Abstract
The soluble (Gs) and membrane-bound (Gm) forms of human respiratory syncytial virus (HRSV) attachment protein were purified by immunoaffinity chromatography from cultures of HEp-2 cells infected with vaccinia virus recombinants expressing either protein. Sucrose gradient centrifugation indicated that Gs, which is secreted into the culture medium, remains monomeric, whereas Gm is an oligomer, probably a homotetramer. Nevertheless, Gs was capable of binding to the surface of cells in vitro, as assessed by a flow cytometry-based binding assay. The attachment of Gs to cells was inhibited by previous heparinase treatment of living cells, and Gs did not bind to CHO cell mutants defective in proteoglycan biosynthesis. Thus, Gs, as previously reported for the G protein of intact virions, binds to glycosaminoglycans presented at the cell surface as proteoglycans. Deletion of a previously reported heparin binding domain from Gs protein substantially inhibited its ability to bind to cells, but the remaining level of binding was still sensitive to heparinase treatment, suggesting that other regions of the Gs molecule may contribute to attachment to proteoglycans. The significance of these results for HRSV infection is discussed.
Human respiratory syncytial virus (HRSV) is the leading cause of hospitalization due to viral respiratory tract infection in infants and young children worldwide (for reviews, see references 4 and 6). Severe disease is characterized by bronchiolitis and pneumonia, which frequently results in long-term abnormal lung function (29, 30). HRSV is a member of the Pneumovirus genus within the Paramyxoviridae family of enveloped, single-stranded, negative-sense RNA viruses. It encodes 10 major mRNAs, which produce 11 viral proteins. Three virally encoded transmembrane glycoproteins are present on the surface of respiratory syncytial virus virions anchored within the envelope; a small hydrophobic protein of unknown function, a fusion (F) protein, and a receptor-binding or attachment protein (G).
Similarly to that with other paramyxoviruses, HRSV infection is thought to be initiated by the attachment of the virion to the target cell, followed by virus entry via fusion of the viral envelope with the host cell plasma membrane. The F protein is responsible for the fusion activity of HRSV, mediating both viral penetration of the host cell (37) and syncytium formation (14). The G protein is thought to represent the attachment protein (23) and mediate most of the interaction between respiratory syncytial virus and target cells by binding cell surface proteoglycans (10, 16, 17, 21, 26, 34).
HRSV G glycoprotein is produced both as a type II transmembrane protein (Gm) that is incorporated into virions and as a soluble form (Gs) lacking the signal/membrane-anchor region that is secreted from infected cells (18, 19). Gs is formed by alternative translation initiation from a second in-frame AUG codon in the G protein open reading frame (M48) (31), followed by N-terminal proteolytic processing (see Fig. 1). Thus, Gs represents the ectodomain of Gm, which is heavily glycosylated by the addition of N- and O-linked oligosaccharides in both forms of the G protein. While the predicted mass of the G protein is approximately 33 kDa, mature G protein migrates on sodium dodecyl sulfate (SDS)-polyacrylamide gels with an estimated mass of 80 to 90 kDa due to the extensive posttranslational glycosylation (32, 38). Glycosylation sites are clustered in two regions of maximal sequence divergence of the G protein primary structure, termed mucin-like domains due to their high percentage of Ser/Thr O-glycosylation acceptor sites and high Pro content. Indeed, the G protein displays the greatest sequence divergence among HRSV gene products between virus isolates (20). The two mucin-like domains are separated by a short, relatively conserved segment located in the central part of the G ectodomain that contains a cluster of four conserved cysteine residues (8, 22).
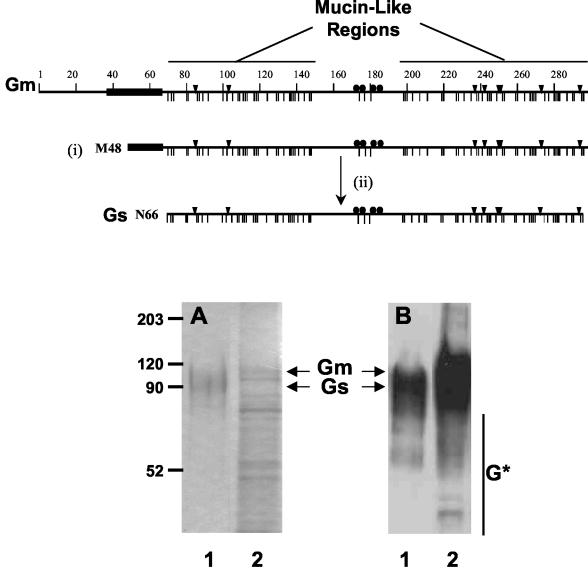
FIG. 1.
SDS-PAGE and Western blotting of Gs and Gm proteins. A scheme of the Gm protein is presented in the upper part of the figure, denoting the hydrophobic region ( ), the potential N- (▾) and O-glycosylation sites (|), and the cysteine residues (•). Also indicated are the two mucin-like regions. Formation of the Gs form occurs by translation initiation (i) at Met48 and subsequent cleavage (ii) after residue 65. HRSV Gs (lanes 1) and Gm (lanes 2) proteins were purified by immunoaffinity chromatography, as described in Materials and Methods, and analyzed by Coomassie staining of an SDS-PAGE gel (A) and by Western blotting with monoclonal antibody 63G (B). Molecular weight markers are shown on the left. G* indicates low-molecular-weight bands reacting with antibody 63G.
), the potential N- (▾) and O-glycosylation sites (|), and the cysteine residues (•). Also indicated are the two mucin-like regions. Formation of the Gs form occurs by translation initiation (i) at Met48 and subsequent cleavage (ii) after residue 65. HRSV Gs (lanes 1) and Gm (lanes 2) proteins were purified by immunoaffinity chromatography, as described in Materials and Methods, and analyzed by Coomassie staining of an SDS-PAGE gel (A) and by Western blotting with monoclonal antibody 63G (B). Molecular weight markers are shown on the left. G* indicates low-molecular-weight bands reacting with antibody 63G.
A specific cellular receptor for HRSV has yet to be identified, although it is well documented that HRSV is capable of binding to cell surface glycosaminoglycans (16, 17, 21, 26, 35). Glycosaminoglycans (GAGs) are long, unbranched polysaccharide chains consisting of repeating disaccharides, one of which is always an aminosugar (N-acetylglucosamine or N-acetylgalactosamine). The second sugar in the disaccharide unit is a hexuronic acid, either glucuronic acid or its epimer, iduronic acid. GAGs are found both within the extracellular matrix and covalently bound to host cell membrane proteins to form proteoglycans. Since GAG chains are generally highly sulfated, they carry a negative charge which is thought to mediate attachment of HRSV to target cells via interactions with basic amino acids in the G protein (10). However, in addition to this nonspecific electrostatic interaction, HRSV preferentially binds to GAGs possessing iduronic acid, N sulfation, and a minimum chain length of 10 saccharides (16, 17). In particular, heparan sulfate, and to a lesser extent chondroitin-4-sulfate, have been implicated in the binding of HRSV to cells (16, 17, 26).
Protein GAG-binding domains are usually positively charged regions containing a high proportion of basic arginine and lysine residues, which interact with the negatively charged sulfate groups of cellular proteoglycans. A GAG-binding domain encompassing a region of positively charged residues was proposed by a study of G protein synthetic peptides (10) and termed the heparin-binding domain (HBD), given its ability to bind to a heparin-agarose affinity column. This segment comprises residues 184 to 198 in HRSV subgroup A and therefore overlaps the conserved cystine cluster and flanks the central conserved region. However, there has been no formal demonstration that the HBD is required for the attachment of G protein to cells. Indeed, it has been suggested that the G protein HBD may extend further than that proposed, given that a recombinant HRSV (rHRSV) expressing G protein lacking the HBD infected cells in a GAG-dependent manner (36).
The oligomeric forms of Gs or Gm are currently unknown; however, it has been suggested that Gm may be a trimer or a tetramer (5, 28). Given that oligomerization is an important structural characteristic that influences protein activity, we sought to determine the oligomeric structure of Gs and Gm by comparison of their sedimentation properties in sucrose gradients. Since it was found that both proteins differ in their oligomeric state, it was of interest to analyze the capacity of purified Gs to bind to cells and to evaluate the relevance of GAGs and the requirement of the HBD region for this binding interaction.
MATERIALS AND METHODS
Cells and viruses.
HEp-2 cells were maintained in Dulbecco's modified eagle's medium (DMEM) supplemented with 2% fetal bovine serum (FBS). Human respiratory syncytial virus (Long strain) was grown in HEp-2 cells and purified from culture supernatants as previously described (12). CV-1 cells were cultured in DMEM supplemented with 10% FBS. CHO (Chinese hamster ovary) cell lines were grown in a 1:1-ratio mixture of DMEM and Ham's F12 medium with 10% FBS. Wild-type CHO K1 cells express a normal complement of cell surface GAGs, whereas mutants of the parental CHO K1 line, pgs D-677 (24) and pgs A-745 (9), are deficient in GAG expression. All continuous cell lines used in this study were incubated at 37°C in an atmosphere of 5% CO2.
Recombinant vaccinia viruses were propagated by infection of CV-1 cells in DMEM supplemented with 2.5% FBS. The recombinant viruses expressing either the HRSV Gm (rVV-Gm) or Gs (rVV-Gs) protein have been described (1). The G insert of rVV-Gm contains a single point mutation that results in a substitution of the methionine at codon 48 for isoleucine (Met48Ile) and inhibits internal translation initiation at that triplet. Thus, rVV-Gm expresses only the membrane-bound form of the G protein. The G insert of rVV-Gs contains a deletion of the first 47 codons, resulting in expression of only the soluble form of G.
Recombinant vaccinia virus expressing Gs protein lacking the heparin binding domain (rVV-GsΔHBD) was prepared by the method of Blasco and Moss (3), as previously described (1). Briefly, the gene encoding the Gs protein (Long strain), previously cloned into plasmid pRB21 (3), was mutated to delete residues 187 to 198 (pRBGsΔHBD) using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions and using the following oligonucleotide primers: forward, 5′ GC AAC AAT CCA ACC TGC TGG GCT ATC TGC ACC ACC AAG CCT ACA AAA AAA CCA ACC 3′; reverse, 5′ GGT TGG TTT TTT TGT AGG CTT GGT GGT GCA GAT AGC CCA GCA GGT TGG ATT GTT GC 3′.
CV-1 monolayers were infected with the vaccinia virus vRB12 (2) and subsequently transfected with pRBGsΔHBD. vRB12 lacks most of the gene encoding vp37, the major protein component of the vaccinia virus envelope, and as a result is unable to form extracellular enveloped virions. Hence, vRB12 is deficient in plaque formation under standard conditions. pRB21-derived plasmids carry a complete copy of vp37 and thus, via homologous recombination with vRB12, restore VP37 function. Recombinant rVV-GsΔHBD was obtained from the supernatant of infected/transfected CV-1 cells and subjected to three consecutive rounds of plaque purification, and a viral stock was grown in this cell line.
Production and purification of G proteins.
HEp-2 cells were infected with the recombinant vaccinia viruses in serum-free DMEM (multiplicity of infection, ≈0.1 PFU/cell). Culture supernatants were collected 48 h postinfection from cells infected with either rVV-Gs or rVV-GsΔHBD and clarified by centrifugation at 15,300 × g for 15 min to remove cell debris. Supernatants were concentrated and equilibrated in phosphate-buffered saline (PBS) by filtration through polyethersulfone membranes (Vivaflow; Sartorius) of 50-kDa exclusion pore size. rVV-Gm-infected cells were also harvested 48 h postinfection and washed twice with PBS, and extracts were prepared by sonication in PBS containing 1% octyl-glucoside. Extracts were subsequently centrifuged at 13,000 × g for 10 min, and the pellets were discarded.
Both Gs- and Gm-containing preparations were loaded separately onto Sepharose columns to which monoclonal antibodies 021/1G and 021/2G (25) had been bound. The columns were washed with 20 volumes of PBS, and the retained material was eluted with 10 volumes of 0.1 M glycine-HCl, pH 2.5. G protein-containing fractions eluted from the columns were neutralized with saturated Tris and concentrated and buffer exchanged to PBS using a Vivaspin (Sartorius) concentrator of 50-kDa exclusion pore size. Gm was purified in the presence of buffers containing 0.1% octyl-glucoside, whereas Gs was maintained in detergent-free buffers at all stages of purification, unless specified.
Purified G proteins were subsequently subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and Coomassie blue staining or Western blotting using anti-G-specific monoclonal antibodies (12).
Sucrose density gradient centrifugation.
Both Gs and Gm proteins were loaded onto preformed 5 to 25% sucrose gradients prepared in PBS in the absence or presence of 0.1% octyl-glucoside. The samples were centrifuged at 39,000 rpm for 15 h at 4°C in an SW40 rotor (Beckman). Twelve 1-ml fractions were collected from the tops of the tubes, and aliquots of each fraction were analyzed by Western blotting using monoclonal antibody 63G (12).
Flow cytometry.
Cell monolayers were detached using 1 mM EDTA in Ca2+- and Mg2+-free PBS and resuspended in DMEM (2% FBS). Cells (3 × 105) were incubated for 45 min at 4°C with 100-μl dilutions of either purified virus or Gs protein, centrifuged at a low speed, washed three times with PBS, and incubated for 75 min with a pool of monoclonal antibodies (25G, 63G, and 021/1G) specific for the G protein (12, 25). Cells were stained by subsequent 30-min incubations with biotin-coupled anti-mouse sheep antibodies and R-Phycoerythrin-conjugated streptavidin, with washing of the cell pellet after each incubation. All incubations and centrifugations were conducted at 4°C. Finally, cells were fixed in 1% paraformaldehyde in PBS, and the fluorescence of 103 cells was determined using a Becton Dickinson FACSCalibur instrument with CellQuest software. In some experiments the cells were pretreated with heparinase I (H2519; Sigma) before being used in the binding assay, as indicated in the figure legends.
RESULTS
Comparison of the sedimentation properties of Gs and Gm.
Purified Gs and Gm proteins were analyzed by SDS-PAGE and Coomassie blue staining (Fig. 1A). While Gs was estimated by densitometry of the gel to be at least 90% pure, the Gm preparation contained several prominent contaminating bands. It should be emphasized that the process of staining the G protein with Coomassie blue (or silver staining, not shown) is extremely inefficient, probably due to the high oligosaccharide content of this molecule. Therefore, the degree of purification of both Gs and Gm may be underestimated by SDS-PAGE. Figure 1B shows a Western blot of the Gs and Gm preparations in which several additional bands are observed to migrate faster than Gs and Gm and may correspond to maturation intermediates and/or degradation products.
Both purified Gs and Gm proteins were subjected to sedimentation in 5 to 25% sucrose gradients (Fig. 2) in the presence or absence of detergent (0.1% octyl-glucoside). Gradient fractions were analyzed by Western blotting using the specific monoclonal antibody 63 G (12). Gs sedimented in the first few fractions near the top of the gradient, and its sedimentation profile was not influenced by the presence or absence of detergent (Fig. 2A and C). This indicates that Gs is indeed a soluble protein with no tendency to aggregate. The low-molecular-weight forms of Gs predominated in the first fraction of the gradient, whereas mature Gs predominated in fractions 3 and 4 of the gradient. Gs sedimented slightly more slowly than bovine serum albumin (BSA), centrifuged in a parallel gradient. Since Gs has an apparent molecular mass greater than that of BSA, as estimated by SDS-PAGE, it is likely that Gs remains monomeric under the experimental conditions used in this study and that it displays an abnormal electrophoretic mobility in SDS-PAGE gels (see Discussion).
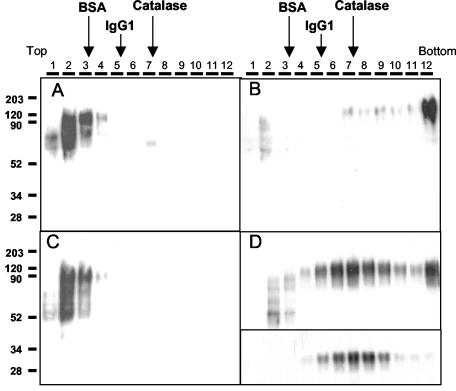
FIG. 2.
Sedimentation of Gs and Gm proteins in sucrose gradients. The purified Gs (A and C) and Gm (B and D) proteins from Fig. 1 were loaded onto preformed 5 to 25% sucrose gradients and then centrifuged and processed as indicated under Materials and Methods. Sedimentation proceeds from left to right. Both samples and gradients shown in panels C and D contained 0.1% octyl-glucoside in PBS, whereas those displayed in panels A and B did not contain detergent. Thirty microliters from each gradient fraction (shown at the top of each panel) were analyzed by Western blotting using antibody 63G. Fractions 5 to 9 of the gradient shown in panel D were pooled together, equilibrated in PBS containing 0.1% octyl-glucoside, and centrifuged a second time in a new 5 to 25% sucrose gradient prepared in PBS containing 0.1% octyl-glucoside. Western blot analysis of fractions obtained from the latter gradient is shown in the inset of panel D. Gradients run in parallel in the absence of detergent were loaded with either BSA (68 kDa), mouse immunoglobulin G1 (IgG1; 150 kDa), or catalase (250 kDa). Fractions of these gradients were analyzed by SDS-PAGE and Coomassie blue staining. The peak of the protein sedimentation profile in each of these gradients is indicated (arrows).
In contrast to Gs, the sedimentation profile displayed by Gm in the sucrose gradient was clearly influenced by the presence of detergent (Fig. 2B and D). In the absence of octyl-glucoside, the majority of Gm sedimented to the bottom of the gradient. Electron microscopy of this material revealed the presence of large vesicles to which Gm was probably bound via its transmembrane region (not shown). However, in the presence of octyl-glucoside detergent, Gm sedimented preferentially to the central fractions (5 to 9) of the gradient, although a small proportion continued to sediment to the bottom of the gradient. It is worth noting that the low-molecular-weight bands of the Gm preparation remained in the first few fractions of the gradient, indicating that they were not associated with mature Gm. When fractions 5 to 9 from the Gm gradient that contained detergent were pooled together and centrifuged a second time in a new gradient containing octyl-glucoside, Gm was found in fractions 5 to 9 and did not sediment to the bottom of the gradient. In comparison with standard proteins of known molecular weight centrifuged in parallel gradients, the peak fractions of Gm in the presence of detergent sedimented as the catalase marker (≈250 kDa). Thus, the mass of Gm estimated by sedimentation analysis is approximately four times higher than the mass of Gs, compatible with Gm possessing an oligomeric, probably tetrameric structure (refer to Discussion).
Gs binds to cell surface GAGs.
It has been reported that HRSV binds to GAGs in the form of cell surface proteoglycans (16, 17, 26) and that this interaction is preferentially mediated by the G protein (35). In spite of the differences we observed between the oligomeric structure of Gs and Gm, since Gs represents the ectodomain of the Gm protein it was of interest to determine whether Gs was capable of binding to cells and if this interaction was mediated by GAGs.
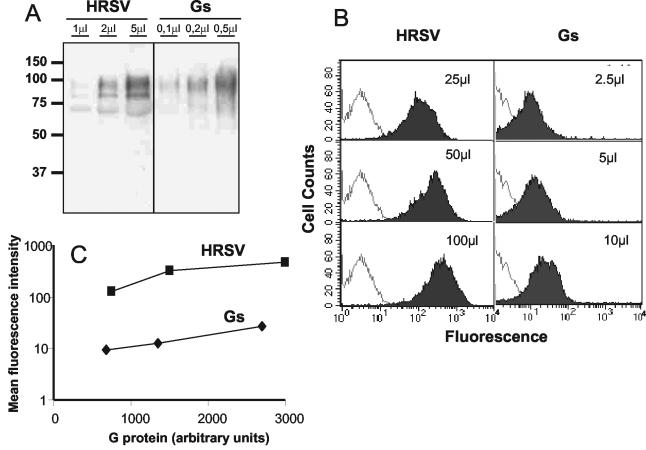
Binding of Gs to HEp-2 cells was tested using a previously described flow cytometry-based binding assay (26). We also attempted to use purified Gm in the same type of assay; however, the aggregating properties of this molecule precluded us from obtaining reproducible results. Thus, for comparative purposes, purified virus (Long strain) was tested in parallel with Gs for binding to HEp-2 cells. The concentration of G protein present in the virus and Gs preparations was estimated by Western blot analysis. As seen in Fig. 3A, the concentration of Gm protein was approximately 10 times lower in the preparation of Long virus than the concentration of Gs present in the preparation of purified protein. Following concentration estimation, dilutions of virus and Gs that contained equivalent amounts of G protein were assayed for their ability to bind to HEp-2 cells (Fig. 3B). There was a dose-dependent increase in the fluorescence of cells as increasing amounts of either virus or Gs were incubated with cells. However, the level of fluorescence produced by Gs bound to cells was significantly lower than that observed with purified virus containing an equivalent amount of Gm protein. Quantitation of these results (Fig. 3C) indicated that cells incubated with Gs emitted 10 to 50 times less fluorescence than cells incubated with equivalent amounts of virion-bound G. Whether this result indicates that Gs binds less efficiently to cells than virus-associated Gm is unknown at present. One should bear in mind that Gs is monomeric; therefore, each molecule of Gs bound to cells would bind only one molecule of each of the three antibodies used in the binding assay. In contrast, each virus particle carries a large number of G molecules that may each bind three antibodies and thus amplify the signal.
FIG. 3.
Binding of Gs protein to HEp-2 cells. (A) The amounts indicated in each lane of either purified HRSV or Gs protein were analyzed by Western blotting with antibody 63G (12). (B) Corresponding amounts of HRSV (Gm) or Gs, as determined by densitometry of the Western blots shown in panel A, were incubated with HEp-2 cells. Following incubation, the binding of virus and Gs to cells was determined by flow cytometry (shaded histograms), as indicated in Materials and Methods. Negative controls of cells incubated without virus or without Gs are shown in each case (unshaded histograms). (C) The amounts of Gs and Gm used in the flow cytometry assay were converted to arbitrary units and plotted against the mean fluorescent intensity (with background fluorescence subtracted) of the histograms shown in panel B.
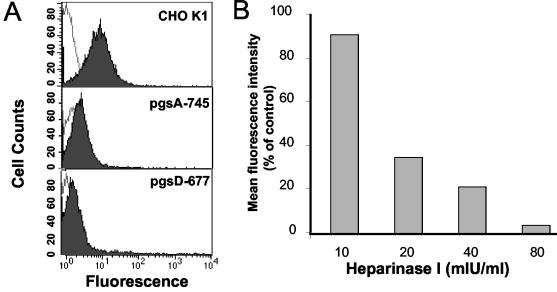
The binding of Gs to proteoglycan-deficient cell lines was investigated in order to determine whether GAGs are involved in the binding of Gs to cells. CHO K1 cells express a full complement of cell surface GAGs, estimated to comprise 70% heparan sulfate and 30% chondroitin-4-sulfate. Gs protein bound to the CHO K1 cell line (Fig. 4A), as previously observed with HEp-2 cells; however, binding to proteoglycan-deficient cell lines was greatly reduced. The mutant cell line pgsA-745 is deficient in xylosyltransferase (9), an enzyme required for the first sugar transfer reaction (transfer of xylose to the core protein), in the formation of all proteoglycans. As a result, pgsA-745 cells express less than 1% of GAGs expressed by the wild-type CHO K1 cell line. Thus, inhibition of Gs attachment to pgsA-745 cells (Fig. 4A) demonstrates that this protein requires proteoglycans for attachment to cells. In order to further specify the particular proteoglycan involved in attachment, binding of Gs to pgsD-677 cells was assessed. The pgsD-677 cell line is deficient in both N-acetylglucosaminyltransferase and glucuronosyltransferase activities required for the synthesis of heparan sulfate (24). Therefore, pgsD-677 cells do not express heparan sulfate but overexpress chondroitin-4-sulfate. There was substantial inhibition of attachment of Gs to pgsD-677 cells in comparison to the case with the parental CHO K1 cell line, implying that Gs specifically binds to cellular heparan sulfate, as has been previously shown for the complete virus (16, 26, 35).
FIG. 4.
Binding of Gs to proteoglycan-deficient CHO cells and effect of heparinase treatment on Gs attachment. (A) Twenty microliters of purified Gs (shown in Fig. 1) was incubated with wild-type CHO K1 cells or with the proteoglycan-deficient cell mutants pgsA-745 or pgsD-677. The attachment of Gs to each cell type was measured by flow cytometry, as indicated in Materials and Methods (shaded histograms). The unshaded histograms correspond to cells incubated without Gs. (B) HEp-2 cells (3 × 105) were incubated for 1 h at 37°C with the indicated amounts of heparinase I before 20 μl of Gs was added to assess attachment by flow cytometry. The results are expressed as the percentage of mean fluorescence intensity relative to that of heparinase mock-treated cells after subtraction of background values (without added Gs).
In order to (i) confirm that attachment of purified Gs to HEp-2 cells is mediated by heparan sulfate and (ii) determine whether Gs protein is capable of attachment by GAG-independent interactions, the effect of heparinase I on attachment was investigated. Heparinase I selectively cleaves glycosidic linkages of heparin and heparan sulfate at highly sulfated regions. Thus, HEp-2 cells were pretreated with heparinase I to remove heparan sulfate from the cell surface before being incubated with purified Gs and subjected to flow cytometry. Quantitation of the results (Fig. 4B) shows that binding of Gs was inhibited in a dose-dependent manner by heparinase I treatment. This indicates that heparan sulfate mediates most if not all of the Gs protein binding to HEp-2 cells. The size and surface topography of heparinase-treated cells (as determined by flow cytometry) were consistent with those of untreated cells, indicating that inhibition of binding was not due to a loss in viability of the cells following heparinase treatment.
Residues 187 to 198 of the proposed HBD are required for efficient binding of Gs to cells.
A GAG-binding domain (heparin binding domain or HBD) encompassing amino acids 184 to 198 was postulated by Feldman et al. (10) following analysis of the ability of G-protein peptides to bind to heparin. However, there is debate as to whether this region is in fact indispensable in order for native G protein to bind to cells during infection (36).
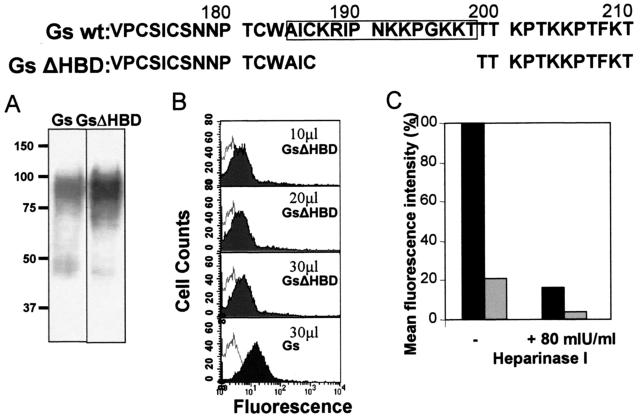
In order to test the relevance of the HBD in the binding of Gs to cells, a recombinant vaccinia virus (rVV-GsΔHBD) was prepared that expresses a Gs deletion mutant lacking residues 187 to 198 of the HBD (Fig. 5). The first three residues of the proposed HBD were retained in the GsΔHBD mutant, since they comprise Cys186, which may contribute to the G protein structure by participating in disulfide bonding that stabilizes a cystine noose motif (8, 15).
FIG. 5.
Binding of Gs lacking the HBD to HEp-2 cells. Partial sequences of the Gs wild-type and GsΔHBD proteins are shown at the top, denoting the proposed HBD (framed) and the amino acids deleted in the latter protein. (A) Western blot analysis (using antibody 63G) of 20 μl of wild-type Gs and mutant GsΔHBD that were concentrated from the supernatants of HEp-2 cells infected with vaccinia virus recombinants expressing the corresponding proteins. (B) The indicated amounts of each protein were assessed for their ability to bind to HEp-2 cells by flow cytometry (shaded histograms). The unshaded histogram is a negative control representing cells incubated without Gs. (C) HEp-2 cells were either incubated with 80 mIU of heparinase I/ml or mock digested as indicated in Fig. 4 before being incubated with 30 μl of Gs (black) or GsΔHBD (grey) and subjected to flow cytometry to assess attachment. The results are expressed as the percentage of mean fluorescence obtained relative to that produced by the binding of Gs to untreated HEp-2 cells after subtraction of background values (without added Gs).
During the course of this study we observed by flow cytometry that Gs that was concentrated from the culture supernatant of vaccinia virus recombinant-infected cells was capable of binding to cells without the need for further purification. Therefore, supernatants of HEp-2 cells infected with either rVV-Gs or rVV-GsΔHBD were concentrated and analyzed by Western blotting (Fig. 5A). Both preparations generated similar band patterns, characterized by a prominent band representing mature Gs and additional faint, faster-migrating bands. Thus, deletion of residues 187 to 198 did not significantly alter the expression and maturation of Gs. In comparison with Gs, GsΔHBD displayed a decreased ability to bind to HEp-2 cells (Fig. 5B). The estimated concentration of GsΔHBD was three times higher than that of Gs (Fig. 5A), yet the mean fluorescence intensity of cells incubated with 30 μl of GsΔHBD was one-fifth of that incubated with 30 μl of Gs (Fig. 5C). However, the residual binding of GsΔHBD to HEp-2 cells could be further reduced by pretreatment of the cells with 80 mIU of heparinase I/ml. Thus, although efficient binding of Gs to GAGs requires residues 187 to 198 of the HBD domain, deletion of this region does not completely eliminate the attachment of Gs to cell surface GAGs.
DISCUSSION
Two different forms of the HRSV G glycoprotein (Gs and Gm) were analyzed in this study. The soluble form (Gs) was obtained from the culture medium of HEp-2 cells infected with a recombinant vaccinia virus expressing this protein. Following immunoaffinity chromatography, a highly purified form of Gs was obtained which remained unaggregated in the absence of detergent and was easily manipulated. A membrane-bound form of the G protein (Gm) was partially purified from the lysates of cells infected with a recombinant vaccinia virus expressing Gm. In contrast to Gs, Gm was difficult to manipulate, since it formed large aggregates in the absence of detergent and had a tendency to aggregate upon storage even in the presence of 0.1% octyl-glucoside.
Sucrose gradient centrifugation of the two forms of the G protein in the presence of detergent revealed that they differ in their sedimentation rates. Gs sedimented at a rate lower than that of BSA (68 kDa), compatible with Gs remaining as a monomer. Given that the mass of Gs estimated by SDS-PAGE (≈90 kDa) exceeds that of BSA, it is likely that Gs has an abnormal electrophoretic mobility due to poor binding to SDS (5). Gm sedimented at a rate similar to that of the catalase marker (250 kDa) but higher than mouse immunoglobulin G1 (150 kDa). Collins and Mottet (5) have previously reported the sedimentation properties of Gm (present in HRSV-infected cell extracts) on sucrose gradients containing 0.1% Triton X-100. They found that Gm sedimented as the F protein trimer (∼190 kDa), although no data were presented for the latter protein. We have carried out a similar analysis of HRSV-infected cell extracts and found that the sedimentation rate of Gm is indistinguishable from the rate observed in the gradient of Fig. 2D (data not shown). These observations are compatible with Gm existing as a homotetramer, given that its sedimentation rate is about four times higher than that of Gs. However, since the preparation of purified Gm still contained other contaminating proteins, we cannot exclude the possibility that Gm associates with some of these proteins to form a hetero-oligomer. It is worth noting that Gm sedimented at the same rate after centrifugation in a second sucrose gradient containing 0.1% octyl-glucoside, while most of the contaminating proteins sedimented in other fractions of the gradient (data not shown). Thus, at present we favor the hypothesis that Gm assembles as a homotetramer. This would resemble the oligomeric structure of the attachment protein of other paramyxoviruses. The recently solved atomic structure of the Newcastle disease virus HN attachment protein revealed that this protein forms homodimers (7) that can also associate into tetramers (27). Oligomerization seems to require the Gm hydrophobic transmembrane region, not present in Gs; however, the differential cellular processing of the secreted and membrane-bound forms of the G protein may also play a role in oligomerization.
We have demonstrated here for the first time that Gs is capable of binding to cells in vitro and that this interaction is mediated mainly, if not entirely, by cellular proteoglycans. Heparan sulfate was found to be the specific GAG involved in attachment, since Gs protein did not bind to a mutant cell line (pgsD-677) which overexpresses chondroitin-4-sulfate but lacks heparan sulfate. This specificity was confirmed by heparinase I treatment of cells. Thus, the specificity of Gs binding to cells mimics the specificity reported for the Gm protein of virions (16, 26), despite their differences in oligomeric structure. Although cells incubated with Gs were 10 to 50 times less fluorescent than cells incubated with equivalent amounts of virion-associated Gm, it is too early to conclude that Gs and Gm possess differing affinities for GAGs until the valency of virion-associated Gm in the cell-binding assay can be correctly calculated.
The contribution of the proposed G protein HBD (residues 184 to 198) to the binding of HRSV to cells has been in question since the infectivity of rHRSV lacking residues 187 to 197 of the G protein was shown to be inhibited by heparin (36). The deletion of residues 187 to 198 markedly reduced the attachment of Gs to cells. However, residual binding of GsΔHBD was inhibited by pretreatment of cells with heparinase I (Fig. 5). Therefore, other GAG-binding regions besides the HBD may exist in the G protein, which would explain both the phenotype of rHRSV lacking residues 187 to 197 and the attachment properties of GsΔHBD. The reasons for which Feldman et al. (10) did not identify other GAG-binding regions are unknown at present, but there are a number of possibilities, including the following: (i) other heparin binding regions, besides amino acids 184 to 197, may have not been represented in the set of peptides used (15-amino-acid-long peptides with a 5-amino-acid overlap and offset); (ii) the presence of multiple oligosaccharide side chains in the native G protein may influence the presentation of protein regions for binding to GAGs.
A recent study (33), in which segments of the G protein were expressed in bacteria and tested for cell binding, also supports the notion that other regions besides the 184-187 region are able to interact with GAGs. However, since protein glycosylation does not occur in bacteria, we believe that testing of cell binding should be done with G protein expressed in mammalian cells, given the highly glycosylated nature of the G protein. Similarly to the manner in which cell-specific glycosylation modifies the antigenic properties of G (13), changes in the glycosylation pattern of the G protein (either Gs or Gm) may influence its interaction with GAGs.
The presence of more than one HBD may represent an advantageous strategy for the virus, since sequence variation of the HBD has been observed among HRSV isolates (11). Multiple GAG-binding domains would allow the G protein to undergo mutation to evade the host defense without losing its capacity to bind to cell surface GAGs. Indeed, if the attachment of G protein to GAGs does in fact involve electrostatic interactions mediated by positively charged residues, multiple clusters of basic amino acids found along the G protein primary structure may contribute to these interactions. The binding assay reported here should allow us to make a more in-depth examination of the contributions of different regions of Gs to its interaction with GAGs.
The precise role of Gs is currently unknown; however, the fact that it is able to bind to cells via GAGs may be an important factor in its function. For instance, since Gs is shed from infected cells before the appearance of progeny virus (19), binding of Gs to cells could make them more susceptible to infection. This would provide a possible explanation for the recent finding that rHRSV expressing only Gs grew efficiently in vitro and resulted in only a modest growth reduction in mice, whereas rHRSV lacking the entire G protein gene grew poorly in certain cell lines and was attenuated in mice (36). Therefore, it is possible that the presence of either form of the G protein bound to target cells is important at some stage of HRSV infection.
Acknowledgments
We are grateful to R. Blasco (INIA, Madrid, Spain) for the kind gift of the pRB21 plasmid and vRB12 vaccinia virus.
This work was supported in part by grants from Ministerio de Ciencia y Tecnología (SAF 2003-08250) and from Instituto de Salud Carlos III (01/24). E. Escribano-Romero was the recipient of a postdoctoral fellowship funded by the European Union (QLK2-CT-1999-00443), and J. Rawling was on a placement year from Imperial College (London, United Kingdom).
REFERENCES
- 1.Bembridge, G. P., R. García-Beato, J. A. Lopez, J. A. Melero, and G. Taylor. 1998. Subcellular site of expression and route of vaccination influence pulmonary eosinophilia following respiratory syncytial virus challenge in BALB/c mice sensitized to the attachment G protein. J. Immunol. 161:2473-2480. [PubMed] [Google Scholar]
- 2.Blasco, R., and B. Moss. 1991. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by the deletion of the gene encoding the 37,000-dalton outer envelope protein. J. Virol. 65:5910-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasco, R., and B. Moss. 1995. Selection of recombinant vaccinia viruses on the basis of plaque formation. Gene 158:157-162. [DOI] [PubMed] [Google Scholar]
- 4.Brandenburg, A. H., H. J. Neijens, and A. D. Osterhaus. 2001. Pathogenesis of RSV lower respiratory tract infection: implications for vaccine development. Vaccine 19:2679-2782. [DOI] [PubMed] [Google Scholar]
- 5.Collins, P. L., and G. Mottet. 1992. Oligomerization and post-translational processing of glycoprotein G of human respiratory syncytial virus: altered O-glycosylation in the presence of brefeldin A. J. Gen. Virol. 73:849-863. [DOI] [PubMed] [Google Scholar]
- 6.Collins, P. L., R. M. Chanock, and B. R. Murphy. 2001. Respiratory syncytial virus, p. 1443-1484. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 7.Crennell, S., T. Takimoto, A. Portner, and G. Taylor. 2000. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 7:1068-1074. [DOI] [PubMed] [Google Scholar]
- 8.Doreleijers, J. F., J. P. M. Langedijk, K. Hard, R. Boelens, J. A. C. Rullmann, W. M. Schaaper, J. T. van Oirschot, and R. Kaptein. 1996. Solution structure of the immunodominant region of protein G of bovine respiratory syncytial virus. Biochemistry 35:14684-14688. [DOI] [PubMed] [Google Scholar]
- 9.Esko, J. D., T. E. Stewart, and W. H. Taylor. 1985. Animal cell mutants defective in Glycosaminoglycan biosynthesis. Proc. Natl. Acad. Sci. USA 82:3197-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feldman, S. A., R. M. Hendry, and J. A. Beeler. 1999. Identification of a linear heparin binding domain for human respiratory syncytial virus attachment glycoprotein G. J. Virol. 73:6610-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García, O., M. Martín, J. Dopazo, J. Rabiza, S. Frabasile, J. Russi, M. Hortal, P. Perez-Breña, I. Martínez, B. García-Barreno, and J. A. Melero. 1994. Evolutionary pattern of human respiratory syncytial virus (subgroup A): cocirculating lineages and correlation of genetic and antigenic changes in the G glycoprotein. J. Virol. 68:5448-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Barreno, B., C. Palomo, C. Peñas, T. Delgado, P. Perez-Breña, and J. A. Melero. 1989. Marked differences in the antigenic structure of human respiratory syncytial virus F and G glycoproteins. J. Virol. 63:925-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Beato, R., I. Martínez, C. Francí, F. X. Real, B. García-Barreno, and J. A. Melero. 1996. Host cell effect upon glycosylation and antigenicity of human respiratory syncytial virus G glycoprotein. Virology 221:301-309. [DOI] [PubMed] [Google Scholar]
- 14.González-Reyes, L., M. B. Ruiz-Argüello, B. García-Barreno, L. Calder, J. A. López, J. P. Albar, J. J. Skehel, D. C. Wiley, and J. A. Melero. 2001. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc. Natl. Acad. Sci. USA 98:9859-9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorman, J. J., J. L. McKimm-Breschkin, B. S. Norton, and K. J. Barnham. 2001. Antiviral activity and structural characteristics of the nonglycosylated central subdomain of human respiratory syncytial virus attachment (G) glycoprotein. J. Biol. Chem. 276:38988-38994. [DOI] [PubMed] [Google Scholar]
- 16.Hallak, L. K., P. L. Collins, W. Knudson, and M. E. Peeples. 2000. Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology 271:264-275. [DOI] [PubMed] [Google Scholar]
- 17.Hallak, L. K., D. Spillmann, P. L. Collins, and M. E. Peeples. 2000. Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection. J. Virol. 74:10508-10513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendricks, D. A., K. Baradaran, K. McIntosh, and J. L. Patterson. 1987. Appearance of a soluble form of the G protein of respiratory syncytial virus in fluids of infected cells. J. Gen. Virol. 68:1705-1714. [DOI] [PubMed] [Google Scholar]
- 19.Hendricks, D. A., K. McIntosh, and J. L. Patterson. 1988. Further characterisation of the soluble form of the G glycoprotein of respiratory syncytial virus. J. Virol. 62:2228-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, P. R., M. K. Spriggs, R. A. Olmsted, and P. L. Collins. 1987. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc. Natl. Acad. Sci. USA 84:5625-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krusat, T., and H. J. Streckert. 1997. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch. Virol. 142:1247-1254. [DOI] [PubMed] [Google Scholar]
- 22.Langedijk, J. P. M., B. L. de Groot, J. C. Berendsen, and J. T. van Oirschot. 1998. Structural homology of the central conserved region of the attachment protein G of respiratory syncytial virus with the fourth subdomain of 55-kDa tumor necrosis factor receptor. Virology 243:293-302. [DOI] [PubMed] [Google Scholar]
- 23.Levine, S., R. Klaiber-Franco, and P. R. Paradiso. 1987. Demonstration that the glycoprotein G is the attachment protein of respiratory syncytial virus. J. Gen. Virol. 68:2521-2524. [DOI] [PubMed] [Google Scholar]
- 24.Lidholt, K., J. L. Weinke, C. S. Kiser, F. N. Lugemwa, K. J. Bame, S. Cheifetz, J. Massagué, U. Lindahl, and J. D. Esko. 1992. A single mutation affects both N-acetylglucosaminyltransferase and glucuronosyltransferase activities in a Chinese hamster ovary cell mutant defective in heparan sulfate biosynthesis. Proc. Natl. Acad. Sci. USA 89:2267-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martínez, I., J. Dopazo, and J. A. Melero. 1997. Antigenic structure of respiratory syncytial virus G glycoprotein and relevance of hypermutation events for the generation of antigenic variants. J. Gen. Virol. 78:2419-2429. [DOI] [PubMed] [Google Scholar]
- 26.Martínez, I., and J. A. Melero. 2000. Binding of human respiratory syncytial virus to cells: implication of sulphated cell surface proteoglycans. J. Gen. Virol. 81:2715-2722. [DOI] [PubMed] [Google Scholar]
- 27.McGinnes, L. W., and T. G. Morrison. 1994. The role of individual cysteine residues in the formation of the mature, antigenic HN protein of Newcastle disease virus. Virology 200:470-483. [DOI] [PubMed] [Google Scholar]
- 28.Melero, J. A., B. García-Barreno, I. Martínez, C. R. Pringle, and P. A. Cane. 1997. Antigenic structure, evolution and immunobiology of human respiratory syncytial virus attachment (G) protein. J. Gen. Virol. 78:2411-2418. [DOI] [PubMed] [Google Scholar]
- 29.Mok, J. Y., and H. Simpson. 1982. Outcome of acute lower respiratory tract infection in infants: preliminary report of seven-year follow-up study. Br. Med. J. 285:333-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pullen, C. R., and E. N. Hey. 1982. Wheezing, asthma and pulmonary dysfunction 10 years after infection with respiratory syncytial virus in infancy. Br. Med. J. 284:1665-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts, S. R., D. Lichtenstein, L. A. Ball, and G. W. Wertz. 1994. The membrane-associated and secreted forms of the respiratory syncytial virus attachment glycoprotein are synthesised from alternative initiation codons. J. Virol. 68:4538-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Satake, M., J. E. Coligan, N. Elango, E. Norrby, and S. Venkatesan. 1985. Respiratory syncytial virus envelope glycoprotein (G) has a novel structure. Nucleic Acids Res. 13:7795-7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shields, B., J. Mills, R. Ghildyal, P. Gooley, and J. Meanger. 2003. Multiple heparin binding domains of respiratory syncytial virus G mediate binding to mammalian cells. Arch. Virol. 148:1987-2003. [DOI] [PubMed] [Google Scholar]
- 34.Techaarpornkul, S., N. Barretto, and M. E. Peeples. 2001. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 75:6825-6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Techaarpornkul, S., P. L. Collins, and M. E. Peeples. 2002. Respiratory syncytial virus with the fusion protein as its only viral glycoprotein is less dependent on cellular glycosaminoglycans for attachment than complete virus. Virology 294:296-304. [DOI] [PubMed] [Google Scholar]
- 36.Teng, M. N., S. S. Whitehead, and P. L. Collins. 2001. Contribution of the respiratory syncytial virus G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology 289:283-296. [DOI] [PubMed] [Google Scholar]
- 37.Walsh, E. E., and J. Hruska. 1983. Monoclonal antibodies to respiratory syncytial virus proteins: identification of the fusion protein. J. Virol. 47:171-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wertz, G. W., P. L. Collins, Y. Hang, C. Gruber, S. Levine, and L. A. Ball. 1985. Nucleotide sequence of the G protein of human respiratory syncytial virus reveals an unusual type of membrane protein. Proc. Natl. Acad. Sci. USA 82:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]