Abstract
Evolution of the domain encoding the V1/V2 variable region of the simian immunodeficiency virus sm (SIVsm) envelope (env) gene was analyzed in relation to route of virus challenge, virus load, and neutralizing antibody (NAb) titers during primary infection of rhesus macaques with the pathogenic SIVsmE660 isolate. In this model system animals are initially infected with multiple viruses as evidenced by the presence of multiple V1/V2 genotypic variants that could be resolved by using a heteroduplex tracking assay (HTA). Overlapping subsets of the multiple variants were established in each animal. There was no selection for the establishment of specific variants in comparing intravenous- and intrarectal-challenged macaques at week 2 postinfection, suggesting that no genotypic selection occurred at the mucosal surface. There was an initial period of significant stability of the V1/V2 variants. Macaques challenged intravenously displayed subsequent V1/V2 diversification significantly earlier than macaques challenged intrarectally and well past the initial resolution of viremia. The time when SIVsmE660-specific NAbs reached a threshold titer of 100 was significantly correlated with the timing of V1/V2 diversification, even though antibodies to the Env protein could be detected much earlier. The time when NAbs reached a titer of 400 was significantly correlated with virus load late in infection. These results show that the route of infection affects the timing of V1/V2 diversification and that this diversification is correlated with the maturation of a specific NAb response. However, prior immunization capable of priming an anamnestic Env antibody response did not accelerate V1/V2 diversification. This result suggests that diversification of the SIV env V1/V2 region is the result of a type-specific antibody response.
Both the human immunodeficiency virus type 1 (HIV-1) and the simian immune virus (SIV) envelope (Env) proteins contain variable regions (12, 34). The V1/V2 variable regions are a multifunctional domain on Env that undergoes a conformational change upon CD4 binding (46, 56), modulates exposure of the coreceptor binding site on Env (55), can contribute to cell tropism and cytopathicity (20, 42), and contains epitopes for antiviral antibodies (22, 31, 49). The V1/V2 region, along with the other variable regions, represents regions of the viral genome that express remarkable capacity to evolve in response to changing selective pressures within the host.
Systemic and mucosal infection represent two major routes of HIV-1 entry into human hosts. Several studies have examined viral evolution in relation to route of infection (29, 48, 50, 51). These results suggested that mucosal barriers act as selective filters for HIV-1 genotypes.
The development of host neutralizing antibodies (NAbs) as part of the humoral response against HIV-1 and SIV most often occurs after the resolution of peak plasma viremia (43, 45). The variable regions on the HIV-1 and SIV Env proteins can serve as linear and conformational epitopes for NAbs (1, 9, 21, 22). Changes in the variable regions enable the virus to escape neutralization by antibodies developed early after infection, allowing virus populations to persist (5, 52). Virus variants that develop after infection are commonly characterized by sequence changes, length changes, and changes in the carbohydrate composition of the V1/V2, namely alterations in N-linked and O-linked glycosylation sites (1, 7, 53, 55). In addition, the route of exposure to antigen in vaccine presentation, and potentially in infection, can affect the nature of the immune response (26, 57).
The analysis of env heterogeneity has yielded and will continue to yield important information about the biology of the viral Env protein. We have focused on the heterogeneity of the V1/V2 variable region to track changes in env gene populations, although other regions of env are amenable to similar analysis. We have examined the effect of site of entry on virus uptake and the rate of evolution as measured by V1/V2 diversification. We found no evidence for sequence selection during entry at a mucosal surface. However, our results show that V1/V2 diversification occurs significantly later in macaques challenged mucosally (intrarectally [i.r.]) than with macaques challenged systemically (intravenously [i.v.]), although there was no significant difference in the total Env antibody response by these two routes. The timing of V1/V2 diversification was correlated with the antiviral NAb titer but was not correlated with peak virus load (VL) or set point viremia levels. The initial changes in V1/V2 amino acid sequences that were observed in macaques challenged i.v. and i.r. were similar and were characterized primarily by changes in a region of potential O-linked glycosylation sites in V1. Prior vaccination primed an anamnestic response, as measured by a neutralization assay specific for SIVsmH-4 and enzyme-linked immunosorbent assay (ELISA) specific for gp120, but did not affect the timing of V1/V2 diversification, suggesting strong type specificity in the V1/V2 immune response. Taken together, these results suggest that the route of virus entry affects the timing of V1/V2 diversification and that the heterogeneity of the SIV env V1/V2 region is correlated with a type-specific antibody response.
MATERIALS AND METHODS
SIV challenge and vaccination.
After passage of SIVsm through a rhesus macaque, the uncloned challenge virus, SIVsmE660, was isolated as previously described (15, 19). Macaques (in groups of six) were challenged either i.v. with 1 ml of a 1:6,000 dilution of the virus stock (∼50% macaque infectious dose) or i.r. with 1 ml of undiluted virus stock of SIVsmE660. However, two of the nonvaccinated macaques challenged i.v. in this study had undetectable plasma VL throughout the postinfection (p.i.) period and were not demonstrated to be infected by any other test and therefore were not included in this analysis. Another group of 8 macaques was vaccinated with SIVsmH4-derived matrix/capsid (MA/CA), gp140 and gp160 genes expressed by using the Venezuelan equine encephalitis virus (VEE) vector system (10). Each VEE vector dose was 107 infectious units given subcutaneously in the right forelimb. Data on the efficacy of the vaccination protocol will be presented separately. Four of these macaques were challenged i.v. and four macaques were challenged i.r. with SIVsmE660 as described above. Another two macaques from a previous study (10) were also examined in a separate analysis to determine the effect of rapid disease progression. These macaques (N2P and W1A) were sacrificed at week 11 p.i. in that previous study due to severe wasting (10). All animal care was performed in accordance with institutional guidelines.
Virus load determination and NAb titers.
Virus load was determined by the bDNA assay (Chiron, Emeryville, Calif.). NAb titers against SIVsmE660 and SIVsmH-4 (18) were determined by CEMx174 and H9 cell-killing assays, respectively, as previously described (35). The values shown are the reciprocal of the dilution of serum needed to protect 50% of the cells from virus-induced killing. SIVsmE660 is a virus stock from an isolate generated from a macaque infected with the SIVsm isolate (18). SIVsmH-4 is an infectious molecular clone from the SIVsm isolate (18) that was used as a source of viral genes for immunization and as the source of the heteroduplex tracking assay (HTA) probe and was also used to generate a virus stock for NAb assays.
ELISA.
Purified SIVsmH-4 gp120 protein was diluted with carbonate buffer (7.5 mM Na2CO3-17.4 mM NaHCO3, pH 9.6) and then added to wells (200 ng/well) and incubated overnight. The plates were washed with phosphate-buffered saline (PBS) and were incubated for 1 h with blocking buffer (3% bovine serum albumin plus 1% normal rabbit sera diluted in PBS). The wells were washed with PBS. Diluted macaque plasma was added to the wells, and the plate was incubated for 1 h at room temperature. The wells were washed, and then rabbit anti-monkey immunoglobulin G-conjugated horseradish peroxidase antibodies (Sigma, St. Louis, Mo.) were added and incubated for 1 h. The wells were washed, and color development was performed by using OPD FAST Tablets (Sigma) per the manufacturer's protocol. The plates were read at an optical density of 450 nm (OD450). Titers are reported as the reciprocal of the highest dilution that yields an OD reading of 0.200.
RNA isolation and RT-PCR.
Viral RNA was isolated from 140 μl of macaque plasma by using the QIAamp viral RNA isolation kit (Qiagen, Valencia, Calif.), resulting in purified RNA in a 50-μl volume. Briefly, the V1/V2 region of the env gene was reverse transcribed by using a modified Titan One Tube RT-PCR System (Roche Applied Science, Indianapolis, Ind.). Primers specific for the V1/V2 region used for reverse transcription-PCR (RT-PCR) were the following: 5′-GTGGAACCTCTTTGAAACATCCATTAAGCC-3′ (smH-4/V1) and 5′-GTGCACAGTATCTAAATCTAATAGCATCCC-3′ (smH-4/V2). RT-PCR was performed as follows. The 20-μl reaction mixture consisted of 5 μl of purified viral RNA, 1× Titan RT-PCR buffer, 5 mM dithiothreitol, 1 mM each deoxynucleoside triphosphate, 10 pmol of V2 primer, and 12 U of avian myeloblastosis virus reverse transcriptase. RT was carried out at 42°C for 30 min. A 30-μl aliquot of PCR mix (1× Titan RT-PCR buffer, 5 mM dithiothreitol, 10 pmol of V1 primer, and 0.5 μl of Titan enzyme mix) was then added to the RT reaction mixture. PCR was carried out in a Robocycler 40 (Stratagene, La Jolla, Calif.) with the following program: 1 cycle of 95°C for 3 min, 54°C for 1 min, and 72°C for 1 min, and 39 cycles of 95°C 1 min, 54°C for 1 min, and 72°C for 1 to 4 min, followed by a final cycle at 72°C for 7 min.
V1/V2 probe construction.
The V1/V2 sequence was amplified from the SIVsmH-4 molecular clone (19) by using primers smH-4/V1 and smH-4/V2 to produce a 430-bp fragment that was inserted into the pT7Blue plasmid by blunt-end ligation (Novagen, Madison, Wis.) to generate the plasmid pSIVsmH-4V1/V2. An aliquot of 5 μg of plasmid DNA was linearized with EcoRI at 37°C for 1 h. The probe in the EcoRI reaction was labeled by filling in the 3′ underhang after adding 10 mM dithiothreitol, 50 μCi of [35S]dATP (1,250 Ci/mmol; NEN Life Science Products), and 10 U of Klenow fragment of DNA polymerase I for 15 min at room temperature. The labeled probe was then released from the vector by digestion with XbaI at 37°C for 1 h. Excess radioactive nucleotide was removed by using a MicroSpin G-50 column (Amersham Pharmacia Biotech, Arlington Heights, Ill.), and the volume was adjusted to 100 μl.
V1/V2 HTA.
V1/V2 heteroduplex annealing mixtures consisted of 1× annealing buffer (0.1 M NaCl, 10 mM Tris-HCl [pH 7.5], 2 mM EDTA), 5 to 10 μl of unpurified RT-PCR product, and 1 μl of labeled V1/V2 smH-4 probe in a total volume of 10 μl. The annealing mixture was incubated at 99°C for 2 min to denature the DNA and then was allowed to anneal at room temperature for 5 min. The heteroduplexes were separated in 6% polyacrylamide (37:5:1 acrylamide-bisacrylamide) gels in 1× Tris-borate-EDTA buffer (0.09 M Tris-borate, 0.002 M EDTA). Heteroduplexes were visualized by autoradiography of dried HTA gels. Dried gels were also exposed to phosphorimaging screens to quantitate the relative abundance of the bands in each sample by using ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.). Two independent RT-PCRs were performed from each viral RNA preparation followed by HTA analysis as described above, which validated adequate sampling in that the same complex patterns were reproducibly observed.
Measure of V1/V2 diversification.
The relative abundance of each variant was calculated as a percent of the total population of genotypic variants at each time point. In an effort to quantitate subsequent diversification, we used the group of genotypic variants present in each animal at week 2 p.i. as the baseline and quantitated the appearance of new genotypic variants in subsequent samples using the following formula: Divx = 100 − Ex, where E represents the variants that were established by week 2 p.i., Ex represents the percent fraction of those variants remaining at week x, and Divx represents the percentage of new variants at week x. Td50 is defined as the time when half of the relative abundance of variants identified by HTA were attributed to variants not found at week 2 p.i. The Mann-Whitney rank sum test was used to determine the statistical significance between values from the i.v.- and the i.r.-challenged groups. Pearson product moment correlation statistics were used to determine the significance of correlations between two variables.
Sequence analysis.
RT-PCR products of interest were purified by using the QIAquick PCR purification kit (Qiagen) and were cloned by using the pT7Blue Perfectly Blunt cloning kit. Inserts in individual clones were amplified by using primers smH-4/V1 and smH-4/V2 by PCR as described above. The products were screened by V1/V2-HTA with the SIVsmH-4V1/V2 probe. Plasmids were sequenced by ABI dye terminator sequencing (Perkin-Elmer Corp., Foster City, Calif.) and were analyzed and aligned with MacVector 7.0 (Genetics Computer Group, Madison, Wis.).
Nucleotide sequence accession numbers.
The sequences described in this study were deposited in GenBank with the accession numbers AY492288 to AY492335.
RESULTS
Characterization of the V1/V2 region of the SIVsmE660 isolate.
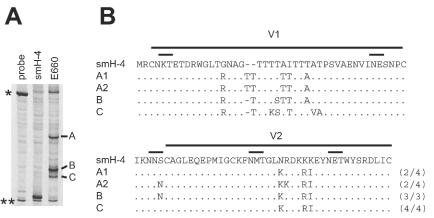
We constructed an HTA probe for the SIVsm V1/V2 region of env from the SIVsmH-4 molecular clone (18). We used this probe to assess the complexity of the SIVsmE660 isolate, which was used as the challenge virus. This analysis showed a heteroduplex pattern consisting of three major V1/V2 variants in the SIVsmE660 stock (Fig. 1A, bands A, B, and C). Band A clones consisted of two distinct V1/V2 populations represented by sequences A1 and A2 (Fig. 1B), which differed by two nucleotide substitutions in V2 but comigrated in the HTA gel. Several changes distinguished the SIVsmH-4 probe from all of the SIVsmE660 variants, but the changes that distinguished the SIVsmE660 variants from each other largely consisted of insertions of threonines and other substitutions in the threonine-rich region of V1. The SIVsmE660 stock was used to challenge macaques through the i.v. or i.r. route, and V1/V2 env diversification was assessed during primary infection. Undiluted virus stock was used in the i.r. challenge versus a 1:6,000 dilution of the virus for the i.v. challenge to compensate for the lower efficiency of infection by a mucosal route compared to that by an i.v. route.
FIG. 1.
Analysis of the V1/V2 region from SIVsmE660. (A) HTA analysis of SIVsmE660 V1/V2 region. Bands A, B, and C represent the three major V1/V2 variants seen in the SIVsmE660 inoculum. Single asterisks represent single-stranded probe, and double asterisks represent double-stranded probe. The PCR product for the SIVsmH-4 control was generated from cloned DNA. In this system, the homoduplex with PCR product is designed to migrate slightly more slowly than the probe reannealed to itself. (B) Amino acid sequence alignments of V1/V2 regions of SIVsmH-4 and the major V1/V2 variants in SIVsmE660. Dots represent identical amino acids. Dashes represent deleted amino acids. Short lines above the sequence highlight N-linked glycosylation sites. A1 and A2 represent different sequences from clones of the PCR product that comigrated with band A in the HTA gel. The frequency of sequences from clones is indicated at the right of each sequence.
Transmission and initial viremia.
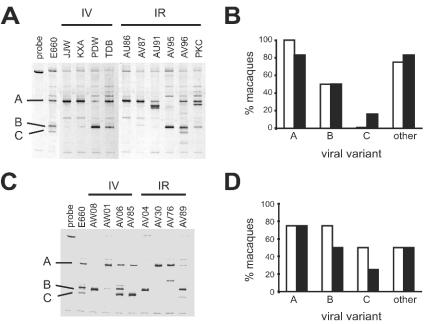
We used the smH-4 V1/V2 probe to analyze V1/V2 diversity to determine which V1/V2 variants were established in each host at week 2 p.i. Only a subset of the variants present in the complex SIVsmE660 stock became established in most animals, while only macaques AV96 (Fig. 2A), AV06, and AV89 (Fig. 2C) acquired all three of the major SIVsmE660 variants. A visual analysis of the V1/V2 HTA patterns at week 2 p.i. did not reveal a correspondence between route of challenge and which V1/V2 variants became established in each host (Fig. 2A and C). The presence of SIVsmE660 V1/V2 viral variants A, B, and C (as shown in Fig. 1) was scored for each animal. The percentage of animals showing the respective SIVsmE660 variants at week 2 p.i. was calculated for each challenge group (Fig. 2B and D). These results showed no significant difference between i.v.- and i.r.-challenged animals with respect to the subset of SIVsmE660 V1/V2 variants established in each animal. These results suggest that the route of challenge, either i.v. or i.r., did not differ with respect to tissue-specific selective determinants that would bias the establishment of a subset of the SIVsmE660 challenge virus when administered at these two sites. All macaques showed typical peak viremia at 2 to 3 weeks p.i., and the levels of peak VL were not significantly different between macaques challenged i.v. or i.r. (Table 1).
FIG. 2.
Established V1/V2 variants at week 2 p.i. (A) HTA analysis of V1/V2 variants detected in plasma at week 2 p.i. from nonvaccinated macaques. (B) Histogram representing the percentage of nonvaccinated macaques scored for the presence of each SIVsmE660 variant shown in Fig. 1. (C) HTA analysis of V1/V2 variants detected in plasma at week 2 p.i. from vaccinated macaques. (D) Histogram representing the percentage of vaccinated macaques scored for the presence of each SIVsmE660 variant shown in Fig. 1. If a viral variant did not correspond with variants A, B, or C in the original SIVsmE660 inoculum, it was scored as other. In panels B and D white bars represent i.v.-challenged macaques and black bars represent i.r.-challenged macaques.
TABLE 1.
VL and NAb dynamics of SIV-naive macaquesc
| Macaque | Group | VL peak (wk p.i.; P = 0.610) | VL peak (copies/ml; P = 0.257) | VL set pointa (copies/ml; P = 0.915) | gp120b ELISA (wk 7 p.i.; P = 0.610) | NAb titerb smH-4 (wk 7 p.i.; P = 0.067) | NAb titerb smE660 (wk 7 p.i.; P = 0.048d) |
|---|---|---|---|---|---|---|---|
| JJW | i.v. | 2 | 1.7 × 107 | 1.5 × 105 | 500 | 1,239 | 35 |
| KXA | i.v. | 3 | 8.7 × 106 | 8.6 × 104 | 2,500 | 3,037 | 79 |
| PDW | i.v. | 2 | 4.0 × 107 | 2.5 × 105 | 1,000 | 2,084 | 20 |
| TDB | i.v. | 2 | 1.5 × 107 | 1.9 × 106 | 4,000 | 7,188 | 26 |
| AU86 | i.r. | 2 | 8.7 × 105 | 4.5 × 105 | 2,000 | 12,728 | <20 |
| AV87 | i.r. | 2 | 3.6 × 107 | 9.4 × 106 | 200 | 6,343 | <20 |
| AU91 | i.r. | 2 | 1.2 × 106 | 4.4 × 104 | 4,000 | 9,676 | <20 |
| AV95 | i.r. | 2 | 8.2 × 106 | 3.3 × 106 | 4,000 | 8,544 | <20 |
| AV96 | i.r. | 2 | 2.6 × 106 | 1.9 × 103 | 1,000 | 2,956 | <20 |
| PKC | i.r. | 2 | 2.2 × 107 | 3.2 × 105 | 8,000 | 8,701 | 61 |
Determined by calculating the geometric mean of bDNA values between wk 24 and 41.
Values represent the inverse of serum dilution.
P values were determined by Mann-Whitney rank sum test.
Determined by Fisher exact test for the presence or absence of NAb against SIV smE660 comparing i.v.- and i.r.-challenged groups.
Resolution of viremia.
The initial decline in viremia was reached by week 6 p.i. in most animals. There was a remarkable level of stability in the composition of the V1/V2 env population through week 8 p.i. HTA analysis showed that 8 out of 10 nonvaccinated macaques maintained stable V1/V2 populations through week 8 p.i., indicating that initial viremia was resolved without detectable changes in V1/V2 sequences regardless of infection route (Fig. 3). This suggests that, during the initial resolution of viremia, there is no apparent competition between these V1/V2 variants and no apparent selective pressure by the host directed at V1/V2. This observation is consistent with a cytotoxic T-lymphocyte (CTL) response resolving the initial viremia between week 2 to 6 p.i (2, 24, 38), although we have not examined other regions of env for early diversification. One example of where a CTL response may have occurred at or near V1/V2 is shown in the HTA analysis of macaque PDW, where a change in relative abundance of two V1/V2 variants appears during the transition between weeks 3 and 4 p.i. followed by stability (Fig. 3A). Macaque AV96 showed a slow outgrowth of a V1/V2 variant between week 3 and week 12 p.i. (Fig. 3B, asterisk). Notably, no new V1/V2 variants were detected in these two macaques beyond those established variants during week 2 to 8 p.i., similar to the cases of the other eight macaques analyzed.
FIG. 3.
V1/V2 evolution in i.v. (A) and i.r. (B) challenge groups. Left panels show the HTA autoradiograms of longitudinal plasma samples using the SIVsmV1/V2 probe. Numbers above lanes indicate the week p.i. Graphs show VL (circle), NAb titers to SIVsmE660 (triangle), and the percent diversity (square) at the indicated week p.i. The designated identifier of the animal is shown above each set of panels.
We examined the initial antibody response by using an ELISA assay to measure the level of anti-gp120 antibodies in the plasma of each animal starting at week 7 p.i. Anti-gp120 antibodies were detected in all animals at week 7 p.i., and the levels of antibodies were not significantly different between i.v.- and i.r.-challenged macaques (Table 1). Neutralization assays were performed to quantitate the subset of effective antibodies present in the plasma of each macaque at week 7 p.i. Detectable NAbs were present in all animals using the SIVsmH-4 variant in neutralization assays, with a trend toward higher NAb titer in the i.r.-challenged animals (P = 0.067) (Table 1). However, neutralization assays against the challenge virus SIVsmE660 showed detectable, albeit low, NAb levels in all of the i.v.-challenged macaques and only one i.r.-challenged macaque (P = 0.048) (Table 1). SIVsmH-4 is easy to neutralize in vitro, whereas the SIVsmE660 mixture is difficult to neutralize, similar to typical primary isolates (D. Montefiori, unpublished data). These results suggest a qualitative, although not quantitative, difference in the early antibody response depending on an i.v. versus a mucosal challenge.
Initial diversification of V1/V2.
In general, there was a dramatic loss of the initially established V1/V2 variants at a point some time after week 8 p.i., with the appearance of new variants indicating a primary phase of V1/V2 diversification. We measured the time at which one-half of the V1/V2 population represented new variants, defined as Td50, as a way to compare the rate of env diversification between animals and between challenge groups. Note that this calculation omits changes in the relative abundance of the transmitted variants (as in AV96 and PDW) to draw attention to the appearance of new variants. These results showed that there was a significant difference between Td50 values in the i.v. group (mean at week 10 p.i.) and the i.r. group (mean at week 18 p.i.) (P = 0.01). The difference in timing of env diversification between the i.v.- and i.r.-challenged groups was apparent even though there was no significant difference in the level or day of the initial peak viremia or in virus set point levels (Table 1). Since VL has been shown to be a predictor of the rate of progression to AIDS (17, 32, 33), we wanted to determine if VL is also associated with the timing of V1/V2 diversity in the macaque cohort. Scatter plots of Td50 values and corresponding peak VL values and set point VL levels for the i.v.- and i.r.-challenged animals showed no significant correlation (data not shown). Together, these data suggest that the timing of peak VL, level of VL peak, and set point viremia do not play a role in the observed differences in the timing of V1/V2 diversity between macaques challenged i.v. and i.r.
Correlation between Td50 and NAb100 titers to the challenge virus.
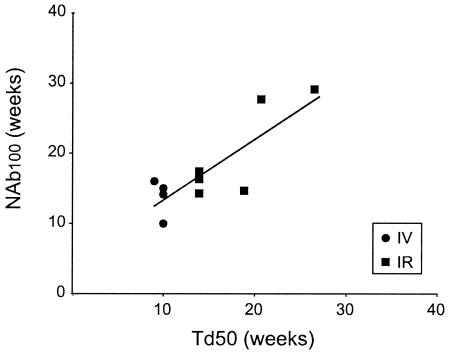
Previous work has shown that the resolution of the initial viremia is correlated with the appearance of CTL activity, which precedes the appearance of NAbs (13, 38). We hypothesized that the delayed diversification of env after the resolution of the initial viremia would reflect the evolution of a NAb response. The time at which NAb to the challenge virus, SIVsmE660, could be detected at a 1:100 dilution (NAb100) was used as a measure of NAb response, because this titer is significantly above the level of detection (a 1:20 dilution) in the assay. Although NAb100 values showed the same trend as Td50 values, the NAb100 values did not significantly differ between the i.v.- and i.r.-challenged macaques (P = 0.171). However, a scatter plot representing the relationship between Td50 and corresponding NAb100 values showed a significant correlation for the nonvaccinated macaques (Fig. 4; r = 0.83, P = 0.003). Thus, env V1/V2 diversification is correlated with the evolution of a host NAb response subsequent to the initial resolution of viremia. Given the surface location of V1/V2 on the Env protein we presume that the selective pressure driving diversification is the host humoral response.
FIG. 4.
Correlation between Td50 and SIVsmE660 NAb titers. Td50 values and NAb100 values are plotted for all of the nonvaccinated animals. The solid line represents the correlation for nonvaccinated animals (r = 0.83, P = 0.003).
Maturation of a NAb response.
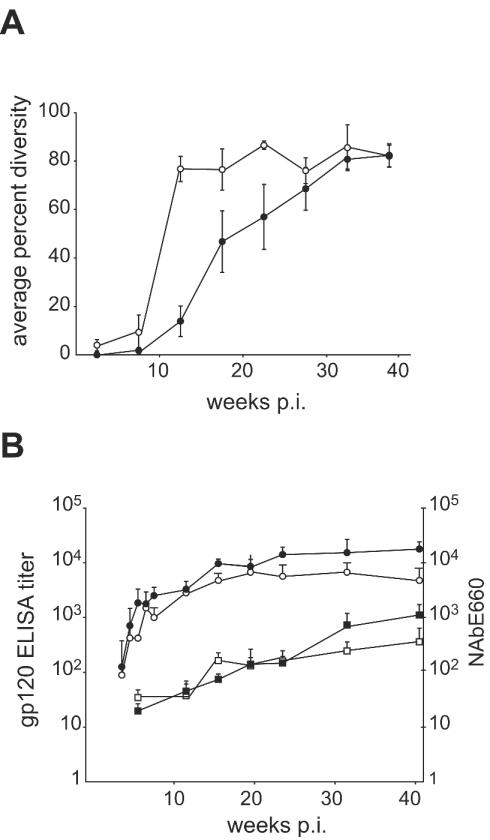
After initial viremia was resolved, V1/V2 diversity continued to increase beyond the Td50 point for a majority of the macaques, as indicated by the HTA patterns and as quantitated by average percent diversity for i.v.- and i.r.-challenged macaques (Fig. 5A). Significant differences in average percent diversity between i.v.- and i.r.-challenged macaques were observed at weeks 12, 16, and 20 p.i. (Fig. 5A). In contrast, there was no difference in the kinetics of the evolution of an antibody response to Env as measured by ELISA, with the i.r.-challenged animals having equivalent or higher titers at most time points (Fig. 5B). Similarly, we measured no consistent difference in the average NAb response to SIVsmE660 in comparing these two groups (Fig. 5B). Thus, V1/V2 diversification may be a more sensitive measure of the presence of selective pressure on this region of the Env protein than the measure of bulk NAb to the challenge virus.
FIG. 5.
Antibody response to SIV infection. Open symbols represent nonvaccinated i.v.-challenged macaques and closed symbols represent nonvaccinated i.r.-challenged macaques. (A) Plots represent average percent diversity during infection. (B) Plots represent averaged ELISA antibody titers against gp120 (circles) and SIVsmE660-specific NAb titers (squares) in plasma. Error bars represent standard errors.
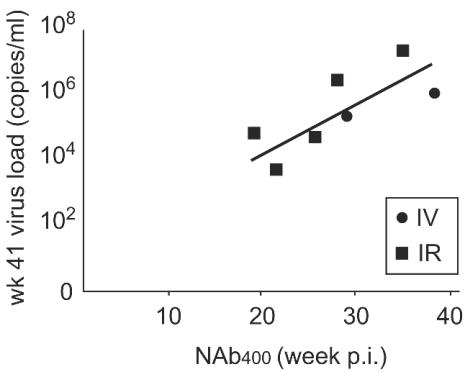
The VL patterns between weeks 24 and 41 p.i. are dynamic in most of the animals (Fig. 3), suggesting that the virus and host have not reached a steady state. Therefore, to begin to assess the long-term effect of a NAb response, we examined the VL at the last time point available (week 41 p.i.) with the time of evolution of a stronger host NAb response to the challenge virus, SIVsmE660; i.e., a titer of 400. A significant correlation between NAb400 and VL at week 41 p.i. is evident (Fig. 6; r = 0.811, P = 0.027). Macaques JJW, KXA, and AU86 were not included in this analysis, because NAb titers to SIVsmE660 did not reach 400 during the p.i. time period. These results suggest that the rate of establishment of a NAb response, as measured by the time required to reach a NAb titer of 400, is in part predictive of VL outcome in nonvaccinated macaques.
FIG. 6.
Correlation between NAb400 and endpoint VL. The week p.i. when NAb titers to SIVsmE660 reached 400 was estimated from the graphs shown in Fig. 3. The VL at week 41 p.i. was determined by bDNA assay. Two of the i.v.-infected animals did not reach a titer of 400 and are not included in this analysis.
V1/V2 sequence evolution.
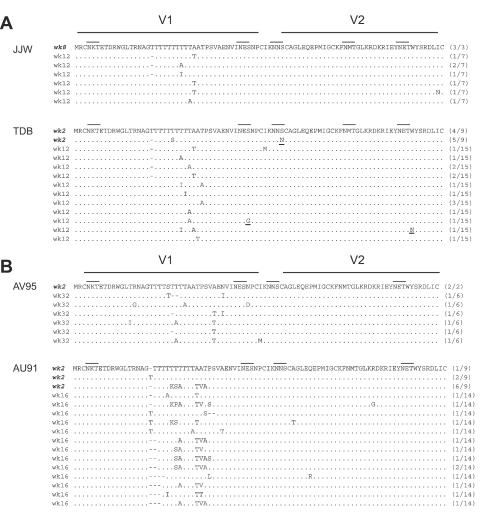
V1/V2 variants from pre- and post-Td50 time points were cloned and sequenced from macaques JJW (i.v.), TDB (i.v.), AV95 (i.r.), and AU91 (i.r.) (Fig. 7). Most of the variability between clones from the pre-Td50 point was confined to the center of the V1 region. In contrast, the V2 region showed no length variation and little sequence variation in the clones analyzed. Comparing pre- and post-Td50 time points, V1/V2 sequence analysis from these four macaques showed numerous amino acid changes in the V1 region in i.v.- and i.r.-challenged animals characterized by serine and threonine changes, suggesting additions and deletions of O-linked glycosylation sites. These results support previous studies showing changes in glycosylation sites in V1 during the time of infection (40, 41). However, there was no discernible difference in the pattern of sequence variability between i.v. and i.r. challenge groups, suggesting that major sequence changes occur in V1 regardless of challenge route but only differ in the timing of when these changes occur. These results suggest that the initial selective pressure on V1/V2, presumably by host antibodies, is largely directed at V1. In this analysis, we do not know if the sequence variants evolved de novo in each host or represented a minor variant present in the transmitting stock that grew out in the face of selective pressure on the major variants.
FIG. 7.
Analysis of V1/V2 sequences before and after corresponding Td50. Italic lettering indicates pre-Td50 sequences. Other sequences are post-Td50 sequences. Dots represent identical amino acids. Dashes represent deleted amino acids. Solid lines above sequences represent N-linked glycosylation sites. The frequency of the specific sequence from clones generated from the PCR product are indicated at the right of each sequence. Inferred amino acid sequence was derived from the nucleotide sequence.
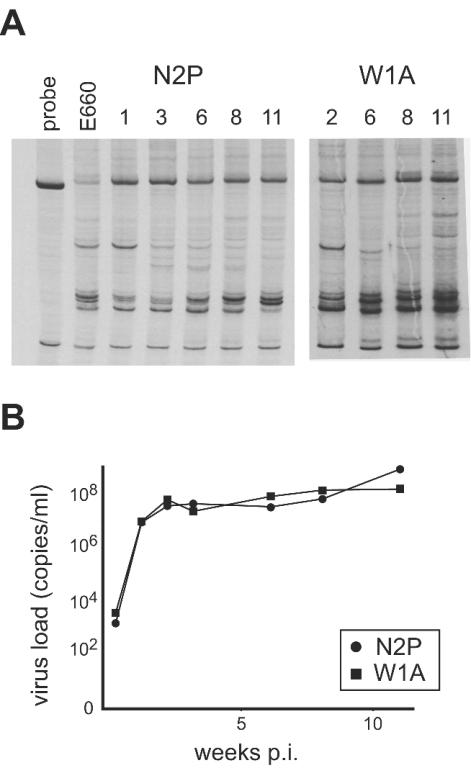
Lack of V1/V2 diversification in rapid progressors.
In a related study (10), two macaques challenged with SIVsmE660 through the i.v. route had a rapid disease course after failing to resolve the initial viremia. The virus population in these animals showed changes only in the relative abundance of established V1/V2 variants, and no new V1/V2 variants were detected (Fig. 8). The NAb titers measured against SIVsmH-4 were below detection at week 8 p.i., and no reactivity to Env was observed in ELISA assays (data not shown). These results suggest that very high VL associated with rapid disease progression blunts the initial humoral response and precludes the establishment of host selective pressures that drive V1/V2 diversification.
FIG. 8.
HTA analysis of the V1/V2 region of macaques that did not resolve initial viremia. (A) HTA autoradiograms of plasma samples using the SIVsmV1/V2 probe. Numbers above lanes indicate week p.i. (B) Graph of VL measurements as determined by bDNA assay. Both macaques were sacrificed at week 11 p.i.
Effect of vaccination on V1/V2 diversification.
As part of a vaccine efficacy study, eight macaques were immunized with an alphavirus vector expressing SIVsmH-4 genes. The SIVsmH-4 molecular clone was generated from the same virus isolate that was used to generate the SIVsmE660 challenge stock. Thus, the cloned genes used in the vaccination are related but not identical to the more complex SIVsmE660 isolate mixture, which was generated from an infected animal and expanded in culture. We used the smH-4 V1/V2 env probe to analyze which V1/V2 variants were established at week 2 p.i. in macaques that were vaccinated with SIVsmH-4-derived genes and challenged with SIVsmE660 via the i.v. or IR route. In most of the vaccinated macaques a subset of the variants present in the complex SIVsmE660 stock became established in each host (Fig. 2C). Scoring for the presence of the SIVsmE660 variants A, B, and C showed similar proportions of each variant in both i.v.- and i.r.-challenged animals after vaccination (Fig. 2D). Prior vaccination primed for an anamnestic response as indicated by the presence of elevated levels of NAbs to SIVsmH-4 at week 2 (Table 2) and in the elevated gp120 ELISA titer (data not shown) in the vaccinated animals. However, HTA analysis of V1/V2 diversification in the vaccinated macaques showed that the V1/V2 HTA pattern was stable between weeks 2 to 8 p.i., and resolution of viremia was not associated with changes in V1/V2 (data not shown). There was no significant difference in the time to V1/V2 diversification between nonvaccinated and vaccinated animals. Differences between Td50 values in i.v.- and i.r.-challenged macaques were not as significant as those seen with nonvaccinated macaques, but similar trends were observed whereby V1/V2 diversification occurred later in i.r.-challenged macaques than in i.v.-challenged macaques (data not shown). Thus, V1/V2 diversification dynamics occurred similarly in both nonvaccinated and vaccinated macaques in spite of the fact that the vaccination primed for an anamnestic response to the SIVsmH-4 Env protein.
TABLE 2.
NAb titers against SIVsmH-4
| Group and vaccination status | Titer at wk p.i.:
|
||
|---|---|---|---|
| 1 | 2 | 7 | |
| Non vaccinated | |||
| i.v. | <20 | <20 | 2,740 ± 1,319a |
| i.r. | <20 | <20 | 7,455 ± 1,341 |
| Vaccinated | |||
| i.v. | 49 ± 11 | 9,657 ± 9,418 | 13,025 ± 2,839 |
| i.r. | 50 ± 7 | 970 ± 807 | 16,240 ± 1,515 |
Error values represent standard errors.
DISCUSSION
The results of this investigation suggest that the route of virus entry into a host affects the timing of the initial evolution of the SIV env V1/V2 region. Detectable changes in the V1/V2 region occurred significantly earlier in animals infected systemically (i.v.) rather than across a mucosal surface (i.r.). In contrast, although different V1/V2 variants became established within each animal among the i.v.- and IR-challenged macaques (Fig. 2), there was no specific selection of variants that could establish an infection via the i.v. or i.r. routes. Thus, specific mutations, at least as linked to V1/V2 genotypic variants, did not seem to play a discriminating role in establishing the initial infection. Previous studies have shown that the route of infection can be a determinant of the complexity of the transmitted variants (16, 48). The results of these studies suggested that mucosal transmission leads to a relatively simple composition of genotypic variants in plasma, perhaps as the result of selection at the mucosal site, whereas a complex virus population becomes established in most of the i.v.-challenged macaques (16, 48). In these studies, the same amount of virus was administered by i.v. and at the mucosal site. In our study, macaques infected by the mucosal route were inoculated with undiluted virus stock to compensate for lower efficiency of mucosal challenge. After compensating for differences in transmission efficiency by these different routes, we did not observe a difference in the complexity or composition of transmitted V1/V2 variants between i.v.- and i.r.-challenged macaques (Fig. 2).
The acute phase of infection (weeks 2 to 8 p.i.) was not accompanied by changes in V1/V2, as was evident in the HTA analysis (Fig. 3 and 8), suggesting that early VL dynamics are not correlated with the initial diversification of V1/V2. In addition, CTL escape by mutation has been measured and shown to occur as early as week 4 p.i., suggesting that the virus is able to escape some cellular immune pressures during the acute phase of infection (38). Thus, the stability of V1/V2 populations during the resolution of viremia suggests that V1/V2 is not an early target of a CTL response. We do note that one animal (PDW; Fig. 3) had a rapid transition between V1/V2 variants at the time of viremia resolution, suggesting a closely linked CTL epitope. These results on the early stability of V1/V2 variants are in contrast to those of Learn et al., who suggested that an initially diverse HIV-1 env population (in humans) undergoes a contraction in complexity, at least within the C2-V5 regions (25).
Induction of antibodies against the HIV-1 and SIV Env protein are an integral (and the relevant) component of the host humoral immune response to viral infection (4, 6, 30, 36). Specifically, NAbs have been suggested to influence V1/V2 evolution (5, 39). We suggest that the diversification of V1/V2 reported here was driven by antibody selection, although some of the data are inconclusive. When using the data from all 10 nonvaccinated macaques, we found a correlation between the evolution of a moderate NAb response and V1/V2 diversification (Fig. 4), with both occurring significantly later for IR-challenged animals. Consistent with this finding, we observed a trend toward a qualitative difference in the antibody response between i.v.- and i.r.-challenged animals at week 7 p.i. (Table 1). However, there was no difference between i.v.- and i.r.-challenged animals in the total antibody response (ELISA), nor was there a consistent difference in the NAb response over the time course (Fig. 5B). Thus, while there is an overall correlation between NAbs and V1/V2 diversification, it is less apparent that differences in the timing of the appearance of NAbs accounts for the differences in the diversification of V1/V2 between i.v.- and i.r.-challenged animals. Studies such as these are limited by the small numbers of animals available. In support of our interpretation is the observation that in a separate vaccinated group of macaques there was a similar trend of early diversification of V1/V2 in i.v.-challenged animals and late diversification in i.r.-challenged animals. We believe this indicates that there is a qualitative difference in the virus-host interaction that is dependent on the route of infection. The correlation between the timing of V1/V2 diversity and a NAb response (Fig. 4) supports the hypothesis that V1/V2 evolution is a sensitive measure of the appearance of a NAb response during SIV infection or at least an antibody response capable of providing selective pressure sufficient to favor the outgrowth of genetic variants.
Although our hypothesis is that NAbs influence V1/V2 diversity, NAb titers apparently trailed V1/V2 diversity in 7 out of 10 nonvaccinated macaques (Fig. 4). A possible explanation for this observation is that NAbs are present prior to V1/V2 diversification but are not detected against the SIVsmE660 isolate used in the neutralization assay. NAbs were detected against SIVsmH-4 at week 7 p.i. in all macaques in this study (Table 1), suggesting that NAbs specific for SIVsmH-4-specific epitopes are present prior to V1/V2 diversification. Previous work has shown that the SIVsmH-4 variant is a sensitive indicator of NAbs, and in vitro neutralization assays using SIVsmH-4 seem to detect epitope specificities that are either absent or not adequately exposed on SIVsmE660 (10). In addition, since only a subset of the SIVsmE660 V1/V2 variants became established within each host (Fig. 2), the in vitro neutralization assay against the complete SIVsm E660 mixture may underestimate the neutralizing potential of antibodies within each animal. The delayed appearance of SIVsmE660-specific NAbs suggests a lengthier maturation of an SIVsmE660-specific NAb response. Others have reported a general phenomenon of lentiviruses characterized by a complex and lengthy maturation of specific NAb development (8, 37, 43, 44, 47) and that evolution of virus-specific antibody responses are independent of the pathogenicity of the virus strain used for infections (8). However, rapid disease progression can be associated with a blunting of the immune response and a reduction of genotypic diversity in both SIVsm-infected macaques (Fig. 8 and reference 27) and HIV-1-infected humans (11, 14, 54).
It was somewhat surprising that vaccination with the SIVsmH-4 env gene was able to prime an anamnestic response for Env antibodies and for SIVsmH-4 NAbs (Table 2) but did not alter the timing of V1/V2 diversification. This implies that the V1/V2 region of the H-4 Env protein was not an effective immunogen for the E660 variants. The likely explanation is that the sequence differences between H-4 V1/V2 and the E660 variants (Fig. 1) represent changes that confer a type-specific antibody response. The production of an anamnestic response for SIVsmH-4 neutralizing antibodies indicates that the E660 complex is displaying these epitopes. If these antibodies apply selective pressure on variants in the E660 mixture it may be possible to identify the target region (epitopes) of the SIVsmH-4 NAbs as having early sequence changes in the vaccinated animals. Testing this hypothesis will require sequencing larger regions of the env genes generated from vaccinated and nonvaccinated animals.
What could be the difference in the virus-host interaction initiated by different routes of infection that would affect the timing of V1/V2 diversification? Virus production after a mucosal infection might occur at sites that are less accessible to selective pressures of antibodies, although the total plasma VL is not different as a function of route. Alternatively, V1/V2 antigen presentation or the capacity/response of B cells and/or T-helper cells may differ by compartment in the face of an infection. Finally, gp120 conformations available to the immune system may differ as a function of compartment, reflecting different proportions of particle-associated gp120, cell surface-associated gp120, or shed soluble gp120. The difference in NAbs to SIVsmH-4 and SIVsmE660 at week 7 p.i. (Table 1) suggests a qualitative difference in the antibody response as a function of route.
The long-term partial control of viremia by the host is likely to be the result of interplay between a number of factors. We found a correlation between the time at which a strong NAb response evolved (titer of 400) and VL at week 41 p.i. (Fig. 6). However, this analysis did not include three animals that had moderate VL but never reached this NAb titer, pointing to the complexity of this system. Schmitz et al. have observed a relationship between NAb and set point (47). These observations support the idea that a strong NAb response is either a contributor to or a marker of more effective control of the VL. However, contradicting this conclusion is the observation that when animals were primed by vaccination for an anamnestic response, there was no correlation between the timing of the appearance of a potent NAb titer and VL at week 41 p.i. (data not shown).
There are numerous glycosylation sites in the V1/V2 region, and changes in glycosylation sites could mask antibody recognition of neutralizing epitopes on Env (6). For three of the animals examined by sequence, substitutions in V1 either added or removed threonines within V1, possibly changing O-linked glycosylations (Fig. 7). In the fourth animal examined more complex substitutions occurred in V1 (AU91; Fig. 7B). Only in one animal did we detect changes related to N-linked glycosylations within this region (TDB; Fig. 7A). The homogeneous runs of threonines on SIVsm differ from the AXU codon motif encoding Thr, Ser, and Asn within V1, V2, and V4 of HIV-1, suggesting a difference in the nature of sequence diversity between SIVsm and HIV-1 (3, 23). It is not clear to what extent these changes in potential O-linked glycosylation sites work in cis or at distal sites as proposed for the HIV-1 glycan shield (53).
Because the SIVsmE660 variant is a heterogeneous viral population, the model presented here approximates exposure of a host to genetically diverse variants at primary infection. Multiple variants have been reported in women recently infected with HIV-1 (28), and we have observed that 50% of subjects in primary HIV-1 infection have multiple V1/V2 populations (K. Ritola and R. Swanstrom, unpublished data). We have also observed the coexistence of multiple V1/V2 variants late in HIV-1 infection (23), in the chronic stages of HIV-1 infection (K. Ritola and R. Swanstrom, unpublished) and after the initial diversification of V1/V2 in macaques (Fig. 3). These coexisting variants may represent the presence of individually evolved type-specific variants independently populating niches determined by an incomplete host immune response. These results show how env diversification is affected by route of virus entry and host immune pressures and suggest that the V1/V2 response may be highly type specific.
Acknowledgments
We thank Mary Connell for assistance with the infections of the macaques. We thank Chad Cecil for providing the gp120 reagent used in the ELISA assay and Nancy Davis for thoughtful discussions and support.
B. J. Rybarczyk and this work were supported by the Seeding Postdoctoral Innovators in Research and Education (SPIRE) program. This study was supported by the MORE Division of NIGMS grant GM000678, NIH grant R01-AI44667 (R.S.), the NIH HIVRAD award PO1-AI46023 (R.S., P.R.J., D.M., R.E.J.), the UNC Center for AIDS Research award (P30-AI50410), and the Carolina Vaccine Institute.
REFERENCES
- 1.Bolmstedt, A., S. Sjolander, J. E. Hansen, L. Akerblom, A. Hemming, S. L. Hu, B. Morein, and S. Olofsson. 1996. Influence of N-linked glycans in V4-V5 region of human immunodeficiency virus type 1 glycoprotein gp160 on induction of a virus-neutralizing humoral response. J. Acquir. Immune Defic. Syndr. 12:213-220. [DOI] [PubMed] [Google Scholar]
- 2.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch, M. L., A. C. Andeweg, R. Schipper, and M. Kenter. 1994. Insertion of N-linked glycosylation sites in the variable regions of the human immunodeficiency virus type 1 surface glycoprotein through AAT triplet reiteration. J. Virol. 68:7566-7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruck, C., C. Thiriart, L. Fabry, M. Francotte, P. Pala, O. Van Opstal, J. Culp, M. Rosenberg, M. De Wilde, and P. Heidt. 1994. HIV-1 envelope-elicited neutralizing antibody titres correlate with protection and virus load in chimpanzees. Vaccine 12:1141-1148. [DOI] [PubMed] [Google Scholar]
- 5.Burns, D. P., and R. C. Desrosiers. 1994. Envelope sequence variation, neutralizing antibodies, and primate lentivirus persistence. Curr. Top. Microbiol. Immunol. 188:185-219. [DOI] [PubMed] [Google Scholar]
- 6.Burton, D. R. 1997. A vaccine for HIV type 1: the antibody perspective. Proc. Natl. Acad. Sci. USA 94:10018-10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chackerian, B., L. M. Rudensey, and J. Overbaugh. 1997. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J. Virol. 71:7719-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole, K. S., M. Murphey-Corb, O. Narayan, S. V. Joag, G. M. Shaw, and R. C. Montelaro. 1998. Common themes of antibody maturation to simian immunodeficiency virus, simian-human immunodeficiency virus, and human immunodeficiency virus type 1 infections. J. Virol. 72:7852-7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connick, E., D. G. Marr, X. Q. Zhang, S. J. Clark, M. S. Saag, R. T. Schooley, and T. J. Curiel. 1996. HIV-specific cellular and humoral immune responses in primary HIV infection. AIDS Res. Hum. Retrovir. 12:1129-1140. [DOI] [PubMed] [Google Scholar]
- 10.Davis, N. L., I. J. Caley, K. W. Brown, M. R. Betts, D. M. Irlbeck, K. M. McGrath, M. J. Connell, D. C. Montefiori, J. A. Frelinger, R. Swanstrom, P. R. Johnson, and R. E. Johnston. 2000. Vaccination of macaques against pathogenic simian immunodeficiency virus with Venezuelan equine encephalitis virus replicon particles. J. Virol. 74:371-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delwart, E. L., H. Pan, H. W. Sheppard, D. Wolpert, A. U. Neumann, B. Korber, and J. I. Mullins. 1997. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J. Virol. 71:7498-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desrosiers, R. C. 1990. The simian immunodeficiency viruses. Annu. Rev. Immunol. 8:557-578. [DOI] [PubMed] [Google Scholar]
- 13.D'Souza, M. P., and B. J. Mathieson. 1996. Early phases of HIV type 1 infection. AIDS Res. Hum. Retrovir. 12:1-9. [DOI] [PubMed] [Google Scholar]
- 14.Ganeshan, S., R. E. Dickover, B. T. Korber, Y. J. Bryson, and S. M. Wolinsky. 1997. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J. Virol. 71:663-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstein, S., W. R. Elkins, W. T. London, A. Hahn, R. Goeken, J. E. Martin, and V. M. Hirsch. 1994. Immunization with whole inactivated vaccine protects from infection by SIV grown in human but not macaque cells. J. Med. Primatol. 23:75-82. [DOI] [PubMed] [Google Scholar]
- 16.Greenier, J. L., C. J. Miller, D. Lu, P. J. Dailey, F. X. Lu, K. J. Kunstman, S. M. Wolinsky, and M. L. Marthas. 2001. Route of simian immunodeficiency virus inoculation determines the complexity but not the identity of viral variant populations that infect rhesus macaques. J. Virol. 75:3753-3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta, P., L. Kingsley, J. Armstrong, M. Ding, M. Cottrill, and C. Rinaldo. 1993. Enhanced expression of human immunodeficiency virus type 1 correlates with development of AIDS. Virology 196:586-595. [DOI] [PubMed] [Google Scholar]
- 18.Hirsch, V. M., R. A. Olmsted, M. Murphey-Corb, R. H. Purcell, and P. R. Johnson. 1989. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339:389-392. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch, V. M., P. M. Zack, and P. R. Johnson. 1990. Molecular characterization of SIV in tissues from experimentally infected macaques. J. Med. Primatol. 19:287-294. [PubMed] [Google Scholar]
- 20.Hoffman, N. G., F. Seillier-Moiseiwitsch, J. Ahn, J. M. Walker, and R. Swanstrom. 2002. Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop. J. Virol. 76:3852-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Javaherian, K., A. J. Langlois, D. C. Montefiori, K. A. Kent, K. A. Ryan, P. D. Wyman, J. Stott, D. P. Bolognesi, M. Murphey-Corb, and G. J. Larosa. 1994. Studies of the conformation-dependent neutralizing epitopes of simian immunodeficiency virus envelope protein. J. Virol. 68:2624-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jurkiewicz, E., G. Hunsmann, J. Schaffner, T. Nisslein, W. Luke, and H. Petry. 1997. Identification of the V1 region as a linear neutralizing epitope of the simian immunodeficiency virus SIVmac envelope glycoprotein. J. Virol. 71:9475-9481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitrinos, K. M., N. G. Hoffman, J. A. Nelson, and R. Swanstrom. 2003. Turnover of env variable region 1 and 2 genotypes in subjects with late-stage human immunodeficiency virus type 1 infection. J. Virol. 77:6811-6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Learn, G. H., D. Muthui, S. J. Brodie, T. Zhu, K. Diem, J. I. Mullins, and L. Corey. 2002. Virus population homogenization following acute human immunodeficiency virus type 1 infection. J. Virol. 76:11953-11959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehner, T., Y. Wang, L. Ping, L. Bergmeier, E. Mitchell, M. Cranage, G. Hall, M. Dennis, N. Cook, C. Doyle, and I. Jones. 1999. The effect of route of immunization on mucosal immunity and protection. J. Infect. Dis. 179:S489-S492. [DOI] [PubMed] [Google Scholar]
- 27.Lifson, J. D., M. A. Nowak, S. Goldstein, J. L. Rossio, A. Kinter, G. Vasquez, T. A. Wiltrout, C. Brown, D. Schneider, L. Wahl, A. L. Lloyd, J. Williams, W. R. Elkins, A. S. Fauci, and V. M. Hirsch. 1997. The extent of early viral replication is a critical determinant of the natural history of simian immunodeficiency virus infection. J. Virol. 71:9508-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long, E. M., H. L. Martin, Jr., J. K. Kreiss, S. M. Rainwater, L. Lavreys, D. J. Jackson, J. Rakwar, K. Mandaliya, and J. Overbaugh. 2000. Gender differences in HIV-1 diversity at time of infection. Nat. Med. 6:71-75. [DOI] [PubMed] [Google Scholar]
- 29.Lu, Y., C. D. Pauza, X. Lu, D. C. Montefiori, and C. J. Miller. 1998. Rhesus macaques that become systemically infected with pathogenic SHIV 89.6-PD after intravenous, rectal, or vaginal inoculation and fail to make an antiviral antibody response rapidly develop AIDS. J. Acquir. Immune Defic. Syndr. 19:6-18. [DOI] [PubMed] [Google Scholar]
- 30.Mascola, J. R., S. W. Snyder, O. S. Weislow, S. M. Belay, R. B. Belshe, D. H. Schwartz, M. L. Clements, R. Dolin, B. S. Graham, G. J. Gorse, M. C. Keefer, M. J. McElrath, M. C. Walker, K. F. Wagner, J. G. McNeil, F. E. McCutchan, and D. S. Burke. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. J. Infect. Dis. 173:340-348. [DOI] [PubMed] [Google Scholar]
- 31.McKeating, J. A., C. Shotton, J. Cordell, S. Graham, P. Balfe, N. Sullivan, M. Charles, M. Page, A. Bolmstedt, and S. Olofsson. 1993. Characterization of neutralizing monoclonal antibodies to linear and conformation-dependent epitopes within the first and second variable domains of human immunodeficiency virus type 1 gp120. J. Virol. 67:4932-4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mellors, J. W., L. A. Kingsley, C. R. Rinaldo, Jr., J. A. Todd, B. S. Hoo, R. P. Kokka, and P. Gupta. 1995. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann. Intern. Med. 122:573-579. [DOI] [PubMed] [Google Scholar]
- 33.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 34.Modrow, S., B. H. Hahn, G. M. Shaw, R. C. Gallo, F. Wong-Staal, and H. Wolf. 1987. Computer-assisted analysis of envelope protein sequences of seven human immunodeficiency virus isolates: prediction of antigenic epitopes in conserved and variable regions. J. Virol. 61:570-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montefiori, D. C., T. W. Baba, A. Li, M. Bilska, and R. M. Ruprecht. 1996. Neutralizing and infection-enhancing antibody responses do not correlate with the differential pathogenicity of SIVmac239delta3 in adult and infant rhesus monkeys. J. Immunol. 157:5528-5535. [PubMed] [Google Scholar]
- 36.Montefiori, D. C., B. S. Graham, J. Zhou, R. A. Bucco, D. H. Schwartz, L. A. Cavacini, and M. R. Posner. 1993. V3-specific neutralizing antibodies in sera from HIV-1 gp160-immunized volunteers block virus fusion and act synergistically with human monoclonal antibody to the conformation-dependent CD4 binding site of gp120. NIH-NIAID AIDS Vaccine Clinical Trials Network. J. Clin. Investig. 92:840-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moog, C., H. J. Fleury, I. Pellegrin, A. Kirn, and A. M. Aubertin. 1997. Autologous and heterologous neutralizing antibody responses following initial seroconversion in human immunodeficiency virus type 1-infected individuals. J. Virol. 71:3734-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493-499. [DOI] [PubMed] [Google Scholar]
- 39.Overbaugh, J., and C. R. Bangham. 2001. Selection forces and constraints on retroviral sequence variation. Science 292:1106-1109. [DOI] [PubMed] [Google Scholar]
- 40.Overbaugh, J., and L. M. Rudensey. 1992. Alterations in potential sites for glycosylation predominate during evolution of the simian immunodeficiency virus envelope gene in macaques. J. Virol. 66:5937-5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Overbaugh, J., L. M. Rudensey, M. D. Papenhausen, R. E. Benveniste, and W. R. Morton. 1991. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J. Virol. 65:7025-7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmer, C., P. Balfe, D. Fox, J. C. May, R. Frederiksson, E. M. Fenyo, and J. A. McKeating. 1996. Functional characterization of the V1V2 region of human immunodeficiency virus type 1. Virology 220:436-449. [DOI] [PubMed] [Google Scholar]
- 43.Pellegrin, I., E. Legrand, D. Neau, P. Bonot, B. Masquelier, J. L. Pellegrin, J. M. Ragnaud, N. Bernard, and H. J. Fleury. 1996. Kinetics of appearance of neutralizing antibodies in 12 patients with primary or recent HIV-1 infection and relationship with plasma and cellular viral loads. J. Acquir. Immune Defic. Syndr. 11:438-447. [DOI] [PubMed] [Google Scholar]
- 44.Pilgrim, A. K., G. Pantaleo, O. J. Cohen, L. M. Fink, J. Y. Zhou, J. T. Zhou, D. P. Bolognesi, A. S. Fauci, and D. C. Montefiori. 1997. Neutralizing antibody responses to human immunodeficiency virus type 1 in primary infection and long-term-nonprogressive infection. J. Infect. Dis. 176:924-932. [DOI] [PubMed] [Google Scholar]
- 45.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ross, T. M., and B. R. Cullen. 1998. The ability of HIV type 1 to use CCR-3 as a coreceptor is controlled by envelope V1/V2 sequences acting in conjunction with a CCR-5 tropic V3 loop. Proc. Natl. Acad. Sci. USA 95:7682-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmitz, J. E., M. J. Kuroda, S. Santra, M. A. Simon, M. A. Lifton, W. Lin, R. Khunkhun, M. Piatak, J. D. Lifson, G. Grosschupff, R. S. Gelman, P. Racz, K. Tenner-Racz, K. A. Mansfield, N. L. Letvin, D. C. Montefiori, and K. A. Reimann. 2003. Effect of humoral immune responses on controlling viremia during primary infection of rhesus monkeys with simian immunodeficiency virus. J. Virol. 77:2165-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sodora, D. L., F. Lee, P. J. Dailey, and P. A. Marx. 1998. A genetic and viral load analysis of the simian immunodeficiency virus during the acute phase in macaques inoculated by the vaginal route. AIDS Res. Hum. Retrovir. 14:171-181. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan, N., M. Thali, C. Furman, D. D. Ho, and J. Sodroski. 1993. Effect of amino acid changes in the V1/V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by a neutralizing antibody. J. Virol. 67:3674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trivedi, P., D. Horejsh, S. B. Hinds, P. W. Hinds II, M. S. Wu, M. S. Salvato, and C. D. Pauza. 1996. Intrarectal transmission of simian immunodeficiency virus in rhesus macaques: selective amplification and host responses to transient or persistent viremia. J. Virol. 70:6876-6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trivedi, P., K. K. Meyer, D. N. Streblow, B. L. Preuninger, K. T. Schultz, and C. D. Pauza. 1994. Selective amplification of simian immunodeficiency virus genotypes after intrarectal inoculation of rhesus monkeys. J. Virol. 68:7649-7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watkins, B. A., M. S. Reitz, Jr., C. A. Wilson, K. Aldrich, A. E. Davis, and M. Robert-Guroff. 1993. Immune escape by human immunodeficiency virus type 1 from neutralizing antibodies: evidence for multiple pathways. J. Virol. 67:7493-7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 54.Wolinsky, S. M., B. T. Korber, A. U. Neumann, M. Daniels, K. J. Kunstman, A. J. Whetsell, M. R. Furtado, Y. Cao, D. D. Ho, and J. T. Safrit. 1996. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537-542. [DOI] [PubMed] [Google Scholar]
- 55.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 56.Wyatt, R., N. Sullivan, M. Thali, H. Repke, D. Ho, J. Robinson, M. Posner, and J. Sodroski. 1993. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J. Virol. 67:4557-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao, Y., X. Dong, and Y. H. Chen. 2002. Neutralizing antibodies mechanism of neutralization and protective activity against HIV-1. Immunol. Res. 25:193-200. [DOI] [PubMed] [Google Scholar]