Abstract
Introduction
Cardiac myosin binding protein-C (cMyBP-C) becomes dephosphorylated in the failing heart and reduced phosphorylation-dependent regulation of cMyBP-C has been implicated in contractile dysfunction. To date, several phosphorylation sites have been identified for human cMyBP-C; however, a comprehensive characterization of the cMyBP-C phosphoproteome is lacking. This study aimed to characterize the cMyBP-C phosphoproteome using two different proteomic-based methods in explanted donor and end-stage failing hearts.
Methods
The first approach used to characterize the cMyBP-C phosphoproteome employed a strong-cation exchange chromatography (SCX) fractionation method (10 pooled samples, technical replicates = 4) and the second employed a sodium dodecylsulfate polyacrylamide gel electrophoresis method (n = 10; technical replicates = 2). Each subsequently underwent titanium dioxide (TiO2) affinity chromatography to enrich for the tryptic phosphopeptides, which were analyzed using an LTQ-Orbitrap mass spectrometer. Moreover, recombinant C0-C2 fragment of mouse cMyBP-C incubated with PKA, PKC, CaMKII and CK2 was analyzed to identify the kinases involved with phosphorylation of cMyBP-C.
Results
Seventeen phosphorylation sites on cMyBP-C were identified in vivo, with the majority localized in the N-terminal domains C0-C2. The three most abundant phosphorylated sites, Ser284, Ser286 and Thr290, are located in the regulatory M-domain of cMyBP-C. Ser284 showed a significant reduction in phosphorylation in HF.
Conclusion
This study demonstrates that cMyBP-C harbours more phosphorylation sites than previously known, with a total of 17 (9 novel) identified phosphorylation sites in vivo. Most sites were primarily located within the N-terminal side of the protein. The most highly phosphorylated site on cMyBP-C was Ser284 and this site showed decreased phosphorylation in the failing heart, which implicate importance for fine-tuning contractility. To date, the functional importance of Ser286 and Thr290 is unknown. In addition, 16 sites were identified after in vitro kinase incubation. The data have been deposited to the ProteomeXchange with identifier PXD000158
Keywords: cardiac myosin binding protein-C, cMyBP-C, phosphorylation, phosphoproteome, mass spectrometry, strong-cation exchange chromatography, titanium dioxide
1. Introduction
Cardiac myosin binding protein-C (cMyBP-C) is a 140-kDa thick-filament associated protein consisting of 11 domains (C0-C10), a proline-alanine rich region between C0-C1 and a unique regulatory sequence, the M-domain, which is located between C1-C2. Cardiac MyBP-C influences cardiac contractility and can regulate acto-myosin interactions and influence the rate of power contraction [1]. Cardiac MyBP-C knock-out mice developed cardiac hypertrophy with contractile dysfunction, indicating that cMyBP-C is necessary for normal cardiac function [2]. Furthermore, multiple mutations in cMyBP-C are responsible for a substantial proportion of genotyped cases of familial hypertrophic cardiomyopathy [3].
The M-domain of cMyBP-C harbors several phosphorylation sites (Ser133, Ser275, Ser284, Ser304 and Ser311) that have been identified as substrates for glycogen synthase kinase 3 beta (GSK3β), protein kinase A (PKA), C (PKC) or Ca2+-dependent calmodulin kinase II (CaMKII) [4-6]. The overall phosphorylation levels of the sites located within the M-domain are reduced in failing hearts [6-8], indicating that reduced phosphorylation-dependent regulation of cMyBP-C may play a role in the underlying mechanism of heart failure (HF). In agreement with this, transgenic mice expressing cMyBP-C ablated for phosphorylation of Ser273, Ser282 and Ser302 (by mutating Serine to Alanine; mouse sequence) resulted in cardiac hypertrophy and contractile dysfunction [9]. Furthermore, PKA phosphorylation of cMyBP-C accelerates cross-bridge kinetics and loss of this regulation leads to cardiac dysfunction [10]. The N-terminus of cMyBP-C interacts with F-actin in a phosphorylation sensitive manner, suggesting that reversible interactions of cMyBP-C with actin could contribute to modulation of cross-bridge kinetics [11].
Although the importance of cMyBP-C phosphorylation for optimal contractile performance has been established, and the few known phosphorylation sites have been intensely investigated, a comprehensive characterization of the cMyBP-C phosphoproteome is lacking. This study aimed to elucidate the phosphorylation sites on cMyBP-C in tissue obtained from human explanted donor and end-stage failing hearts. We used a strong-cation exchange chromatography (SCX) fractionation method [12] coupled with titanium dioxide (TiO2) [13] to enrich for phosphopeptides and identified 17 phosphorylation sites (10 novel sites) on cMyBP-C. A second method using sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) to separate cMyBP-C allowed us to quantify the most abundant phosphorylation sites. This method demonstrates that the most abundantly phosphorylated site on cMyBP-C, Ser284, is reduced in the failing heart. Moreover, we identified phosphorylation sites of CaMKII, CK2 and PKA in vitro on the C0-C2 fragment of cMyBP-C.
2. Methods
Tissue was used from explanted donor and end-stage HF hearts from patients with idiopathic dilated cardiomyopathy (IDCM). All tissues used in this study were obtained after informed consent and with approval of the local Ethical Committees (Human Research Ethics approval from St. Vincent's Hospital (H03/118) and from The University of Sydney (#7326). Overall cMyBP-C phosphorylation levels in explanted donor (n=8) and end-stage failing hearts (n=8) (and dephosphorylated sample as negative control) untreated and treated with PKA were analyzed using phos-tag acrylamide gels (FMS Laboratory; Hiroshima University, Japan).
A comprehensive analysis of the cMyBP-C phosphoproteome was performed using SCX-fractionation and SDS-PAGE gel electrophoresis fractionation of cMyBP-C. The SCX-based method (10 pooled samples, technical replicates = 4) provided high coverage of the cMyBP-C phosphoproteome, but required large protein amounts, whereas the SDS-PAGE method (n = 10; technical replicates = 2) allowed us to quantify the most abundant sites but provided low coverage of the cMyBP-C phosphoproteome. TiO2 was used to enrich for phosphopeptides for both methods. All enriched tryptic peptides were analyzed by LC-MS/MS on an Agilent 1200 nano-LC system in front of an LTQ-Orbitrap mass spectrometer (Thermo Scientific). Each MS1 scan was followed by collision induced dissociation (CID, acquired in the LTQ) of the 5 most abundant precursor ions. Only MS1 signals exceeding 1000 counts triggered the MS2 scans. Raw MS/MS data were batch searched against the Human IPI database version 3.79 using the Sorcerer 2™-SEQUEST® algorithm (Sage-N Research, Milpitas, CA, USA). Data were further analyzed using Scaffold PTM version 1.1.2. The data is available in Pride (http://www.ebi.ac.uk/pride/) through accession number 24805 (Reviewer account: Username = review78541, Password = aNz2Rwf8).
Recombinant mouse cMyBP-C fragment C0-C2 (2 μg) was incubated with 200U protein kinase A (PKA), 2000U Calcium calmodulin kinase II (CaMKII), 1000U Casein kinase 2 (CK2) or 0.2U of protein kinase C (PKC) for 3hrs at 30°C. Protein was digested using trypsin and peptides were analyzed in triplicate on an EASY-nLC 1000 connected to an Orbitrap Elite (Thermo) equipped with a nanoelectrospray ion source. Database searching and processing was performed as described above using the mouse IPI database version 3.80.
A detailed expanded methods description is available in the online data supplement.
3. Results and Discussion
3.1 Assessment of cMyBP-C phosphorylation from healthy and failing tissue using phos-tag acrylamide gel electrophoresis
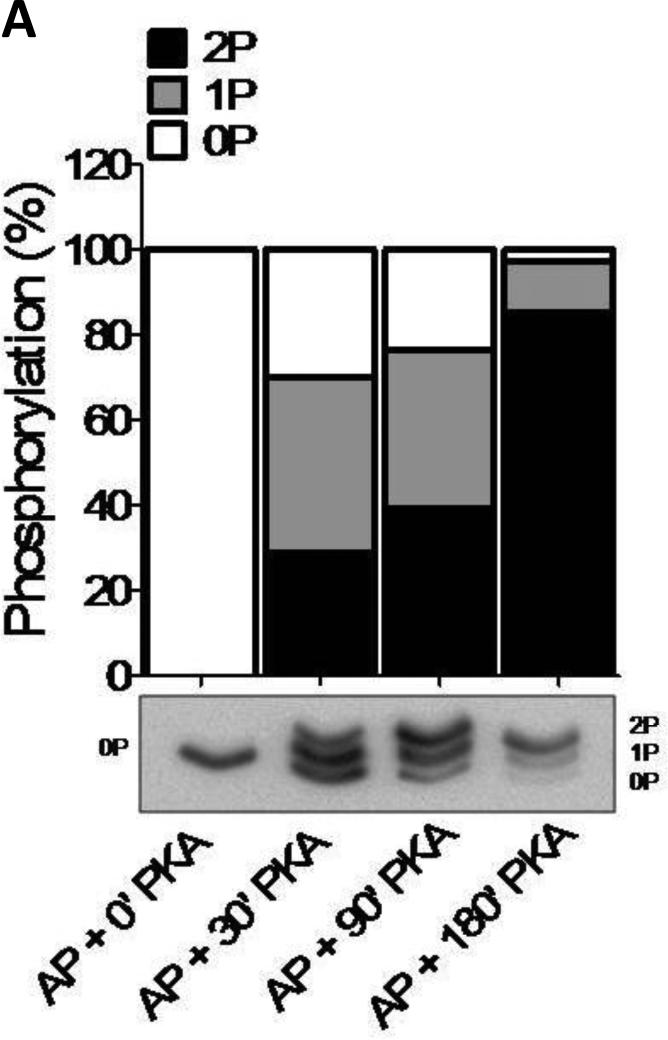
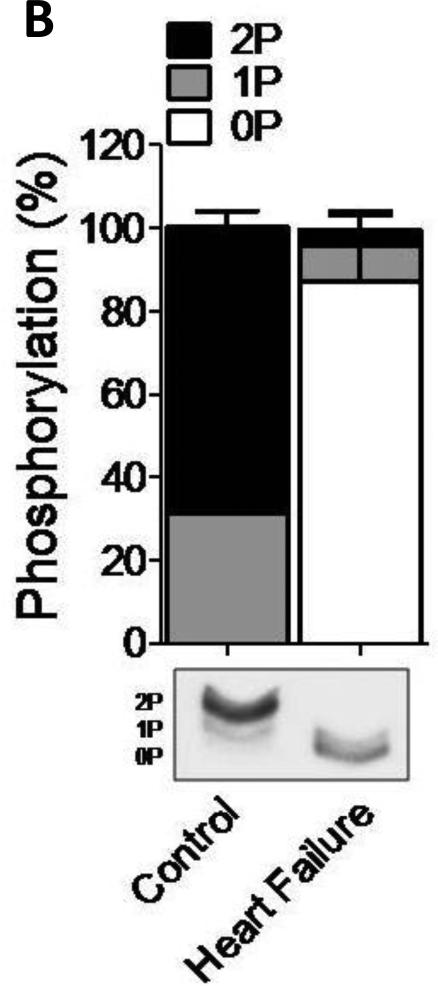
Phos-tag gel-based analysis was carried out to establish the level of phosphorylation of the human samples (n=8) treated with PKA for up to 180 minutes (Fig. 1). To validate the use of phos-tag gels for cMyBP-C, myofilament proteins were dephosphorylated using alkaline phosphatase (AP) alone or subsequently incubated with PKA (Fig. 1A). An antibody against cMyBP-C amino acid residues 2-14 was used as it recognizes “total” cMyBP-C [14;15]. The results show dephosphorylated cMyBP-C at time point ‘0’ and increased phosphorylation after 30, 90 and 180 minutes of incubation with PKA (Fig. 1A). Although four sites on cMyBP-C (Ser275, Ser284, Ser304 and Ser311, based on human sequence (Uniprot #Q14896)) have been identified as PKA substrates [5], the phos-tag gel only showed 3 bands (Fig.1B). These most likely correspond to the un-, mono- and di-phosphorylated forms of cMyBP-C based on the migration of the AP treated (dephosphorylated) sample. This reduced number of phosphorylated species of cMyBP-C compared to the potential known (and any unknown sites) might be due to the low abundance of phosphorylation at residues, which could contribute to higher phosphorylation levels. These low abundant forms could, therefore, be below the detection level of the phos-tag gels. Alternatively, it is feasible that there can only be one or two sites phosphorylated at any given time. Regardless, the cardiac samples derived from patients with end-stage HF (n=8) showed significantly reduced phosphorylation compared to the samples derived from donor (n=8) (two-way ANOVA, *p<0.05) using phos-tag acrylamide gels (Fig. 1B).
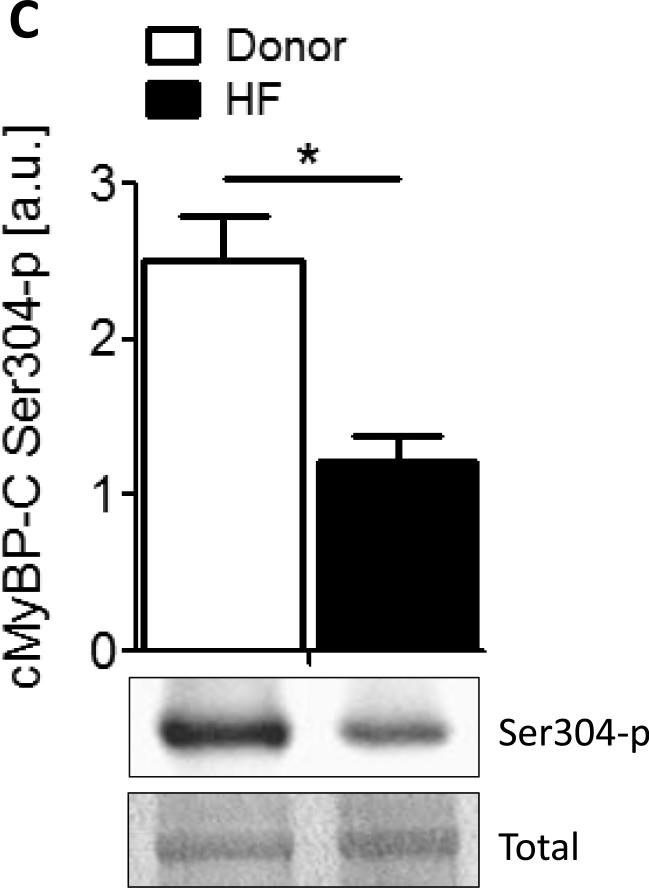
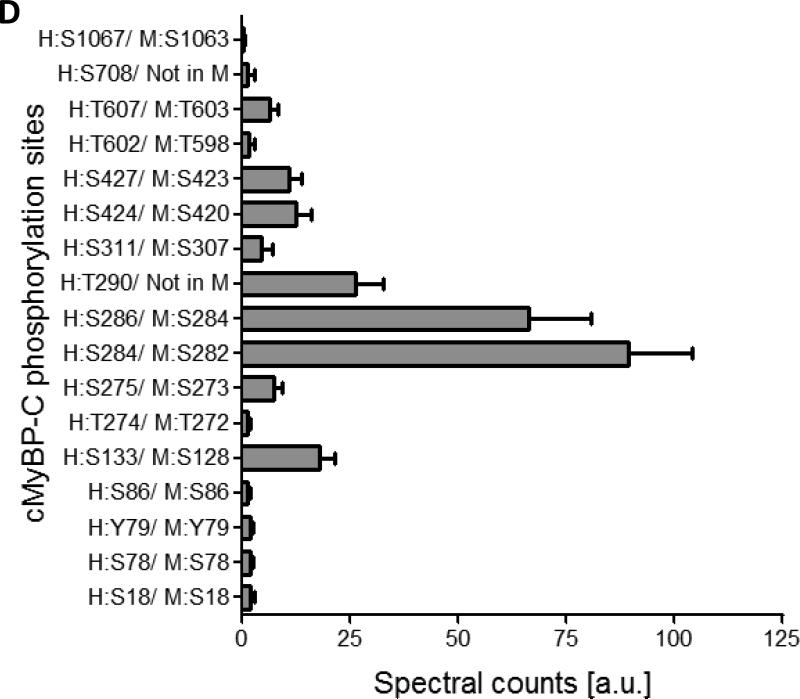
Figure 1. Characterization of the cMyBP-C phosphoproteome.
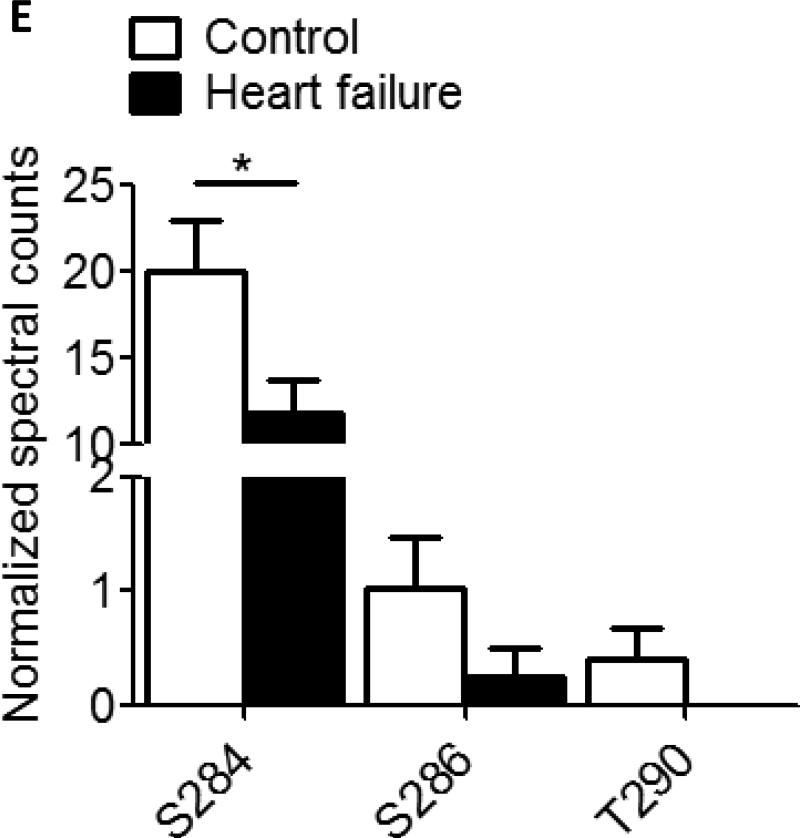
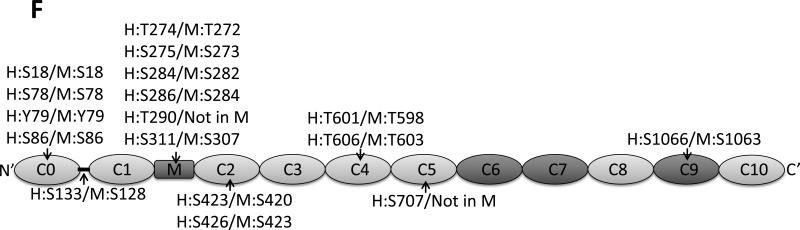
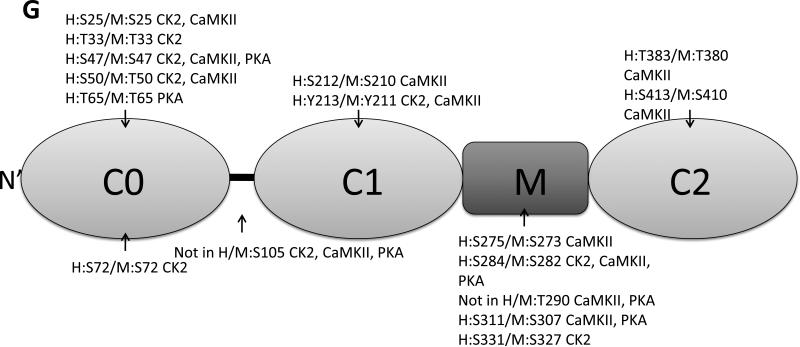
A. Phos-tag gel analysis of healthy donor tissue dephosphorylated using AP and subsequently phosphorylated using PKA. B. Reduced phosphorylation is shown by phos-tag gel analysis of samples derived from donor (n=8) and HF (n=8) hearts. C. Myofilament enriched pooled donor (tech. rep.=4) and HF (tech. rep.=4) samples were SCX-fractionated and TiO2 enriched for phosphopeptides. Samples were analyzed by LC-MS/MS on an LTQ-Orbitrap mass spectrometer. Seventeen phosphorylation sites were identified on cMyBP-C. Sites Ser284, Ser286 and Thr290 were observed as the most abundantly phosphorylated. D. Western blot of human donor and HF tissue (n=5) immunostained for phosphorylated Ser304 and corrected for total protein staining. Phosphorylation of Ser304 was significantly decreased in the HF group (*p<0.05 in a t-test). E. SDS-PAGE-based separation of cMyBP-C of n=10 healthy donor and n=10 HF heart samples (technical replicate = 2). Phosphorylation of Ser284 was significantly decreased (*p<0.05 in a t-test). F. Schematic representation of the domains of cMyBP-C and the identified phosphorylation sites. The residue numbers of human (uniprot reference # Q14896), and mouse (uniprot reference # O70468) are listed. Between C1 and C2 domains is a proline-alanine rich linker, between C1 and C2 donains is the M-domain and the dark grey boxes represent the Fn type II domains. Supplemental table 1 lists the predicted substrate kinase motifs for the identified sites and the phosphorylation sites listed on PhosphoSitePlus (www.phosphosite.org). All phosphopeptide spectral counts (for both method 1 and method 2) were corrected for total acquired spectra of each MS run and all spectra were manually validated. G. Schematic representation of the C0-C2 domains of cMyBP-C and the identified phosphorylation sites. Phosphorylation sites of CaMKII, CK2 and PKA are listed and the residue numbers of human (uniprot reference # Q14896), and mouse (uniprot reference # O70468) are listed.
3.2 Characterization of the cMyBP-C phosphoproteome using a SCX-based method
Samples from 10 pooled healthy (tech. rep.=4) and 10 pooled HF hearts (tech. rep.=4) were trypsin digested, SCX-fractionated, TiO2 enriched and analyzed by LC-MS/MS on an LTQ-Orbitrap mass spectrometer. All MS spectra were manually inspected on peak assignment and the localization of the phosphorylation sites. The SCX-based method was effective at characterizing phosphorylation sites on cMyBP-C and identified a total of 17 phosphorylated residues in vivo. The known sites, Ser133, Ser275, Ser284, and Ser311 were identified; however, site Ser304 was not detected in our analysis, most likely because this residue lies on a very short tryptic peptide, which may result in ion suppression or impaired fragmentation of the peptide. Possibly an alternate digestion method would produce a peptide containing the Ser304 site that is more amenable to mass spectrometry. Figure 1C shows reduced phosphorylation of Ser304 in the HF group (n=5) (t-test, *<0.05). The reduced phosphorylation observed in the failing group is in line with results from previous studies [7;8].
Nine of the 17 sites that were identified are novel (Supplemental Table 2). Specifically, the N-terminus side of cMyBP-C was extensively phosphorylated: sites Ser18, Ser78, Y79 and Ser86 were observed in the C0-domain; Thr274, Ser275, Ser284, Ser286, Thr290 and Ser311 in the M-domain; and Ser424 and Ser427 in the C2 domain. Thr602 and Thr607 were observed in domain C4, Ser708 in C5, and Ser1067 at the C-terminal domain C9 (Table 1, Fig. 1C/E). Shaffer et al. showed that the domains C0-C1-m-C2 bind to F-actin and that phosphorylation of the M-domain reduced binding to actin [11]. Interestingly, the three most abundantly phosphorylated sites (Ser284, Ser286 and Thr290) are all present in close proximity of each other in the regulatory M-domain of cMyBP-C, indicating that phosphorylation of these sites is important for regulating binding to actin. In addition, Ser275 is conserved in the skeletal muscle fast isoform (MYBPC2), which is an indicates presence of this site in MYBPC2.
Overview of identified phosphorylation sites on cMyBP-C
| Site | Peptide sequence | Peptide start-end | Best A-score | Loc. prob. | Charge | C/ HF |
|---|---|---|---|---|---|---|
| ‘Identification’ method 1: SCX- based | ||||||
| S18 | sVEVAAGSPAVFEAETER | 18-35 | 61.7 | 100% | 2+ | D |
| S78 | EVGPADQGsYAVIAGSSK | 70-87 | 7.2 | 71% | 2+ | HF |
| Y79 | EVGPADQGSyAVIAGSSK | 70-87 | 20.0 | 98% | 2+ | D/ HF |
| S86 | EVGPADQGSYAVIAGSsK | 70-87 | 8.1 | 13% | 2+ | D/ HF |
| S133 | VIEAEKAEPmLAPAPAPAEATGAPGEAPAPAAELGESAPsPK | 94-135 | 41.9 | 100% | 3+ | D/ HF |
| T274 | tSLAGGGR | 274-281 | 21.9 | 100% | 2+ | D/ HF |
| S275 | RTsLAGGGR | 273-281 | 34.0 | 100% | 2+ | D/ HF |
| S284 | RIsDSHEDTGILDFSSLLKK | 283-300 | 29.3 | 100% | 2+ | D/ HF |
| S286 | RISDsHEDTGILDFSSLLK | 282-300 | 12.6 | 82% | 4+ | D/ HF |
| T290 | ISDSHEDtGILDFSSLLK | 283-300 | 16.2 | 93% | 3+ | D/ HF |
| S311 | TPRDsKLEAPAEEDVWEILR | 307-326 | 38.2 | 100% | 2+ | D/ HF |
| S424 | RTLTIsQcSLADDAAYQcVVGGEK | 419-442 | 8.3 | 68% | 3+ | D/ HF |
| S427 | TLTISQcsLADDAAYQcVVGGEK | 420-442 | 28.1 | 100% | 2+ | D/ HF |
| T602 | LtIDDVTPADEADYSFVPEGFAcNLSAK | 601-628 | 8.8 | 71% | 3+ | D/ HF |
| T607 | VHKLTIDDVtPADEADYSFVPEGFAcNLSAK | 598-628 | 44.5 | 100% | 3+ | D/ HF |
| S708 | APARPAPDAPEDTGDsDEWVFDKK | 693-716 | 28.1 | 100% | 3+ | HF |
| S1067 | ATLVLQVVDKPsPPQDLR | 1056-1073 | 51.9 | 100% | 3+ | HF |
| ‘Quantification’ method 2: SDS-PAGE-based | ||||||
| S284 | RIsDSHEDTGILDFSSLLK | 282-300 | 45.01 | 100% | 3+ | D/HF |
| S286 | RISDsHEDTGILDFSSLLK | 282-300 | 18.7 | 96% | 3+ | D/HF |
| T290 | ISDSHEDtGILDFSSLLK | 283-300 | 6.04 | 55% | 3+ | D |
‘Identification’ SCX-based method 1: 10 donor and 10 failing heart samples were pooled. Each healthy donor and failing heart sample were analyzed in quadruplicate.
‘Quantification’ SDS-PAGE-based method 2: 10 healthy donor and 10 failing heart samples were all separately analyzed in duplicate.
Loc. Prob.; Localization probability, TiO2; Titanium dioxide, SCX; strong-cation exchange chromatography, C; donor, HF; heart failure, SDS-PAGE; dodecylsulfate polyacrylamide gel electrophoresis, A-score; post-translational modification scoring algorithm used by Scaffold-PTM [16]. Phosphorylated amino acids are indicated in lower case bold, modified C and M residues indicated in lower case.
In Supplemental Table 2 we list the predicted kinase substrate motifs (human protein reference database, www.hrpd.org) for these newly identified phosphorylation sites. The kinases PKA and PKC have additional predicted sites (Ser18, Ser86, Thr274, Thr602 and Ser1067), which may also be altered in heart failure, since PKA and PKC activity is changed in the failing heart. Other kinases were predicted to phosphorylate cMyBP-C at multiple sites, indicating that other signaling pathways, underappreciated in the myofilaments, may be involved. All these sites are based on the human cMyBP-C sequence (Uniprot #Q14896).
3.3 Reduced phosphorylation in the M-domain of cMyBP-C in the failing heart
Only three sites (Ser284, Ser286, Thr290, human cMyBP-C sequence (Uniprot #Q14896)) on cMyBP-C were identified using the SDS-PAGE-based method, and these correlated to the three most abundant sites identified using the SCX-based method. All three sites show a reduced phosphorylation in the HF group compared to the donor group based on spectral counts, however, only Ser284 was significantly reduced (*p<0.05,t-test ) (Table 1, Fig. 1D). This is in agreement with the phos-tag analysis of figure 1A, where we visualize only the most abundant phosphorylation sites. To date, the functional role of Ser286 or Thr290 has been entirely unexplored, although these sites, together with Ser284, likely play an important role in cardiac contractility.
The difference in phosphorylation yield between the two methods is likely due to the different protein amounts of the starting material. For the SCX-based method, 1 mg of pooled myofilament enriched protein was used, while the SDS-PAGE-based method only used 30 μg for each sample. The large amounts of protein necessary for the SCX-based method prevented the analysis of biological replicates, which was not the case for the SDS-PAGE-based method.
3.4 In vitro phosphorylation of the C0-C2 domain of mouse cMyBP-C
Ten novel phosphorylation sites were identified on full-length human cMyBP-C (figure 1D), the majority located in the N-terminal domain of cMyBP-C. In the online supplemental table 2, we performed a kinase substrate motif prediction (human protein reference database) to predict which kinases might be involved with phosphorylation of the novel sites on cMyBP-C. The majority of the novel sites were predicted to be phosphorylated by PKA and PKC, two kinases whose expression levels are reduced in the failing heart. Moreover, CK2 a serine/threonine selective kinase is predicted to phosphorylate numerous novel sites. Two of the known sites that show reduced phosphorylation during HF are predicted to be phosphorylated by CaMKII. These four kinases (PKA, PKC, CaMKII and CK2) were used in an in vitro phosphorylation assay in combination with MS to identify the phosphorylation sites of each kinase.
The recombinant C0-C2 fragment of mouse cMyBP-C was incubated with 200U PKA, 2000U CaMKII, 1000U CK2 or 0.2U PKC. Three sites, Ser273 (human Ser275), Ser282 (human Ser284) and Ser307 (human Ser311) were identified, which overlap with sites observed in the human tissue. Ser273 was phosphorylated by CaMKII, Ser282 by CK2, CaMKII and PKA, and Ser307 by CaMKII and PKA. All three sites are located in the M-domain of cMyBP-C (Fig. 1G). We did not observe phosphorylation of Ser273 by PKA even though this site is a known PKA substrate. The most likely reason for this is that the phosphorylation level at site Ser275 was at a stoichiometry to low to detect using mass spectrometry. Thirteen novel sites were identified that were not observed in the human tissues. In the in vivo situation there is interplay between kinases and phosphatases, meaning that not all phosphorylatable residues were necessarily phosphorylated at time of tissue procurement. Also, it can be reasoned that the sites phosphorylated in the fragment protein are normally buried in the complete structure of cMyBPC, indicating that proteolysis can influence the phosphorylation state of the protein. Additionally, association with other proteins and formation of protein complexes can make potential phosphorylation sites inaccessible to the kinase in vivo. Therefore, more research is needed to confirm whether the sites observed on the recombinant C0-C2 fragment are physiological relevant. Eight of the novel sites identified on the recombinant C0-C2 fragment were phosphorylated by CK2, 9 sites by CaMKII and 4 sites by PKA (Fig. 1G). We did not identify any phosphorylation sites for PKC, which might have been a methodological problem since multiple PKC sites on cMyBP-C have been previously reported [9]. To our knowledge, this is the first time that CK2 has been implicated with phosphorylation of cMyBP-C or any other cardiac myofilament protein, and more research is warranted to identify whether CK2 is targeted to the myofilament and the implications of cMyBP-C phosphorylation by CK2 in vivo.
4. Limitations
In mass spectrometry, spectral counting is a generally accepted method for analysis of peptide abundance. However, caution should be exerted when comparing spectral counts of different peptides, because ionization of each peptide may vary. Spectral counts may therefore not always exactly reflect the relative abundance of phosphorylation sites on different peptides. The phosphorylation sites of Ser284, Ser286 and Thr290 in our results are located on the same peptide and can therefore be compared without hindrance. We should, however, be aware when comparing these sites to the other observed phosphorylation sites.
5. Conclusion
This study characterizes the phosphoproteome of human cMyBP-C. Seventeen phosphorylation sites were observed on cMyBP-C, 10 of which have not been previously reported (Supplemental Table 2). Moreover, we show that phosphorylation of the three most highly phosphorylated sites, Ser284, Ser286 and Thr290, are reduced in the failing heart. These sites likely play an important role in cardiac contractility and more research is needed to determine the functional effects of phosphorylation at these sites to increase our understanding of cMyBP-C's role in the failing heart. In addition, we identified phosphorylation sites for CaMKII, CK2 and PKA on the C0-C2 fragment of cMyBP-C.
Supplementary Material
Highlights.
We analysed the phosphoproteome of cMyBP-C in human explanted donor and end-stage failing hearts.
Seventeen phosphorylation sites on cMyBP-C were identified in vivo.
Nine phosphorylation sites are novel identified phosphorylation sites.
The majority of these sites are localized in the M-domain of cMyBP-C.
Sixteen sites were identified after in vitro kinase incubation using different kinases.
Acknowledgement
We would like to thank Dr. S. Sadayappan for his kind gift of the cMyBP-C antibodies and the recombinant C0-C2 cMyBP-C fragment and Dr. C dos Remedios for providing us with the banked human explanted hearts. This work was in part supported by National Institutes of Health Grant R01 HL63038 (A.M.M.) and in part by an American Heart Association postdoctoral fellowship (V.K., 12POST11520006). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD000158.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None
Reference List
- 1.Stelzer JE, Fitzsimons DP, Moss RL. Ablation of myosin-binding protein-C accelerates force development in mouse myocardium. Biophys J. 2006;90:4119–27. doi: 10.1529/biophysj.105.078147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris SP, Bartley CR, Hacker TA, McDonald KS, Douglas PS, Greaser ML, et al. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ Res. 2002;90:594–601. doi: 10.1161/01.res.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- 3.James J, Robbins J. Signaling and Myosin-binding Protein C. Journal of Biological Chemistry. 2011;286:9913–9. doi: 10.1074/jbc.R110.171801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohamed AS, Dignam JD, Schlender KK. Cardiac myosin-binding protein C (MyBP-C): identification of protein kinase A and protein kinase C phosphorylation sites. Arch Biochem Biophys. 1998;358:313–9. doi: 10.1006/abbi.1998.0857. [DOI] [PubMed] [Google Scholar]
- 5.Barefield D, Sadayappan S. Phosphorylation and function of cardiac myosin binding protein-C in health and disease. J Mol Cell Cardiol. 2010;48:866–75. doi: 10.1016/j.yjmcc.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuster DW, Sequeira V, Najafi A, Boontje NM, Wijnker PJ, Witjas-Paalberends ER, et al. GSK3beta phosphorylates newly identified site in the proline-alanine-rich region of cardiac myosin-binding protein C and alters cross-bridge cycling kinetics in human: short communication. Circ Res. 2013;112:633–9. doi: 10.1161/CIRCRESAHA.112.275602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Copeland O, Sadayappan S, Messer AE, Steinen GJM, van der Velden J, Marston SB. Analysis of cardiac myosin binding protein-C phosphorylation in human heart muscle. Journal of Molecular and Cellular Cardiology. 2010;49:1003–11. doi: 10.1016/j.yjmcc.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 8.El Armouche A, Pohlmann L, Schlossarek S, Starbatty J, Yeh YH, Nattel S, et al. Decreased phosphorylation levels of cardiac myosin-binding protein-C in human and experimental heart failure. J Mol Cell Cardiol. 2007;43:223–9. doi: 10.1016/j.yjmcc.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Sadayappan S, Gulick J, Osinska H, Martin LA, Hahn HS, Dorn GW, et al. Cardiac myosin-binding protein-C phosphorylation and cardiac function. Circ Res. 2005;97:1156–63. doi: 10.1161/01.RES.0000190605.79013.4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stelzer JE, Patel JR, Moss RL. Protein kinase A-mediated acceleration of the stretch activation response in murine skinned myocardium is eliminated by ablation of cMyBP-C. Circ Res. 2006;99:884–90. doi: 10.1161/01.RES.0000245191.34690.66. [DOI] [PubMed] [Google Scholar]
- 11.Shaffer JF, Kensler RW, Harris SP. The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner. J Biol Chem. 2009;284:12318–27. doi: 10.1074/jbc.M808850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dephoure N, Gygi SP. A solid phase extraction-based platform for rapid phosphoproteomic analysis. Methods. 2011;54:379–86. doi: 10.1016/j.ymeth.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics. 2005;4:873–86. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Govindan S, Sarkey J, Ji X, Sundaresan NR, Gupta MP, de Tombe PP, et al. Pathogenic properties of the N-terminal region of cardiac myosin binding protein-C in vitro. J Muscle Res Cell Motil. 2012;33:17–30. doi: 10.1007/s10974-012-9292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006;5:749–57. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–92. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.