(The American Journal of Human Genetics 93, 54–66; July 11, 2013)
In the version of this paper published online June 13, 2013, two authors were unfortunately omitted from the author list. Ping An and Cheryl A. Winkler have now been added here, online, and in the full article that appears in the print issue. Both added authors are at the Basic Research Laboratory, Center for Cancer Research, National Cancer Institute, Frederick National Laboratory, Science Applications International Corporation-Frederick, Frederick, MD 21702, USA.
Within the Material and Methods and within the Acknowledgments, references to primers and suggestions provided by Cheryl Ann Winkler have been removed given that she is now included in the author list.
Additionally, the rs number of the G2 allele has been corrected to rs71785313 in Figure 2 and in the main text. In the first paragraph of the Results section, site 1,079 has been corrected to 1,097. In Figure 2, rs11613667 has been corrected to rs116136671, and the genomic position of the G2 allele has been corrected to 34991993. A corrected Figure 2 appears here.
Figure 2.
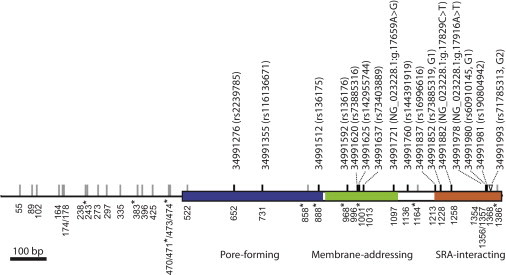
Spatial Distribution of Genetic Variants at the Functional-Domain-Encoding Region of APOL1
The APOL1 genetic region, which codes for the pore-forming (blue), membrane-addressing (green), and SRA-interacting (orange) domains, and the adjacent intron are marked by genetic variants identified in this study. Each synonymous and intronic SNP is labeled by a gray line. Each nonsynonymous SNP is labeled by a black line with its genomic position (based on NCBI Genome browser build 36.1) and reference SNP ID if it is described in dbSNP. For those nonsynonymous SNPs that are not present in dbSNP (build 137), the changes of nucleotide state are given according to RefSeq NG_023228.1. The indel variant (i.e., G2 allele) is labeled by an inverted triangle. The nucleotide site of each variant at our sequence alignment is labeled at the bottom of the gene. The nucleotide sites that define the G3 haplotype are labeled with asterisks.
In Table S1, in the "Name" column the slash in "apol1_ex7_s2/" has been removed so that it reads "apol1_ex7_s2."
Finally, the following text has been added to the Acknowledgments:
This project was funded in part with Federal funds from the Frederick National Laboratory for Cancer Research, NIH, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the United States Government. This research was supported (in part) by the Intramural Research Program of the NIH, Frederick National Lab, Center for Cancer Research.
The authors regret these errors.


