Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) infection of in vitro target cells is characterized by the expression of the latency-associated open reading frame (ORF) 73 gene (LANA-1) and the absence of progeny virus production. This default latent infection can be switched into lytic cycle by phorbol ester and by the lytic cycle ORF 50 (RTA) protein. In this study, the kinetics of latent and lytic gene expression immediately following KSHV infection of primary human dermal microvascular endothelial (HMVEC-d) and foreskin fibroblast (HFF) cells were examined by real-time reverse transcriptase PCR and whole-genome array. Within 2 h postinfection (p.i.), high levels of ORF 50 transcripts were detected in both cell types, which declined sharply by 24 h p.i. In contrast, comparatively low levels of ORF 73 expression were detected within 2 h p.i., increased subsequently, were maintained at a steady state, and declined slowly by 120 h p.i. The RTA and LANA-1 proteins were detected in the majority of infected cells by immunoperoxidase assays. In genome array, only 29 of 94 (31%) KSHV genes were expressed, which included 11 immediate-early/early, 8 early, and 5 late lytic genes and 4 latency-associated genes. While the expression of latent ORF 72, 73, and K13 genes continued, nearly all of the lytic genes declined or were undetectable by 8 and 24 h p.i. in HMVEC-d and HFF cells, respectively. Only a limited number of RTA-activated KSHV genes were expressed briefly, and the majority of KSHV genes involved in viral DNA synthesis and structural proteins were not expressed. However, early during infection, the lytic K2, K4, K5, K6, and vIRF2 genes with immune modulation functions and the K7 gene with antiapoptotic function were expressed. Expression of K5 was detected for up to 5 days of observation, and vIRF2 was expressed up to 24 h p.i. The full complement of lytic cycle genes were expressed when 12-O-tetradecanoylphorbol-13-acetate was added to the HMVEC-d cells after 48 h p.i. These data suggest that in contrast to alpha- and betaherpesviruses and some members of gammaherpesviruses, gamma-2 KSHV in vitro infection is characterized by the concurrent expression of latent and a limited number of lytic genes immediately following infection and a subsequent decline and/or absence of lytic gene expression with the persistence of latent genes. Expression of its limited lytic cycle genes could be a “strategy” that evolved in KSHV allowing it to evade the immune system and to provide the necessary factors and time to establish and/or maintain latency during the initial phases of infection. These are unique observations among in vitro herpesvirus infections and may have important implications in KSHV biology and pathogenesis.
The hallmark of infection by members of the Herpesviridae family is the establishment of latent infection and the maintenance of their genome in the host with the expression of a limited number of viral genes in the latently infected cells (43). Upon entry into the in vitro target cells, members of alpha-, beta-, and gammaherpesviruses have been shown to enter the lytic replicative cycle, which is characterized by a cascade of gene expression (24, 28, 42, 43). The genes expressed in the initial stages of lytic replication are called α or immediate-early genes, products of which activate the cascade of β or early gene expression essential for viral DNA replication and for the regulation of late or γ gene expression (24, 28, 42, 43). Even though the late genes can be detected before viral DNA replication, substantially higher levels are expressed after viral DNA replication. The lytic infection leads to the production of infectious viral progeny. Unlike many members of alpha-, beta-, and gammaherpesviruses, lytic gamma-1 Epstein Barr virus (EBV) replication has not been demonstrated during in vitro infection of susceptible primary human B cells. Instead, EBV establishes a latent infection in these cells, resulting in the transformation and the establishment of B-lymphoblastoid cell lines (24).
Kaposi's sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8, is associated with the endothelial tumor Kaposi's sarcoma (KS) and with two lymphoproliferative disorders, body cavity-based B-cell lymphoma (BCBL) or primary effusion lymphoma (PEL), and a subset of multicentric Castleman's disease (9, 14, 18, 29, 46, 47). Gamma-2-KSHV DNA sequence analyses demonstrate that a large region of the KSHV genome is conserved among herpesviruses and is colinear with gamma-1 EBV and gamma-2-herpesvirus saimiri (34, 44). KSHV encodes more than 90 open reading frames (ORFs), which are designated as ORFs 4 to 75 by their homology to herpesvirus saimiri ORFs (34, 44). Divergent regions in between the conserved gene blocks contain more than 20 KSHV unique genes, which are designated with the prefix K. Several KSHV-encoded proteins are homologs of host proteins, probably captured by the virus from the host over the course of evolution by molecular piracy. These genes include K2 (v-interleukin-6 [vIL-6]), K4 (v-macrophage inhibitory protein II [vMIP-II]), K3 and K5 (MIR-1 and MIR-2; immunomodulatory proteins), K6 (vMIP-I), K7 (antiapoptotic protein), K9 (v-interferon regulatory factor [vIRF]), vIRF2 (K11.1), ORF 16 (vBcl-2), K13 (v-FLICE-inhibitory protein [vFLIP]), K14 (vOX-2), ORF 72 (v-cyclin D), and ORF 74 (v-G protein-coupled receptor) (14, 18, 29, 35, 44, 46, 47).
Similar to EBV-associated lymphomas, cell lines with B-cell characteristics established from BCBL carry KSHV in a latent form, and a lytic cycle can be induced by 12-O-tetradecanoylphorbol-13-acetate (TPA) (14, 18, 29, 40, 46, 47). In vivo, KSHV DNA and transcripts have been detected in human B cells, macrophages, keratinocytes, endothelial cells, and epithelial cells (14, 18, 29, 48, 56). In vitro, KSHV has been shown to infect human B, endothelial, epithelial, and fibroblast cells (1, 2, 3, 7, 13, 19, 25, 30, 39, 52, 53). In addition, KSHV also infects a variety of animal cells, such as owl monkey kidney cells, baby hamster kidney fibroblast cells, Chinese hamster ovary cells, and primary embryonic mouse fibroblast cells (1, 2, 5, 14, 29, 32). If in vitro permissiveness of a cell type were judged by the initiation of a lytic-cycle gene expression cascade and formation of progeny virus from the input KSHV, as yet no suitable cell culture system exists. However, if in vitro permissiveness were judged by the expression of KSHV latency genes and the ability to support KSHV lytic replication after activation by agents, a variety of human and animal cells, including human endothelial and fibroblast cells, are permissive, as evidenced by the retention of viral genome in a latent form, by the expression of KSHV latency-associated ORF 73 protein (LANA-1), and by the ability to support lytic replication upon activation by TPA and the KSHV switch gene, ORF 50 (RTA) (1, 2, 5, 11, 14, 19, 25, 29, 30, 52, 53). Several studies have also reported the detection of lytic cycle in about 1 to 5% of infected cells after 48 h postinfection (p.i.) (5, 11, 19, 25, 30, 53). It should be noted that the in vitro latent KSHV infection in primary endothelial or fibroblast cells or in nonadherent B cells is unstable, and the viral DNA is not maintained efficiently and is usually lost in subsequent culturing of infected cells (2, 5, 25, 52).
In all of the infected BCBL cell lines studied so far, KSHV exists in a latent state and expresses only a few genes, such as ORF 73, ORF 72, K13, K12, K15, and ORF 10.5 (14, 18, 29, 45, 46, 47). Northern hybridization, RNase protection assay, real-time reverse transcriptase PCR (RT-PCR), and gene array analyses have been used to demonstrate the temporal pattern of KSHV latent and lytic gene expression in uninduced cells as well as following induction by TPA or RTA (14, 18, 29, 45, 50, 55). The KSHV RTA (regulator of transcription and activation), like its functional homolog ZTA in EBV, plays crucial roles in switching from latency to lytic replication. Studies have shown that RTA can activate the KSHV immediate-early/early or primary (K8, K5, K2, K12, Nut1, and ORF 6, 57, and 74), early or secondary (K9, ORF 59, ORF 65, and K3), and late or tertiary (K1, K8.1A, and ORF 21) lytic cycle genes, either alone or in synergy with other viral regulatory genes (14, 18, 29, 46, 50, 55). KSHV RTA is also shown to interact with host cellular proteins, resulting in the modulation of cellular as well as viral gene expression (14, 18, 29, 50, 55). Even though transcription profiling of KSHV genes in both latent and lytic infections has been performed, most of these studies were conducted in latently infected B cells, and KSHV gene expression immediately following a primary infection of target cells was not analyzed.
Using real-time RT-PCR and KSHV whole-genome array, we examined the kinetics of KSHV latent and lytic gene expression during primary infection of primary human dermal microvascular endothelial (HMVEC-d) and foreskin fibroblast (HFF) cells. Here, we demonstrate that early during infection, KSHV expresses the lytic ORF 50 gene and the latent ORF 73 gene concurrently, with initial higher levels of ORF 50 expression followed by a rapid decline. In contrast, ORF 73 expression was low at the initial time points, increased subsequently, was maintained at a steady level for a while, and then declined slowly. Analyses of all KSHV latent and lytic gene expression by gene arrays at two different time points demonstrated that in addition to the latent genes, KSHV also expresses a limited number of lytic genes immediately after infection, most of which decline sharply during the course of infection. These findings suggest that KSHV initiates concurrent latent and limited lytic gene expression immediately following infection. Many of the expressed KSHV lytic genes have been shown to possess immune modulation and antiapoptotic functions, suggesting that they may be providing survival time and sufficient factors and advantage necessary for the coexistence with the host cell during the initial time of infection.
MATERIALS AND METHODS
Cells.
KSHV-carrying BCBL-1 were cultured in RPMI 1640 (Gibco BRL, Grand Island, N.Y.) medium with 10% heat-inactivated fetal bovine serum (HyClone, Logan, Utah), l-glutamine, and antibiotics (1, 2, 3, 32). HFF cells (Clonetics, Walkersville, Md.) were cultured in Dulbecco's modified Eagle's medium (Gibco BRL), and HMVEC-d cells (CC-2543; Clonetics) were maintained in endothelial basal media 2 in the presence of necessary growth factors (32). The HMVEC-d cells from Clonetics used here are probably the same as the dermal microvascular endothelial cells (DMVEC) used by other investigators.
Virus.
The KSHV lytic cycle was induced from BCBL-1 cells by 20 ng of TPA/ml. After 96 h, virus from the supernatant was collected, and different batches were pooled and purified as per procedures described before (32). KSHV DNA was extracted from the virus, and copy numbers were quantitated by real-time DNA-PCR using primers amplifying the KSHV ORF 73 gene as described below.
Isolation of DNA.
HMVEC-d and HFF cells were grown to confluence in six-well plates (3 × 105 to 4 × 105 cells/well), washed to remove the serum and growth factors, and infected with KSHV. At different time points, cells were washed twice with 1× phosphate-buffered saline (PBS) to remove the unbound virus. To remove the virus bound to the cell but not internalized, cells were treated with 0.25% trypsin-EDTA for 5 min, harvested, and washed twice in serum-free medium. Total DNA from infected and mock-infected cells was prepared using a Qiagen DNeasy tissue kit (Qiagen, Valencia, Calif.), quantitated spectrophotometrically, and stored at −20°C.
Real-time DNA-PCR.
Real-time PCR was performed to quantify the internalized viral DNA in the infected cells. A 100-ng aliquot of DNA sample and KSHV ORF 73 gene Taqman probe (Applied Biosystems, Foster City, Calif.) (Table 1) and Quantitect PCR mix were used. We designed primer pairs and dual-labeled fluorogenic probes (Taqman) based on the ORF 73 and 50 sequences. The highly repetitive central region of the orf73 gene was removed from consideration, as well as a low-complexity region (high in proline and serine in the amino acid sequence) near the 5′ end and a small region high in proline at the C-terminal end. The primer-probe set was designed from a highly homologous region at the 3′ end using the ABI Primer Express 2.0 software. As this set was to be used for DNA real-time PCR on cellular DNA, we verified that the primers had no significant homology with Alu repetitive sequences. For ORF 50, the KSHV unique mRNA sequence region of nucleotides 340 to 1460 (GenBank accession no. AF091350) not overlapping the splice site was used for the design of the primer-probe set.
TABLE 1.
Primer and probe sequences used for real-time PCR
| Gene | ORF 73 (68 bp)a | ORF 50 (73 bp)a |
|---|---|---|
| Forward primer | 5′-CGCGAATACCGCTATGTACTCA-3′ | 5′-CGCAATGCGTTACGTTGTTG-3′ |
| Reverse primer | 5-GGAACGCGCCTCATACGA-3′ | 5′-GCCCGGACTGTTGAATCG-3′ |
| Taqman probe | 6FAM-ACATCACCACCCCACAGACCTGGAG-TAMRA | 6FAM-ACCTGTGCCCCCTCTTCGACACC-TAMRA |
Product length.
The KSHV ORF 73 gene cloned in the pGEM-T vector (Promega, Madison, Wis.) was used for the external standard. Known amounts of ORF 73 plasmid (106, 105, 104, 103, 102, 101, and 100 copies) were used in the amplification reaction mixtures along with the test samples. The reaction conditions were as follows: initial preheating at 95°C for 10 min followed by 44 cycles of 95°C for 15 s and 60°C for 30 s. The lower limit of ORF 73 gene detection was 10 to 100 copies, and the most accurate detection was from 100 to 106 copies. Special care was taken to keep the slope of the standard curve close to 3.3, so that the amplification efficiency of the cycles was 2 (16). The Ct values were used to plot the standard graph and to calculate the relative copy numbers of viral DNA in the samples.
Isolation of total RNA.
KSHV-infected cells were washed twice with 1× PBS to remove the unbound virus and lysed with RLT buffer (Qiagen), and the monolayer was scraped to collect the lysate. Total RNA was isolated from the lysate using RNeasy kits (Qiagen) according to the manufacturer's protocols, quantified spectrophotometrically, and stored at −80°C.
Real-time RT-PCR.
The ORF 50 and ORF 73 transcripts were detected by real time RT-PCR using gene-specific Taqman probes (Applied Biosystems). Prior to reverse transcription, the RNA samples were subjected to DNase I (amplification grade; Invitrogen, Carlsbad, Calif.) treatment to remove contaminating DNA according to the manufacturer's protocol. The DNase I-treated RNA samples were reverse transcribed and amplified in the presence of Taqman probe (Table 1) using Taqman EZ RT-PCR reagents (Applied Biosystems), transcript-specific primers, and Taqman probe. For the preparation of external control samples, the genes were cloned into the pGEM-T vectors and transcribed in vitro using the MEGAScript T7 high-yield transcription kit (Ambion Inc., Austin, Tex.). The transcripts were quantitated by spectrophotometer, and different dilutions of known concentration of gene-specific transcripts (106, 105, 104, 103, 102, 101, and 100 copies) were prepared. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal control. A 250-ng aliquot of DNase I-treated total RNA from each sample was used in duplicate reactions for real-time RT-PCR. Extreme care was taken to make sure that the values obtained for reactions were not contributed from contaminating DNA. All samples were used in separate reactions of real-time DNA-PCR (without RT) to confirm the absence of contaminating DNA. The reaction conditions used for ORF 50 and ORF 73 gene amplification consisted of four stages: 50°C for 2 min, 60°C for 30 s, and 95°C for 10 min, followed by 44 cycles of 95°C for 15 s and 60°C for 30 s. GAPDH conditions varied slightly in the amplification stage, with denaturation of the strands at 95°C for 20 s and 62°C for 1 min, and were repeated for 40 cycles.
The relative copy numbers of the transcripts were calculated from the standard graph plotted using the Ct values for different dilutions of in vitro-transcribed transcripts. These values were normalized to each other using the values of the GAPDH control reactions. The number of GAPDH transcripts in each sample was calculated from the standard graph plotted using copy numbers from different dilutions of the known concentrations of human RNA and Taqman GAPDH reagents (Applied Biosystems). The standard graph of the external control reactions was used to determine the relative exact copy numbers of respective transcripts in each sample, while the GAPDH control reaction was used to normalize the amount of initial template in each reaction mixture. The lower limit of detection in the standard samples was 10 to 100 copies of transcripts for both the ORF 50 and ORF 73 genes, while copies in the range of 100 to 106 were detected with more accuracy. Similar to real-time DNA-PCR, maximum care was taken to keep the slope of the standard curve close to 3.3.
Analyses of KSHV gene expression by genome microarray.
Commercially available KSHV gene arrays (Celonex Inc., Edmonton, Canada) spotted in triplicate on amino-modified ceramic DNA slides were used. Cellular transcripts like tubulin, actin, GAPDH, and ribosomal S9 represented the positive controls and were used for the normalization of chip-to-chip variations. Control plasmids and human immunodeficiency virus genes represented the negative controls. Each gene was represented by oligos of 110 nucleotides in length. Spots were 100 to 120 μm in diameter, with each spot separated from the nearest spot by 350 μm. The specificity and the sensitivity were checked prior to the experiment by hybridizing with labeled cDNA prepared from BCBL cells induced with TPA for 72 h.
The RNAs from uninfected and KSHV-infected HMVEC-d and HFF cells at different time p.i. were prepared and quantified, and integrity was checked on formaldehyde-agarose gels. For each experiment, 15 μg of RNA was used. The RNA transcripts from the infected and uninfected samples were converted into cDNA using Superscript II (Invitrogen) in the presence of the fluorescent dyes Cy-3 and Cy-5, respectively. The reaction was carried out in a 30-μl total volume. The deoxynucleoside triphosphates were used at a final concentration of 500 μM except for dTTP, which was used at a final concentration of 100 μM. Three millimoles of Cy-3 or Cy-5 dUTPs was used in each 30-μl reaction mixture, and the cDNA populations were generated using virus-specific primer mix as well as primer mix for the control genes. The reaction was performed at 42°C for 2 h and stopped by adding 3 μl of 25 mM EDTA. The RNA left in the reaction mixture was degraded by adding 1.5 μl of 500 mM NaOH at 70°C for 10 min, and the pH was neutralized by adding 1.5 μl of 500 mM HCl.
The fluorescent-labeled cDNA was purified from the unincorporated free nucleotides by using QIAquick PCR purification spin columns (Qiagen), denatured at 94°C for 5 min followed by chilling on ice before mixing with 2× hybridization buffer containing 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 50% formamide, 0.4% sodium dodecyl sulfate (SDS), and 40 μg of salmon sperm DNA/μl. The mixture was preheated to 50°C and applied over the chip. Prior to hybridization, the slides were pretreated by boiling in sterile water for 3 min, followed by incubation with prehybridization buffer for 1 to 4 h at 42°C. The slides were rinsed in sterile water, washed in isopropyl alcohol, dried by centrifugation, and preheated to 50°C before hybridization to minimize the postwash background. The hybridization mix was applied over the spotted arrays, covered with coverslips (HybriSlips; Grace Bio-labs, Bend, Oreg.), placed inside the hybridization chamber (Ambion), and incubated at 42°C for 16 to 20 h. After hybridization, slides were washed in low-stringency buffer containing 1% SSC and 0.2% SDS for 5 min three times at 42°C. This was followed by two washes for 5 min each in low-stringency buffer containing 0.1% SSC and 0.2% SDS at 42°C and a single wash in buffer containing 0.1% SSC alone at room temperature. The slides were rinsed in sterile water at room temperature, dried by centrifugation, and scanned immediately.
The slides were scanned with an Affymetrix scanner 428 (Santa Clara, Calif.), and the Cy-3 and Cy-5 spot signal images were analyzed by using Affymetrix Jaguar software (version 4). A custom-designed KSHV gene array list file compatible with the Jaguar software generated at the University of Kansas Medical Center Biotechnology Core Facility was used to import the intensities of the individual spots and to calculate the ratios between different samples. The background intensities were also measured similarly. The variation among different slides was negated by normalizing the intensities of the spots corresponding to the cellular housekeeping genes and applying the same normalization factor to all the spots. Using the housekeeping genes, the Cy-3 and Cy-5 intensity values were also normalized to each other. The background intensities were subtracted from the average intensity for three different spots of each individual gene, and the standard deviations were calculated. The calibrated Cy-3/Cy-5 expression ratios of the normalized, background-subtracted intensities were calculated and imported to the Cluster program (15). The genes and the ratios were clustered by using a self-organizing map algorithm by the average linkage clustering, and the cluster results were visualized by using the Treeview program (15). Cy-3/Cy-5 expression ratios below 2.0 were considered not significant for further analyses.
Immunoperoxidase assay.
HMVEC-d and HFF cells grown in eight-well glass chamber slides were infected with KSHV, incubated for different periods, washed with 1× PBS, and fixed with ice-cold acetone. Rabbit polyclonal antibodies against KSHV LANA (2, 30, 53) and RTA (a kind gift from Tetsutaro Sata, National Institute of Infectious Diseases, Tokyo, Japan) (23), and monoclonal antibodies against K5 (a kind gift from K. Yamanishi and K. Ueda, Osaka University, Osaka, Japan) (20, 35) were used. Prestandardized dilutions of the primary antibodies in 1× Tris-buffered saline containing 1% bovine serum albumin were added to the slides and incubated at 4°C overnight. After the incubation, the slides were washed with 1× PBS, incubated with biotinylated anti-rabbit or anti-mouse antibodies (Vectastain ABC kit; Vector Laboratories, Burlingame, Calif.), and detected using avidin-biotin complex carrying horseradish peroxidase. Diamino benzidine was used as the substrate for the peroxidation reaction, and the reaction was stopped after sufficient color development. The slides were dehydrated in a series of 70 to 90% and absolute ethanol followed by xylene, sealed permanently by cytoseal, and stored at room temperature. Slides were examined under a Nikon light microscope with digital imaging system.
RT-PCR.
The total RNA isolated from different uninfected and infected cells was treated with DNase I (Invitrogen) and converted into cDNA using random hexamers. The reaction was performed using the Thermoscript RT-PCR system (Invitrogen) at 55°C according to the manufacturer's instructions. The primer sequences used in the RT-PCR analyses are listed in Table 2. The RT-PCR products amplified for 40 cycles from 250 ng of cDNA by KSHV gene-specific primers were electrophoresed on a 2% agarose gel and analyzed using an AlphaImager 2200 (Alpha Innotech Corporation, San Leandro, Calif.) gel documentation system.
TABLE 2.
Primer sequences used for RT-PCR
| Gene | Forward primer | Reverse primer | Product size (bp) |
|---|---|---|---|
| ORF7 | 5′-ACTGTGGGGGTCTGTCATCT-3′ | 5′-GGTTGAAGTTGGGCTCTATC-3′ | 377 |
| ORF8 | 5′-CTGTATCCGCCAAGTTCG-3′ | 5′-GGTCAAACACGCTACTCG-3′ | 432 |
| ORF9 | 5′-AGGAAAAGAGCAGTCAGGTC-3′ | 5′-CGTAGGGGATTCTGTCGTGT-3′ | 324 |
| ORFK2 | 5′-AAAACACGCACCGCTTGACCTG-3′ | 5′-TTCACTGCTGGTATCTGGAACG-3′ | 534 |
| ORFK4 | 5′-GCATCCTGCTCGTCGCTGTG-3′ | 5′-GCTTCTTCACCCAGTCTTTC-3′ | 240 |
| ORFK5 | 5′-GCAGAGAAAACCGAGCAC-3′ | 5′-GGGAAGAGGTGGGGAACG-3′ | 332 |
| ORFK6 | 5′-GCTGCCTAACCCAGTTTTTG-3′ | 5′-TTACCTGACGCCCACAGAAA-3′ | 209 |
| ORF25 | 5′-GTCCACCCCTTCTTTGATTTTT-3′ | 5′-TTTCCCGAGTTGACCCAGTAGG-3′ | 335 |
| ORF50 | 5′-GCCCTCTGCCTTTTGGTT-3′ | 5′-GATGATGCTGACGGTGTG-3′ | 351 |
| ORFK8 | 5′-ACTGACGGTGGGGAAAAC-3′ | 5′-GCGTGGGAGAATGTGACT-3′ | 535 |
| ORFK13 | 5′-ATTGACATTAGGGCATCC-3′ | 5′-AAAGGAGGAGGGCAGGTT-3′ | 327 |
| ORF72 | 5′-GATAATAGAGGCGGGCAATG-3′ | 5′-TAAAGCAGGTGTCCAAAGAA-3′ | 338 |
| ORF73 | 5′-GAAGTGGATTACCCTGTTGTTAGC-3′ | 5′-TTGGATCTCGTCTTCCATCC-3′ | 307 |
| vIRF2 | 5′-CGGAATGGCTCACGGACTTTAT-3′ | 5′-AGACATCCTTCACATCCCTTGT-3′ | 346 |
RESULTS
Kinetics of KSHV entry in primary human endothelial and fibroblast cells.
As an initial step to examine the kinetics of KSHV gene expression early during infection, we first determined the kinetics of viral entry into HMVEC-d and HFF cells. HMVEC-d cells represent the major in vivo target of KSHV infection, and HFF cells were used as an additional in vitro adherent target cell (1-3, 5, 32). KSHV DNA was extracted from the purified virus, and copy numbers were quantitated by real-time DNA-PCR using primers amplifying the KSHV ORF 73 gene. Only enveloped virus particles were seen in these electron microscopy examinations, indicating the purity of virus preparations, and electron microscopy of virus-infected target cells showed only enveloped virus particles (1, 3, 32). Target cells were infected with KSHV with a multiplicity of infection (MOI) of 100 viral DNA copies per cell for different time periods at 37°C. Real-time DNA-PCR was carried out to amplify the ORF 73 genes, and external copy standards were used to obtain the copy numbers of the amplified ORF 73 gene. Since every viral genome contains a single copy of the ORF 73 gene, the number of viral DNA molecules could be calculated from the corresponding copy number of the ORF 73 gene. Internalized viral DNA could be detected in HFF and HMVEC-d cells as early as 5 min p.i. This rapid entry into the target cells was supported by our laboratory's earlier electron microscopic observation of KSHV internalization via endocytic vesicles in the target cells within 5 min p.i. (1). Internalized viral copy numbers increased rapidly during the first 60 to 90 min of infection and reached a plateau around 90 to 120 min p.i., with no further significant increase in the internalized DNA copy numbers (Fig. 1). Similar levels of internalized viral DNA were detected in the HMVEC-d and HFF cells. These results were reproducible and consistent, with only minor variations between experiments and batches of viruses. Preincubation of virus with 100 μg of heparin/ml blocked more than 90% of KSHV DNA entry (data not shown). Together with the steady increase in the internalized viral DNA, these results demonstrated the specificity of the real-time DNA-PCR assay.
FIG. 1.
Kinetics of KSHV entry into HMVEC-d and HFF cells. HMVEC-d and HFF cells were infected at an MOI of 100 KSHV DNA copies per cell. At different time points, unbound KSHV was removed by washing and the bound, noninternalized virus was removed by treating the cells with trypsin-EDTA for 5 min. Infected and mock-infected cells were washed, and internalized viral DNA was isolated and quantified by real-time DNA-PCR with primers and Taqman probe specific for ORF 73 (Table 1). Viral DNA copy numbers were calculated from the standard graph generated by real-time PCR of known concentrations of a cloned ORF 73 gene. Each reaction was done in duplicate, and each point represents the average ± standard deviation of three experiments.
Early during primary infection of HMVEC-d and HFF cells, the KSHV immediate-early lytic ORF 50 gene was expressed along with the latent ORF 73 gene.
KSHV ORF 73-encoded LANA-1 has been shown to be expressed in vivo and in vitro in latently infected cells and to play roles in the maintenance of latency, tethering viral DNA to the host chromosome, and latent viral DNA replication along with the host chromosome (14, 29). The KSHV ORF 50-encoded regulatory protein RTA is an immediate-early gene which activates many KSHV genes, leading to the expression of lytic genes in a cascade manner, thus facilitating a complicated but systematic lytic replication process (14, 18, 29, 46, 47, 50, 55). Since the expression of both these proteins plays major roles in the KSHV life cycle, we examined their expression patterns by quantitative real-time RNA-PCR after primary infection of HMVEC-d and HFF cells. Extreme care was taken to make sure that the values obtained in these reactions were not contributed from contaminating DNA, and all samples were tested by real-time DNA-PCR for the detection of any trace amount of DNA.
As early as 30 min p.i., nearly equal copy numbers of KSHV ORF 73 transcripts were detected in both cell types (Fig. 2). In HMVEC-d cells, ORF 73 expression steadily increased, reached a peak around 24 h (1,440 min), and declined very slowly thereafter, with levels almost in a steady state in the observed period of 5 days p.i. (Fig. 2A). The level of ORF 73 transcript in HFF cells at 24 h p.i. was about a log lower than that in HMVEC-d cells, with the peak level reached around 1,440 min (24 h) and subsequently declining very slowly (Fig. 2B).
FIG. 2.
Kinetics of KSHV ORF 73 and 50 gene expression during primary infection of HMVEC-d (A) and HFF (B) cells. Cells were infected at an MOI of 100 KSHV DNA copies per cell. At different time points p.i. RNA was isolated and treated with DNase I, and 250 ng of DNase-treated RNA was subjected to real-time RT-PCR with ORF 73 and 50 gene-specific primers and Taqman probes. Known concentrations of DNase-treated in vitro-transcribed ORF 50 and ORF 73 transcripts were used in a real-time RT-PCR to construct a standard graph from which the relative copy numbers of viral transcripts were calculated and normalized, with GAPDH used as the internal control. Each reaction was done in duplicate, and each point represents the average ± standard deviation of three experiments.
Employing PCR, immunofluorescence, and immunoperoxidase assays, previous studies have examined ORF 73 expression after more than 2 days p.i. (5, 11, 13, 19, 25, 30, 53). Although the expression of ORF 50 following a primary infection was not measured systematically it was presumed to be absent, and establishment of latent infection was considered as the default KSHV infection (5, 11, 13, 14, 18, 19, 25, 29, 30). However, as early as 30 min p.i., we detected a robust level of ORF 50 gene expression in HMVEC-d and HFF cells, with slightly higher levels of expression in HFF cells (Fig. 2). During the early stages of infection, ORF 50 gene expression levels in HMVEC-d cell were about 1 log higher than that of ORF 73 expression and 2 logs higher in HFF cells (Fig. 2). After reaching a peak, ORF 50 expression declined sharply at later time points. In HMVEC-d cells, the peak level of ORF 50 expression was around 2 to 8 h p.i. with a sharp steady decline thereafter, with levels below that of ORF 73 after 24 h p.i. and almost undetectable by 120 h (Fig. 2A). In HFF cells, the same pattern of ORF 50 gene expression was observed, with a peak level at 480 min (8 h) p.i. followed by a sharp decline, with its level almost a log lower than that of ORF 73 by 72 h p.i. (Fig. 2B). The specificity of these real-time RT-PCRs was demonstrated by the absence of contaminating DNA in any of the DNase-treated RNA samples used in this study. Even though we observed slight differences in the copy number of transcripts in different experiments and batches of viruses, the patterns of gene expression were closely similar and highly reproducible.
It is very evident from these results that the expression patterns of ORF 50 and ORF 73 follow a similar pattern during a primary infection of adherent HMVEC-d and HFF cells. These findings suggest that KSHV enters the latent cycle concurrently with the activation of the lytic cycle ORF 50 gene in the initial stages of infection.
KSHV early lytic RTA and latent LANA-1 proteins were expressed in the majority of infected cells.
To determine whether ORF 50 mRNA detection represented expression in a limited number of infected cells or in all the infected cells and to determine whether the detection of a high level of ORF 50 message also resulted in protein expression, an immunoperoxidase assay was performed using rabbit anti-RTA and LANA antibodies. As early as 2 h p.i., RTA was detected in the nuclei of more than 90% of HMVEC-d cells (Fig. 3A), and after 8 h p.i. it was detected in HFF cells (Fig. 3F). The stained nuclei exhibited the ORF 50 characteristic diffused and grainy appearance (23). This expression was also detected after 24 h p.i. (Fig. 3B and G), a time when the ORF 50 messages were in decline (Fig. 2). Similar to ORF 50 expression, the characteristic spicular appearance of LANA-1 was also detected in more than 90% of infected cell nuclei, and results at 48 h p.i. are shown in Fig. 3D and I. The specificity of these reactions was demonstrated by the absence of reactivities of anti-ORF 50 and ORF 73 antibodies with the uninfected cells (Fig. 3C, E, H, and J) and by the absence of reactivities of normal rabbit antibodies with the infected cells (data not shown). These results demonstrated the expression of RTA and LANA-1 in most of the infected cells and suggested that the detection of ORF 50 and 73 transcripts probably represents these infected cells.
FIG. 3.
Immunoperoxidase assay detecting KSHV ORF 73 (LANA-1) and ORF 50 (RTA) proteins in HMVEC-d and HFF cells. Cells in eight-well chamber slides were infected with KSHV for 2 h, washed, and incubated at 37°C. At different time points, cells were washed, fixed with cold acetone, incubated at 4°C overnight with a predetermined dilution of rabbit antibodies against LANA-1 or RTA or with preimmune antibodies, processed for immunostaining as described in Materials and Methods, and examined under a light microscope. (A) ORF 50 expression in HMVEC-d cells 2 h p.i. (B) ORF 50 expression in HMVEC-d cells 24 h p.i. (C) Uninfected HMVEC-d cells with anti-ORF 50 antibodies. (D) ORF 73 expression in HMVEC-d cells 48 h p.i. (E) Uninfected HMVEC-d cells with anti-ORF 73 antibodies. (F and G) ORF 50 expression in HFF cells at 8 and 24 h p.i., respectively. (H) Uninfected HFF cells with anti-ORF 50 antibodies. (I) ORF 73 expression in HFF cells 48 h p.i. (J) Uninfected HFF cells with anti-ORF 73 antibodies. Magnification: ×40 (A, C, E, F, H, and J); ×100 (B, D, and G).
KSHV initiated the lytic cycle soon after infection of HMVEC-d and HFF cells, which was not sustained subsequently.
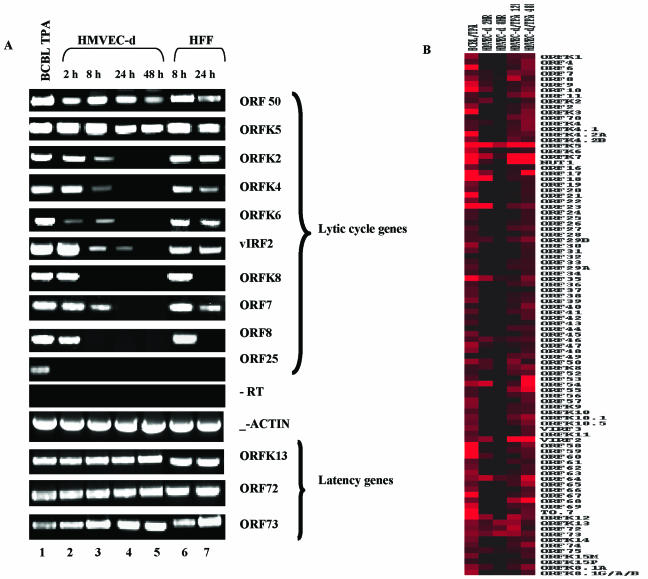
Overexpression of ORF 50 in cells which were already latently infected with KSHV has been shown to activate the lytic cycle, resulting in the production of virus progeny (5, 14, 18, 29, 46, 47, 50, 55). To determine whether the high-level expression of ORF 50 during a primary infection also results in the induction of KSHV lytic cycle gene expression, a KSHV gene array was used to examine the expression profiles of all KSHV genes in the infected HMVEC-d and HFF cells. We selected two time points p.i., with one time point representing the maximum ORF 50 expression and another representing the time when ORF 50 expression was at a decline (Fig. 2). For HMVEC-d cells we selected 2 and 8 h p.i., and 8 and 24 h p.i. were selected for HFF cells. Total RNA from the infected or uninfected cells was converted into cDNA and subjected to gene array analyses as described in Materials and Methods. The resulting normalized, calibrated ratios of expression of each gene were used to construct the cluster diagrams (Fig. 4 and Table 3).
FIG. 4.
Gene array analyses of KSHV latent and lytic genes in HMVEC-d and HFF cells. (A) Cluster diagram representing the expression of KSHV latent and lytic genes. HMVEC-d cells were infected with KSHV for 2 and 8 h, and HFF cells were infected for 8 and 24 h. KSHV-positive BCBL-1 cells induced with TPA for 72 h were used as a control for the gene array analyses. The RNA transcripts from the infected and uninfected samples were converted into cDNA in the presence of fluorescent dyes Cy-3 and Cy-5 dUTPs, respectively. All samples were used in separate reactions of real-time DNA-PCR (without RT) to confirm the absence of contaminating DNA. The fluorescently labeled cDNAs were hybridized with KSHV arrays spotted on ceramic chips and scanned using an Affymetrix scanner. The average intensity of three different spots of each individual gene was calculated, and background intensities were subtracted and normalized across different arrays using cellular housekeeping gene transcripts. The normalized infected Cy-3/uninfected Cy-5 expression intensity ratios were used to construct the cluster diagrams, and the intensity ratios closer to 1.0 and below were taken as an indication of nonexpressed genes. KSHV genes detected at higher levels during infection are shown in progressively darker shades of red. (B) KSHV genes expressed in HMVEC-d and HFF cells during early times after primary infection are represented in a genomic view. The KSHV genes are designated per the methods of Jenner et al. (22) and color coded to represent the latency and lytic (primary, secondary, and tertiary) cycle-associated genes.
TABLE 3.
Genome array analyses: intensity ratios of KSHV latent and lytic genes expressed early during infection of HMVEC-d and HFF cellsa
HMVEC-d cells were infected with KSHV for 2 and 8 h, and HFF cells were infected for 8 and 24 h. KSHV carrying BCBL-1 cells induced with TPA for 72 h were used as a control for the gene array analyses. Affymetrix analyses of KSHV genes in TPA-induced BCBL-1 cells (control) and infected HMVEC-d and HFF cells described in the legend for 4A were used to calculate the intensity ratios of expressed genes and to construct the table. The normalized infected Cy-3/ uninfected Cy-5 expression intensity ratios were used to construct the cluster diagrams. Intensity ratios closer to 1.0 and below were taken as an indication of nonexpressed genes, and intensity ratios less than 2.0 and below were considered as nonsignificant expression. KSHV genes were categorized as per Jenner et al. (22) and color coded to represent the latency and lytic cycle-associated genes as described for Fig. 4. Green, latency-associated genes; blue, purple, and orange; primary, secondary, and tertiary lytic genes, respectively; white, uncharacterized.
In the BCBL-1 cells induced with TPA for 72 h, expression of almost all the viral genes was detected (Fig. 4A). In contrast, though lytic cycle gene expression was observed in the infected HMVEC-d and HFF cells, there was a dramatic absence of a number of genes (Fig. 4 and Table 3). The validity of the viral chip analysis was confirmed by RT-PCR, and all the genes selected showed the presence or absence of the corresponding mRNAs by RT-PCR according to what was observed in the gene array experiments (see Fig. 5). The number of viral genes expressed in HMVEC-d and HFF cells was remarkably similar, and almost identical genes were expressed in both cells with similar kinetics. Of the total 94 KSHV genes described by Jenner et al. (22), 31% (29) were expressed in both cell types, which included all four KS lesion-associated latency genes and 11 primary, 8 secondary, 5 tertiary, and 1 uncharacterized lytic genes (Fig. 4 and Table 3). High gene expression was observed at a time point when ORF 50 expression was at the maximum, with considerable reduction at the time point when ORF 50 expression was much below that at the early time points. At 2 h p.i. in HMVEC-d cells and at 8 h p.i. in HFF cells, when ORF 50 expression was the maximum, only 32 and 31% of the total 94 genes was expressed in HMVEC-d and HFF cells, respectively. At 8 h p.i. in HMVEC-d cells and 24 h p.i. in HFF cells, this was reduced to 12 and 14%, respectively. The patterns of gene expression were closely similar and highly reproducible between experiments.
FIG. 5.
(A) RT-PCR confirmation of KSHV transcription in HMVEC-d and HFF cells. Total RNA prepared from the infected or uninfected cells was treated with DNase I and amplified by RT-PCR. The products were separated by gel electrophoresis, visualized, and quantified. Lane 1, TPA-induced BCBL-1 cells; lanes 2 to 5, HMVEC-d cells after 2, 8, 24, and 48 h p.i., respectively; lanes 6 and 7, HFF cells after 8 and 24 h p.i., respectively. −RT, all samples were used in separate reactions of DNA-PCR (without RT) to confirm the absence of contaminating DNA, and an example is shown. (B) Cluster diagram representing KSHV latent and lytic gene transcription in the HMVEC-d cells after TPA stimulation. HMVEC-d cells were infected with KSHV and, after 48 h, cells were stimulated with 20 ng of TPA/ml for 12 or 48 h and analyzed by KSHV gene array as per the procedures described for Fig. 4A.
In contrast to the activation of the KSHV lytic cycle and progeny virus production after the overexpression of ORF 50 in latently infected cells (5, 14, 18, 29, 46, 47, 50, 55), it was evident from these results that even though high levels of ORF 50 were expressed following primary infection, not all lytic KSHV genes were activated. Moreover, even the initially activated genes either declined or were not detected subsequently in significant levels. Since there appeared to be a block in the ORF 50-mediated lytic cycle activation, to determine the stage of the block we analyzed the temporal patterns of KSHV latent and lytic cycle gene expression and their predicted putative functions as well as reported functions.
Expression of latency-associated KSHV genes was initiated early during infection of HMVEC-d and HFF cells, and they continued to be synthesized subsequently.
In the KS lesion endothelial cells, expression of KSHV ORF 73, ORF 72, K13, and K12 genes was detected (14, 18, 29, 46, 47, 48, 56). In the infected HMVEC-d and HFF cells, ORF 73, ORF 72, and K13 genes were expressed from earlier times p.i. and increased at later time points (Table 4). RT-PCRs amplifying the ORF 72 and 73 and K13 genes confirmed the specificity of the gene array (Fig. 5A). The ORFs 73, 72, and K13 are expressed from a common promoter (14, 29), and K12 is expressed from a separate promoter (14, 29). K12 was classified as a primary lytic gene (immediate-early/early), since it was activated by RTA and the synthesis increased after the induction of the lytic cycle (14, 29, 31, 45, 50). Although K12 transcription was detected at a moderately higher level in HMVEC-d cells at 2 h p.i. and in HFF cells at 8 h p.i., it was undetectable at 8 h p.i. in HMVEC-d cells and was greatly reduced in HFF cells at 24 h p.i. (Table 4). LANA-2, an ORF 10.5-encoded latency-associated protein, can be detected in the B cells of BCBL and multicentric Castleman's disease tissue and in BCBL cell lines, but not in KS tissue (14, 29, 41). It is interesting that the expression of ORF 10.5 was detected in the TPA-induced BCBL-1 cells but not in the infected HMVEC-d and HFF cells (Table 4). This finding also confirmed the specificity of the gene array and the adherent cell infection analyses. These data demonstrated that the latency-associated ORF 73, ORF 72, and K13 genes were expressed even during the early stages of primary infection and continued to be synthesized subsequently.
TABLE 4.
Expression of latency- and RTA-associated KSHV gene expression early during infection of HMVEC-d and HFF cells
| Gene | Expression in cell typea
|
||||
|---|---|---|---|---|---|
| BCBL/TPA | HMVEC-d
|
HFF
|
|||
| 2 h | 8 h | 8 h | 24 h | ||
| Latency associated | |||||
| ORF K10.5 | ++ | − | − | − | − |
| ORF K13 | ++ | + | ++ | ++ | ++ |
| ORF 72 | ++ | ++ | ++ | + | ++ |
| ORF 73 | ++ | ++ | +++ | ++ | +++ |
| ORF K12 | ++++ | ++ | − | ++ | + |
| RTA activatedb | |||||
| ORF K8 (PL) | + | ++ | (+) | ++ | − |
| ORF K5 (PL) | ++++ | +++ | +++ | +++ | + |
| ORF K2 (PL) | + | ++ | − | ++ | − |
| ORF K12 (PL/Lat) | ++++ | ++ | − | + | + |
| Nut-1 (PL) | ++++ | + | − | − | − |
| ORF 57 (PL) | ++ | − | − | − | − |
| ORF 6 (PL) | +++ | − | − | − | − |
| ORF K9 (SL) | ++ | − | − | − | − |
| ORF 59 (SL) | ++++ | + | − | − | − |
| ORF 65 (SL) | ++ | − | − | − | − |
| ORF K3 (SL) | +++ | − | − | − | − |
| ORF K1 (TL) | ++ | − | − | − | − |
| ORF K8.1A (TL) | + | ++ | − | ++ | − |
| ORF 21 (TL) | +++ | − | − | − | − |
| Total % expressed RTA-activated genes | 100 | 43 | 7 | 36 | 14 |
For presentation purposes, the expression ratios of KSHV genes in TPA-induced BCBL-1 cells (BCBL/TPA) and infected HMVEC-d and HFF cells described for Fig. 4A and Table 3 were used to assign arbitrary values. Symbols: −, <2; +, <5; ++, >5; +++, >10; ++++, >25 gene expression intensity ratio; (+), detected by RT-PCR but not by gene array. Ratios less than 2 were not considered significant.
For calculating the percentage of expressed RTA-activated genes in HMVEC-d and HFF cells, expression in TPA-induced BCBL cells was considered 100%. PL, primary lytic gene; SL, secondary lytic gene; TL, tertiary lytic gene; PL/Lat, primary lytic/latency associated.
Only a limited number of KSHV primary (immediate-early/early), secondary (early), and tertiary (late) lytic cycle genes were expressed early during infection of HMVEC-d and HFF cells.
Including ORF 50 genes, 23 (100%) primary lytic genes were expressed in the TPA-induced BCBL cells (Fig. 4; Table 3) (22). In contrast, in the HMVEC-d cells 12 (52%) and 4 (17%) primary lytic genes were expressed, at 2 h and 8 h p.i., respectively. In the HFF cells, 11 (48%) and 4 (17%) primary lytic genes were expressed at 8 and 24 h p.i., respectively. Similar genes were expressed in both cell types with almost similar intensities, with the exception of the Nut1 gene (49) absent in HFF cells (Fig. 4; Table 3). In the TPA-induced BCBL cells, 29 (97%) of the secondary lytic genes were expressed, while only 7 (23%) and 2 (10%) were detected in both HMVEC-d and HFF cells at early as well as late times p.i. (Fig. 4; Table 3). These results demonstrated a drastic reduction in the expression of KSHV secondary lytic genes compared to that of the primary lytic genes. Expression of tertiary lytic genes was almost comparable to that of secondary lytic genes, with 93% of genes expressed in the TPA-induced BCBL cells while only 19 and 4% of tertiary lytic genes were expressed at 2 h and 8 h p.i. in HMVEC-d cells (Fig. 4; Table 3). At 8 and 24 h p.i., HFF cells expressed only 19 and 7% of the tertiary lytic genes (Fig. 4; Table 3).
Only a limited number of ORF 50-activated KSHV genes were expressed in infected HMVEC-d and HFF cells.
After the induction of the KSHV lytic cycle from the latently infected cells by TPA and histone deacetylase inhibitors, the ORF 50 gene was rapidly expressed within 2 to 4 h, well before many other mRNAs, and the expression was resistant to inhibitors of protein synthesis (14, 29, 45, 50, 55). These characteristics classified ORF 50 as an immediate-early gene. When plasmids constitutively expressing the ORF 50 protein were transfected into the BCBL cell lines harboring latent KSHV, activation of ORF 50 as well as kinetically appropriate expression of downstream early- and late-stage lytic KSHV mRNAs and proteins were observed (14, 29, 50, 55). Various studies have shown that ORF 50 can activate KSHV primary (immediate-early/early) genes (K8, K5, K2, K12, Nut1, ORF 57, and ORF 6), secondary (early) genes (K9, ORF 59, ORF 65, and K3), and tertiary (late) lytic cycle genes (K1, K8.1A, and ORF 21) (14, 29, 31, 45, 50, 55). Whether ORF 50 protein stimulates each of these genes by the same mechanism or different mechanisms is not known.
The gene array data were further analyzed to determine whether the high level of ORF 50 expression in these cells early during infection also results in the activation of its target genes (Table 4). The differential expression of ORF 50 shown in Fig. 2 was also confirmed by the gene array (Fig. 4A) and by the RT-PCRs (Fig. 5A). Even though the maximum expression of ORF 50 was seen between 2 and 8 h p.i. in HMVEC-d and HFF cells, of the 14 genes that have been shown to be activated by ORF 50 in HMVEC-d cells only 6 genes at 2 h p.i. (43%) and 1 gene at 8 h p.i. (7%) were expressed (Table 4). Similarly, in HFF cells, only five genes (36%) and two genes (14%) were expressed at 8 and 24 h p.i., respectively (Table 4). Among the seven primary (immediate-early/early) lytic target genes of ORF 50, expression of five genes in HMVEC-d cells (K8, K5, K2, K12, and Nut1) and four genes in HFF cells (K8, K5, K2, and K12) were observed (Table 4). In contrast, only a very low level of ORF 59 was observed at early time points, and none of other three secondary (early) target genes of ORF 50 was activated in both the cells. Among the three tertiary (late) target genes of ORF 50, only the K8.1A gene was expressed in both the cells at the earlier time points (Table 4).
The immediate-early gene ORF K8 encoding K-bZip is activated immediately downstream of RTA in the lytic cycle cascade activation (14, 29). However, though K8 was expressed at a moderately high level at 2 and 8 h p.i. in HMVEC-d and HFF cells, respectively (Table 3), it was not detected at later time points (Table 4), which was also confirmed by the RT-PCR (Fig. 5A). In contrast, the K5 gene (IE-1; BHV4-IE-1 homolog), also an immediate-early gene, was expressed throughout the time of observation at higher levels (Fig. 4A; Tables 3 and 4), which was also confirmed by the RT-PCRs (Fig. 5A). These data suggest that only a subset of ORF 50 target genes was activated during the early stage of infection and was not sustained at later times of infection. Together with the expression of only a limited set of KSHV immediate-early genes, these results suggest that the arrest in lytic gene expression occurs at early stages downstream of ORF 50 expression.
Overall, 52% of primary, 23% of secondary, and 19% of tertiary lytic genes were expressed during the initial stages of HMVEC-d and HFF cell infection. These results suggested that the lytic cycle was initiated soon after infection in the HMVEC-d and HFF cells, probably induced by the high level of expression of RTA. However, this was not sufficient for the complete lytic cycle to proceed, which was evident from the reduction in the expression levels and especially in the number of lytic genes at later stages of infection, with a sustained expression of latency-associated genes. To understand the stage of viral lytic cycle block, we further analyzed the expressed and nonexpressed KSHV lytic cycle genes, based on their demonstrated functions as well as putative functions that were predicted based on analogy with other herpesviruses (22, 29, 34, 44).
A majority of KSHV genes involved in viral DNA synthesis were not expressed in the HMVEC-d and HFF cells.
After infection of target cells, herpes viral lytic cycle DNA replication has been shown to occur downstream of immediate-early/early gene expression and is dependent upon several virally encoded gene products (24, 28, 42). At least 14 KSHV genes have been categorized as involved in DNA synthesis and nucleic acid metabolism (22, 29, 34, 44). However, only three of these genes were synthesized during primary infection. The KSHV primary lytic DNA polymerase gene encoded by ORF 9 has been shown to be involved in KSHV DNA synthesis (14, 29). Moderate levels of ORF 9 synthesis were observed at 2 and 8 h p.i. in HMVEC-d and HFF cells, respectively, and were not detected subsequently (Fig. 6A and Table 3). The secondary lytic ORF 46 gene encoding uracil DNA nucleosidase was expressed at moderate levels at 2 and 8 h p.i. in HMVEC-d cells and only at 8 h p.i. in HFF cells. The tertiary lytic ORF 54 gene encoding dUTPase was expressed at only 2 h p.i. in HMVEC-d cells and at 8 and 24 h p.i. in HFF cells (Fig. 6A and Table 3). Only a low level of secondary lytic ORF 59 (a processivity factor critical for viral DNA synthesis) was expressed in these cells (Table 3). In contrast, KSHV primary lytic ORF 6 (single-stranded DNA binding protein), ORF 56 (DNA replication protein), ORFs 60 and 61 (RNase reductase), ORF 70 (thymidylate synthase), and tertiary lytic ORF 21 (thymidine kinase), ORF 37 (alkaline exonuclease), and ORFs 40, 41, and 42 (helicase/primase) necessary for viral DNA synthesis were not detected (Fig. 6A and Table 3). These results suggest that lytic cycle-associated KSHV DNA replication may not occur during the initial stages of HMVEC-d and HFF cell infection.
FIG. 6.
Clustering of KSHV genes expressed in HMVEC-d and HFF cells according to the predicted putative functions and demonstrated functions. The cluster diagram representing the expression of KSHV genes and the experimental procedures were as described for Fig. 4A. KSHV genes detected at higher levels are shown in progressively darker shades of red. The designations of KSHV genes are as per Jenner et al. (22).
The majority of KSHV genes encoding structural and membrane-associated proteins were not expressed in the HMVEC-d and HFF cells.
Among at least 23 genes predicted to encode KSHV structural and membrane-associated proteins, only 6 were expressed during HMVEC-d and HFF cell infections (Fig. 6B and Table 3). However, sustained expression was observed only with ORFs 64 and 75, predicted to encode the tegument proteins. Expression of ORF 8 (glycoprotein B), ORF 29B (packaging protein), ORF 47 (glycoprotein gL), and gpK8.1A was observed only during the earlier time p.i. (Fig. 6B and Table 3).
KSHV lytic genes with immunomodulation and antiapoptotic functions were expressed early during primary infection.
The immediate-early (primary lytic) gene K5 was continuously synthesized at higher levels during both early and late time points (Fig. 6C and 5A; Table 3). The K5 gene product (MIR-2) has been shown to be involved in the downregulation of major histocompatibility complex (MHC) class I, ICAM-1, and B7-2 molecules, which are essential in the elimination of virus-infected cells by cytotoxic T and NK cells and for T-cell stimulation (10, 20, 21, 27, 52). In contrast, the K3 gene, also known to encode a protein (MIR-1) involved in the downregulation of MHC class I molecule, was not detected (Fig. 6C and Table 3). The primary lytic K4 (vMIP-II) and K6 (vMIP I-α) were expressed during the early time of KSHV infection in both cells, and these MIPs are predicted to protect the virus-infected cells (14, 29, 31). The primary lytic K2 gene encoding vIL-6 and the secondary lytic vIRF2, both with immunomodulatory functions and protecting cells from interferon action (4, 8, 12, 26, 35, 38, 57) were also synthesized during the earlier time points of infection (Fig. 6C and 5A and Table 3). Similarly, the primary lytic K7 antiapoptotic gene (17, 51, 54) was synthesized in both cell types during the early time of infection, along with the latent K13 protein encoding the antiapoptotic vFLIP protein (6, 29) (Fig. 6D and Table 3). These data suggested that even though the full complement of KSHV lytic genes was not expressed in HMVEC-d and HFF cells, some of the genes encoding proteins with immunomodulatory functions and antiapoptotic function were synthesized during the initial stages of infection (Fig. 6C and Table 3).
RT-PCR confirmation of gene array data.
To confirm the gene array results by an independent method, RT-PCR analyses were carried out for 13 selected KSHV genes with differential gene array expression (Table 3). In addition to the RNA samples used for the array analysis, we also determined their expression levels in the independently derived RNA samples of infected and mock-infected cells (Fig. 5A). Since several studies have reported the detection of the lytic cycle in about 1 to 5% of infected cells after 48 h p.i. (5, 11, 19, 25, 30, 53), to rule out the influence of spontaneous reactivation on the transcription by the primary infection RNA isolated at 2, 8, 24, and 48 h p.i. of HMVEC-d cells was examined by RT-PCR (Fig. 5A).
Consistent with the gene array data, latency-associated ORF K13, 72, and 73 transcripts were consistently detected at high levels (Fig. 5A, lanes 2 to 7), even at 48 h p.i. Similar to the results from real-time RT-PCR and gene array, we observed a more than 50% reduction in the ORF 50 transcript level by 48 h p.i. in HMVEC-d cells (Fig. 5A, lanes 2 to 5). Among the immunomodulatory genes detected by gene array, expression of K5 transcript was detected at high levels even at 48 h p.i. (Fig. 5A, lanes 2 to 5). In HMVEC-d cells, expression levels of K2 (vIL-6), K4 (vMIP-II), and K6 (vMIP-I) were detected until 8 h p.i. and undetectable by 24 h p.i., and the vIRF2 transcript was detected until 24 h p.i. (Fig. 5A, lanes 2 to 5). Expression levels of ORFs K8, 7, and 8 were not detected at 24 or 48 h p.i., and ORF 25 was not detected even at 48 h p.i. (Fig. 5A and Table 3). Our RT-PCR confirmation was not strictly quantitative, since we were comparing individual cell analyses with results on mass cultures. Hence, our data do not indicate whether the detection of KSHV transcripts represented expression in all the infected cells and the variations in the expression between cells. Nevertheless, these results suggested that the observed limited lytic cycle gene expressions under our conditions of infection were probably not influenced by spontaneous reactivation. We have also observed similar results with an input MOI of 10 viral DNA copies per cell. We observed only minor differences between the array and RT-PCR assays, and the differences in the magnitudes of induction detected could be due to the increased sensitivity of the RT-PCR and/or the potential for chip saturation. However, importantly, RT-PCR confirmed the expression of the genes detected by the arrays, in the RNA samples used for array analyses as well as in the independently derived set of RNA samples.
KSHV early lytic immunomodulatory K5 protein was detected at later time points of infection in the majority of infected cells.
Among the limited lytic KSHV genes expressed during primary infection, the K5 gene was expressed at sustained higher levels. To determine the extent of K5 expression, infected cells were examined by immunoperoxidase assay with rabbit anti-K5 antibodies (20, 35), along with antibodies to RTA and LANA-1 (Fig. 7). LANA-1 was detected in more than 90% of the infected cell nuclei, and results at 5 days p.i. are shown in Fig. 7A. Similarly, ORF 50 was detected in the nuclei of >90% of all infected cells at 5 days p.i. (Fig. 7B). Expression of K5 was also detected in >90% of the entire infected cell population as early as 2 h p.i. (data not shown) and at 24 h and 5 days p.i. (Fig. 7C, D, and E). The specificity of these reactions was demonstrated by the absence of reactivities of anti-K5 antibodies with the uninfected cells (Fig. 7F) and by the absence of reactivities of normal rabbit antibodies with the infected cells (data not shown). Comparable to the previous studies (20), K5 was detected predominantly in the cytoplasm and in the perinuclear region of infected HMVEC-d cells (Fig. 7C to E). The K5 staining was comparatively weaker, which may have been due to the use of monoclonal antibodies and diffuse cytoplasmic distribution.
FIG. 7.
Immunoperoxidase assay detecting KSHV K5, LANA1, and RTA proteins in HMVEC-d cells. Cells in eight-well chamber slides were infected with KSHV for 2 h, washed, and incubated at 37°C. At different time points, cells were washed, fixed with cold acetone, incubated at 4°C overnight with predetermined dilutions of antibodies against K5, ORF 73, or ORF 50 or with preimmune antibodies, processed for immunostaining as described in Materials and Methods, and examined under a light microscope. (A) LANA-1 expression in HMVEC-d cells after 5 days p.i. (B) RTA expression in HMVEC-d cells after 5 days p.i. (C to E) K5 expression in HMVEC-d cells after 24 h (C) and 5 days (D and E) p.i. (F) Uninfected HMVEC-d cells with anti-K5 antibodies. Magnification: ×20 (A); ×40 (B, C, D, and F); ×100 (E). The arrows indicate the cytoplasmic and perinuclear staining seen with anti-K5 antibodies.
HMVEC-d cells after primary infection could be induced by TPA to express the full range of KSHV lytic cycle genes.
KSHV enters the latency cycle soon after infection of endothelial cells in vitro, and the lytic cycle can be activated by TPA, ORF 50, or other inducing factors (5). Addition of 20 ng of TPA/ml along with virus during primary infection resulted in the virtual absence of KSHV gene expression (data not shown). This was not unexpected, due to the fact that TPA is a powerful inhibitor of endocytosis and blocks KSHV entry into the target cells (1). To determine whether the primary infections in our experiments can also lead to a latent infection, HMVEC-d cells were infected with KSHV and after 48 h, when most of the lytic cycle genes were not detected (Fig. 5A), cells were incubated with 20 ng of TPA/ml for 12 or 48 h and the gene expression profiles were examined by viral array hybridization.
After 12 h of TPA induction of latently infected HMVEC-d cells, 73 out of 94 (77%) KSHV transcripts could be detected (Fig. 5B). This is in contrast to the 30 transcripts detected in the early stages of primary infection, which declined to 10 genes at 8 h p.i. (Fig. 4A). Approximately 87 and 80% of primary and secondary lytic genes, respectively, were activated after 12 h of TPA treatment, whereas only 59% of the tertiary lytic genes could be detected at the same time point. After 48 h of TPA induction, 93 out of 94 (99%) KSHV transcripts could be detected by the array. Most of the lytic genes not detected during primary infection were expressed at higher levels after TPA induction. The detection of nearly 100% of viral transcripts after TPA induction clearly demonstrated that KSHV infection of HMVEC-d cells in our system resulted in a latent infection, and the latency could be switched subsequently to express the full complement of lytic cycle genes. Whether this reactivation represents all infected cells or few cells was not determined. However, since a recent study showed that TPA or ORF 50 can activate the lytic cycle in >80% of LANA-1-expressing endothelial and other cells after 48 h p.i. with BCBL-1-derived virus (5), similar to the virus used in the present study, we presume that our results may also represent reactivation from the majority of the infected cells.
DISCUSSION
In this report, we have described the extensive and thorough analyses of KSHV gene expression events immediately following infection of two different primary cells. These results represent a significant advance over our current knowledge regarding KSHV gene expression and biology. Nylon and glass slide arrays have been used to describe the KSHV transcriptome in uninduced and TPA-induced PEL cells as well as the expression pattern differences between TPA-induced and RTA-induced PEL cells (22, 31, 37). RT-PCR was also used to detect the latency-associated transcripts in BCBL-1 cells and the KSHV transcription profile in the KS lesion (16). However, all these studies concentrated around the latently infected PEL cells, and the present study is the first of its kind documenting the KSHV gene expression profile in primary permissive cells immediately after de novo infection.
The validity of the gene array was shown by the detection of limited lytic cycle gene transcription, their decline at later time points, and transcription of all the lytic cycle genes by TPA after 48 h p.i. The consistency of RT-PCR results with the array data and the absence of transcription of the type I-B latency gene ORF 10.5 (LANA-2/vIRF3) (14, 22, 29, 41, 45), shown to be transcribed only in KSHV-associated B-cell malignancies and not in endothelial-derived KS tumors, also validate the array data. Furthermore, even though RT-PCR is more sensitive and accurate than the arrays, similar to the array data, RT-PCR detected the K8 and ORF 8 gene only during earlier times p.i. and not at later times p.i. and did not detect ORF 25 at the observed time points. These results also validated the array data. We could not detect the expression of ORFs 32, 44, and 49 in the TPA-induced BCBL cell array; however, these were detected when the latently infected HMVEC-d cells were induced with TPA. In contrast, ORF 37 encoding the KSHV alkaline exonuclease was detected in the BCBL cells but not in the TPA-induced HMVEC-d cells. This discrepancy could be due to cell type variation, expression levels, time of detection, and/or our criterion of considering gene expression intensity ratios of less than 2 as not significant. Apart from this, there were no other significant array data discrepancies.
The initiation of latent and lytic cycle genes concurrently during KSHV infection of primary human adherent HMVEC-d and HFF cells is one of the important observations in this study. Whether this phenomenon also occurs in other in vitro target cells, such as human B cells, keratinocytes, and epithelial cells and animal cells, remains to be elucidated. Similarly, whether wild-type KSHV from the saliva of immunocompetent and immunodeficient individuals (14, 29, 47) and from KS tissues (29, 47) behaves in a similar fashion is not known at present. We have also observed similar results with an input MOI of 10 viral DNA copies per cell. Although we have optimized our infection procedures and obtained nearly synchronized infection by using input purified virus with equal DNA copy numbers and we have made consistent observations, we interpret our results with caution. Some variations in the kinetics might be expected, since KSHV infection procedures are influenced by several factors, such as the use of TPA-induced BCBL cells as the source of virus stock, method of virus stock preparation, batch-to-batch variations in the virus stocks, and possible defective-genome-containing viruses.
Our experimental design, including the examination of KSHV gene expression kinetics at very early time points after infection, and utilization of sensitive real-time RT-PCR and gene arrays representing the whole range of latent and lytic cycle KSHV genes, differs considerably from those of the previous KSHV in vitro infection studies. Earlier studies preceded the availability of sensitive techniques and examined limited KSHV gene expression in the context of testing the ability of target cells to support a lytic infection, as measured by the production of serially passable infectious virus. These studies were done by infecting a variety of human and animal cells, including primary endothelial cells as well as endothelial cells immortalized by telomerase (TIME) or E6/E7 proteins of human papillomavirus (5, 11, 13, 19, 25, 30, 53). All of these studies differed from our study, as they were conducted at a later time point of infection, which ranged from 48 h to several days. These studies included 48 h p.i. in primary human umbilical vein endothelial cells with the recombinant BAC36 KSHV (19), 48 h p.i. in telomerase-transformed endothelial (TIME) cells with the BCBL-1 virus (25), 24 to 48 h p.i. in a variety of human and animal target cells including TIME cells with the BCBL-1 virus (5, 25), 48 h p.i. in primary DMVEC (identical to the HMVEC-d cells used here) and TIME-DMVEC cells with BCBL-1 virus (52), 6 days p.i. in primary human keratinocytes with BCBL-1 virus (14, 29), 25 days p.i. in primary DMVEC with virus from JSC cells (11), and 7 to 14 days p.i. in human papillomavirus E6/E7-transformed human dermal endothelial cells with BCBL-1 virus (30).
Since these previous in vitro studies utilized later time points of infection, the detection of LANA-1 coupled with the absence of infectious virus and lytic cycle ORF 59 and gpK8.1 lead to the notion that the primary KSHV infection leads to the establishment of latency without any apparent lytic cycle initiation. However, several studies also reported the detection of lytic cycle markers such as ORF 59 and gpK8.1A in <5% of cells after 48 h p.i. (5, 11, 25, 52), and these were considered cells entering spontaneous activation of the lytic cycle. In a recent study using recombinant BAC36 KSHV, Gao et al. (19) detected LANA-1 in >90% of the infected primary human umbilical vein endothelial cells and KSHV ORF 65 (minor capsid protein) in <5% of cells after 2 days p.i. The reason for only the small percentage of cells entering the lytic cycle is not known at present. Our study shows that lytic gene expression of KSHV is initiated soon after the infection. However, this initial lytic activation is not successful enough to proceed to a full-fledged lytic replication and progeny virus development. Rather, the lytic cycle was less aggressive, with the expression of only a subset of lytic cycle genes and subsiding rapidly thereafter. Results presented here suggest that, depending upon the time of examination, many of the lytic cycle genes may not be detectable at later time points. However, these may become detectable if there is a spontaneous reactivation of the lytic cycle in a certain percentage of infected cells at later time points. Since low levels of ORF 59 transcripts in HMVEC-d cells and gpK8.1A transcripts in both cell types were also detected at the initial time of infection in our experiments, detection of these proteins at earlier time points by the above-mentioned studies may represent an outcome of primary infection, and detection at later time points may represent entry into the lytic cycle.
The present study revealed a number of unique observations as well as raised several important questions regarding the biology of KSHV infection.
Among the well-characterized in vitro infections of other herpesviruses, concurrent KSHV lytic and latent gene expression is a unique observation. The in vitro infection of human B cells by the gamma-1-EBV results in transformation and establishment of latency (24). Whether EBV also initiates a limited lytic cycle is not known. In vitro infection by alpha- and betaherpesviruses results in the rapid initiation of lytic cycle, virus progeny formation, and cell death (28, 42). This is probably not unexpected, since the majority of cells used in these studies do not represent the in vivo target cells in which these viruses enter latency (28, 42). For the alpha- and betaherpesviruses there is no clear-cut in vitro model for latency, and there is no good reproducible in vitro neuronal cell model in which the alphaherpesviruses can establish a latent infection. Whether concurrent in vitro latent and lytic infections also occur in other herpesvirus-infected cells in which these viruses are shown to be in a latent state in vivo needs to be examined.
Another important observation in this study was that, despite KSHV transactivator ORF 50 gene expression at high levels soon after infection, only a subset of ORF 50 target genes were activated during the early stage of infection, and these were not sustained at later times of infection. This implies that expression of ORF 50 alone is not sufficient to fully activate the lytic cycle during the early time of infection of primary endothelial and fibroblast cells. Since providing ORF 50 in trans has been shown to activate the KSHV lytic cycle cascade in latently infected cells (5, 14, 29, 50, 55), several important questions arise from the above observation: why doesn't the expression of ORF 50 during primary infection result in the completion of the lytic cycle cascade? What factor(s) is responsible for the initiation of the latent and lytic cycles of KSHV during primary infection? What factor(s) governs the outcome of infection?
Several possible explanations can be speculated for these questions. Activation of a subset of ORF 50 target genes, expression of only a limited set of KSHV immediate-early genes, and the absence of a true lytic cascade suggest that the arrest in lytic gene expression occurs at early stages downstream of ORF 50 expression. There are several factors that can potentially have an influence over the ORF 50 functions during a primary infection: for example, (i) the nature of a virion-associated genome entering the nucleus and its modifications, such as methylation, and (ii) posttranslational modifications of ORF 50 synthesized from the input DNA and their potential functional differences from the ORF 50 provided in trans (5, 14, 29, 50, 55). Modification of protein influencing the different functions has been well documented for the herpes simplex virus type 1 transcriptional transactivator protein ICP-4, which undergoes modifications such as extensive phosphorylation, poly-ADP ribosylation, and nucleotidylation (42). Whether the ORF 50 protein undergoes such modifications and whether they exert influence on the ORF 50 activation cascade remain to be studied. The functions of ORF 50 can also potentially be influenced by the newly synthesized ORF 73 protein (11, 29) and by a KSHV virion-associated factor(s). Further studies are required to analyze these possibilities.
Furthermore, as depicted in the model shown in Fig. 8, there are several important multistep complex events that precede the initiation of KSHV gene expression in the nucleus. It is critical not to overlook the influence of host cell signal pathways and host cell gene expression induced by KSHV interactions with the host cell surface receptor molecules, since our studies suggest that the above early events may have a strong influence over host cell gene expression (2, 32, 33). For example, in an integrin-dependent manner, KSHV induced the mitogenic ERK signal pathways within 5 min p.i., and this induction was dependent upon viral gene expression (32). In addition, within 2 h p.i. in endothelial cells, KSHV modulated the transcription of several host cell molecules that have hitherto not been linked to KSHV biology, including a variety of genes involved in major cellular processes such as transcription, apoptosis, cell cycle, angiogenesis, tumorigenesis, microtubular dynamics, structural proteins, transport, and signal transduction (33). Since ORF 50 must be interacting with several host-cell transcriptional and other proteins to mediate its transactivating functions and the induction of the lytic cycle cascade, the inability of ORF 50 to activate the lytic cycle cascade during a primary infection may be also due to the negative and/or positive regulatory control over such functions by the early events of infection. The ability of ORF 50 to activate KSHV after primary infection of endothelial cells was shown by providing ORF 50 in trans to the cells usually after 48 h p.i. (5), a time point at which the influence of early events of infection may have subsided.
FIG. 8.
Model for KSHV gene expression early during in vitro infection. Early events of KSHV infection and gene expression in the target cells are depicted in overlapping dynamic phases. KSHV binds to the cell surface via its interactions with heparan sulfate (HS) (3), integrins (2), and possibly another yet-to-be-identified molecule(s) (phase 1). In phase 2, virus enters into the target cells (1-3, 32), probably overlapping with the induction of host cell signal pathways (32). In phase 3, viral capsid and tegument moves in the cytoplasm, probably facilitated by the induced signal pathways, and probably overlaps with the signal pathways induced during phase 4 host cell gene transcription and expression (33). In phase 5, viral DNA enters into the nucleus, followed by concurrent latent and lytic cycle gene expression as shown in this study, which is probably influenced by the KSHV-induced signal pathways and expressed host cell genes. Phase 6 involves the overlapping viral gene-induced host cell gene expression, which also may exert an influence on subsequent viral gene expression. During the early time of primary infection of endothelial and fibroblast cells, KSHV expresses the lytic ORF 50 gene and the latent ORF 73 gene concurrently, with initially higher levels of ORF 50 expression followed by a rapid decline. Nearly all latency-associated and limited lytic genes are transcribed concurrently. While the expression of latent ORF 72, 73, and K13 genes continues, nearly all of the lytic genes decline or are undetectable by 24 h p.i. Only a limited number of RTA-activated KSHV genes are expressed briefly, and the majority of KSHV genes involved in viral DNA synthesis and structural proteins are not expressed. Early during infection, the lytic K2, K4, K5, K6, and vIRF2 genes with immune modulation functions and the K7 gene with antiapoptotic function are expressed, which may be providing sufficient survival time for the infected cells from the host immune surveillance apparatus and apoptosis and the sufficient advantage necessary for the establishment of latent infection during the initial time of infection.
The factor(s) governing the activation of ORF 50 gene expression during primary infection and its subsequent decline is also not known at present. Following primary infection of infected endothelial cells, a lytic cycle is detected in about 1 to 5% of cells after 48 h p.i. (5, 11, 14, 19, 25). Even though the ORF 50 transcript levels declined after 24 h, detection of the protein up to 5 days p.i., albeit at low levels, suggests that the spontaneous reactivation of the lytic cycle could be also due to the action by the ORF 50 protein after the cessation of the influence exerted by the early events of infection. Why only a small percentage of cells enter a spontaneous lytic cycle is not known, which suggests a complex interaction between KSHV latency and lytic cycle-associated proteins and host cell factors. We surmise from our observations that the regulation, function, and influence of ORF 50 protein during a primary infection are much more complex than the simple activation of the lytic cycle.
Other important questions that arise from our studies are (i) why KSHV expresses a limited number of lytic cycle genes along with latency genes, and (ii) what possible role this may play in the biology of KSHV. Initially, we considered the possibility that the initial limited lytic cycle gene expression could be due to the random transcription of newly entered viral DNA in the nucleus by the host cell transcriptional factors. However, examination of functions attributed to the transcribed genes suggests that this may not be a random process but probably is an infectious process adapted by KSHV, perhaps selected by the protection afforded to the virus to escape from apoptosis and immune responses.
There are several obstacles that viruses have to overcome during infection of target cells in the host, such as the external threats to infected cells by constant immune surveillance by cytotoxic T cells and NK cells, interferons (IFN-α, -β, and -γ), tumor necrosis factor alpha, complement, antibodies, ADCC, NK cells, cytotoxic T lymphocytes, and phagocytic cells, as well as internal obstacles such as the transcriptional block and cellular apoptosis probably triggered by the virus binding and entry processes. To establish a successful infection, KSHV must have developed many ways to manipulate and overcome these obstacles. KSHV expresses its K2, K4, K5, K6, and vIRF2 genes with immunomodulatory functions and K7 with antiapoptotic function during the initial stages of infection. Extrapolation of functions attributed to these expressed limited KSHV lytic genes suggests that they may be providing a pivotal survival advantage during the initial stages of infection (Fig. 8).
For example, the K5 gene was expressed at a sustained high level, which has been shown to be involved in the downregulation of MHC class I, ICAM-1, and B7-2 molecules, which are the most important elements necessary for the elimination of virus-infected cells by cytotoxic T and NK cells and for T-cell stimulation (10, 20, 21, 27, 52). Recent studies by Tomescu et al. (52) demonstrated the downregulation of cell surface MHC class I, ICAM-1 (CD54), and PE-CAM (CD31) in the pDMVEC after infection through coculture with induced PEL cells and in DMVEC-TIME cells infected directly with cell-free virus. These studies were done at 48 h p.i., and the downregulation was predominantly seen in the KSHV-infected LANA-positive cells. Less than 1% of cells expressed the lytic RTA and K5 (MIR-2) proteins by immunofluorescence assay examination. However, since K5 gene expression has also been shown to be independent of ORF 50 expression (35), those authors suggested that they cannot rule out the observed downregulation in part mediated by the expression of extremely low levels of lytic gene products, including K3 (MIR-1) and K5 (MIR-2), remaining below the detection levels of the used immunofluorescence assay method (52). Our demonstration of a high level of K5 expression in the primary infection is exciting, since together with the findings by Tomescu et al. (52) these results suggest that expression of K5 protein probably provides an important avenue for the in vivo-infected cells to escape immune surveillance.
We also detected the transient expression of the KSHV vIRF2 gene up to 24 h p.i. in HMVEC-d cells, whose product has been shown to have profound inhibitory effects on IFN-α and -β gene transcription, probably by its ability to bind specifically to several transcription factors, such as IRF1, IRF2, IRF3, IRF7, IRF8, Rel/p65, and CBP/p300 (4, 8, 29). These data, together with the expression of K4 (vMIP-II), K6 (vMIP-Iα), and K2 (v-IL-6) gene products, which have been shown to possess immunomodulatory functions and to protect cells from IFN action (12, 26, 29, 35, 38, 44, 47, 57), and the K7 gene encoding a protein with antiapoptotic function (17, 51, 54), suggest that these gene products probably also provide a survival advantage for the infected cells (38). KSHV ORF 9 encoding viral DNA polymerase, ORF 46 encoding uracil DNA nucleosidase, ORF 54 gene encoding dUTPase, and ORF 75 (an FGARAT homolog with a probable role in the biosynthesis of purines) were also expressed. Whether these gene products provide any functional help for the initial establishment of latency (latent viral DNA replication?) is not known at the present time. The advantage provided by these and other expressed lytic cycle proteins needs to be studied further. Though only a limited number of lytic cycle KSHV genes were expressed during a primary infection, these genes may be providing sufficient survival time for the infected cells from the host immune surveillance apparatus and from apoptosis, time probably necessary to establish the latent KSHV infection.
KS is an enigma among the known human tumors, due to its multifocal nature and the variety of cell types, such as the spindle-shaped cells and inflammatory cells, in the lesions. The ORFs 73, 72, K13, and K12 were detected in vascular endothelial and spindle cells of KS lesions, and the lytic cycle proteins were detected in about 1 to 10% of infiltrating monocytic cells of KS lesions (14, 18, 29, 46-48, 56). If in vivo KSHV infection of endothelial cells also results in concurrent latent and limited lytic gene expression as we observed here, it is possible that KS lesion endothelial cells may also represent a primary infection by KSHV. In this scenario, depending upon the time of infection prior to the lesion development, the time when the biopsy was taken for examination, and the sensitivity of the assay used, one would expect to detect a limited number of lytic gene transcripts, with varying levels in the KS spindle and endothelial cells. The finding by Staskus et al. (48), who detected the nuclear KSHV lytic Nut-1 RNA (T.1.1) in about 10% of spindle tumor cells expressing the KSHV latency-associated T0.7 RNA (K12), support such a notion. Less than 1% of Nut-1 RNA-containing KS lesion cells also expressed the KSHV major capsid ORF 25 gene (48), and enveloped herpesvirus particles also have been observed in a small percentage of KS spindle cells by electron microscopy (36), which may represent spontaneous lytic activation. Additional studies are required to examine the expression of other lytic cycle genes in KS cells to correlate the relevance of in vitro gene expression presented here with the in vivo infection of KSHV. If a similar limited number of KSHV lytic cycle genes were to be detected in the early KS lesion endothelial cells, then KS lesions may represent a stage induced by the primary KSHV infection of endothelial cells.
Acknowledgments
We thank T. Sata, NIID, Tokyo, Japan, for RTA antibodies and K. Yamanishi and K. Ueda, NIID, Tokyo, Japan, for ORF50 antibodies.
This study was supported in part by Public Health Service grants CA 75911 and CA 82056 to B.C. and by a University of Kansas Medical Center Biomedical Research Training program postdoctoral fellowship to P.P.N.
REFERENCES
- 1.Akula, S. M., P. P. Naranatt, N. S. Walia, F. Z. Wang, B. Fegley, and B. Chandran. 2003. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) infection of human fibroblast cells occurs through endocytosis. J. Virol. 77:7978-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akula, S. M., N. P. Pramod, F.-Z. Wang, and B. Chandran. 2002. Integrin α3β1 (CD49c/28) is a cellular receptor for Kaposi's sarcoma associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell 108:407-419. [DOI] [PubMed] [Google Scholar]
- 3.Akula, S. M., F.-Z. Wang, J. Vieira, and B. Chandran. 2001. Human herpesvirus 8 interaction with target cells involves heparan sulfate. Virology 282:245-255. [DOI] [PubMed] [Google Scholar]
- 4.Barnes, B., B. Lubyova, and P. M. Pitha. 2002. On the role of IRF in host defense. J. Interferon Cytokine Res. 22:59-71. [DOI] [PubMed] [Google Scholar]
- 5.Bechtel, J. T., Y. Liang, J. Hvidding, and D Ganem. 2003. Host range of Kaposi's sarcoma-associated herpesvirus in cultured cells. J. Virol. 77:6474-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belanger, C., A. Gravel, A. Tomoiu, M. E. Janelle, J. Gosselin, M. J. Tremblay, and L. Flamand. 2001. Human herpesvirus 8 viral FLICE-inhibitory protein inhibits Fas-mediated apoptosis through binding and prevention of procaspase-8 maturation. J. Hum. Virol. 4:62-73. [PubMed] [Google Scholar]
- 7.Blackbourn, D. J., E. Lennette, B. Klencke, A. Moses, B. Chandran, M. Weinstein, R. G. Glogau, M. H. Witte, D. L. Way, T. Kutzkey, B. Herndier, and J. A. Levy. 2000. The restricted cellular host range of human herpesvirus 8. AIDS 14:1123-1133. [DOI] [PubMed] [Google Scholar]
- 8.Burysek, L., and P. M. Pitha. 2001. Latently expressed human herpesvirus 8-encoded interferon regulatory factor 2 inhibits double-stranded RNA-activated protein kinase. J. Virol. 75:2345-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 10.Coscoy, L., and D. Ganem. 2001. A viral protein that selectively downregulates ICAM-1 and B7-2 and modulates T cell costimulation. J. Clin. Investig. 107:1599-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciufo, D. M., J. S. Cannon, L. J. Poole, F. Y. Wu, P. Murray, R. F. Ambinder, and G. S. Hayward. 2001. Spindle cell conversion by Kaposi's sarcoma-associated herpesvirus: formation of colonies and plaques with mixed lytic and latent gene expression in infected primary dermal microvascular endothelial cell cultures. J. Virol. 75:5614-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng, H., M. J. Song, J. T. Chu, and R. Sun. 2002. Transcriptional regulation of the interleukin-6 gene of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 76:8252-8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dezube, B. J., M. Zambela, D. R. Sage, J.-F. Wang, and J. D. Fingeroth. 2002. Characterization of Kaposi sarcoma-associated herpesvirus/human herpesvirus-8 infection of human vascular endothelial cells: early events. Blood 100:888-896. [DOI] [PubMed] [Google Scholar]
- 14.Dourmishev, L. A., A. L. Dourmishev, D. Palmeri, R. A. Schwartz, and D. M. Lukac. 2003. Molecular genetics of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) epidemiology and pathogenesis. Microbiol. Mol. Biol. Rev. 67:175-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fakhari, F. D., and D. P. Dittmer. 2002. Charting latency transcripts in Kaposi's sarcoma-associated herpesvirus by whole-genome real-time quantitative PCR. J. Virol. 76:6213-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng, P., J. Park, B. S. Lee, S. H. Lee, R. J. Bram, and J. U. Jung. 2002. Kaposi's sarcoma-associated herpesvirus mitochondrial K7 protein targets a cellular calcium-modulating cyclophilin ligand to modulate intracellular calcium concentration and inhibit apoptosis. J. Virol. 76:11491-11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganem, D. 1997. KSHV and Kaposi's sarcoma: the end of the beginning? Cell 91:157-160. [DOI] [PubMed] [Google Scholar]
- 19.Gao, S. J., J. H. Deng, and F. C. Zhou. 2003. Productive lytic replication of a recombinant Kaposi's sarcoma-associated herpesvirus in efficient primary infection of primary human endothelial cells. J. Virol. 77:9738-9749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haque, M., K. Ueda, K. Nakano, Y. Hirata, C. Parravicini, M. Corbellino, and K. Yamanishi. 2001. Major histocompatibility complex class I molecules are down-regulated at the cell surface by the K5 protein encoded by Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8. J. Gen. Virol. 82:1175-1180. [DOI] [PubMed] [Google Scholar]
- 21.Ishido, S., J. K. Choi, B. S. Lee, C. Wang, M. DeMaria, R. P. Johnson, G. B. Cohen, and J. U. Jung. 2000. Inhibition of natural killer cell-mediated cytotoxicity by Kaposi's sarcoma-associated herpesvirus K5 protein. Immunity 13:365-374. [DOI] [PubMed] [Google Scholar]
- 22.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katano, H., Y. Sato, H. Itoh, and T. Sata. 2001. Expression of human herpesvirus 8 (HHV-8)-encoded immediate early protein, open reading frame 50, in HHV-8-associated diseases. J. Hum. Virol. 4:96-102. [PubMed] [Google Scholar]
- 24.Kieff, E., and A. B. Rickinson. 2002. Epstein-Barr virus and its replication, p. 2511-2573. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 25.Lagunoff, M., J. Bechtel, E. Venetsanakos, A. M. Roy, N. Abbey, B. Herndier, M. McMahon, and D. Ganem. 2002. De novo infection and serial transmission of Kaposi's sarcoma-associated herpesvirus in cultured endothelial cells. J. Virol. 76:2440-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, M., H. Lee, J. Guo, F. Neipel, B. Fleckenstein, K. Ozato, and J. U. Jung. 1998. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor. J. Virol. 72:5433-5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Means, R. E., S. Ishido, X. Alvarez, and J. U. Jung. 2002. Multiple endocytic trafficking pathways of MHC class I molecules induced by a herpesvirus protein. EMBO J. 21:1638-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mocarski, E. S., and C. M. Courcell. 2002. Cytomegalovirus and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 29.Moore, P. S., and Y. Chang. 2002. Kaposi's sarcoma associated herpesvirus, p. 2803-2833. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 30.Moses, A. V., K. N. Fish, R. Ruhl, P. P. Smith, J. G. Strussenberg, L. Zhu, B. Chandran, and J. A. Nelson. 1999. Long-term infection and transformation of dermal microvascular endothelial cells by human herpesvirus 8. J. Virol. 73:6892-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura, H., M. Lu, Y. Gwack, J. Souvlis, S. L. Zeichner, and J. U. Jung. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible RTA transactivator. J. Virol. 77:4205-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naranatt, P. P., S. M. Akula, C. A. Zien, H. H. Krishnan, and B. Chandran. 2003. Kaposi's sarcoma-associated herpesvirus induces phosphatidylinositol 3-kinase-PKC-ζ-MEK-ERK signaling pathway in target cells early during infection: implications for infectivity. J. Virol. 77:1524-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naranatt, P. P., H. H. Krishnan, S. R. Svojanovsky, C. Bloomer, S. Mathur, and B. Chandran. 2004. Host gene induction and transcriptional reprogramming in Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) infected endothelial, fibroblast and B cells: insights into modulation events early during infection. Cancer Res. 64:72-84. [DOI] [PubMed] [Google Scholar]
- 34.Neipel, F., J. C. Albrecht, and B. Fleckenstein. 1997. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J. Virol. 71:4187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okuno, T., Y. B. Jiang, K. Ueda, K. Nishimura, T. Tamura, and K. Yamanishi. 2002. Activation of human herpesvirus 8 open reading frame K5 independent of ORF50 expression. Virus Res. 90:77-89. [DOI] [PubMed] [Google Scholar]
- 36.Orenstein, J. M., S. Alkan, A. Blauvelt, K. T. Jeang, M. D. Weinstein, D. Ganem, and B. Herndier. 1997. Visualization of human herpesvirus type 8 in Kaposi's sarcoma by light and transmission electron microscopy. AIDS 11:35-45. [DOI] [PubMed] [Google Scholar]
- 37.Paulose-Murphy, M., N. K. Ha, C. Xiang, Y. Chen, L. Gillim, R. Yarchoan, P. Meltzer, M. Bittner, J. Trent, and S. Zeichner. 2001. Transcription program of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 75:4843-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ploegh, H. L. 1998. Viral strategies of immune evasion. Science 280:248-253. [DOI] [PubMed] [Google Scholar]
- 39.Renne, R., D. Blackbourn, D. Whitby, J. Levy, and D. Ganem. 1998. Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J. Virol. 72:5182-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 41.Rivas, C., A. E. Thlick, C. Parravicini, P. S. Moore, and Y. Chang. 2001. Kaposi's sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J. Virol. 75:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roizman, B., and D. M. Knipe. 2002. Herpes simplex viruses and their replication, p. 2339-2359. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 43.Roizman, B., and P. E. Pellett. 2002. The family Herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 44.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (HHV-8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schulz, T. F., Y. Chang, and P. S. Moore. 1998. Kaposi's sarcoma associated herpesvirus (human herpesvirus 8), p. 87-134. In D. J. McCance (ed.), Human tumor viruses. American Society for Microbiology, Washington, D.C.
- 47.Schulz, T. F., J. Sheldon, and J. Greensill. 2002. Kaposi's sarcoma associated herpesvirus (KSHV) or human herpesvirus 8 (HHV-8). Virus Res. 82:115-126. [DOI] [PubMed] [Google Scholar]
- 48.Staskus, K. A., A. W. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. D. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun, R., S. F. Lin, L. Gradoville, and G. Miller. 1996. Polyadenylylated nuclear RNA encoded by Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 93:11883-11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thome, M., P. Schneider, K. Hofmann, H. Fickenscher, E. Meinl, F. Neipel, C. Mattmann, K. Burns, J. L. Bodmer, M. Schroter, C. Scaffidi, P. H. Krammer, M. E. Peter, and J. Tschopp. 1997. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386:517-521. [DOI] [PubMed] [Google Scholar]
- 52.Tomescu, C., W. K. Law, and D. H Kedes. 2003. Surface downregulation of major histocompatibility complex class I, PE-CAM, and ICAM-1 following de novo infection of endothelial cells with Kaposi's sarcoma-associated herpesvirus. J. Virol. 77:9669-9684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vieira, J., P. O'Hearn., L. Kimball., B. Chandran, and L. Corey. 2001. Activation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) lytic replication by human cytomegalovirus. J. Virol. 75:1378-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, H. W., T. V. Sharp, A. Koumi, G. Koentges, and C. Boshoff. 2002. Characterization of an anti-apoptotic glycoprotein encoded by Kaposi's sarcoma-associated herpesvirus which resembles a spliced variant of human survivin. EMBO J. 21:2602-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.West, J. T., and C. Wood. 2003. The role of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 regulator of transcription activation (RTA) in control of gene expression. Oncogene 22:5150-5163. [DOI] [PubMed] [Google Scholar]
- 56.Zhong, W., H. Wang, B. Herndier, and D. Ganem. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. USA 93:6641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zimring, J. C., S. Goodbourn, and M. K. Offermann. 1998. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF1-mediated transcription. J. Virol. 72:701-707. [DOI] [PMC free article] [PubMed] [Google Scholar]