001;83:301–310. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- 40.Davies KJA, Goldberg AL. Proteins damaged by oxygen radicals are rapidly degraded in extracts of red blood cells. J Biol Chem. 1987;262:8227–8234. [PubMed] [Google Scholar]
- 41.Pickering AM, Koop AL, Teoh CY, Ermak G, Grune T, Davies KJA. The Immunoproteasome, the 20S Proteasome and the PA28alphabeta Proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem J. 2010;432:585–594. doi: 10.1042/BJ20100878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S Proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 43.Ciechanover A. The ubiquitin-Proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 44.Reinheckel T, Sitte N, Ullrich O, Kuckelkorn U, Davies KJA, Grune T. Comparative resistance of the 20S and 26S Proteasome to oxidative stress. Biochem J. 1998;335(Pt 3):637–642. doi: 10.1042/bj3350637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reinheckel T, Ullrich O, Sitte N, Grune T. Differential impairment of 20S and 26S Proteasome activities in human hematopoietic K562 cells during oxidative stress. Arch Biochem Biophys. 2000;377:65–68. doi: 10.1006/abbi.2000.1717. [DOI] [PubMed] [Google Scholar]
- 46.Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, Tsukihara T. Structure determination of the constitutive 20S Proteasome from bovine liver at 2.75 A resolution. J Biochem. 2002;131:171–173. doi: 10.1093/oxfordjournals.jbchem.a003084. [DOI] [PubMed] [Google Scholar]
- 47.Liu CW, Corboy MJ, DeMartino GN, Thomas PJ. Endoproteolytic activity of the Proteasome. Science. 2003;299:408–411. doi: 10.1126/science.1079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Navon A, Goldberg AL. Proteins are unfolded on the surface of the ATPase ring before transport into the Proteasome. Mol Cell. 2001;8:1339–1349. doi: 10.1016/s1097-2765(01)00407-5. [DOI] [PubMed] [Google Scholar]
- 49.Rattan SI. Synthesis, modifications, and turnover of proteins during aging. Exp Gerontol. 1996;31:33–47. doi: 10.1016/0531-5565(95)02022-5. [DOI] [PubMed] [Google Scholar]
- 50.Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 51.Davies KJA. Oxidative stress, antioxidant defenses, and damage removal, repair, and replacement systems. IUBMB Life. 2000;50:279–289. doi: 10.1080/713803728. [DOI] [PubMed] [Google Scholar]
- 52.Stadtman ER. Protein oxidation and aging. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- 53.Sohal RS, Dubey A. Mitochondrial oxidative damage, hydrogen peroxide release, and aging. Free Radic Biol Med. 1994;16:621–626. doi: 10.1016/0891-5849(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 54.Chondrogianni N, Gonos ES. Proteasome dysfunction in mammalian aging: steps and factors involved. Exp Gerontol. 2005;40:931–938. doi: 10.1016/j.exger.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 55.Ferrington DA, Husom AD, Thompson LV. Altered Proteasome structure, function, and oxidation in aged muscle. Faseb J. 2005;19:644–646. doi: 10.1096/fj.04-2578fje. [DOI] [PubMed] [Google Scholar]
- 56.Husom AD, Peters EA, Kolling EA, Fugere NA, Thompson LV, Ferrington DA. Altered Proteasome function and subunit composition in aged muscle. Arch Biochem Biophys. 2004;421:67–76. doi: 10.1016/j.abb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 57.Viteri G, Carrard G, Birlouez-Aragon I, Silva E, Friguet B. Age-dependent protein modifications and declining Proteasome activity in the human lens. Arch Biochem Biophys. 2004;427:197–203. doi: 10.1016/j.abb.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 58.Ponnappan U, Zhong M, Trebilcock GU. Decreased Proteasome-mediated degradation in T cells from the elderly: A role in immune senescence. Cell Immunol. 1999;192:167–174. doi: 10.1006/cimm.1998.1418. [DOI] [PubMed] [Google Scholar]
- 59.Carrard G, Dieu M, Raes M, Toussaint O, Friguet B. Impact of ageing on Proteasome structure and function in human lymphocytes. Int J Biochem Cell Biol. 2003;35:728–739. doi: 10.1016/s1357-2725(02)00356-4. [DOI] [PubMed] [Google Scholar]
- 60.Keller JN, Huang FF, Markesbery WR. Decreased levels of Proteasome activity and Proteasome expression in aging spinal cord. Neuroscience. 2000;98:149–156. doi: 10.1016/s0306-4522(00)00067-1. [DOI] [PubMed] [Google Scholar]
- 61.Bulteau AL, Petropoulos I, Friguet B. Age-related alterations of Proteasome structure and function in aging epidermis. Exp Gerontol. 2000;35:767–777. doi: 10.1016/s0531-5565(00)00136-4. [DOI] [PubMed] [Google Scholar]
- 62.Petropoulos I, Conconi M, Wang X, Hoenel B, Bregegere F, Milner Y, Friguet B. Increase of oxidatively modified protein is associated with a decrease of Proteasome activity and content in aging epidermal cells. J Gerontol A Biol Sci Med Sci. 2000;55:B220–227. doi: 10.1093/gerona/55.5.b220. [DOI] [PubMed] [Google Scholar]
- 63.Conconi M, Friguet B. Proteasome inactivation upon aging and on oxidation-effect of HSP 90. Mol Biol Rep. 1997;24:45–50. doi: 10.1023/a:1006852506884. [DOI] [PubMed] [Google Scholar]
- 64.Salmon AB, Leonard S, Masamsetti V, Pierce A, Podlutsky AJ, Podlutskaya N, Richardson A, Austad SN, Chaudhuri AR. The long lifespan of two bat species is correlated with resistance to protein oxidation and enhanced protein homeostasis. Faseb J. 2009;23:2317–2326. doi: 10.1096/fj.08-122523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chondrogianni N, Gonos ES. Proteasome function determines cellular homeostasis and the rate of aging. Adv Exp Med Biol. 2011;694:38–46. doi: 10.1007/978-1-4419-7002-2_4. [DOI] [PubMed] [Google Scholar]
- 66.Perez VI, Buffenstein R, Masamsetti V, Leonard S, Salmon AB, Mele J, Andziak B, Yang T, Edrey Y, Friguet B, et al. Protein stability and resistance to oxidative stress are determinants of longevity in the longest-living rodent, the naked mole-rat. Proc Natl Acad Sci U S A. 2009;106:3059–3064. doi: 10.1073/pnas.0809620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grune T, Catalgol B, Licht A, Ermak G, Pickering AM, Ngo JK, Davies KJA. HSP70 mediates dissociation and reassociation of the 26S Proteasome during adaptation to oxidative stress. Free Radic Biol Med. 2011 doi: 10.1016/j.freeradbiomed.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang X, Yen J, Kaiser P, Huang L. Regulation of the 26S Proteasome complex during oxidative stress. Sci Signal. 2010;3:ra88. doi: 10.1126/scisignal.2001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wiese AG, Pacifici RE, Davies KJA. Transient adaptation of oxidative stress in mammalian cells. Arch Biochem Biophys. 1995;318:231–240. doi: 10.1006/abbi.1995.1225. [DOI] [PubMed] [Google Scholar]
- 70.Ullrich O, Reinheckel T, Sitte N, Hass R, Grune T, Davies KJA. Poly-ADP ribose polymerase activates nuclear Proteasome to degrade oxidatively damaged histones. Proc Natl Acad Sci U S A. 1999;96:6223–6228. doi: 10.1073/pnas.96.11.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 72.Banasik M, Komura H, Shimoyama M, Ueda K. Specific inhibitors of poly(ADP-ribose) synthetase and mono(ADP-ribosyl)transferase. J Biol Chem. 1992;267:1569–1575. [PubMed] [Google Scholar]
- 73.Piskunova TS, Yurova MN, Ovsyannikov AI, Semenchenko AV, Zabezhinski MA, Popovich IG, Wang ZQ, Anisimov VN. Deficiency in Poly(ADP-ribose) Polymerase-1 (PARP-1) Accelerates Aging and Spontaneous Carcinogenesis in Mice. Curr Gerontol Geriatr Res. 2008:754190. doi: 10.1155/2008/754190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ustrell V, Hoffman L, Pratt G, Rechsteiner M. PA200, a nuclear Proteasome activator involved in DNA repair. Embo J. 2002;21:3516–3525. doi: 10.1093/emboj/cdf333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baumeister W, Walz J, Zuhl F, Seemuller E. The Proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 76.Schmidt M, Haas W, Crosas B, Santamaria PG, Gygi SP, Walz T, Finley D. The HEAT repeat protein Blm10 regulates the yeast Proteasome by capping the core particle. Nat Struct Mol Biol. 2005;12:294–303. doi: 10.1038/nsmb914. [DOI] [PubMed] [Google Scholar]
- 77.Blickwedehl J, Agarwal M, Seong C, Pandita RK, Melendy T, Sung P, Pandita TK, Bangia N. Role for Proteasome activator PA200 and postglutamyl Proteasome activity in genomic stability. Proc Natl Acad Sci U S A. 2008;105:16165–16170. doi: 10.1073/pnas.0803145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li J, Rechsteiner M. Molecular dissection of the 11S REG (PA28) Proteasome activators. Biochimie. 2001;83:373–383. doi: 10.1016/s0300-9084(01)01236-6. [DOI] [PubMed] [Google Scholar]
- 79.Rechsteiner M, Hill CP. Mobilizing the proteolytic machine: cell biological roles of Proteasome activators and inhibitors. Trends Cell Biol. 2005;15:27–33. doi: 10.1016/j.tcb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 80.Zhang Z, Zhang R. Proteasome activator PA28 gamma regulates p53 by enhancing its MDM2-mediated degradation. Embo J. 2008;27:852–864. doi: 10.1038/emboj.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fruh K, Gossen M, Wang K, Bujard H, Peterson PA, Yang Y. Displacement of housekeeping Proteasome subunits by MHC-encoded LMPs: a newly discovered mechanism for modulating the multicatalytic proteinase complex. Embo J. 1994;13:3236–3244. doi: 10.1002/j.1460-2075.1994.tb06625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Belich MP, Glynne RJ, Senger G, Sheer D, Trowsdale J. Proteasome components with reciprocal expression to that of the MHC-encoded LMP proteins. Curr Biol. 1994;4:769–776. doi: 10.1016/s0960-9822(00)00174-3. [DOI] [PubMed] [Google Scholar]
- 83.Teoh CY, Davies KJA. Potential roles of protein oxidation and the Immunoproteasome in MHC class I antigen presentation: the ‘PrOxI’ hypothesis. Arch Biochem Biophys. 2004;423:88–96. doi: 10.1016/j.abb.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 84.Tanaka K, Kasahara M. The MHC class I ligand-generating system: roles of immunoProteasomes and the interferon-gamma-inducible Proteasome activator PA28. Immunol Rev. 1998;163:161–176. doi: 10.1111/j.1600-065x.1998.tb01195.x. [DOI] [PubMed] [Google Scholar]
- 85.Fruh K, Yang Y. Antigen presentation by MHC class I and its regulation by interferon gamma. Curr Opin Immunol. 1999;11:76–81. doi: 10.1016/s0952-7915(99)80014-4. [DOI] [PubMed] [Google Scholar]
- 86.Watanabe Y, Suzuki O, Haruyama T, Akaike T. Interferon-gamma induces reactive oxygen species and endoplasmic reticulum stress at the hepatic apoptosis. J Cell Biochem. 2003;89:244–253. doi: 10.1002/jcb.10501. [DOI] [PubMed] [Google Scholar]
- 87.Ferrington DA, Hussong SA, Roehrich H, Kapphahn RJ, Kavanaugh SM, Heuss ND, Gregerson DS. ImmunoProteasome responds to injury in the retina and brain. J Neurochem. 2008;106:158–169. doi: 10.1111/j.1471-4159.2008.05345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kotamraju S, Matalon S, Matsunaga T, Shang T, Hickman-Davis JM, Kalyanaraman B. Upregulation of immunoProteasomes by nitric oxide: potential antioxidative mechanism in endothelial cells. Free Radic Biol Med. 2006;40:1034–1044. doi: 10.1016/j.freeradbiomed.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 89.Ethen CM, Hussong SA, Reilly C, Feng X, Olsen TW, Ferrington DA. Transformation of the Proteasome with age-related macular degeneration. FEBS Lett. 2007;581:885–890. doi: 10.1016/j.febslet.2007.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hussong SA, Kapphahn RJ, Phillips SL, Maldonado M, Ferrington DA. ImmunoProteasome deficiency alters retinal Proteasome's response to stress. J Neurochem. 113:1481–1490. doi: 10.1111/j.1471-4159.2010.06688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang Z, Krutchinsky A, Endicott S, Realini C, Rechsteiner M, Standing KG. Proteasome activator 11S REG or PA28: recombinant REG alpha/REG beta hetero oligomers are heptamers. Biochemistry. 1999;38:5651–5658. doi: 10.1021/bi990056+. [DOI] [PubMed] [Google Scholar]
- 92.Fabunmi RP, Wigley WC, Thomas PJ, DeMartino GN. Interferon gamma regulates accumulation of the Proteasome activator PA28 and immunoProteasomes at nuclear PML bodies. J Cell Sci. 2001;114:29–36. doi: 10.1242/jcs.114.1.29. [DOI] [PubMed] [Google Scholar]
- 93.Groettrup M, Soza A, Eggers M, Kuehn L, Dick TP, Schild H, Rammensee HG, Koszinowski UH, Kloetzel PM. A role for the Proteasome regulator PA28alpha in antigen presentation. Nature. 1996;381:166–168. doi: 10.1038/381166a0. [DOI] [PubMed] [Google Scholar]
- 94.McNaught KS, Olanow CW, Halliwell B, Isacson O, Jenner P. Failure of the ubiquitin-proteasome system in Parkinson's disease. Nat Rev Neurosci. 2001;2:589–594. doi: 10.1038/35086067. [DOI] [PubMed] [Google Scholar]