Abstract
Hmx1 is a variant homeodomain transcription factor expressed in the developing sensory nervous system, retina, and craniofacial mesenchyme. Recently, mutations at the Hmx1 locus have been linked to craniofacial defects in humans, rats, and mice, but its role in nervous system development is largely unknown. Here we show that Hmx1 is expressed in a subset of sensory neurons in the cranial and dorsal root ganglia which does not correspond to any specific sensory modality. Sensory neurons in the dorsal root and trigeminal ganglia of Hmx1dm/dm mouse embryos have no detectable Hmx1 protein, yet they undergo neurogenesis and express sensory subtype markers normally, demonstrating that Hmx1 is not globally required for the specification of sensory neurons from neural crest precursors. Loss of Hmx1 expression has no obvious effect on the early development of the trigeminal (V), superior (IX/X), or dorsal root ganglia neurons in which it is expressed, but results in marked defects in the geniculate (VII) ganglion. Hmx1dm/dm mouse embryos possess only a vestigial posterior auricular nerve, and general somatosensory neurons in the geniculate ganglion are greatly reduced by mid-gestation. Although Hmx1 is expressed in geniculate neurons prior to cell cycle exit, it does not appear to be required for neurogenesis, and the loss of geniculate neurons is likely to be the result of increased cell death. Fate mapping of neural crest-derived tissues indicates that Hmx1-expressing somatosensory neurons at different axial levels may be derived from either the neural crest or the neurogenic placodes.
Keywords: Hmx1, homeodomain, geniculate ganglion, trigeminal ganglion, dorsal root ganglion, posterior auricular nerve, neural crest
INTRODUCTION
In mammals, general somatic sensation to the face and head is supplied by multiple cranial ganglia. The most prominent of these is the trigeminal ganglion (TG), which innervates the anterior face, jaw, and orbital region, but general sensory fibers are also supplied to the lateral face and head by the geniculate (facial), superior (dorsal IX/X) and cervical dorsal root ganglia (DRG). The pinna, or external ear, lies at the intersection of the cutaneous domains served by these ganglia and receives sensory innervation from all of these sources (Folan-Curran and Cooke, 2001).
Significant progress has been made in identifying the key transcriptional regulators of somatosensory neurogenesis, including an early requirement for bHLH transcription factors of the neurogenin family, and the later role of homeodomain transcription factors such as Brn3a, Islet1, and Klf7 (Dykes et al., 2011; Lei et al., 2006; Marmigere and Ernfors, 2007; Sun et al., 2008). The regulation of the development of the special sensory neurons of the vestibulocochlear system is also well-studied (Fritzsch et al., 2006; Kelley, 2006). In contrast, very little is known about genetic programs that distinguish the hindbrain sensory ganglia serving general somatic sensation. Similarly, less is known about distinct markers or mechanisms governing the development of the two classes of sensory neurons that reside in the geniculate ganglion: the general somatosensory neurons that contribute to the posterior auricular nerve, and the special viscerosensory neurons that contribute to the chorda tympani and the greater superficial petrosal nerve and innervate taste receptors (Yamout et al., 2005).
Hmx1 is a variant homeodomain protein expressed predominantly the developing craniofacial mesenchyme, retina, and sensory nervous system (Wang et al., 2000; Yoshiura et al., 1998). Genetic approaches have identified Hmx1 (human NKX5-3) alleles causing ear deformities in multiple species, including the dumbo (dmbo, Hmx1dm) and misplaced ears (mpe) mutations in mice (Munroe et al., 2009), the dumbo (dmbo) strain of “fancy” rat (Kuramoto et al., 2010), and the oculo-auricular syndrome in humans (Schorderet et al., 2008). Homozygous individuals of all three species exhibit related phenotypes, including ventrally displaced ears and often associated eye deformities. The ocular defects observed in these disorders are broadly consistent with the known role of Hmx1 homologues in the development of the chick retina (Schulte and Cepko, 2000). Although studies of sensory development in the Dumbo mouse and rat have not been described, it has been reported that Hmx1 is essential for sensory differentiation in chick embryos, and that in its absence neural-crest derived sensory precursors adopt a melanocyte fate (Adameyko et al., 2009).
Here we show that Hmx1 is expressed in a subset of sensory neurons in the cranial and dorsal root ganglia. In the cranial ganglia, Hmx1 expression is restricted to the mandibular lobe of the TG, general somatosensory neurons in the geniculate ganglion, and some neurons of the developing statoacoustic (vestibulocochlear, VIII) and superior (IX/X) ganglion complexes. Expression is also detected in some sympathetic neurons. DRG and TG neurons in Hmx1dm/dm embryos have no detectable Hmx1 protein, yet express markers of general sensory differentiation and subtype specification normally. Loss of Hmx1 function results in marked cell loss in the somatosensory component of the geniculate ganglion, but not in the other sensory ganglia in which it is expressed. The selective requirement for Hmx1 in the geniculate ganglion may be due to its early expression in that ganglion compared to other somatosensory neurons, but it is not required for geniculate neurogenesis. Genetic fate-mapping studies indicate that the Hmx1-expressing sensory neurons may arise from either neural crest or neurogenic placodes, depending on location.
MATERIALS AND METHODS
Mice and genotyping
Hmx1 mice bearing the dmbo allele, strain B6;C3Fe-Hmx1dmbo/Rw/JcsJKjn, were obtained from Jackson Laboratories (Stock #008677). The Hmx1dm allele was originally the result of ENU mutagenesis on a C57BL/6 genetic background (Wilson et al., 2005), and mice were crossed to C57BL/6N (Charles River) for 2–5 additional generations prior to the experiments described. Experiments were performed with littermate controls. The Brn3atauLacZ (Brn3atlz) mouse strain, targeting a bovine tau/LacZ fusion expression cassette to the Pou4f1 gene locus, has been previously described (Quina et al., 2005). The mouse strain Tg(Wnt1-Cre)11Rth/MileJ (Danielian et al., 1998) was obtained from Jackson Laboratories (Stock #007807). The mouse strain B6.Cg-Gt(Rosa)26Sortm6(CAG-ZsGreen1)Hze/J (Ai6, Madisen et al., 2010), which inducibly expresses ZsGreen from the Rosa26 locus, was obtained from Hongkui Zeng, Allen Institute for Brain Science (Seattle).
Genotyping of the dmbo and wild-type Hmx1 alleles was performed by real-time PCR using oligonucleotide primers that have 3' termini at the point mutation which characterizes the dmbo mutation. The 54bp amplicon extends from position 157 to position 210 of the Hmx1 reference mRNA sequence, NM_010445 (Figure S1). The oligonucleotide primer sequences are: Common 5', gaggacgaggatccggagc; Hmx1 WT 3', ctgctgtcgccgccgctg; dmbo 3', ctgctgtcgccgccgcta. The qPCR efficiency of the matched and mismatched primers differ by 5–10 amplification cycles (32–1024 fold differential product generation at threshold), allowing the alleles to be easily distinguished. Real-time PCR was performed using Power SYBR Green Master Mix (Life Technologies), according to manufacturer's instructions, with an annealing temperature of 65°C. The Brn3atlz allele was genotyped as previously described (Quina et al., 2005).
Antibody generation
Because the GC-rich Hmx1 open reading frame was not amenable to PCR, a codon-optimized synthetic version of the coding sequence was generated by de novo synthesis (Figure S1). From this sequence we generated a bacterial expression construct for a glutathione-S-transferase (GST)/Hmx1 fusion protein containing amino acids 2–188 of the Hmx1 protein, excluding the conserved homeodomain, in the vector pGEX-KG (Smith and Johnson, 1988). The protein was isolated from bacterial lysates by affinity chromatography, and rabbits were immunized with the uncleaved GST fusion product.
Optical projection tomography
Whole mount embryo staining for βgalactosidase activity was performed with X-gal as previously described (Eng et al., 2001). Samples for optical projection tomography (OPT, Sharpe et al., 2002) were stained with X-gal, dehydrated in methanol, then cleared in 1:2 benzyl alcohol/benzyl benzoate. OPT was performed using a Bioptonics 3100M scanner, with data acquired for each specimen through 400 rotational positions.
BrdU labeling and immunofluorescence
Staged embryos were generated for analysis by interbreeding mice of the appropriate genetic background, with the assignment of noon on the day after the appearance of a mucous plug in mated animals as embryonic day 0.5 (E0.5). Embryos were further staged by size and developmental landmarks as described by Theiler (Theiler, 1972). Harvested embryos were fixed 1–3 hours in 4% paraformaldehyde in phosphate-buffered saline, cryoprotected in sucrose, embedded in OCT, and sectioned at 15–20μm in a cryostat. Cumulative BrdU labeling was performed by injection of pregnant dams with 2–3 serial injections of BrdU timed at 2–3 hour intervals (intraperitoneal, 50mg/Kg in 0.9% saline). Labeling was cumulative because the short time period before embryo harvest did not allow sufficent time for the dilution of the BrdU signal in cells which continued to divide. Thus unlabeled cells represent only those which were postmitotic or quiescent at the start of the labeling period. Embryo stage at the start of the labeling period was estimated based on the stage at harvest (Theiler, 1972) and the elapsed time from the start of labeling. Sections stained for BrdU were treated with DNase simultaneously with the primary antibodies.
Polyclonal rabbit and guinea pig antisera against Brn3a have been previously described (Quina et al., 2005). Other antisera included: rabbit anti-Islet1 (Abcam), rabbit monoclonal anti-caspase-3 (Cell Signaling Tech), rabbit anti-peripherin (Millipore), rabbit anti-Phox2b (gift of J-F Brunet, Pattyn et al., 1997), rabbit anti-Neurofilament 200 Ab (Sigma-Aldrich), guinea pig anti-Sox 10 (gift of M. Wegner, Maka et al., 2005), goat anti-TrkA, anti-TrkB and anti-TrkC (R&D Systems), goat anti-RET (Fitzgerald), goat anti-βgalactosidase (Biogenesis), rat monoclonal anti-BrdU (Abcam), and mouse monoclonal anti-Histone H3 (phospho S10) antibody (H3-P, Abcam ab7031). Secondary antibodies labeled with Alexa fluorophores were obtained from Life Technologies.
RESULTS
Hmx1 is expressed in diverse sensory neurons
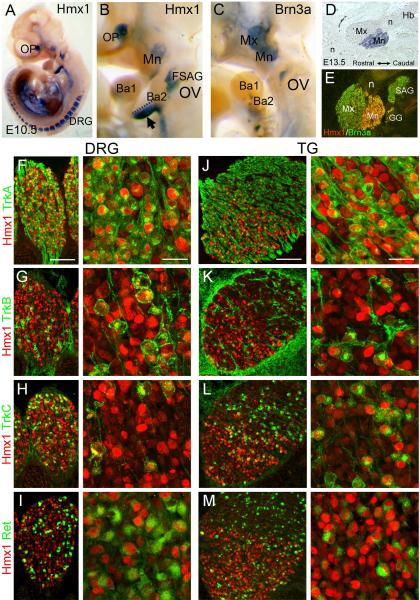
At E10.5, in situ hybridization revealed Hmx1 mRNA expression in the TG, DRG, the optic vesicle, the faciostatoacoustic ganglion complex adjacent to the otic vesicle, and the ventral portion of branchial arch 2 (Figure 1A,B). Comparison with the expression pattern of the pan-sensory marker Brn3a (Figure 1C) demonstrated that Hmx1 expression is restricted to the caudal part of the TG encompassing the mandibular trigeminal lobe (mnTG). This expression pattern of Hmx1 mRNA is consistent with prior reports (Munroe et al., 2009), but the cell-specific expression of the Hmx1 protein has not been examined. For this reason we generated an Hmx1 antibody to the amino terminal portion of the Hmx1 protein (Figure S1, Methods), which does not include the conserved homeodomain region, and is not predicted to cross-react with the related gene products Hmx2 and Hmx3. Hmx1 immunostaining accurately reproduced the expression pattern of Hmx1 mRNA at all stages, including the restriction of Hmx1 expression to the mnTG (Figure 1D,E). No immunoreactivity was observed in the previously described expression domains of Hmx2 and Hmx3, such as the otic vesicle (Wang and Lufkin, 2005).
Figure 1. Hmx1 expression in sensory neurons.
(A–C) Whole-mount in situ hybridization for Hmx1 and Brn3a mRNA in E10.5 embryos. (D) In situ hybridization for Hmx1 at E13.5, sagittal section. (E) Immunofluorescence for Hmx1 and Brn3a at E13.5 shows restriction of Hmx1 protein to the mandibular lobe of the TG, as well as expression in the geniculate and statoacoustic ganglia. (F–M) Co-expression of Hmx1 with the early markers of sensory subtypes TrkA, TrkB, TrkC and Ret in the DRG and TG of E13.5 embryos. Ba1, 2, branchial arch 1,2; DRG, dorsal root ganglion; FSAG, faciostatoacoustic ganglion complex; GG, geniculate ganglion; Hb, hindbrain; Mn, mandibular lobe, trigeminal ganglion; Mx, maxillary lobe, trigeminal ganglion; n, trigeminal nerve (proximal and distal branches); OP, optic vesicle; OV, otic vesicle; SAG, statoacoustic ganglion. Scale F,J, 100μm, 50μm.
In the mnTG and the DRG, Hmx1 is expressed in the developmental interval during which sensory neurons differentiate into their major subtypes, including nociceptors, mechanoreceptors and proprioceptors. Differentiation of these functional classes is associated with expression of specific neurotrophin receptors including TrkA, TrkB, TrkC and Ret (Marmigere and Ernfors, 2007). Hmx1 expression was observed in some but not all of differentiating DRG neurons, suggesting that it might be restricted to one or more of these functional sensory classes. However, co-expression of Hmx1 was observed with all four neurotrophins in both the DRG and TG at E13.5 (Figure 1F–M), essentially ruling out a role for Hmx1 in the specification of these recognized sensory subtypes.
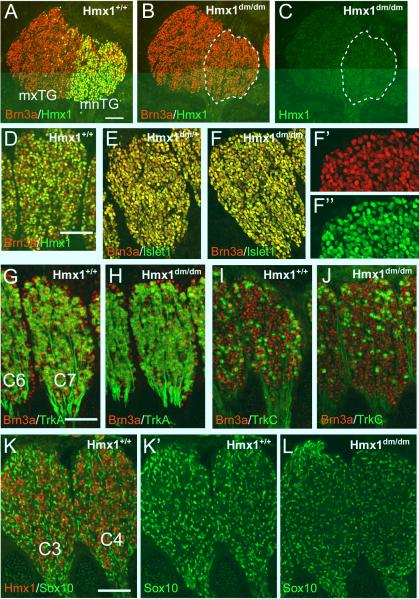
The Hmx1 dmbo allele consists of a nonsense mutation that lies amino-terminal to the Hmx1 homeodomain, and thus it should result in loss-of-function for a DNA binding transcription factor. However, the amino-terminal antisera generated to Hmx1 would be expected to recognize the truncated Hmx1 protein if it is present in Hmx1dm/dm mice. To determine whether a truncated Hmx1 gene product persists in mutant embryos, we examined the expression of Hmx1 protein in the TG of E13.5 Hmx1dm/dm embryos and controls (Figure 2A–C). Hmx1 immunoreactivity was undetectable in the Hmx1dm/dm embryos, probably due to rapid cellular clearance of the truncated mutant protein, thus demonstrating that the dmbo allele is a functionally null mutation.
Figure 2. Hmx1 is not required for sensory neurogenesis or subtype specification.
(A–C) Immunofluorescence staining for Hmx1 and Brn3a in sagittal sections of the TG of control and Hmx1dm/dm E13.5 embryos showing that the dmbo allele is a functional null. (D) Co-localization of Hmx1 and Brn3a in E13.5 DRG. At this stage Hmx1 is expressed in a subset of the Brn3a+ neurons. Sagittal sections, cervical level ganglia. (E,F) Co-localization of the sensory neurogenesis markers Brn3a and Islet1 in DRG of control and Hmx1dm/dm E12.5 embryos. (G–J) Normal expression of TrkA and TrkC in DRG of control and Hmx1dm/dm E13.5 embryos. (K–L) Expression of Hmx1 and Sox10 in DRG of control and Hmx1dm/dm E13.5 embryos. Hmx1 is not co-expressed with Sox10, and Sox10 expression is unchanged in Hmx1dm/dm ganglia. C3–C7, dorsal root ganglion of C3–C7; mnTG, mandibular lobe, trigeminal ganglion; mxTG, maxillary lobe, trigeminal ganglion. Scale A, 100μm; D,G,K, 50μm.
In the initial study linking the mouse dmbo allele to the Hmx1 locus, the dmbo allele was carried as a heterozygote in trans with a balancer allele, Rw, which consists of a ~50MB inversion on Chromosome 5 containing the Hmx1 locus (wild-type), and which precludes recombination of the genomic region containing Hmx1 (Munroe et al., 2009; Stephenson et al., 1994). In this study, partially penetrant exencephaly and subsequent neonatal lethality were associated with the Hmx1dm/dm genotype, such that surviving Hmx1dm/dm animals were observed at 41% lower than expected frequency by the third postnatal week in dmbo/Rw × dmbo/dmbo intercrosses. However, a majority of Hmx1dm/dm mice were viable, and the characteristic misplaced ear phenotype was 100% penetrant in these animals. For the present study, we obtained the strain B6;C3Fe-Hmx1dmbo/Rw/JcsJKjn and crossed these Hmx1dm mice with C57BL/6N (Charles River), and used real-time PCR to directly genotype the Hmx1dm and Hmx1+ alleles (Methods), thus eliminating the Rw allele. We genotyped 85 embryos resulting from 10 crosses using Hmx1dm/+ parental mice that had been backcrossed to C57BL/6 for 2–5 generations. Embryos were harvested between E10.5 and E14.5. The observed frequency of the resulting genotypes was 21 Hmx1dm/dm, 44 Hmx1dm/+, and 20 Hmx1+/+ (expected frequency 21.25/42.5/21.25), and none of these embryos exhibited exencephaly. From these results it can be concluded that exencephaly is not a highly penetrant phenotype associated with the Hmx1dm/dm genotype, and that the previously observed exencephaly may have resulted from other alleles that co-segregated with Hmx1dm in the Hmx1dmbo/Rw mice, or from a genetic interaction between dmbo and other alleles associated with the Rw or dmbo genetic backgrounds.
Hmx1 is not required for sensory neurogenesis
Previously it has been reported that Hmx1 expression is required for sensory neurogenesis, and that in the absence of Hmx1, neural crest precursors fated to differentiate into sensory neurons will instead adopt a glial/melanocyte fate (Adameyko et al., 2009). These conclusions were based on siRNA knockdowns of Hmx1 expression in chick DRG precursors, and a resulting switch from neural to melanocyte marker expression. Given that Hmx1dm/dm mice exhibit no detectable Hmx1 protein, the sensory ganglia in these mice provide a further test of a general role for Hmx1 in sensory neurogenesis. We used the transcription factors Brn3a and Islet1 as markers of sensory neurogenesis, because co-expression of these factors characterizes sensory neurons at all levels of the neural axis, and they are not expressed in glia, melanocytes, or their committed precursors (Adameyko et al., 2009; Dykes et al., 2011; Fedtsova et al., 2003; Lanier et al., 2007). First, we noted that in the TG and in the DRG, Hmx1 is not expressed in all sensory neurons, but only in a subset of the Brn3a+ cells (Figures 2A,D). In the TG, the Hmx1+ neurons are restricted to the mnTG at all stages, whereas in the DRG they are distributed throughout the ganglion. At E13.5 Hmx1 was also detected in a cluster of neurons in the statoacoustic ganglion, and in a subset of neurons in the superior ganglion of the IX/X complex (Figure S2A–D). The presence of Hmx1-negative sensory neurons in the cranial ganglia is not consistent with a universal Hmx1 requirement for sensory neurogenesis. Hmx1 protein could also be detected at E13.5 in sympathetic neurons of the sympathetic chain and stellate ganglia, in accordance with in situ hybridization data available in databases (Figure S2E,F; http://developingmouse.brain-map.org/).
We then examined markers of sensory neurogenesis in Hmx1dm/dm mice and controls. Brn3a and Islet1 were co-expressed at E12.5, in all DRG neurons in Hmx1dm/dm mice, indistinguishably from controls (Figure 2E,F). TrkA and TrkC were also unchanged at E13.5 in the Hmx1dm/dm DRG (Figure 2G–J) and in the TG (Figure S3). Finally, if sensory neurons failed to undergo neurogenesis and adopted a non-neural fate, they would be predicted to show abnormal or persistent expression of the neural crest marker Sox10, which in mid-gestation sensory ganglia is a marker for glial precursors, not neurons (Kuhlbrodt et al., 1998; Lanier et al., 2007; Sun et al., 2008). Sox10 was not co-expressed with Hmx1 in wild-type embryos, and its expression was not increased at E13.5 in Hmx1dm/dm DRG (Figure 2K,L) or TG (Figure S4). We conclude that Hmx1 is not essential for the selection of neural over non-neural fates among neural crest-derived cells, for sensory neurogenesis, or for early sensory subtype specification.
Hmx1 is required for the normal development of the somatic sensory component of the geniculate ganglion
We next used a strain of mice bearing a sensory-targeted axonal tauLacZ reporter, Brn3atlz, to examine the growth and targeting of sensory axons in Hmx1dm/dm mice. Brn3atlz is a null allele of Brn3a, but heterozygous animals have been shown to have no detectable sensory phenotype (Eng et al., 2001) and nearly normal expression of Brn3a and its downstream targets (Eng et al., 2004), due to gene dosage compensation at the Brn3a locus (Trieu et al., 2003). Thus Brn3atlz is used here only as a reporter allele.
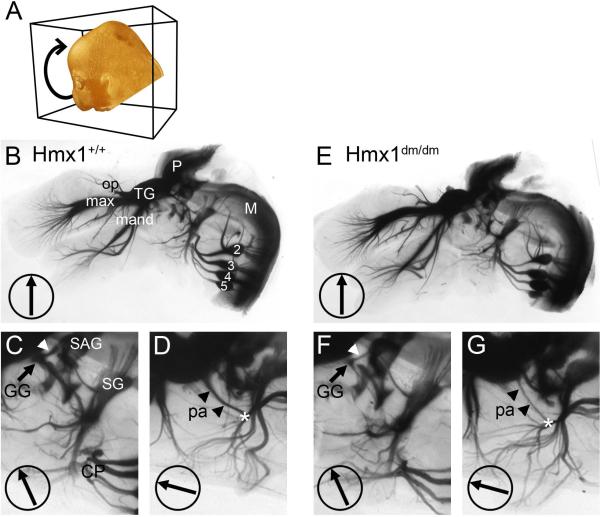
In order to visualize the developing cranial ganglia and their projections in Hmx1dm/dm embryos and controls, we used optical projection tomography (OPT) in which 400 rotational images of cleared embryos are captured and used to generate videos or are digitally reconstructed into a three dimensional dataset (Sharpe et al., 2002). Figure 3 shows 2-D projections of E14.5 embryos derived from the OPT data set, and Video 1 is a composite generated from images of E14.5 Hmx1dm/dm embryos and controls through 360 degrees of rotation. The three major components of the trigeminal system, the ophthalmic, maxillary, and mandibular divisions, were all present in Hmx1dm/dm embryos. Although the restricted expression of Hmx1 to the mnTG suggested that loss of Hmx1 activity would result in a major defect or rerouting of axons in the mandibular division of the nerve, all of the branches innervating the lower face and jaw appeared grossly unchanged in Hmx1dm/dm embryos (Figure 3B,E; Video 1). Examination of the cervical DRG at C2–C5 levels, and their projections to the cervical plexus, also did not reveal obvious abnormalities in mutant embryos (Figure 3C,F).
Figure 3. OPT imaging of developing sensory cranial ganglia.
E14.5 Hmx1+/+ and Hmx1dm/dm embryos bearing a Brn3atlz reporter allele were hemisected and stained for βgalactosidase activity, cleared, and imaged through 400 rotational positions. See also Supplemental Video. (A) Schematic of rotational imaging of embryo. (B–D) Rotational images of Hmx1+/+ embryo taken at 0, 24, and 75 degrees of rotation. Arrowheads indicate the course of nerve fibers which originate from the geniculate ganglion and project centrally (white arrowheads C,F), and into the periphery, forming the posterior auricular nerve (black arrowheads D,G). The other nerves originating in the geniculate ganglion, the chorda tympani and the greater superficial petrosal nerve, do not convey general somatic sensation and are not labeled by the Brn3atlz reporter. Asterisks indicate the intersection of the posterior auricular nerve and fibers from the superior ganglion which also innervate the auricle. (E–G) Rotational images of Hmx1dm/dm embryo taken at 0, 24, and 75 degrees of rotation. 2–5, DRG of corresponding spinal segments; CP, cervical plexus; GG, geniculate ganglion; M, medulla; max, maxillary branch, trigeminal nerve; mand, mandibular branch, trigeminal nerve; op, ophthalmic branch, trigeminal nerve; P, pons; pa, posterior auricular nerve; SAG, statoacoustic ganglion; SG, superior ganglion (of IX/X ganglion complex); TG, trigeminal ganglion.
In contrast, marked abnormalities were observed in the geniculate ganglion. The size of the ganglion was significantly decreased in Hmx1dm/dm embryos (Figure 3C,F), and only vestigial fibers were observed in its distal projection, the posterior auricular nerve (Yamout et al., 2005), which joined fibers from the superior ganglion to innervate the pinna (Figure 3D,G). Likewise its proximal projection, which joined fibers from the statoacoustic complex to enter the brainstem, was greatly diminished. The other nerves originating in the geniculate ganglion, the chorda tympani and the greater superficial petrosal nerve, innervate taste buds and thus do not convey general somatic sensation (Yamout et al., 2005). These nerves were not labeled by the Brn3atlz reporter.
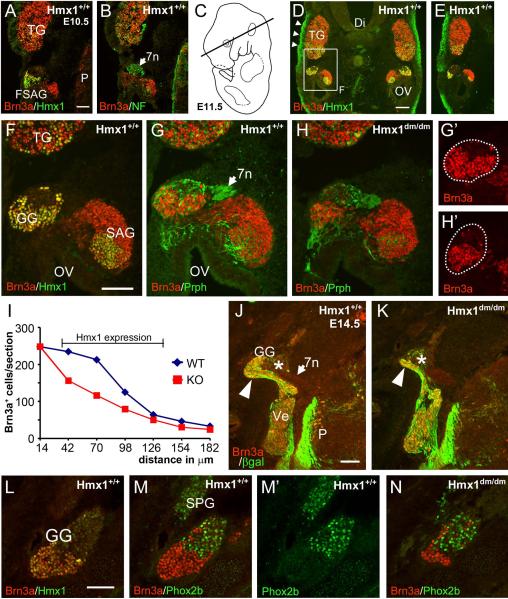
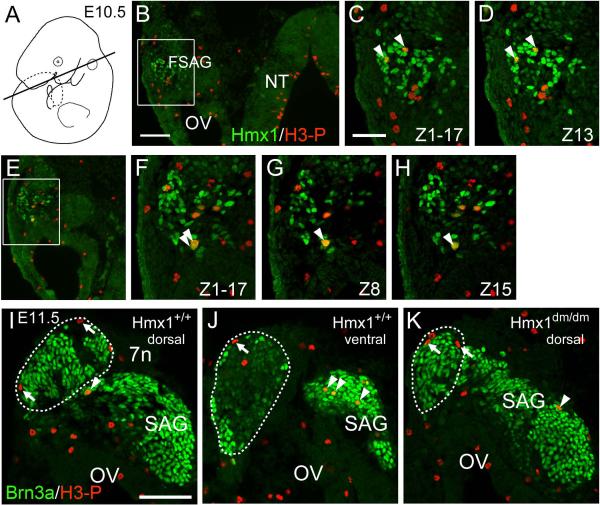
To determine whether the loss of the geniculate ganglion in Hmx1dm/dm embryos was likely to be a cell-autonomous effect of the loss of Hmx1 function, we examined Hmx1 expression in the faciostatoacoustic ganglion complex (FSAG, which gives rise to the geniculate ganglion), and the TG from the time of the first appearance of differentiated neurons in these ganglia. Hmx1 protein was first detected at E10.5 in a region of the FSAG complex immediately adjacent to the facial nerve (Figure 4A,B). We noted that in the mnTG Brn3a expression precedes that of Hmx1 by about one developmental day (compare 4A,4D), and that Hmx1 is barely detectable and Brn3a is strongly expressed at E10.5. In contrast, in geniculate ganglion precursors in the FSAG complex the temporal order of expression is reversed, in that Hmx1 is strongly expressed and Brn3a is weakly expressed at E10.5.
Figure 4. Early loss of developing geniculate neurons in Hmx1dm/dm embryos.
Horizontal sections, rostral is at the top in all views. (A,B) First detectable expression of Hmx1 in the faciostatoacoustic ganglion complex at E10.5. Hmx1 is strongly expressed in precursors of the geniculate ganglion adjacent to the facial nerve (arrow). (C) Schematic of E11.5 embryo showing plane of section for views D–H. (D,E) Expression of Hmx1 in the dorsal (D) and ventral (E) geniculate ganglion, the statoacoustic ganglion, and the mnTG at E11.5. In this embryo, alternate sections were stained for Brn3a/Hmx1 and Brn3a/Peripherin. All Hmx1+ neurons are also Brn3a+ at this stage. The boxed area is enlarged in (F). (F–H) Enlarged view of geniculate and statoacoustic ganglia at E11.5. Comparison of Brn3a expression in (G) and (H) shows first detectable loss of Brn3+ neurons in the geniculate ganglion of Hmx1dm/dm embryos. (I) Brn3a+ cell number in E11.5 geniculate ganglia of Hmx1+/+ and Hmx1dm/dm embryos, counted in the area shown in (G', H'). Cells were counted in every other section, immunostained for Brn3a and peripherin, at 28μm intervals across the extent of the Hmx1-expressing part of the ganglion. The sections shown in G,G' and H.H' are at the 42μM level. Cell loss occurs only in the region of the ganglion normally expressing Hmx1 (bracket). Paired T test for comparison of cell number in 11 matched sections on right and left sides of embryo, P=0.002. Total cell counts: 1273, 850. (J,K) Expression of Brn3a and βgalactosidase in Hmx1+/+ and Hmx1dm/dm embryos bearing a Brn3atlz reporter allele at E14.5. The βgalactosidase marker allows staining of the peripheral and central projections of general somatic sensory neurons. There is a profound loss of sensory neurons in the Brn3a+/Hmx1+ part of the ganglion (arrowhead). The viscerosensory component of the geniculate ganglion (asterisk), which is Hmx1 negative and largely Brn3a negative, is unaffected. (L–N) Hmx1 and Phox2b discriminate somatosensory and viscerosensory neurons in the geniculate ganglion. In (L) Hmx1/Brn3a expression identifies the somatosensory component of the geniculate ganglion in a horizontal section at E14.5. In (M) an adjacent section shows Phox2b expression in the viscerosensory geniculate ganglion, non-overlapping with Brn3a. Phox2b, but not Hmx1 or Brn3a, is also expressed in the adjacent parasympathetic sphenopalatine ganglion. The viscerosensory neurons are not affected in a Hmx1dm/dm embryo (N) showing marked reduction of the somatosensory component. 7n, facial nerve; Di, diencephalon; FSAG, faciostatoacoustic ganglion complex; GG, geniculate ganglion; OV, otic vesicle; P, pons; SAG, statoacoustic ganglion; SPG, sphenopalatine ganglion; TG, trigeminal ganglion. Scale A, F, J, 100μm; D, 200μm.
At E11.5, the developing geniculate ganglion and statoacoustic ganglion complex could be clearly distinguished (Figure 4C–G). Hmx1 was detected in the mnTG, the geniculate ganglion, and a subset of statoacoustic neurons. At this and later stages, all of the Hmx1+ neurons were seen to co-express Brn3a. Robust expression of Hmx1 was also observed in regions of cranial mesenchyme, such as that directly overlying the TG (Figure 4D). We assessed whether the number of geniculate ganglion neurons is diminished at this stage in Hmx1dm/dm embryos relative to controls by counting all Brn3a+ neurons in aligned sections from both genotypes. This showed a significant reduction in cell number in the mutant embryo throughout the region of the geniculate that normally expresses Hmx1 (Figure 4G–I). At E14.5, only a vestigial geniculate ganglion is evident in Hmx1dm/dm embryos (Figure 4J,K). Cell counts at this stage were greatly diminished in the geniculate ganglion of the mutant (Figure S5).
In addition to providing general somatic sensation to the ear, special viscerosensory neurons, which are anatomically part of the geniculate ganglion complex, also innervate the taste buds (Oakley and Witt, 2004). Because Hmx1+ neurons are a subset of Brn3a+ neurons in the geniculate ganglion, all of the Hmx1+ neurons are identified by the Brn3atlz reporter. We did not observe Brn3a+ neurons projecting from the geniculate ganglion to the tongue (Figure 3; Video 1), demonstrating that the Hmx1/Brn3a expression was restricted to the somatosensory part of the ganglion. We also also noted that the geniculate ganglion contains a significant compartment of Hmx1−/Brn3a− neurons that were unaffected in the knockout (Figure 4J,K). In order to clearly determine whether these unaffected geniculate neurons comprised the viscerosensory component of the ganglion, we examined the expression of Phox2b, a homeodomain transcription factor that is expressed in autonomic ganglia and cranial viscerosensory neurons derived from the epibranchial placodes, but not cranial somatosensory neurons (Coppola et al., 2010; Dauger et al., 2003; Pattyn et al., 1997). Phox2b was not co-expressed with Brn3a in the Hmx1+/Brn3a+ region of the ganglion (Figure 4 L,M), and the Phox2b-expressing cells were still present in Hmx1dm/dm embryos (Figure 4N). Thus we conclude that Hmx1 is required only for the development of the general somatosensory component of the geniculate ganglion.
Relationship of Hmx1 expression to neurogenesis and apoptosis in the geniculate ganglion
The extensive cell loss observed in the geniculate ganglion of Hmx1dm/dm embryos by E14.5 could be due to increased cell death, a failure of neurogenesis, or both. In order to determine the extent of apoptotic cell death in the geniculate ganglion during the interval when reduced cell numbers first become apparent, we examined E11.5 embryos for activated caspase-3 expression. Caspase-3+ cells were rare in the Hmx1-expressing domain of the geniculate ganglion in both Hmx1dm/dm embryos and controls (Figure 5A–D), and the number of caspase-3 reactive cells was similar in the part of the statoacoustic ganglion that does not express Hmx1. We performed a complete count of caspase-3+ cells in two matched series of sections of E11.5 Hmx1dm/dm embryos and controls, and no significant difference was detected in apoptotic cell number (21 sections counted in each genotype; Hmx1+/+, range 0–5 caspase+ cells/section, total caspase-3+ cells, 23; Hmx1dm/dm, range 0–6, total 22; paired t-test p=0.908). We also examined activated caspase-3 expression in the geniculate ganglion at E12.5 (data not shown), but at this stage the significantly lower cell numbers in the geniculate ganglion of Hmx1dm/dm embryos did not permit a meaningful comparison with controls. From these data we conclude that there is no precipitous apoptosis of geniculate ganglion neurons shortly after neurogenesis in Hmx1dm/dm embryos, but we cannot exclude gradual attrition of this cell population in the E11.5–E14.5 interval.
Figure 5. Apoptosis in the developing geniculate ganglion.
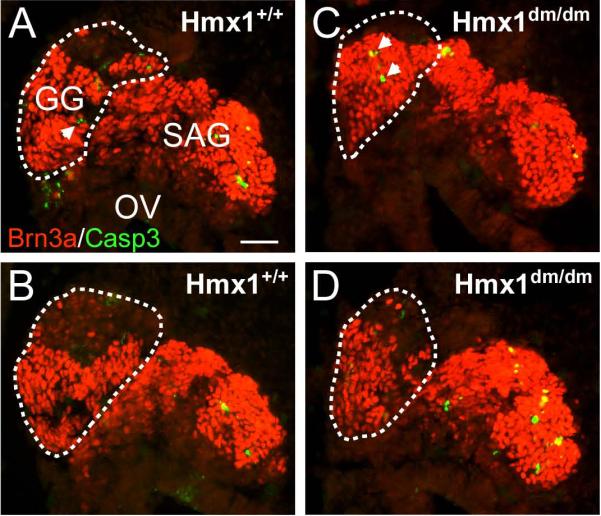
Immunofluorescence for activated Caspase 3 was used to identify apoptotic neurons in the developing faciostatoacoustic complex in E11.5 mice. (A,B) Hmx1+/+ embryo. (C,D) Hmx1dm/dm embryo. The planes of section in A, C are dorsal to B,D. The area of the geniculate ganglion is circled. Apoptotic neurons (arrows) were rare at this stage and no significant difference was observed between genotypes. GG, geniculate ganglion; OV, otic vesicle; SAG, statoacoustic ganglion. Scale, 50μm.
If the loss of Hmx1 function affects geniculate neurogenesis, then Hmx1 expression must be initiated prior to cell cycle exit, i.e. before the “birthdate” of the Hmx1+ geniculate neurons. For this reason we examined co-expression of Hmx1 with the mitotic marker phospho-S10 Histone H3 (H3-P). Hmx1 was clearly co-expressed with H3-P in dividing geniculate ganglion precursors at E10.5 (Figure 6A–H). We then examined the geniculate ganglion and statoacoustic ganglion of Hmx1dm/dm and control embryos for mitotic neural precursors at E11.5 (Figure 6I–K). Few, if any, mitotic cells could be identified in the region of the geniculate ganglion that expresses Hmx1 at this stage in either genotype, but ongoing neurogenesis was detected in the statoacoustic ganglion. Thus the Hmx1-expressing geniculate neurons are an early-born population in which neurogenesis is largely complete by E11.5.
Figure 6. Hmx1 expression in dividing geniculate ganglion precursors.
(A–H) E10.5 control embryo immunostained for Hmx1 and H3-P. H3-P immunoreactive cells are sparse because the marker is only expressed during a portion of M phase in dividing cells. (B,E) Low magnification epifluorescence images showing the location of dividing cells in the neural tube and otic region. (C,D) Z-stack incorporating 17 optical sections and a single 1μm section showing co-localization of Hmx1 and H3-P co-localization in two neuroblasts. (F–H) Z-stack and two selected optical sections 7μm apart showing two neuroblasts which are likely to be the daughter cells of a single recent mitosis. (I–K) E11.5 Hmx1+/+ and Hmx1dm/dm embryos immunostained for Brn3a and H3-P. The geniculate ganglion is circled; Brn3a-expressing neurons in this region also express Hmx1 in control embryos. Mitotic cells on the periphery of the geniculate ganglion (arrows) are consistently Brn3a negative. No Brn3a+ mitotic cells were observed in the geniculate at this stage (every section examined in 2 ganglia of each genotype). In contrast, mitotic Brn3a+ cells were observed frequently in the statoacoustic ganglion (arrowheads). 7n, facial nerve; FSAG, faciostatoacoustic ganglion complex; NT, neural tube; OV, otic vesicle; SAG, statoacoustic ganglion. Scale B, 100μm; C, I, 50μm.
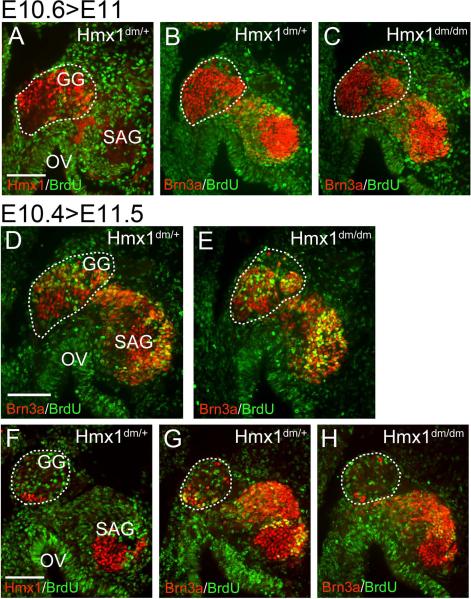
To further assess the relationship of Hmx1 expression to cell cycle exit, we performed two series of cumulative BrdU labeling beginning early and late on E10.5 (designated “E10.4” and “E10.6” respectively; the developmental stage at the onset of labeling must be inferred from the observed stage at harvest and the elapsed time). In these embryos, only neurons which have exited the cell cycle prior to the start of labeling should remain unlabeled by BrdU. Analysis of the “E10.6” embryo series demonstrated that nearly all Hmx1-expressing geniculate neurons exit the cell cycle prior to this stage (Figure 7A–C), indicating that the mitotic cells detected at E10.5 (Figure 6) are probably undergoing their last cell division. Analysis of the “E10.4” labeled embryos showed BrdU labeling of a significant fraction of the Hmx1+ neurons in the dorsal part of the geniculate ganglion (Figure 7D). The pattern of BrdU labeling was similar in Hmx1dm/dm and control embryos (Figure 7D,E). The small complement of Hmx1+ neurons in the ventral geniculate ganglion had mostly completed neurogenesis by this stage (Figure 7F).
Figure 7. Timing of cell cycle exit in the geniculate ganglion.
Cumulative BrdU labeling was performed in two litters of embryos, beginning early and late on gestational day E10.5 (designated “E10.4” and “E10.6”, see text). (A–C) In the later-labeled embryos, nearly all Hmx1-expressing neurons in the geniculate ganglion (circled) have exited the cell cycle by the start of the labeling period in both control and Hmx1dm/dm embryos. (D,E) In the dorsal geniculate ganglion (circled) of earlier-labeled embryos, some geniculate neurons are still undergoing (a presumably final round) of DNA synthesis at the onset of labeling. A similar fraction of BrdU-positive neurons are observed in the geniculate ganglion of control and Hmx1dm/dm embryos. (F–H) In the ventral geniculate ganglion (circled) of earlier-labeled embryos, only a few Hmx1-expressing somatosensory neurons are present; most of the ganglion at this level consists of Hmx1-negative viscerosensory neurons. Hmx1 neurons at this level have mostly exited the cell cycle by the stage of labeling. Hmx1 expression also identifies a population of early-born neurons in the statoacoustic ganglion. Neither population shows altered incorporation of BrdU in Hmx1dm/dm embryos. GG, geniculate ganglion; OV, otic vesicle; SAG, statoacoustic ganglion. Scale, 100μm.
We also used BrdU labeling to assess the timing of neurogenesis in Hmx1-expressing neurons in the statoacoustic ganglion and the trigeminal ganglion. The pan-sensory marker Brn3a allowed assessment of BrdU uptake in a comparison group of neurons, including those not expressing Hmx1. In the statoacoustic ganglion of “E10.4” labeled embryos Hmx1 expression identified a group of BrdU-negative early-born neurons, whilst Brn3a-expressing neurons in the remainder of the ganglion were still in the cell cycle at this stage (Figure 7D–F). As in the geniculate ganglion, cell cycle exit in the statoacoustic ganglion was not affected in the Hmx1 knockout.
We initially considered the possibility that premature exit of geniculate precursors from the cell cycle in Hmx1dm/dm mice might account for the reduced number of geniculate neurons. However we conclude that the timing of neurogenesis is not affected in the geniculate ganglion of Hmx1dm/dm mice, and that Hmx1 is unlikely to function either in maintaining a pool of precurors in the cell cycle, or promoting cell cycle exit. Instead, there is a brief developmental interval (~11.0) in which geniculate ganglion neurogenesis is complete, and cell number is nearly normal. Thus we infer that cell loss in the geniculate ganglion takes place by excess cell death in Hmx1dm/dm ganglia between E11.0 and E14.5. Several factors may have contributed to our failure to detect a statistically significant increase in dying neurons using Caspase-3 staining, including: 1) the significant background of apoptosis in the normal ganglion, 2) the brief expression of the Caspase-3 marker, which thus labels very few cells in any given sample, 3) the protracted period of cell death from E11.5 to E14.5, which yields only a few apoptotic events at any time point, and 4) the small total number of geniculate neurons available for sampling.
Distinct subsets of Hmx1 sensory neurons originate in the neural crest and neurogenic placodes
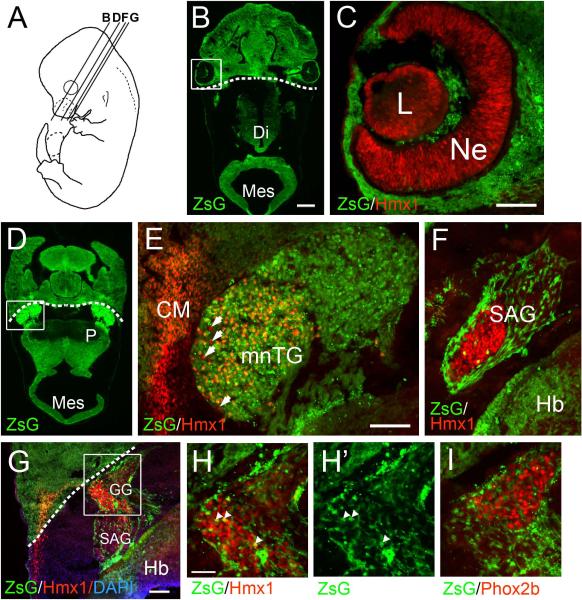
Work principally in avian embryos and lower vertebrates has demonstrated the dual embryological origin of the cranial sensory ganglia, in which components of the trigeminal, geniculate, and IX/X ganglion complexes originate in either the neural crest or ectodermal placodes (Baker and Bronner-Fraser, 2001; Le Douarin and Smith, 1988). In this context, we used a transgenic fate-mapping strategy to determine the embryological origin of the Hmx1-expressing neurons in the cranial sensory ganglia. A Wnt1-Cre transgenic line, in combination with Cre-inducible reporters, has been widely used to fate-map derivatives of the dorsal neural tube and neural crest (Danielian et al., 1998). In order to determine the developmental origin of Hmx1-expressing cranial sensory neurons we crossed Wnt1-Cre with the mouse strain Ai6, which inducibly expresses the reporter ZsGreen from the Rosa26 locus (Madisen et al., 2010). Consistent with prior studies (Yoshida et al., 2008), in Wnt1-Cre/Ai6 embryos examined at E12.5 we observed a clear boundary of expression in the craniofacial mesenchyme, just posterior to the eye, between frontal structures which are neural crest-derived and ZsGreen positive, and parietal structures which are mesoderm-derived and ZsGreen-negative (dashed lines in low power views in Figure 8). However, the migration of the neurons and associated glia of the cranial ganglia do not obey these anatomical boundaries.
Figure 8. Transgenic fate mapping of cranial sensory neurons.
A Wnt1-Cre driver line was crossed with the reporter Ai6, which expresses ZsGreen under the control of a modified Rosa26 locus (Methods). Embryos were examined at E12.5. The endogenous Zs-green fluorescence and immunofluorescence for the stated transcription factor is shown. Dashed lines in low power views indicate boundary between frontal (neural crest) and parietal (mesodermal) mesenchyme (Yoshida et al., 2008). (A) Schematic showing planes of section in subsequent views. (B,C) Expression of Hmx1 in the developing lens and retina. (D–E) Expression of Hmx1 in the TG. Overall, a large majority of both Hmx1+ and Hmx1− TG neurons express the ZsGreen marker. Arrows indicate rare Hmx1+ neurons which do not co-express ZsGreen. Prominent Hmx1 expression is also seen in the migrating cranial mesenchyme overlying the TG. (F) Expression of Hmx1 in the statoacoustic ganglion. Few if any Hmx1+ neurons express ZsGreen. The ganglion is surrounded by ZsGreen-expressing glial precursors. (G–I) Views of the geniculate ganglion, in which few Hmx1+ somatosensory or Phox2b+ viscerosensory neurons express ZsGreen, although ZsGreen+ glial precursors surround and invade the ganglion, sometimes overlapping neurons. Arrowheads in (H) indicate 3/130 Hmx1+ neurons which are potentially neural crest-derived. CM, cranial mesenchyme; Di, diencephalon; GG, geniculate ganglion; Hb, hindbrain; L, lens; Mes, mesencephalon; mnTG, mandibular lobe, trigeminal ganglion; mxTG, maxillary lobe, trigeminal ganglion; Ne, neuroepithelium (of retina); P, pons; SAG, statoacoustic ganglion. Fine punctate green signal in some views is an artifact of the aggregation of the marker protein in neurons (Madisen et al., 2010). Scale B, 400μm; C, E, G, 100μm; H, 50μm.
We then examined the expression of Hmx1 in the sensory ganglia of fate-mapped embryos at E12.5. In the eye, Hmx1-expressing cells in the lens and retinal neuroepithelium were ZsGreen-negative, consistent with their origin in the lens placode and ventral telencephalon (Figure 8B–C). In the mnTG, the vast majority of Hmx1-immunoreactive neurons were ZsGreen-positive, although a few Hmx1+/ZsGreen-negative cells could be clearly distinguished (Figure 8D–E). Indeed throughout the TG, the large majority of the cells were labeled. As expected, Hmx1+ neurons in the statoacoustic ganglion did not express ZsGreen, consistent with their origin in the otic placode (Figure 8F). Finally, in the geniculate ganglion, both the somatosensory Hmx1+ neurons and the viscerosensory Phox2b+ neuron populations were largely ZsGreen-negative, although the associated glial precursors in both ganglia expressed ZsGreen as expected (Figure 8G–I). Together these results indicate that in rodents the neural crest contribution to the neuronal population of the geniculate ganglion may be less than that reported in birds and amphibians, whereas the neural crest contribution to the TG may be proportionately greater.
DISCUSSION
In the present study we have shown that the variant homeodomain factor Hmx1 is required for development of somatic sensory neurons in the geniculate ganglion, which give rise to the posterior auricular nerve. In contrast, although Hmx1 is specifically expressed in the posterior TG and also in the superior ganglion, embryonic cell loss was not detected in these ganglia. Early expression of Hmx1 in dividing geniculate neuroblasts, particularly with respect to the marker of definitive sensory neurogenesis Brn3a, may make the geniculate ganglion specifically sensitive to the loss of Hmx1 function.
In contrast, the results presented here effectively exclude the existing hypothesis that Hmx1 or other Hmx factors are globally required for the selection of sensory neural over glial/melanocyte fates in the neural crest (Adameyko et al., 2009). It is clear that sensory neurogenesis and the initial specification of sensory subtypes proceed unimpeded in the DRG and TG of Hmx1dm/dm embryos. Two other members of the Hmx-class have been identified in mice, Hmx2 and Hmx3, which have known and partially overlapping roles in the development of the vestibulocochlear system (Chatterjee et al., 2010). However, Hmx2 and Hmx3 are not expressed in the early DRG or TG (Wang et al., 2000), and thus cannot compensate for the loss of Hmx1 in Hmx1dm/dm embryos.
The Hmx1 dependence of somatosensory geniculate neurons underscores the distinct nature of these cells from the cranial viscerosensory neurons serving taste sensation. Co-expression of Hmx1 and Brn3a provides a molecular link between the somatosensory geniculate neurons and other caudal somatosensory neurons in the mandibular TG, the superior ganglion of the IX/X complex, and the DRG. In contrast, Hmx1 is excluded from the viscerosensory neurons of the geniculate ganglion and the nodose and petrosal ganglia of the distal IX/X complex. Most of the neurons in these ganglia are also Brn3a-negative. Thus co-expression of Hmx1 and Brn3a uniquely defines a set of somatosensory neurons that extends from the mandibular trigeminal through the entire caudal neural axis. Hmx1 and Brn3a in the hindbrain somatosensory ganglia are expressed in a “mirror image” to the paired-like homeobox gene Phox2b, which is expressed in and required for the development/survival of the viscerosensory hindbrain ganglia but not somatosensory neurons (Coppola et al., 2010; Dauger et al., 2003).
Classic studies in avian embryos have identified a proximal or “root” part of the geniculate (VII) ganglion that lies close to the statoacoustic ganglion, and is of neural crest origin (Ayer-Le Lievre and Le Douarin, 1982; D'Amico-Martel and Noden, 1983; Le Douarin and Smith, 1988), while neurons of the distal portion of the geniculate are of placodal origin. However, the evidence for dual origin of the IX/X ganglion system in the literature is much more extensive than for the geniculate (Narayanan and Narayanan, 1980). D'Amico-Martel and Noden (1983) for instance, reported only a few crest-derived neurons in the proximal geniculate in just 1/7 of chick-quail chimeric transplanted embryos. Our results indicate that in the mouse geniculate ganglion most or all the Hmx1+ somatosensory neurons, like the Phox2b+ viscerosensory neurons, are not derived from neural crest and are thus likely to be of placodal origin. We cannot rule out a population of sensory neurons derived from neural crest in the geniculate that does not express either marker, but the major known somatosensory and viscerosensory functions of the geniculate appear to be accounted for by the Hmx1- and Phox2b-expressing cells, respectively. Recent studies using different marker systems are in accord with these findings. Fate mapping of rhombomere 4-derived neural crest using a Hoxb1-cre driver labels very few neurons of the geniculate ganglion, as identified by NeuN (Arenkiel et al., 2003) or Islet1/2 (Yang et al., 2008) expression. Remarkably, in Hoxa2 null mice, there is a marked increase in neural differentiation in neural crest-derived cells in the vicinity of the geniculate (Yang et al., 2008). In another study, Ap2α-Cre fate mapping of placode-derived cells frequently labeled geniculate neurons. In contrast, Wnt1-Cre fate-mapped neurons co-expressing the neural marker Islet1/2 were rarely observed in the geniculate, and these cells instead expressed glial markers (Harlow et al., 2011).
Studies of the embryological origin of the cranial sensory neurons may also need to consider species differences in the anatomical arrangement of functional populations of sensory neurons. A recent study combining fate mapping and tract-tracing in the salamander clearly shows a population of neural-crest derived somatosensory neurons located in the geniculate ganglion (Harlow and Barlow, 2007). However, these neurons innervate the oral cavity. Our tract-tracing experiments did not reveal somatosensory projections from the geniculate to the oral cavity (Video 1), and in mammals somatic sensation in the oral area is served by neurons of the TG (Arvidsson et al., 1995), which probably contains the rodent equivalents of the orally-projecting neural crest-derived neurons in the salamander geniculate. Our data thus do not support a model in which the neurons serving somatosensory and viscerosensory function in the rodent geniculate ganglion are determined by their respective derivation from the neural crest or placodes. Finally, in contrast to the mixed embryological origin of the neuronal component of the geniculate, our Wnt1-Cre/ZsGreen fate mapping experiments confirm that the glial precursors of the geniculate and statoacoustic ganglia are derived from neural crest, in accordance with many prior studies.
Fate mapping using a Wnt1-Cre driven ZsGreen reporter also allowed determination of the embryological origin of the Hmx1-expressing neurons in the mnTG. Classic studies in chick-quail chimeric embryos have determined that the neuronal component of the TG is derived in part from neural crest, and in part from the trigeminal placodes (D'Amico-Martel and Noden, 1983). In birds, neural-crest derived cells predominate close to the hindbrain, and placode-derived neurons populate the peripheral part of the ganglion. Our results show that in the mouse the large majority of the Hmx1+ neurons in the mnTG are of neural crest origin, but ZsGreen-negative Hmx1+ cells were also observed, indicating that Hmx1 expression is not determined by embryological origin. In general, Wnt1-Cre fate mapping labeled the majority of neurons throughout the TG, demonstrating that the mouse TG as a whole is predominantly of neural crest origin. Most of the cells which were not labeled by Wnt1-Cre are probably of placodal origin, but it must also be considered that Cre-mediated activation of the ZsGreen reporter may not be 100% efficient, and the unlabeled cells in the TG represent an upper limit for the placodal contribution to the ganglion. The ZsGreen-negative cells were not restricted to the periphery of the ganglion, as observed for placode-derived neurons in the chick. A strikingly similar, but reversed, pattern of labeling has been obtained in the mouse TG with the placodal fate mapping Cre-driver Ap2a-Cre (Harlow et al., 2011). We conclude that the mouse TG has a mixed embryological origin, but unlike in birds, the neural crest component predominates, and cells within the ganglion are not segregated by origin.
The only functional role previously identified for Hmx1 is in the chick retina, where two Hmx1 paralogs, SOHo1 and Hmx1/GH6, are co-expressed from adjacent genetic loci. These genes are expressed in an overlapping nasal high/temporal low gradient (Stadler and Solursh, 1994; Wang et al., 2000; Yoshiura et al., 1998), and have the ability to repress the axial patterning signal EphA3 (Schulte and Cepko, 2000). The Hmx1 factors in mouse and chick are expressed beginning at the optic cup stage (Deitcher et al., 1994; Schulte and Cepko, 2000), but our immunofluorescence results also show expression later, in differentiating retinal ganglion cells, suggesting a dual role for Hmx1 in retinal development. Hmx1 loss-of-function mutations in mouse and human result in microphthalmia and/or coloboma, consistent with a role in early eye patterning (Munroe et al., 2009; Schorderet et al., 2008), but oculoauricular syndrome patients also develop progressive rod-cone dystrophy (Vaclavik et al., 2011). The role of Hmx1 in eye development may overlap that of the Vax homeodomain family of transcription factors which help to determine both axial patterning and the segregation of the retinal precursors from adjacent structures (Mui et al., 2005). A full understanding of the role of Hmx1 in retinal development may require a conditional strategy to separate early development roles from possible later effects on retinal ganglion cell differentiation.
Still to be addressed mechanistically is the role of Hmx1 in craniofacial development. Neural tube closure defects, initially reported in Hmx1dm/dm mice, disappeared after two generations of crosses to C57bl/6 mice, but the characteristic recessive Dumbo ear phenotype remained fully penetrant. Geniculate somatosensory neurons, via the posterior auricular nerve, innervate target tissues in the pinna that are also affected by loss of Hmx1 expression, at first suggesting that the effect on geniculate neurons could be non-cell autonomous, perhaps due to the loss of some trophic factor in the mesenchyme. However, the expression of Hmx1 in the geniculate ganglion precedes expression in the mesenchyme, and cell loss in the geniculate precedes target innervation. Thus it is likely that the effects of Hmx1 null mutations in the geniculate ganglion are cell autonomous. It will be interesting to see if the role of Hmx1 in sensory neurons, eye, and mesenchyme are linked by conserved downstream target genes or mechanisms.
CONCLUSIONS
Hmx1 is widely expressed in peripheral somatosensory neurons, but not viscerosensory neurons. The mouse Dumbo allele is a complete loss-of-function for Hmx1. Hmx1 is not generally required for determination of sensory cell fate in the neural crest, and sensory precursors lacking Hmx1 do not adopt non-neural fates. Among somatosensory neurons, Hmx1 is uniquely required for early development of the somatosensory component of the geniculate ganglion. Transgenic fate mapping of the neural crest shows that Hmx1-expressing neurons in the cranial sensory ganglia may arise either from neural crest or neurogenic placodes.
Supplementary Material
HIGHLIGHTS
Hmx1 is a transcription factor which has been linked to ear malformations in humans, rats, and mice.
Hmx1 is expressed in cranial and DRG somatosensory neurons, including all sensory modalities.
The mouse “Dumbo” (dm) mutation is a complete loss of function for Hmx1.
DRG neurons in Hmx1dm/dm embryos undergo neurogenesis and express sensory markers normally.
Hmx1dm/dm embryos show marked defects in the geniculate ganglion/posterior auricular nerve.
Acknowledgements
We would like to thank J.-F. Brunet and M. Wegner for generous gifts of antibodies. We would also like to thank Dr. Bernd Fritzsch for helpful discussions and Dr. Clare Baker for helpful comments on the manuscript. Supported in part by Department of Veterans Affairs MERIT funding, and NIH awards HD33442, MH065496, and NS064933 (E.E.T). E.E.T. is a NARSAD Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adameyko I, Lallemend F, Aquino JB, Pereira JA, Topilko P, Muller T, Fritz N, Beljajeva A, Mochii M, Liste I, Usoskin D, Suter U, Birchmeier C, Ernfors P. Schwann cell precursors from nerve innervation are a cellular origin of melanocytes in skin. Cell. 2009;139:366–379. doi: 10.1016/j.cell.2009.07.049. [DOI] [PubMed] [Google Scholar]
- Arenkiel BR, Gaufo GO, Capecchi MR. Hoxb1 neural crest preferentially form glia of the PNS. Dev Dyn. 2003;227:379–386. doi: 10.1002/dvdy.10323. [DOI] [PubMed] [Google Scholar]
- Arvidsson J, Fundin BT, Pfaller K. Innervation of the hard palate in the rat studied by anterograde transport of horseradish peroxidase conjugates. J Comp Neurol. 1995;351:489–498. doi: 10.1002/cne.903510402. [DOI] [PubMed] [Google Scholar]
- Ayer-Le Lievre CS, Le Douarin NM. The early development of cranial sensory ganglia and the potentialities of their component cells studied in quail-chick chimeras. Dev Biol. 1982;94:291–310. doi: 10.1016/0012-1606(82)90349-9. [DOI] [PubMed] [Google Scholar]
- Baker CV, Bronner-Fraser M. Vertebrate cranial placodes I. Embryonic induction. Dev Biol. 2001;232:1–61. doi: 10.1006/dbio.2001.0156. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Kraus P, Lufkin T. A symphony of inner ear developmental control genes. BMC Genet. 2010;11:68. doi: 10.1186/1471-2156-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola E, Rallu M, Richard J, Dufour S, Riethmacher D, Guillemot F, Goridis C, Brunet JF. Epibranchial ganglia orchestrate the development of the cranial neurogenic crest. Proc Natl Acad Sci U S A. 2010;107:2066–2071. doi: 10.1073/pnas.0910213107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amico-Martel A, Noden DM. Contributions of placodal and neural crest cells to avian cranial peripheral ganglia. Am J Anat. 1983;166:445–468. doi: 10.1002/aja.1001660406. [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Dauger S, Pattyn A, Lofaso F, Gaultier C, Goridis C, Gallego J, Brunet JF. Phox2b controls the development of peripheral chemoreceptors and afferent visceral pathways. Development. 2003;130:6635–6642. doi: 10.1242/dev.00866. [DOI] [PubMed] [Google Scholar]
- Deitcher DL, Fekete DM, Cepko CL. Asymmetric expression of a novel homeobox gene in vertebrate sensory organs. J Neurosci. 1994;14:486–498. doi: 10.1523/JNEUROSCI.14-02-00486.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykes IM, Tempest L, Lee SI, Turner EE. Brn3a and islet1 act epistatically to regulate the gene expression program of sensory differentiation. J Neurosci. 2011;31:9789–9799. doi: 10.1523/JNEUROSCI.0901-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng SR, Gratwick K, Rhee JM, Fedtsova N, Gan L, Turner EE. Defects in sensory axon growth precede neuronal death in Brn3a-deficient mice. J Neurosci. 2001;21:541–549. doi: 10.1523/JNEUROSCI.21-02-00541.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng SR, Lanier J, Fedtsova N, Turner EE. Coordinated regulation of gene expression by Brn3a in developing sensory ganglia. Development. 2004;131:3859–3870. doi: 10.1242/dev.01260. [DOI] [PubMed] [Google Scholar]
- Fedtsova N, Perris R, Turner EE. Sonic hedgehog regulates the position of the trigeminal ganglia. Dev Biol. 2003;261:456–469. doi: 10.1016/s0012-1606(03)00316-6. [DOI] [PubMed] [Google Scholar]
- Folan-Curran J, Cooke FJ. Contribution of cranial nerve ganglia to innervation of the walls of the rat external acoustic meatus. J Peripher Nerv Syst. 2001;6:28–32. doi: 10.1046/j.1529-8027.2001.006001028.x. [DOI] [PubMed] [Google Scholar]
- Fritzsch B, Beisel KW, Hansen LA. The molecular basis of neurosensory cell formation in ear development: a blueprint for hair cell and sensory neuron regeneration? Bioessays. 2006;28:1181–1193. doi: 10.1002/bies.20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow DE, Barlow LA. Embryonic origin of gustatory cranial sensory neurons. Dev Biol. 2007;310:317–328. doi: 10.1016/j.ydbio.2007.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow DE, Yang H, Williams T, Barlow LA. Epibranchial placode-derived neurons produce BDNF required for early sensory neuron development. Dev Dyn. 2011 doi: 10.1002/dvdy.22527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley MW. Regulation of cell fate in the sensory epithelia of the inner ear. Nat Rev Neurosci. 2006;7:837–849. doi: 10.1038/nrn1987. [DOI] [PubMed] [Google Scholar]
- Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramoto T, Yokoe M, Yagasaki K, Kawaguchi T, Kumafuji K, Serikawa T. Genetic analyses of fancy rat-derived mutations. Exp Anim. 2010;59:147–155. doi: 10.1538/expanim.59.147. [DOI] [PubMed] [Google Scholar]
- Lanier J, Quina LA, Eng SR, Cox E, Turner EE. Brn3a target gene recognition in embryonic sensory neurons. Dev Biol. 2007;302:703–716. doi: 10.1016/j.ydbio.2006.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin NM, Smith J. Development of the peripheral nervous system from the neural crest. Annu Rev Cell Biol. 1988;4:375–404. doi: 10.1146/annurev.cb.04.110188.002111. [DOI] [PubMed] [Google Scholar]
- Lei L, Zhou J, Lin L, Parada LF. Brn3a and Klf7 cooperate to control TrkA expression in sensory neurons. Dev Biol. 2006;300:758–769. doi: 10.1016/j.ydbio.2006.08.062. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maka M, Stolt CC, Wegner M. Identification of Sox8 as a modifier gene in a mouse model of Hirschsprung disease reveals underlying molecular defect. Dev Biol. 2005;277:155–169. doi: 10.1016/j.ydbio.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Marmigere F, Ernfors P. Specification and connectivity of neuronal subtypes in the sensory lineage. Nat Rev Neurosci. 2007;8:114–127. doi: 10.1038/nrn2057. [DOI] [PubMed] [Google Scholar]
- Mui SH, Kim JW, Lemke G, Bertuzzi S. Vax genes ventralize the embryonic eye. Genes Dev. 2005;19:1249–1259. doi: 10.1101/gad.1276605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe RJ, Prabhu V, Acland GM, Johnson KR, Harris BS, O'Brien TP, Welsh IC, Noden DM, Schimenti JC. Mouse H6 Homeobox 1 (Hmx1) mutations cause cranial abnormalities and reduced body mass. BMC Dev Biol. 2009;9:27. doi: 10.1186/1471-213X-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan CH, Narayanan Y. Neural crest and placodal contributions in the development of the glossopharyngeal-vagal complex in the chick. Anat Rec. 1980;196:71–82. doi: 10.1002/ar.1091960108. [DOI] [PubMed] [Google Scholar]
- Oakley B, Witt M. Building sensory receptors on the tongue. J Neurocytol. 2004;33:631–646. doi: 10.1007/s11068-005-3332-0. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. Expression and interactions of the two closely related homeobox genes Phox2a and Phox2b during neurogenesis. Development. 1997;124:4065–4075. doi: 10.1242/dev.124.20.4065. [DOI] [PubMed] [Google Scholar]
- Quina LA, Pak W, Lanier J, Banwait P, Gratwick K, Liu Y, Velasquez T, O'Leary DD, Goulding M, Turner EE. Brn3a-expressing retinal ganglion cells project specifically to thalamocortical and collicular visual pathways. J Neurosci. 2005;25:11595–11604. doi: 10.1523/JNEUROSCI.2837-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorderet DF, Nichini O, Boisset G, Polok B, Tiab L, Mayeur H, Raji B, de la Houssaye G, Abitbol MM, Munier FL. Mutation in the human homeobox gene NKX5-3 causes an oculo-auricular syndrome. Am J Hum Genet. 2008;82:1178–1184. doi: 10.1016/j.ajhg.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte D, Cepko CL. Two homeobox genes define the domain of EphA3 expression in the developing chick retina. Development. 2000;127:5033–5045. doi: 10.1242/dev.127.23.5033. [DOI] [PubMed] [Google Scholar]
- Sharpe J, Ahlgren U, Perry P, Hill B, Ross A, Hecksher-Sorensen J, Baldock R, Davidson D. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296:541–545. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Stadler HS, Solursh M. Characterization of the homeobox-containing gene GH6 identifies novel regions of homeobox gene expression in the developing chick embryo. Dev Biol. 1994;161:251–262. doi: 10.1006/dbio.1994.1025. [DOI] [PubMed] [Google Scholar]
- Stephenson DA, Lee KH, Nagle DL, Yen CH, Morrow A, Miller D, Chapman VM, Bucan M. Mouse rump-white mutation associated with an inversion of chromosome 5. Mamm Genome. 1994;5:342–348. doi: 10.1007/BF00356552. [DOI] [PubMed] [Google Scholar]
- Sun Y, Dykes IM, Liang X, Eng SR, Evans SM, Turner EE. A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat Neurosci. 2008;11:1283–1293. doi: 10.1038/nn.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theiler K. The house mouse; development and normal stages from fertilization to 4 weeks of age. Springer-Verlag; Berlin, New York: 1972. [Google Scholar]
- Trieu M, Ma A, Eng SR, Fedtsova N, Turner EE. Direct autoregulation and gene dosage compensation by POU-domain transcription factor Brn3a. Development. 2003;130:111–121. doi: 10.1242/dev.00194. [DOI] [PubMed] [Google Scholar]
- Vaclavik V, Schorderet DF, Borruat FX, Munier FL. Retinal dystrophy in the oculo-auricular syndrome due to HMX1 mutation. Ophthalmic Genet. 2011;32:114–117. doi: 10.3109/13816810.2011.562955. [DOI] [PubMed] [Google Scholar]
- Wang W, Lo P, Frasch M, Lufkin T. Hmx: an evolutionary conserved homeobox gene family expressed in the developing nervous system in mice and Drosophila. Mech Dev. 2000;99:123–137. doi: 10.1016/s0925-4773(00)00488-3. [DOI] [PubMed] [Google Scholar]
- Wang W, Lufkin T. Hmx homeobox gene function in inner ear and nervous system cell-type specification and development. Exp Cell Res. 2005;306:373–379. doi: 10.1016/j.yexcr.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Wilson L, Ching YH, Farias M, Hartford SA, Howell G, Shao H, Bucan M, Schimenti JC. Random mutagenesis of proximal mouse chromosome 5 uncovers predominantly embryonic lethal mutations. Genome Res. 2005;15:1095–1105. doi: 10.1101/gr.3826505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamout A, Spec A, Cosmano J, Kashyap M, Rochlin MW. Neurotrophic factor receptor expression and in vitro nerve growth of geniculate ganglion neurons that supply divergent nerves. Dev Neurosci. 2005;27:288–298. doi: 10.1159/000086708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zhou Y, Barcarse EA, O'Gorman S. Altered neuronal lineages in the facial ganglia of Hoxa2 mutant mice. Dev Biol. 2008;314:171–188. doi: 10.1016/j.ydbio.2007.11.032. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Vivatbutsiri P, Morriss-Kay G, Saga Y, Iseki S. Cell lineage in mammalian craniofacial mesenchyme. Mech Dev. 2008;125:797–808. doi: 10.1016/j.mod.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Yoshiura K, Leysens NJ, Reiter RS, Murray JC. Cloning, characterization, and mapping of the mouse homeobox gene Hmx1. Genomics. 1998;50:61–68. doi: 10.1006/geno.1998.5284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.