Abstract
Chemicals that activate nuclear factor-E2-related factor-2 (Nrf2) often increase multidrug resistance-associated protein expression in liver. Hepatocyte-specific deletion of Kelch-like ECH-associated protein 1 (Keap1) activates Nrf2. Use of hepatocyte-specific Keap1 deletion represents a non-pharmacological method to determine whether constitutive Nrf2 activation upregulates liver transporter expression in vivo. The mRNA, protein expression and localization of several biotransformation and transporters was determined in livers of wild-type and hepatocyte-specific Keap1-null mice. Sulfotransferase 2a1/2, NADP(H):quinone oxidoreductase 1, Cytochrome P450 2b10, 3a11, and glutamate-cysteine ligase catalytic subunit expression was increased in livers of Keap1-null mice. Oatp1a1 expression was nearly abolished, as compared to that detected in livers of wild-type mice. By contrast, Mrp 1-5 mRNA and protein levels were increased in Keap1-null mouse livers, with Mrp4 expression being more than 15-fold higher than wild-types. In summary, Nrf2 has a significant role in affecting expression of Oatp and Mrp expression.
Keywords: Keap1, Nrf2, nfe2l2, Organic anion transporting polypeptide (Oatp), Multidrug resistance-associated protein (Mrp), Abcc4
Introduction
The nuclear factor-E2-related factor-2 (Nrf2; nfe2l2) transcription factor coordinately regulates gene expression of antioxidant and phase-II drug metabolizing enzymes (DMEs) by binding to antioxidant/electrophile responsive elements (ARE/EpRE) found in the promoters of target genes [1-3]. The cytoplasmic protein Keap1 suppresses Nrf2 transcriptional activity under basal conditions through direct binding to Nrf2 and promoting proteasomal degradation [4-6]. In cells under stress, Nrf2 dissociates from Keap1 leading to decreased Nrf2 degradation. As a result, Nrf2 accumulates in the nucleus and recruits to the ARE/EpRE, where it initiates transcription of target genes. Mice with hepatocyte-specific deletion of Keap1 have constitutive nuclear accumulation of Nrf2 and activation of ARE-dependent gene transcription [7]. In this study, hepatocyte-specific Keap1-null mice were generated because whole-body Keap1-deletion results in severe growth retardation and lethality by approximately three weeks [7].
Antioxidants and DMEs are important for protecting cells against oxidative stress, and play a central role to the detoxification of xenobiotics. High expression of Nrf2-dependent antioxidant stress genes and DMEs in hepatocyte-specific Keap1-null mutant mice confer potent resistance to drug toxicity [7]. However, hepatobiliary transporters, which mediate hepatic uptake and efflux processes, also act to prevent tissue injury through enhanced efflux activity. Recent studies demonstrate that Nrf2 regulates gene expression of transporters including Multidrug resistance-associated proteins (Mrp) 1-4 [8-10], in addition to phase-II enzymes and DMEs. Therefore, constitutive Nrf2 activation may influence expression of hepatobiliary transporters in mice with hepatocyte-specific deletion of Keap1, which in turn could contribute to altered chemical disposition in liver and increased resistance to chemical-induced hepatotoxicity.
Hepatic uptake transporters are localized to the basolateral membrane and transport xenobiotics and bile acids into hepatocytes. They include organic anion-transporting polypeptides (Oatps) (mainly Oatp1a1, 1a4, and 1b2), sodium/taurocholate-cotransporting polypeptide (Ntcp), and organic cation transporters. Efflux transporters are localized to both the basolateral and canalicular membranes. Basolateral efflux transporters, including Mrp 1, 3-6, mediate efflux of chemicals from hepatocytes into blood. The canalicular efflux transporters Mrp2, multidrug resistance proteins (Mdrs), bile salt export pump (Bsep), and breast cancer resistance protein (Bcrp) mediate excretion of multiple chemicals and their metabolites from hepatocytes into bile. In the current study we determined the mRNA and protein expression of several DMEs and transporters in livers of wild-type and hepatocyte-specific Keap1-null mice. The results indicate that constitutive activation of Nrf2 alters gene and protein expression of hepatic uptake and efflux transporters as well as drug metabolizing enzymes, and thus Nrf2 likely plays a large role in cellular resistance to toxic chemicals by affecting xenobiotic disposition.
Materials and Methods
Animals
Mice with hepatocyte-specific deletion of Keap1 gene (Alb-Cre∷Keap1flox/−) were generated by Dr. Yamamoto's research group and the generation method was previously described [7]. Corresponding wild-type controls were Keap1+/+ mice Livers were harvested from mice at age of 9-13 week, snap frozen with liquid nitrogen, and stored in −80°C for future analysis.
RNA Extraction
Total RNA from liver was extracted using the RNA Bee reagent (Tel-Test Inc., Friendswood, TX) according to the manufacturer's protocol. RNA integrity was confirmed by formaldehyde-agarose gel electrophoresis and concentration was determined by UV absorbance at 260 nm.
Branched DNA Signal Amplification (bDNA) Assay
Probe sets for mouse Cyp4a14, 2b10, 3a11, 2e1, Ho-1, Nqo1, Sult2a1/2, Mrp1-6, Oatp1a1, 1a4, 1b2, 1a6, 2b1, Bsep, Bcrp, Ntcp, and Mdr2 have been described previously [11-17]. The method for the bDNA assay has been described in detail previously [18].
Membrane, Cytosol, and Nuclear Fraction Preparation
Livers (∼50 mg) were homogenized in sucrose-Tris (ST) buffer (10 × volume of sample amount, 0.25 mol/L sucrose, 10 mmol/L Tris–HCl, pH 7.4) containing protease inhibitor cocktail (2 μL/mL, Sigma Chemical Co. P8340) and centrifuged at 100,000 × g for 60 min at 4°C. The resulting supernatant was the cytosolic fraction and the pellet contained the membrane fraction. ST buffer was used to resuspend the resulting pellet (membrane fraction). Nuclear proteins were prepared from liver using the NE-PER kit (Pierce, Rockford, IL) according to the manufacturer's instructions. Membrane, cytosolic, and nuclear protein concentrations were determined by the method of Lowry using Bio-Rad protein assay reagents (Bio-Rad Laboratories, Hercules, CA). Equal protein loading for SDS-PAGE was confirmed by Coomassie Brilliant Blue staining.
Western Analysis of Mouse Liver Fractions
Methods for western blot analysis of transport and enzyme proteins were according to previous reports [18]. Detailed methods for detecting Oatp, Mrp, and Nrf2 protein levels have been previously described [18]. β-actin was used as a loading control and measured as a housekeeping gene. Proteins (50 μg of protein/lane) were electrophoretically resolved using polyacrylamide gels (8, 10, or 12% resolving, 4% stacking) and transblotted overnight at 4°C onto PVDF membrane (Millipore, Bedford, MA). The membranes were blocked with 2% nonfat dry milk in PBS with 0.1% Tween (PBS/T) for 1 h and then incubated for 2 h with the primary antibody diluted in blocking buffer (PBS/T with 2% milk) at room temperature. After washing, the membranes were incubated for 1 h with a species-appropriate peroxidase-labeled secondary antibody (Sigma-Aldrich, St. Louis, MO) diluted in 2% milk-PBS/T at room temperature. After incubation with the secondary antibody, membranes were washed with PBS, incubated with ECL chemiluminescent kit (Amersham Life Science, Arlington Heights, IL), and exposed to Fuji medical X-ray film (Fisher Scientific, Springfield, NJ). The intensity of the protein bands was quantified using Kodak Molecular Imaging Software (Vision 4.0.4, Eastman Kodak Company).The intensity of the protein bands was quantified using Image J 1.38× software download from National Institute of Health, USA (http://rsb.info.nih.gov/ij/). Background was subtracted before quantification. Mrp4 protein expression could not be quantified because Mrp4 protein was not detected in liver fractions of wild-type mice.
Immunofluorescent Staining and Epifluorescent Microscopy
Frozen mouse livers were embedded in optimal cutting temperature compound and brought to −20°C. Cryosections (5 μm) were thaw-mounted onto Superfrost glass slides (Fisher Scientific) and stored at −80°C under dehumidified conditions until use. Tissue sections were fixed with 4% paraformaldehyde for 5 min. Sections were blocked with 5% goat or donkey serum/phosphate-buffered saline (PBS) with 0.1% Triton X-100 for 1 hr and then incubated with BXP-53 (Bcrp), Mrp2, M3II-2 (Mrp3) or M4I-10 primary antibodies (donated from Drs. George Scheffer and Bruno Steiger, VU Medical Center, The Netherlands) diluted 1:100 in 5% goat serum/PBS with Triton X-100 for 2 hrs at room temperature. All antibody solutions were filtered through 0.22 μm membrane syringe-driven filter units (Osmonics Inc., Minnetonka, MN) prior to use. After incubation with primary antibody, the sections were washed three times in PBS with Triton X-100 and incubated for 1 hr with either goat anti-rat or donkey anti-rabbit IgG AlexaFluor 488 IgG (Invitrogen Corporation, Carlsbad, California) diluted 1:200 in 5% goat serum/ PBS with Triton X-100. Slides were washed in PBS and then rinsed twice in distilled deionized water. Sections were air dried and mounted in Prolong Gold with DAPI (Invitrogen Corp.). Sections were visualized and images were captured using a Zeiss Axiocam Epifluorescent Microscope at an excitation wavelength of 409 and 488 nm with AxioVision LE Rel.4.5. Negative control staining was performed by incubating cryosections without primary antibody (data not shown).
Statistical Analysis
Statistical analyses of differences were performed by Student's t test. P < 0.05 was considered statistically significant. Unless otherwise stated, all data were presented as mean ± SE of five animals.
Results
Liver Expression of Drug Metabolizing and Antioxidant Enzymes in Hepatocyte-Specific Keap1-null Mice
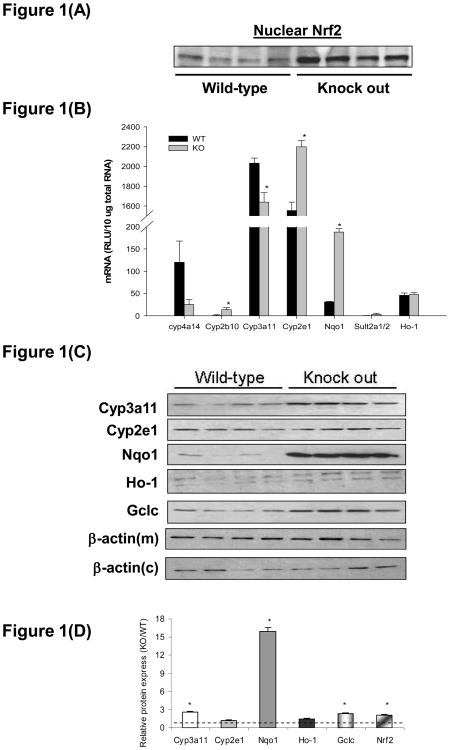
mRNA expression of antioxidant stress and drug metabolizing enzymes is illustrated in Figure 1(A). Hepatocyte-specific deletion of Keap1 decreased mRNA expression of Cyp4a14 and 3a11 in mouse livers to 20% and 80% of wild type mice, respectively. Cyp2b10, 2e1, Nqo1, and Sult2a1/2 mRNA was induced in livers of hepatocyte-specific Keap1-null mice, by 17.0, 1.4, 6.0, and 16.0-fold, respectively, compared to wild-types. Hepatic Ho-1 mRNA expression was similar between knockout and wild type mice. Western blotting showed that Cyp3a11, Nqo1, Gclc, and Nrf2 protein expression in livers was significantly increased in hepatocyte-specific Keap1-null mice, being 2.6-, 16.0-, 2.3-, and 2.1-fold of that detected in livers of wild type mice, respectively (Figure 1(B) and (C)). Hepatic Cyp2e1 and Ho-1 protein levels in knockout mice were not significantly different from wild types. Cyp4a14, 2b10, and Sult2a1/2 protein expression was not analyzed. β-actin protein expression in cytosolic and membrane fractions did not differ between wild types and Keap1-null mice.
Figure 1.
Drug metabolizing and antioxidant enzyme Cytochrome P450 (Cyp) 4a14, 2b10, 3a11, 2e1, NAD(P)H:quinone oxidoreductase 1 (Nqo1), Sulfotransferase (Sult) 2a1/2, Heme oxygenase-1 (Ho-1), and nuclear factor-E2-related factor-2 (Nrf2) mRNA and protein expression in livers of wild-type (WT) and hepatocyte-specific Keap1-null (KO) mice. (A). Western blot of Nrf2 proteins in nuclear fractions isolated from livers from WT and KO mice (protein loading amount 50 μg/lane, n = 4). (B) Cyp4a14, 2b10, 3a11, 2e1, Nqo1, Sult2a1/2, and Ho-1 mRNA expression. Total RNA was isolated from liver and mRNA levels were quantified using branched DNA signal amplification assay. mRNA expression level is presented as mean RLU ± SEM (n = 4 animals). Asterisks (*) represent a statistical difference between WT and KO (p < 0.05). (c). Western blots of Cyp3a11, 2e1, Nqo1, Ho-1, Gclc, and membrane fraction B-actin [β-actin(m)] and cytosolic B-actin [β-actin(c)] protein expression in liver fractions from WT and KO mice (protein loading amount 50 μg/lane, n = 4). (C). Quantification of Cyp3a11, 2e1, Nqo1, Ho-1, Gclc, and Nrf2 relative protein expressions in KO mice compared to WT. Asterisks (*) represent a statistical difference from corresponding WT (p < 0.05).
Liver Expression of Uptake Transporters in Hepatocyte-Specific Keap1-null Mice
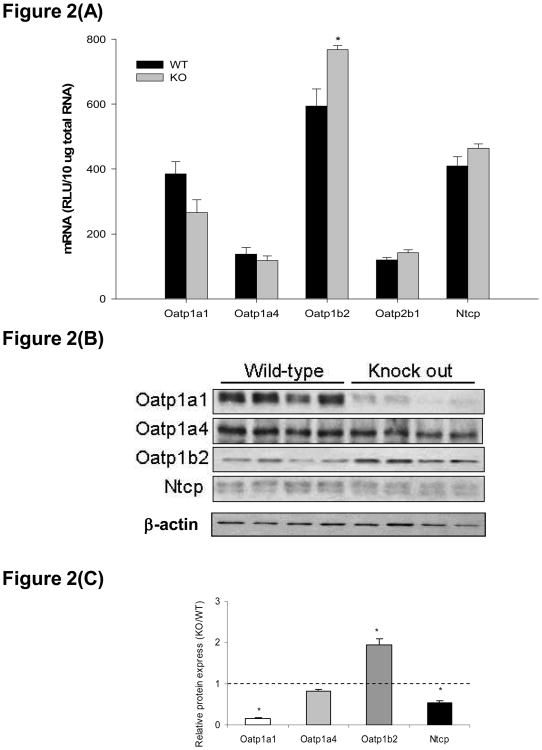
As shown in Figure 2(A), hepatic Oatp1b2 mRNA expression was increased 1.3-fold hepatocyte-specific Keap1-null mice compared to wild type mice. Expression of Oatp1a1, 1a4, 2b1, and Ntcp mRNA was not significantly different between hepatocyte-specific Keap1-null mice and wild types, while Oatp1a1 mRNA expression in knockout mice was 70% that in wild types. Figures 2(B) and (C) show protein expression of hepatic uptake transporters in wild-type and hepatocyte-specific Keap1-null mice. Consistent with its mRNA expression level, Oatp1b2 protein expression was induced 2.0-fold in livers from hepatocyte-specific Keap1-null mice. In contrast to its high hepatic expression in wild-type mice, Oatp1a1 protein expression was almost completely abolished in hepatocyte-specific Keap1-null mice; levels of Oatp1a1 protein in knockout mice were only 15% of wild types. Deletion of Keap1 decreased Ntcp protein expression to 53% of wild types. No difference in Oatp1a4 protein expression was observed between genotypes.
Figure 2.
Organic anion transporting polypeptide (Oatp) 1a1, 1a4, 1b2, 2b1, and sodium/taurocholate cotransporting polypeptide (Ntcp) mRNA and protein expression in livers of wild-type (WT) and hepatocyte-specific Keap1-null (KO) mice. (A). mRNA expression of Oatp1a1, 1a4, 1b2, 2b1, and Ntcp. Total RNA was isolated from liver and mRNA levels were quantified using branched DNA signal amplification assay. mRNA expression level is presented as mean RLU ± SEM (n = 4 animals). Asterisks (*) represent a statistical difference between WT and KO (p < 0.05). (B). Individual Western blots of Oatp1a1, 1a4, 1b2, and Ntcp in livers from WT and KO mice (protein loading amount 50 μg/lane, n = 4). (C). Quantification of Oatp1a1, 1a4, 1b2, and Ntcp relative protein expressions in KO mice compared to WT. Asterisks (*) represent a statistical difference from corresponding WT (p < 0.05).
Liver Expression of Efflux Transporters in Hepatocyte-Specific Keap1-null Mice
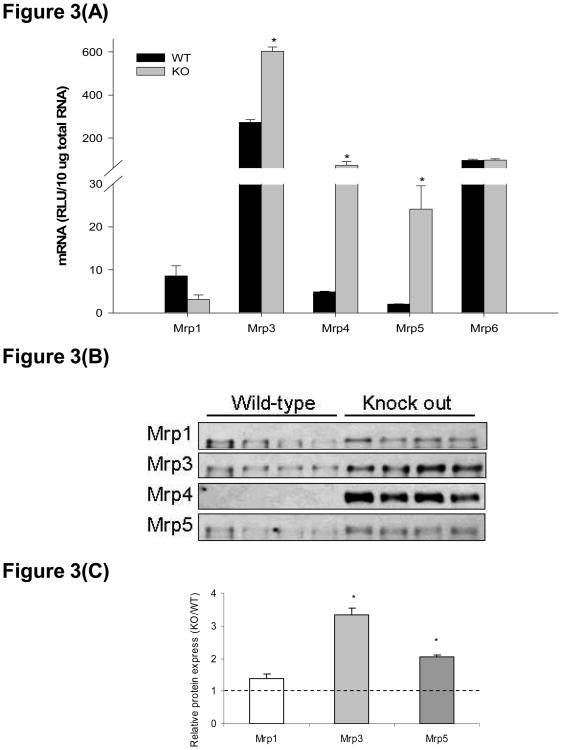
Basolateral efflux transporter Mrp3, 4, and 5 mRNA were elevated in livers of hepatocyte-specific Keap1-null mice as compared to wild types, with 2.2, 15.1, and 12.0-fold increases, respectively (Figure 3(A)). Hepatic Mrp1 mRNA in hepatocyte-specific Keap1-null mice was decreased to 36% of wild types, and Mrp6 mRNA expression in livers was similar between genotypes. As illustrated in Figures 3 (B) and (C), the basolateral efflux transporters, namely Mrp1, 3, 4, and 5, display induced protein expression in livers of hepatocyte-specific Keap1-null mice. Western blotting assay did not detect Mrp4 protein in livers of wild type mice, whereas marked expression was observed in hepatocyte-specific Keap1-null mice. Quantification of the western blots showed that Mrp1, 3, and 5 protein expression was 1.4-, 3.3- and 2.0-fold higher, respectively, in livers of the knockout mice compared to wild types.
Figure 3.
Multidrug resistance-associated protein (Mrp) 1, 3-6 mRNA and protein expression in livers of wild-type (WT) and hepatocyte-specific Keap1-null (KO) mice. (A). mRNA expression of Mrp1, 3-6. Total RNA was isolated from liver and mRNA levels were quantified using branched DNA signal amplification assay. mRNA expression level is presented as mean RLU ± SEM (n = 4 animals). Asterisks (*) represent a statistical difference between WT and KO (p < 0.05). (B). Individual Western blots of Mrp1, 3, 4, and 5 in livers from WT and KO mice (protein loading amount 50 μg/lane, n = 4). (C). Quantification of Mrp1, 3, and 5 relative protein expressions in KO mice compared to WT. No quantification for Mrp4 because it is undetectable in WT livers. Asterisks (*) represent a statistical difference from corresponding WT (p < 0.05).
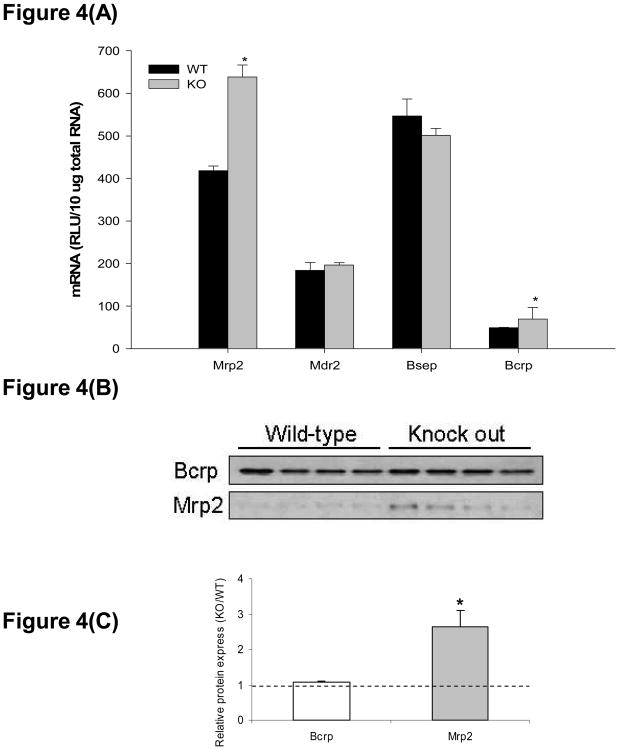
Liver mRNA expression of canalicular efflux transporters is shown in Figure 4(A). Mrp2 and Bcrp mRNA were increased approximately 1.5-fold in hepatocyte-specific Keap1-null mice, comparing to wild types, whereas Mdr2 and Bsep mRNA levels were similar between genotypes. Mrp2 protein was 2.6-fold higher in livers from hepatocyte-specific Keap1-null mice compared to wild types, whereas no difference in Bcrp protein was observed (Figures 4(B) and (C)).
Figure 4.
Multidrug resistance-associated protein 2 (Mrp2), Multidrug resistance protein 2 (Mdr2), bile salt export pump (Bsep), breast cancer resistance protein (Bcrp) mRNA and protein expression in livers of wild-type (WT) and hepatocyte-specific Keap1-null (KO) mice. (A). mRNA expression of Mrp2, Mdr2, Bsep, and Bcrp. Total RNA was isolated from liver and mRNA levels were quantified using branched DNA signal amplification assay. mRNA expression level is presented as mean RLU ± SEM (n = 4 animals). Asterisks (*) represent a statistical difference between WT and KO (p < 0.05). (B). Individual Western blots of Bcrp and Mrp2 in livers from WT and KO mice (protein loading amount 50 μg/lane, n = 4). (C). Quantification of Bcrp and Mrp2 relative protein expressions in KO mice compared to WT. Asterisks (*) represent a statistical difference from corresponding WT (p < 0.05).
Localization of Efflux transporters in Livers of Hepatocyte-Specific Keap1-null Mice
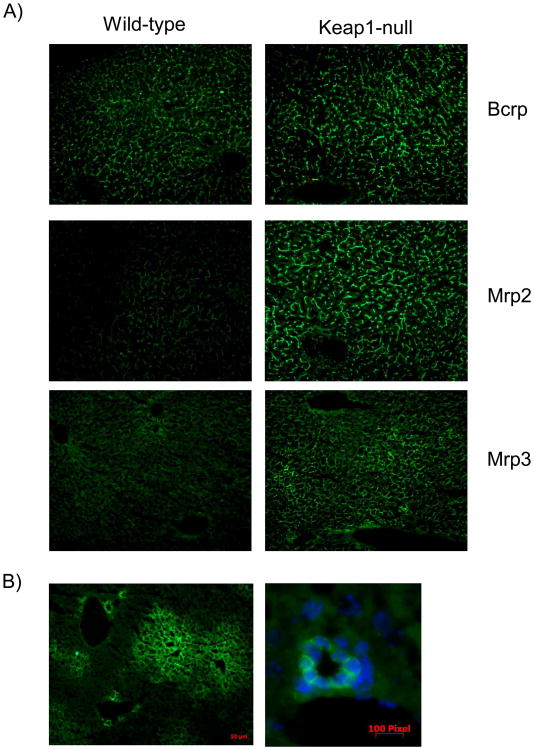
Immunohistochemical staining was performed on frozen liver sections from wild-type and hepatocyte specific null-mice. Canalicular Bcrp and Mrp2 staining was punctuate and branch-like in both genotypes (Figure 5). The intensity of Bcrp and Mrp2 staining was higher in liver sections from Keap1-null mice. Mrp3 staining was basolateral, and significantly higher in both centrilobular and periportal regions of livers of Keap1-null mice. Basal Mrp4 staining was barely detectable in liver sections from wild-type mice, whereas Mrp4 staining was markedly increased in livers of Keap1-null mice (Figure 5). Specifically, Mrp4 protein was upregulated in centrilobular hepatocytes and bile-duct epithelial cells in mice with a hepatocyte-specific deletion of Keap1.
Figure 5.
Immunofluorescent detection of Bcrp, Mrp2-4 in livers of wild-type and hepatocyte-specific Keap1-null (Keap1-null) mice. (A) Representative photomicrographs depict fluorescent immunohistochemical localization of Bcrp, Mrp2, and Mrp3 in liver. Cryosections (4-5 μm) of livers from WT and Keap1-null mice were stained with BXP-53 (Bcrp), Mrp2, M3II-2 (Mrp3), and M4I-10 antibodies followed by incubation with goat or donkey anti-rat or –rabbit IgG AlexaFluor 488 IgG. Photomicrographs of Bcrp and Mrp2 staining in canaliculi, and Mrp3 staining in the basolateral membrane are shown. (Original magnification ×200). (B) Representative photomicrographs depict fluorescent immunohistochemical localization of Mrp4 in liver. Mrp4 staining was upregulated in hepatocytes surrounding the central vein (left panel) and bile-duct epithelia (right panel) (Original magnifications ×100, 400, 630).
Discussion
Deletion of the Keap1 gene results in disruption between Keap1 and Nrf2 resulting in constitutive Nrf2 activation [7]. As expected, Nqo1 expression was increased in livers Keap1-null mice compared to wild types. Additionally, Keap1 deletion increased liver Cyp2b10, 3a11, and Sult2a1/2 mRNA and/or protein expression. Increased enzyme expression in hepatocyte-specific Keap1-null mice suggests that Nrf2 has a role in Cyp2b10, 3a11, and Sult2a1/2 up-regulation. Studies have demonstrated that many metabolizing enzymes are regulated by several transcription factors. First, Cyp3a is up-regulated via Pregnane-x-receptor (PXR), Constitutive Androstane Receptor (CAR), the glucocorticoid receptor (GR), or Nrf2 activation [15; 20; 21]. Second, Aryl hydrocarbon receptor (AhR) activation can cause Nrf2 activation [22] – via Nrf2 as a target gene of the AhR, indirectly via oxidative stress from Cyp1a1-generated reactive oxygen species, or direct cross-interaction of AhR/XRE (xenobiotic response elements). Nrf2 also regulates AhR expression and subsequently modulates AhR-target genes expression (e.g. Cyp1a1 and 1b1) [23]. Third, crosstalk between CAR and Nrf2 transcriptional pathways likely exists because compounds that activate the ARE/EpRE also activate CAR [24; 25].
Oatp1a1 and Ntcp mRNA expression was not significantly changed in Keap1-null mice, whereas Oatp1b2 was induced. In contrast, Oatp1a1 and Ntcp protein expression was decreased. Oatp1a1 is often down-regulated by microsomal enzyme inducers (MEIs) such as AhR and PXR ligands, as well as, CAR peroxisome proliferator-activated receptor α (PPARα), and Nrf2 activators [15]. Oatp1b2 mRNA expression is not affected by treatment with typical MEIs [15]. However, Oatp1b2 mRNA and protein expression was elevated in livers of hepatocyte-specific Keap1-null mice.
In the current study, Mrp transporters (Mrp1-5) were increased by hepatocyte-specific Keap1 deletion. Nrf2 activating compounds, such as butylated hydroxyanisole, oltipraz, and ethoxyquin increase Mrp expression [26]. Nrf2 regulates constitutive and inducible mRNA and protein expression of mouse Mrp1 in embryo fibroblasts [9]. An ARE-like sequence is present in the Mrp2 promoter [27], Nrf2 binds to AREs in the promoter regions of mouse Mrp2, and activation of the Nrf2, caused by hepatocyte-specific deletion of GCLC or Nrf2 activators, stimulates induction of hepatic Mrp2-4 [8]. In addition to Nrf2, other nuclear receptors may also be involved in regulation of Mrps expression including AhR (for Mrp2, 3, 5, and 6), CAR (for Mrp2-6), PXR and PPARα (for Mrp3) [26; 28-30]. Data in the present study illustrate that Mrp2-4 and Bcrp expression are increased with Keap1 deletion, which are consistent with previous findings in Keap1 knockdown mice [31].
The expression of several gene targets was differentially regulated at the mRNA and protein level – namely Cyp3a11, Cyp2e1, Oatp1a1, and Ntcp. The discordance between mRNA and protein levels indicate that the role for Keap1 in modulating DME and transporter protein expression is complex and could occurs through affecting pathways that modulate post-translational processes, such as proteasomal degradation via ubiquitination. It is known that Nrf2 activity is regulated through the modulation of the Cullin3 (Cul3)-containing E3-ligase complex [32], but it is not well described about how Nrf2 activity affects the levels of other proteins through indirect or non-transcriptional mechanisms.
Hepatobiliary transporter expression was increased livers of hepatocyte specific Keap1-null mice, with Mrp4 mRNA expression in livers of Keap1-null mice being more than 15-fold higher than wild-type mice and is consistent with previous findings [31, 33]. The data presented demonstrate a non-pharmacological induction of Multidrug resistance-associated proteins via modulation of the Keap1-Nrf2 pathway.
Acknowledgments
The authors would like to thank Drs. Curtis Klaassen and George Scheffer for their generous antibody contributions. The authors would also like to thank Sarah Campion for her technical assistance with immunohistochemical staining. This work was supported by RI-INBRE Grant # P20RR016457 from NCRR, NIH and K22ES013782 from NIEHS, NIH.
Footnotes
This work was presented, in part, at the Annual Society of Toxicology meeting held March 16-20, 2008, in Seattle, Washington.
References
- 1.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–22. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 2.Kwak MK, Itoh K, Yamamoto M, Sutter TR, Kensler TW. Role of transcription factor Nrf2 in the induction of hepatic phase 2 and antioxidative enzymes in vivo by the cancer chemoprotective agent 3H-1, 2-dimethiole-3-thione. Mol Med. 2001;7:135–45. [PMC free article] [PubMed] [Google Scholar]
- 3.Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem J. 2003;374:337–48. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem. 2003;278:21592–600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 5.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itoh K, Wakabayashi N, Katoh Y, Ishii T, O'Connor T, Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells. 2003;8:379–91. doi: 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- 7.Okawa H, Motohashi H, Kobayashi A, Aburatani H, Kensler TW, Yamamoto M. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem Biophys Res Commun. 2006;339:79–88. doi: 10.1016/j.bbrc.2005.10.185. [DOI] [PubMed] [Google Scholar]
- 8.Maher JM, Dieter MZ, Aleksunes LM, Slitt AL, Guo G, Tanaka Y, Scheffer GL, Chan JY, Manautou JE, Chen Y, Dalton T, Yamamoto M, Klaassen CD. Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology. 2007;46:1597–610. doi: 10.1002/hep.21831. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi A, Suzuki H, Itoh H, Yamamoto M, Sugiyama Y. Transcription factor Nrf2 is required for the constitutive and inducible expression of multidrug resistance-associated protein 1 in mouse embryo fibroblasts. Biochem Biophys Res Commun. 2003;310:824–9. doi: 10.1016/j.bbrc.2003.09.086. [DOI] [PubMed] [Google Scholar]
- 10.Aleksunes LM, Slitt AL, Maher JM, Augustine LM, Goedken M, Chan JY, Cherrington NJ, Klaassen CD, Manautou JE. Induction of Mrp3 and Mrp4 transporters during acetaminophen hepatotoxicity is dependent on Nrf2. Toxicol Appl Pharmacol. 2008;226:74–83. doi: 10.1016/j.taap.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aleksunes LM, Slitt AL, Cherrington NJ, Thibodeau MS, Klaassen CD, Manautou JE. Differential expression of mouse hepatic transporter genes in response to acetaminophen and carbon tetrachloride. Toxicol Sci. 2005;83:44–52. doi: 10.1093/toxsci/kfi013. [DOI] [PubMed] [Google Scholar]
- 12.Alnouti Y, Klaassen CD. Tissue distribution and ontogeny of sulfotransferase enzymes in mice. Toxicol Sci. 2006;93:242–55. doi: 10.1093/toxsci/kfl050. [DOI] [PubMed] [Google Scholar]
- 13.Buist SCN, Klaassen CD. Rat and mouse differences in gender-predominant expression of organic anion transporter (Oat1-3; Slc22a6-8) mRNA levels. Drug Metab Dispos. 2004;32:620–5. doi: 10.1124/dmd.32.6.620. [DOI] [PubMed] [Google Scholar]
- 14.Cheng X, Maher J, Chen C, Klaassen CD. Tissue distribution and ontogeny of mouse organic anion transporting polypeptides (Oatps) Drug Metab Dispos. 2005;33:1062–73. doi: 10.1124/dmd.105.003640. [DOI] [PubMed] [Google Scholar]
- 15.Cheng X, Maher J, Dieter MZ, Klaassen CD. Regulation of mouse organic anion-transporting polypeptides (Oatps) in liver by prototypical microsomal enzyme inducers that activate distinct transcription factor pathways. Drug Metab Dispos. 2005;331:276–82. doi: 10.1124/dmd.105.003988. [DOI] [PubMed] [Google Scholar]
- 16.Cherrington NJ, Slitt AL, Maher JM, Zhang XX, Zhang J, Huang W, Wan YJ, Moore DD, Klaassen CD. Induction of multidrug resistance protein 3 (mrp3) in vivo is independent of constitutive androstane receptor. Drug Metab Dispos. 2003;31:1315–9. doi: 10.1124/dmd.31.11.1315. [DOI] [PubMed] [Google Scholar]
- 17.Hartley DP, Klaassen CD. Detection of chemical-induced differential expression of rat hepatic cytochrome P450 mRNA transcripts using branched DNA signal amplification technology. Drug Metab Dispos. 2000;28:608–16. [PubMed] [Google Scholar]
- 18.Cheng Q, Aleksunes LM, Manautou JE, Cherrington NJ, Scheffer GL, Yamasaki H, Slitt AL. Drug metabolizing enzyme and transporter expression in a mouse model of diabetes and obesity. Mol Pharm. 2008;5:77–91. doi: 10.1021/mp700114j. [DOI] [PubMed] [Google Scholar]
- 19.Aleksunes LM, Scheffer GM, Jakowski AB, Pruimboom-Brees IM, Manautou JE. Coordinated expression of multidrug resistance-associated proteins (Mrps) in mouse liver during toxicant-induced injury. Toxicol Sci. 2006;89:370–9. doi: 10.1093/toxsci/kfi332. [DOI] [PubMed] [Google Scholar]
- 20.Xie W, Barwick JL, Simon CM, Pierce AM, Safe S, Blumberg B, Guzelian PS, Evans RM. Reciprocal activation of xenobiotic response genes by nuclear receptors SXR/PXR and CAR. Genes Dev. 2000;14:3014–23. doi: 10.1101/gad.846800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pascussi JM, Drocourt L, Gerbal-Chaloin S, Fabre JM, Maurel P, Vilarem MJ. Dual effect of dexamethasone on CYP3A4 gene expression in human hepatocytes Sequential role of glucocorticoid receptor and pregnane × receptor. Eur J Biochem. 2001;268:6346–58. doi: 10.1046/j.0014-2956.2001.02540.x. [DOI] [PubMed] [Google Scholar]
- 22.Kohle C, Bock KW. Coordinate regulation of Phase I and II xenobiotic metabolisms by the Ah receptor and Nrf2. Biochem Pharmacol. 2007;73:1853–62. doi: 10.1016/j.bcp.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 23.Shin S, Wakabayashi N, Misra V, Biswal S, Lee GH, Agoston E, Yamamoto M, Kensler TW. NRF2 modulates aryl hydrocarbon receptor signaling: influence on adipogenesis. Mol Cell Biol. 2007;27:7188–97. doi: 10.1128/MCB.00915-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slitt AL, Cherrington NJ, Fisher CD, Negishi M, Klaassen CD. Induction of genes for metabolism and transport by trans-stilbene oxide in livers of Sprague-Dawley and Wistar-Kyoto rats. Drug Metab Dispos. 2006;24:1190–7. doi: 10.1124/dmd.105.007542. [DOI] [PubMed] [Google Scholar]
- 25.Slitt AL, Cherrington NJ, Dieter MZ, Aleksunes LM, Scheffer GL, Huang W, Moore DD, Klaassen CD. trans-Stilbene oxide induces expression of genes involved in metabolism and transport in mouse liver via CAR and Nrf2 transcription factors. Mol Pharmacol. 2006;69:1554–63. doi: 10.1124/mol.105.014571. [DOI] [PubMed] [Google Scholar]
- 26.Maher JM, Cheng X, Slitt AL, Dieter MZ, Klaassen CD. Induction of the multidrug resistance-associated protein family of transporters by chemical activators of receptor-mediated pathways in mouse liver. Drug Metab Dispos. 2005;33:956–62. doi: 10.1124/dmd.105.003798. [DOI] [PubMed] [Google Scholar]
- 27.Vollrath V, Wielandt AM, Iruretagoyena I, Chianale J. Role of Nrf2 in the regulation of the Mrp2 (ABCC2) gene. Biochem J. 2006;395:599–609. doi: 10.1042/BJ20051518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Assem M, Schuetz EG, Leggas M, Sun D, Yasuda K, Reid G, Zelcer N, Adachi M, Strom S, Evans RM, Moore DD, Borst P, Schuetz JD. Interactions between hepatic Mrp4 and Sult2a as revealed by the constitutive androstane receptor and Mrp4 knockout mice. J Biol Chem. 2004;279:22250–7. doi: 10.1074/jbc.M314111200. [DOI] [PubMed] [Google Scholar]
- 29.Chen C, Klaassen CD. Rat multidrug resistance protein 4 (Mrp4, Abcc4): molecular cloning, organ distribution, postnatal renal expression, and chemical inducibility. Biochem Biophys Res Commun. 2004;317:46–53. doi: 10.1016/j.bbrc.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 30.Moffit JS, Aleksunes LM, Maher JM, Scheffer GL, Klaassen CD, Manautou JE. Induction of hepatic transporters multidrug resistance-associated proteins (Mrp) 3 and 4 by clofibrate is regulated by peroxisome proliferator-activated receptor alpha. J Pharmacol Exp Ther. 2006;317:537–45. doi: 10.1124/jpet.105.093765. [DOI] [PubMed] [Google Scholar]
- 31.Reisman SA, Csanaky IL, Aleksunes LM, Klaassen CD. Altered disposition of acetaminophen in Nrf2-null and Keap1-knockdown mice. Toxicol Sci. 2009;109:31–40. doi: 10.1093/toxsci/kfp047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villeneuve NF, Lau A, Zhang DD. Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases. Antioxid Redox Signal. 2010;13:1699–712. doi: 10.1089/ars.2010.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada K, Shoda J, Taguchi K, Maher JM, Ishizaki K, Inoue Y, Ohtsuki M, Goto N, Takeda K, Utsunomiya H, Oda K, Warabi E, Ishii T, Osaka K, Hyodo I, Yamamoto M. Ursodeoxycholic acid stimulates Nrf2-mediated hepatocellular transport, detoxification, and antioxidative stress systems in mice. Am J Physiol Gastrointest Liver Physiol. 2008;295:G735–47. doi: 10.1152/ajpgi.90321.2008. [DOI] [PubMed] [Google Scholar]