Abstract
Infectious bursal disease virus (IBDV) is a nonenveloped virus with an icosahedral capsid composed of two proteins, VP2 and VP3, that derive from the processing of the polyprotein NH2-pVP2-VP4-VP3-COOH. The virion contains VP1, the viral polymerase, which is both free and covalently linked to the two double-stranded RNA (dsRNA) genomic segments. In this study, the virus assembly process was studied further with the baculovirus expression system. While expression of the wild-type polyprotein was not found to be self-sufficient to give rise to virus-like particles (VLPs), deletion or replacement of the five C-terminal residues of VP3 was observed to promote capsid assembly. Indeed, the single deletion of the C-terminal glutamic acid was sufficient to induce VLP formation. Moreover, fusion of various peptides or small proteins (a green fluorescent protein or a truncated form of ovalbumin) at the C terminus of VP3 also promoted capsid assembly, suggesting that assembly required screening of the negative charges at the C terminus of VP3. The fused polypeptides mimicked the effect of VP1, which interacts with VP3 to promote VLP assembly. The C-terminal segment of VP3 was found to contain two functional domains. While the very last five residues of VP3 mainly controlled both assembly and capsid architecture, the five preceding residues constituted the VP1 (and possibly the pVP2/VP2) binding domain. Finally, we showed that capsid formation is associated with VP2 maturation, demonstrating that the protease VP4 is involved in the virus assembly process.
Infectious bursal disease virus (IBDV) causes a disease highly contagious to young chickens, namely infectious bursal disease or Gumboro disease (5). Infection by this virus leads to a severe immunosuppression characterized by the destruction of B cells present in the bursa of Fabricius. The induced immunodepression increases the susceptibility to other pathogens. As a consequence, IBDV represents a large economic burden that calls for better understanding of the virus cycle in order to develop new antiviral drugs and better vaccines. In particular, knowledge of the viral assembly process should provide a structural basis for the rational design of new antiviral molecules.
IBDV belongs to the Birnaviridae family, which is composed of nonenveloped viruses containing two segments of double-stranded RNAs (A and B) (10). While the B segment (IBDB) encodes the VP1 protein, the putative viral RNA-dependent RNA polymerase, segment A (IBDA), contains two partially overlapping open reading frames (ORFs). The smaller ORF encodes VP5, a nonstructural protein of 17 kDa. The larger ORF encodes a polyprotein precursor in the order NH2-pVP2-VP4-VP3-COOH. The polyprotein appears to be cotranslationally processed through the proteolytic activity of VP4 to generate pVP2 (amino acids [aa] 1 to 512), VP4 (aa 513 to 755), and VP3 (aa 756 to 1012) (1, 9, 16). pVP2, the VP2 precursor, is further processed at its C terminus to become VP2, through the cleavage of three alanine-alanine bonds (positions 487 to 488, 494 to 495, and 501 to 502) and an alanine-phenylalanine bond (positions 441 to 442) (6). The resulting peptides have been found associated with the viral particle. VP2 and VP3 are believed to form, respectively, the outer and inner layers of the virion (2). VP1 is contained within the viral particle, both free and genome linked (10). VP3 interacts with both VP1 (11, 17) and genomic RNAs (18). The VP1 binding motif of VP3 was shown to be constituted by at least the last 10 aa of the VP3 C terminus (12, 18).
Different types of viral assemblies can be isolated from IBDV-infected cells. In addition to IBDV virions, tubes with diameters of about 55 nm (type I) and 24 to 26 nm (type II) are formed during infection (7, 8, 15). While type II tubes contain VP4, type I tubes are made of pVP2. Electron cryomicroscopy studies revealed that the structure of the virion is based on a T = 13 lattice formed by trimer-clustered subunits (2).
Recombinant expression of capsid proteins of nonenveloped viruses leads to capsid assembly processes characterized by variable efficiencies. In the case of IBDV, using the baculovirus-insect cell expression system, IBDA polyprotein expression does not give rise to virus-like particles (VLPs), but to pVP2-containing type I tubes (4, 14). The lack of pVP2-to-VP2 maturation on these tubes indicates that, under these conditions, an important factor for VLP assembly was missing. The fusion of an exogenous sequence at the C terminus of VP3 was shown to strongly promote the assembly of VLPs (4). When the VLPs are formed, the final maturation process converting pVP2 into VP2 is observed. It has recently been shown that VLPs are also formed when VP1 is coexpressed with the polyprotein (13). Thus, similar effects on capsid morphogenesis were observed when coexpressing IBDA with the VP1 gene or through expression of an extended IBDA segment with an exogenous sequence fused at the 3′ end of the large ORF. These observations suggest that the electrostatic potential associated with the charges of C terminus of VP3 needs screening by the presence of another protein. To test this hypothesis and to better characterize the interactions between VP3 and VP1, we engineered VP3 mutants and studied the effect of the mutation on capsid morphogenesis. We show that deletion or replacement of the last five charged residues of VP3 suppresses the need for VP1 or a fused protein to promote capsid assembly. Even the single deletion of the last C-terminal residue of VP3, glutamic acid 257, leads to efficient capsid formation. The coexpression of such deleted or substituted polyproteins and VP1 results in VLPs that contain VP1, demonstrating that the C terminus of VP3 displays two functional regions: an assembly controlling domain constituted of the last five residues of VP3 and a VP1 (and possibly pVP2/VP2) binding domain made by the upstream contiguous residues. The C terminus of VP3 appears to be a molecular switch that triggers capsid assembly upon VP1 binding.
MATERIALS AND METHODS
Plasmid and recombinant baculovirus constructs.
Plasmids pFBΔIBDA and pFBΔIBDA-GFP (4) were used as templates for the generation of all of the constructed pFastBac derivates. An ovalbumin cDNA stretch was PCR amplified with the primers GCAGCAGCTAGCGGGCTCCATCGGCGCAGCAAGCATGG and GCAGCAGGTACCTTAAGGGGAAACACATCTGCCAAAG and with truncated ovalbumin cDNA as a template (3). The luciferase gene was PCR amplified with the primers CGGTACTGTGCTAGCAGCCACCATGGAAGACGCC and ATGTCTGCTCGAAGCGGTACCCCGCCCCGACTC and plasmid pGL3 (Promega) as template. Both PCR products were digested with NheI and KpnI and cloned into pFBΔIBDA-GFP previously cut by the same restriction enzymes. The resulting plasmids were named pFBΔIBDA-OVA and pFBΔIBDA-LUC. Plasmids coding for IBDA polyproteins modified at their C termini were constructed. The modifications were introduced by using the Pfu DNA polymerase with the QuikChange site-directed mutagenesis kit (Stratagene) as described by the manufacturer. Nucleotide sequence analyses were carried out to confirm nucleotide substitutions. Thus, plasmids coding for truncated IBDA polyproteins (−10, −5, and −1 aa) were constructed and named pFBΔIBDA − 10, pFBΔIBDA − 5, and pFBΔIBDA − 1, respectively. A plasmid encoding an IBDA polyprotein in which the last five residues were replaced by five alanines was constructed and named pFBΔIBDADEDLE5A. Plasmids coding for elongated IBDA polyproteins (+10, +5, and +1 aa) were constructed and named pFBΔIBDA + 10, pFBΔIBDA + 5, and pFBΔIBDA + 1, respectively. The plasmid pT7-B-HDR (6) was used as a template to PCR amplify the VP1 gene by using the primers CGCGAATTCTAATACGACTCACTATAGGATGAGTGACATTTTCAACAGTCCAC and CGCGAATTCTTAGCGGCTCTCCTTTTGGCGTTGCCG. The PCR product was EcoRI restricted and subcloned into pFastBac1 (Gibco BRL) to generate pFBIBDB.
The resulting plasmids, pFBΔIBDA-OVA, pFBΔIBDA-LUC, pFBΔIBDA − 10, pFBΔIBDA − 5, pFBΔIBDA − 1, pFBΔIBDA + 1, pFBΔIBDA + 5, pFBΔIBDA + 10, pFBΔIBDA-DEDLE5A, and pFBIBDB were used to generate the recombinant baculoviruses BacΔIBDA-OVA, BacΔIBDA-LUC, BacΔIBDA − 10, BacΔIBDA − 5, BacΔIBDA − 1, BacΔIBDA + 1, BacΔIBDA + 5, BacΔIBDA + 10, BacΔIBDADEDLE5A, and BacIBDB, respectively. Briefly, the pFastBac derivates were transformed into DH10Bac competent cells, which contain the bacmid. Colonies containing recombinant bacmids were identified by disruption of the lacZα gene. High-molecular-weight DNA was prepared from selected colonies and used to transfect Sf9 cells with Lipofectin. Recombinant baculoviruses were generated by standard procedures, and high-titer viral stocks of the recombinant baculoviruses (108 PFU/ml) were prepared.
Preparation of protein assembly specimens.
Sf9 cells (2 × 108) were infected at a multiplicity of infection higher than 5 PFU/ml in the presence of protease inhibitors leupeptin (0.5 μg/ml) and aprotinin (1 μg/ml), collected with the cell culture medium, and frozen 100 h postinfection. After thawing, cell lysates were clarified and a protease inhibitor cocktail tablet (Boehringer Mannheim, Mannheim, Germany) was resuspended at the dilution recommended by the manufacturer. The supernatants were centrifuged at 40,000 rpm in a 45 Ti rotor (Beckman Coulter, Villepinte, France) for 1 h at 4°C. The pellets were resuspended in 4.5 ml of a mixture of Tris (10 mM; pH 8), 250 mM NaCl, 5 mM EDTA, and 10 mM β-mercaptoethanol completed with a protease inhibitor cocktail tablet and were then treated with Freon 113. Purification was carried out by density gradient centrifugation in a CsCl solution. The concentration of protein in the purified suspension was estimated by the method of Bradford with bovine serum albumin as a standard and UV spectrophotometry at 280 nm.
Electron microscopy.
Specimens were prepared from the appropriate CsCl gradient fraction containing the different assembled forms by desalting through Micro Bio-Spin chromatography columns (Bio-Rad) equilibrated with a buffer containing 50 mM Tris (pH 7.4) and 150 mM NaCl. Samples of the suspensions were applied to an air-glow-discharged carbon-coated grid and stained with a 2% uranyl acetate aqueous solution. Grids were observed in a CM12 electron microscope (Philips) operated at 80 kV.
Protein analysis. (i) Immunoprecipitations.
Sf9 cells (3 × 106) cells were infected (or mock infected) at a multiplicity of 10 PFU per cell and maintained in 2 ml of Hinks medium with 10% fetal calf serum. At 48 h postinfection, the medium was discarded, and cells were lysed in extraction buffer (50 mM Tris [pH 8], 150 mM NaCl, 2% Triton X-100) with a protease inhibitor cocktail (Boehringer). This material was centrifuged for 30 min at 13,000 × g. Aliquots of the supernatants were incubated for 2 h at room temperature under gentle agitation with 1 μl of ascites fluid of hybridomas specific for pVP2/VP2 and for VP3 and 35 μl of a 1:1 protein A-Sepharose bead solution (Pharmacia). The beads were pelleted and washed four times with 1 ml of extraction buffer and then treated for 2 min at 100°C in Laemmli denaturation buffer plus 5% 2-mercaptoethanol and centrifuged. The resulting supernatants were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (10 or 12.5% polyacrylamide, 0.1% SDS) and proteins were detected by Coomassie blue staining. Protein molecular weight standards (Pharmacia) were used.
(ii) Western blot analyses.
Gels were transferred on Immobilon membranes (Millipore) for 1 h at 50 V. Proteins were visualized by using dilutions of ascites fluid and relevant secondary antibodies conjugated to alkaline phosphatase (Biosys) and nitroblue tetrazolium-5-bromo-4-chloro-3-indolyl phosphate as substrate.
(iii) Trypsin digestion and MALDI-TOF analysis.
Trypsin digestions were achieved directly in the gel matrix. The excised gel plugs were washed in 50% CH3CN in 50 mM NH4CO3 (vol/vol) and then transferred to Eppendorf tubes. After desiccation of the gel in a SpeedVac for 30 min, the digestion was performed in 25 μl of 50 mM ammonium bicarbonate (pH 8.0) and 0.5 μg of modified trypsin (Promega, sequencing grade) for 18 h in an Eppendorf thermomixer at 37°C with vortexing at 500 rpm. A 0.5-μl aliquot of sample was applied as spots directly to the stainless steel matrix-assisted laser desorption ionization (MALDI) plate for MALDI-time-of-flight (TOF) mass spectrometry. The sample was then allowed to dry at room temperature before addition of 0.5 μl of the matrix solution. This dried-droplet sampling method was employed using a freshly prepared solution at 3-mg/ml 1-cyano-4-hydroxycinnamic acid matrix in 50% (vol/vol) acetonitrile and 0.1% (vol/vol) trifluoroacetic acid. For acquisition, the accelerating voltage used was 20 kV. Peptide spectra were recorded in positive reflector mode and with a delayed extraction of 130 ns and a 62% grid voltage. To analyze some peptides, spectra were recorded by the positive linear method with a delayed extraction of 160 ns and a 62% grid voltage. The spectra were calibrated by using an external calibration sample composed of Des-Arg 1 bradykinin (M+ H)+ = 904.468 Da, human angiotensin I (M+ H)+ = 1,296.685 Da, neurotensin (M+ H)+ = 1,672.917 Da, melltin from bee venom (M+ H)+ = 2,845.762 Da and bovine insulin B chain disulfonate (M+ H)+ = 3,494.651 Da. Samples digested with trypsin were calibrated by internal calibration with autolysis of trypsin: (M+ H)+ = 2,211.104 and 842.509 Da.
RESULTS
Capsid assembly is regulated by nonspecific association of the C terminus of VP3 with various proteins.
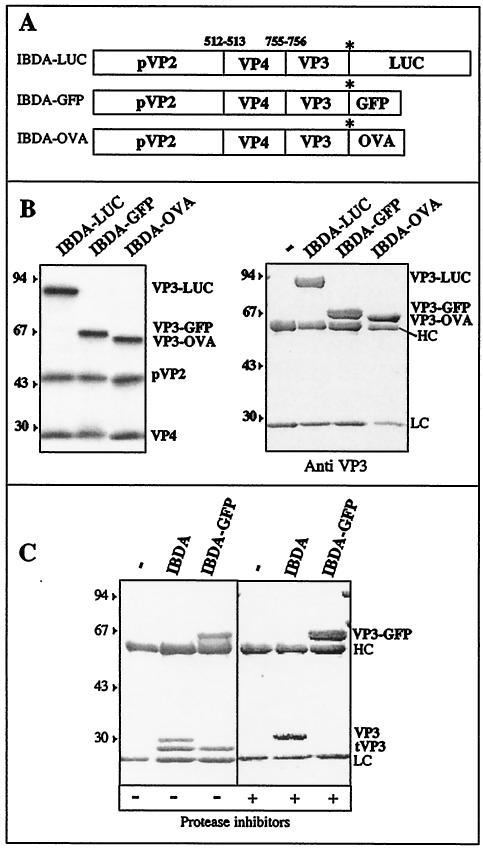
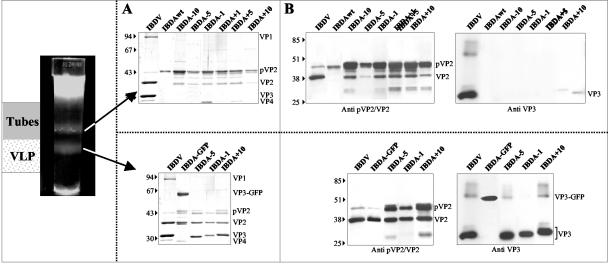
Recombinant expression of the wild-type IBDA polyprotein in the baculovirus-Sf9 cell system mainly leads to the formation of type I tubes (4, 14). Fusion of the green fluorescent protein (GFP; 238 aa) to the C terminus of VP3 provides for the correct capsid assembly signal. Under these conditions, VLPs are efficiently produced and are composed of VP2 and the VP3-GFP fusion protein (4). To determine the importance of the specificity and the length of the fused sequence on the assembly process, we constructed two recombinant baculoviruses, BacΔIBDA-OVA and BacΔIBDA-LUC (Fig. 1A). These recombinants drove the expression of the complete IBDA polyprotein fused at its C terminus with either a truncated 259-aa-long form of ovalbumin (OVA) or the entire 550-aa-long luciferase (LUC), respectively. These proteins were chosen because they exhibit no homology with GFP. In addition, while luciferase was used to further investigate the maximal size of a protein that can be accommodated within the VLPs, the truncated ovalbumin was used because as a glycoprotein, it is expected to fold randomly when synthesized in the cytosol. If VLP assembly were indeed induced by this last construct, it would suggest that the assembly signal is not sequence specific. Validation of the constructs was carried out by using an in vitro expression system and by immunoprecipitation of Sf9-infected cells and SDS-PAGE. Figure 1B (left panel) showed that the in vitro expression and processing of the fusion polyproteins yielded the expected cleavage products pVP2 (46 kDa) and VP4 (30 kDa) as well as a polypeptide corresponding to the expected size for the chimeric VP3-GFP, VP3-OVA, and VP3-LUC fusion proteins. Immunoprecipitations with an anti-VP3 antibody of Sf9 cell extracts infected with BacΔIBDA-GFP, BacΔIBDA-OVA, and BacΔIBDA-LUC revealed a single protein species having in all cases the expected relative molecular mass for each chimerical fusion protein (VP3-GFP, VP3-OVA, and VP3-LUC) (Fig. 1B, right panel). All of the components of the fusion proteins, pVP2, VP4, and the different VP3 chimeric forms, were thus expressed in the constructs. These results were obtained when protease inhibitors were added at all steps of preparation: leupeptin and aprotinin during infection and a manufactured protease inhibitor cocktail during extraction. In the absence of protease inhibitors, VP3 was cleaved (Fig. 1C). Indeed, when VP3 was expressed as a component of the wild-type polyprotein, protease digestion gave rise to an additional lower-molecular-mass form of VP3. When fused to the GFP, in addition to the fused protein, the additional band was still present, demonstrating the existence of a cleavage site near the C terminus of VP3. All further experiments have thus been carried out with protease inhibitors.
FIG. 1.
Construction of recombinant baculoviruses expressing the fusion polyproteins IBDA-LUC, IBDA-GFP, and IBDA-OVA. (A) Schematic representation of the three constructs expressing the IBDA polyproteins fused at their C terminus with luciferase (LUC), GFP, and ovalbumin (OVA). (B) Expression of the different chimerical polyproteins. (Left) The constructs were expressed in the rabbit reticulocyte expression system (Promega), and expression products were analyzed by SDS-PAGE. The different fusion proteins are cotranslationally processed to give rise to pVP2, VP4, and the VP3 proteins fused at their C terminus to luciferase, GFP, and ovalbumin. (Right) Immunoprecipitation analyses using an anti-VP3 antibody. Sf9 cells were infected with recombinant baculoviruses. Immune complexes were analyzed by SDS-PAGE under reducing conditions. The gel was stained with Coomassie blue. The relative molecular weights (shown in thousands) were determined by reference to marker proteins. HC and LC indicate the positions of the heavy and light chains of the immunoglobulins, respectively. Note the presence of a unique immunoprecipitated band in each lane, suggesting that no cleavage occurs inside the fusion proteins in infected cells incubated with an antiprotease inhibitor cocktail. (C) Protease susceptibility of VP3 and VP3-GFP visualized by SDS-PAGE. VP3 and VP3-GFP were expressed, extracted in the absence or presence of protease inhibitors, and immunoprecipitated. For each case, the first lane is the control immunoprecipitation without antigens.
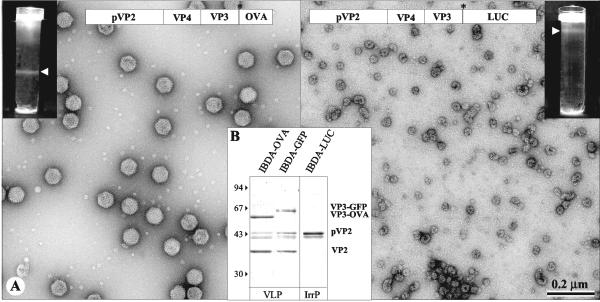
To analyze the type of assembly produced by the two chimerical molecules IBDA-OVA and IBDA-LUC, we expressed the two polyproteins and extracted and purified the resulting assemblies by density gradient centrifugation. In parallel, IBDA-GFP VLPs were purified by the same procedure. As shown in Fig. 2, a band located at different positions in the gradient was visible for both IBDA-OVA and IBDA-LUC extractions. Electron microscopy of negatively stained preparations of the IBDA-OVA band revealed the presence of a large number of VLPs with a diameter of about 60 nm (Fig. 2A, left panel). These particles were similar to VLP-GFP and viral particles (4). In contrast, observation of the IBDA-LUC band showed irregular, rather isometric, particles with a diameter of about 30 nm (Fig. 2A, right panel). SDS-PAGE associated with MALDI-TOF analysis showed that the major components of the VLPs-OVA were the VP3-OVA fusion protein and VP2 (Fig. 2B). pVP2 and cleaved intermediates were also identified. These results were similar to those obtained when GFP was fused (Fig. 2B, lane 3) (4). In the case of IBDA-LUC, a single 48-Da protein identified as pVP2 formed the irregular particles.
FIG. 2.
Analysis of the structures produced by BacΔIBDA-OVA and BacΔIBDA-LUC. (A) Electron microscopy of the samples purified by CsCl gradient centrifugation. The gradients were illuminated and photographed (inserts). (Left) BacΔIBDA-OVA. (Right) BacΔIBDA-LUC. Material collected from the band present in the gradients was negatively stained with 2% uranyl acetate. Large numbers of VLP-OVA proteins were observed, whereas small irregular particles were obtained with BacΔIBDA-LUC. (B) Polypeptide identification. SDS-PAGE analysis and Coomassie blue staining of the purified VLP-OVA and VLP-GFP and of the irregular particles (IrrP) collected from the IBDA-LUC band. Protein assignment was carried out by tryptic digestion and MALDI-TOF analysis. VLP-OVA proteins were found containing VP3-OVA, VP2, and small amounts of the pVP2 and pVP2 cleaved forms. The relative molecular weights (in thousands), determined by reference to marker proteins, are indicated on the left.
We could thus conclude that VLP assembly was promoted by the fusion at the C terminus of VP3 of different exogenous sequences, showing that the required signal for VLP assembly was not sequence specific. The absence of VLPs with IBDA-LUC suggested that steric hindrance prevented large proteins from being incorporated within the VLPs.
Characterization of the VP3 domain controlling the VLP assembly.
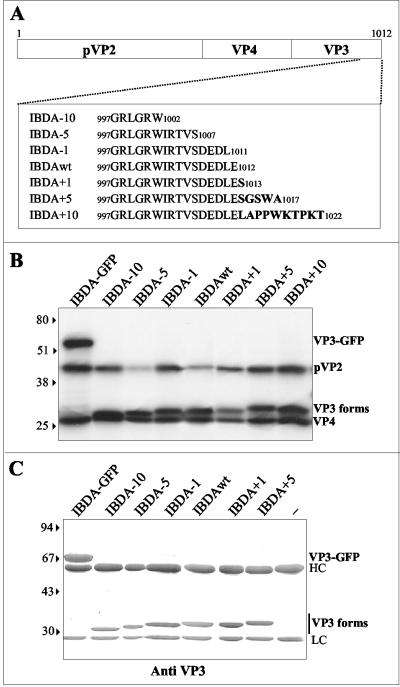
The fact that VLP assembly was promoted by a nonspecific sequence fused at the C terminus of VP3 suggested that the regulation domain was located close to the fusion site. We thus decided to investigate the assemblies generated by several mutants characterized by deletions (−10, −5, and −1 aa) and additions (+1, +5, and +10 aa) at the C terminus of the polyprotein (Fig. 3A). The baculoviruses BacΔIBDA − 10, BacΔIBDA − 5, BacΔIBDA − 1, BacΔIBDA + 1, BacΔIBDA + 5, and BacΔIBDA + 10 expressing the corresponding deletion and addition mutants were engineered. Validation of the constructs was carried out in vitro and in vivo. In vitro expression (Fig. 3B) and cell extract immunoprecipitation (Fig. 3C) showed that the processing of the wild-type and mutated polyproteins yielded the expected cleavage products pVP2 (46 kDa) and VP4 (30 kDa) as well as the different forms of VP3.
FIG. 3.
Expression of IBDA polyproteins modified at the C terminus of VP3. (A) Schematic representation of the constructs expressing the different IBDA polyprotein forms. Deletions of 10, 5, or 1 aa or fusions of 1, 5, or 10 aa were engineered at the C terminus of VP3. The sequence is reported according to the single-letter code. (B) In vitro expression of the different chimeric polyproteins. The constructs were expressed in the rabbit reticulocyte expression system (Promega), and expression products were analyzed by SDS-PAGE. The different fusion proteins were cotranslationally processed to give rise to pVP2, VP4, and the different forms of VP3. (C) Immunoprecipitation analyses using an anti-VP3 antibody. Sf9 cells were infected with recombinant baculoviruses. Immune complexes were analyzed by SDS-PAGE under reducing conditions. The gel was stained with Coomassie blue. The relative molecular weights (shown in thousands) were determined by reference to marker proteins. HC and LC indicate the positions of the heavy and light chains of immunoglobulins, respectively. The last lane is the immunoprecipitation control without antigens. Note the presence of a unique immunoprecipitated band in each lane, suggesting that no cleavage occurred inside the VP3 in infected cells incubated with a protease inhibitor cocktail.
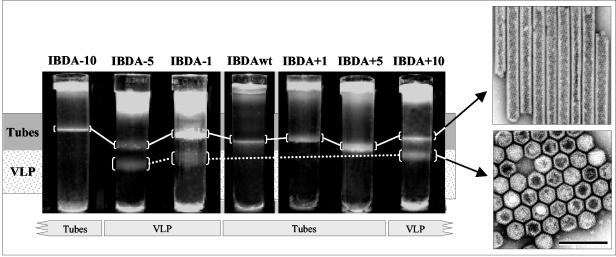
Assemblies resulting from the expression of the different polyproteins were extracted and subjected to density gradient centrifugation. In parallel, the wild-type IBDA was expressed and submitted to the same procedure (Fig. 4). Two types of band profiles in the density gradients were obtained. Whereas a single band characterized wild-type IBDA, ΔIBDA − 10, ΔIBDA + 1, and ΔIBDA + 5, two bands were visible with ΔIBDA − 5, ΔIBDA − 1, and ΔIBDA + 10. Electron microscopy of negatively stained preparations revealed that for preparations displaying two bands, the upper band contained 55-nm diameter tubes, whereas the lower band was made by VLPs. When a single band was present in the gradient, it was composed of tubes. The protein composition of each band was investigated by SDS-PAGE and Western blot analysis (Fig. 5). The major components of the VLPs (the lower band) were VP2 and the different VP3s. The main component of the tubes (the upper band) was pVP2. In all cases, minor components were present: VP2 in the tubes and pVP2 in the VLPs. A faint 28-kDa protein, assigned as the viral protease VP4 by MALDI-TOF analysis, was present in some preparations of tubes, VLPs, and virus (Fig. 5).
FIG. 4.
Electron microscopy analysis of the structures expressed by the recombinant baculoviruses expressing IBDA polyproteins modified at the C terminus of VP3. After an 18-h centrifugation in CsCl at 100,000 × g, the gradients were photographed. Gradients contain one or two bands. The upper bands contain tubes, whereas the lower bands contain VLPs. To promote VLP assembly, one to five residues have to be removed from the C terminus of VP3 or more than five residues have to be added to the C terminus. This demonstrates that an exogenous peptide has to interact with the last 5 aa of VP3 to promote assembly. Concentrated samples giving rise to paracrystalline structures have been negatively stained with 2% uranyl acetate solution. The bar represents 0.2 μm.
FIG. 5.
Biochemical analysis of the structures expressed by the recombinant baculoviruses expressing IBDA polyproteins modified at the C terminus of VP3. Material extracted from each band shown in Fig. 4 was analyzed by SDS-PAGE followed by Coomassie blue staining (A) or Western blot analyses with an anti-pVP2/VP2 antibody or with an anti-VP3 antibody (B). The upper bands, the tubes, mainly contain pVP2, whereas the lower bands, the VLPs, contain VP2 and VP3. The relative molecular weights (in thousands), determined by reference to marker proteins, are indicated on the left.
The efficient assembly of IBDA − 1 and IBDA − 5 mutants into VLPs showed that the last residue of VP3 prevented the VLP assembly process when the wild-type IBDA polyprotein was expressed alone. A large deletion (−10 aa) allowed tube formation, but not VLP assembly, suggesting that the [−10, −6] C-terminal domain of VP3 also played an important role in capsid assembly (to simplify the notation of the amino acid stretches, we defined the position of the amino acid C terminus −1). Addition of more than 5 aa at the C terminus of VP3 also promoted the VLP assembly. These results showed that, in order to induce the formation of VLPs, the [−10, −6] C-terminal domain of VP3 had to be present and the last residue of VP3, glutamic acid 257, had to be deleted, or its electrostatic potential had to be screened through interactions with exogenous peptides. The deletion of the last 10 residues of VP3 only induced pVP2 tube formation, suggesting that the [−10, −6] C-terminal domain of VP3 participated in the pVP2-VP3 interaction.
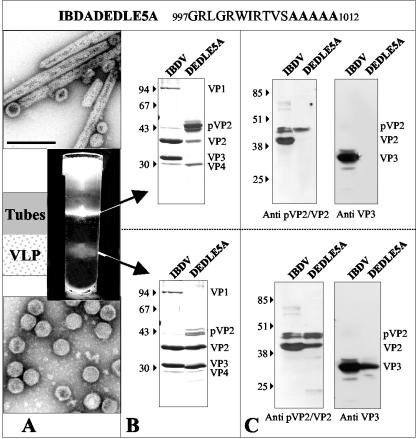
To analyze the role of the negative charges located at the C terminus of VP3 in the assembly process, we engineered a new recombinant baculovirus (BacΔIBDA-DEDLE5A) expressing the IBDA polyprotein in which the last five residues were replaced with alanines. Assemblies of BacΔIBDA-DEDLE5A-infected cells were extracted and subjected to density gradient centrifugation (Fig. 6A). Two bands were observed in the gradient. Electron microscopy of negatively stained preparations revealed tubes as the major component of the upper band and VLPs for the lower band. SDS-PAGE (Fig. 6B) and Western blot analysis (Fig. 6C) showed again that VLPs were mainly made of VP2 and the mutated VP3, whereas tubes were constituted of pVP2. These results suggested that in the absence of exogenous proteins or peptides, the negative charges of the last glutamic and aspartic residues at the C terminus of VP3 prevented the VLP assembly. In conclusion, VLP assembly required these charged residues to be deleted, replaced, or shielded by an exogenous protein. Because the single deletion of the glutamic acid 257 was sufficient for efficient capsid assembly, we could conclude that the last residue of VP3 played an important assembly control role during capsid morphogenesis.
FIG. 6.
Analysis of the structures produced by BacΔIBDA-DEDLE5A. Sf9 cells were infected with BacΔIBDA-DEDLE5A for treatment with Freon 113 as described in Materials and Methods. After 18 h of centrifugation in a CsCl density of 1.30 at 35,000 rpm, the gradient were illuminated with white light and photographed (A). Material collected from the bands present in the gradients was negatively stained with 2% uranyl acetate. Large numbers of VLPs were observed in the lower band, whereas tubes were mainly observed in the upper band. The bar represents 0.2 μm. (B) SDS-PAGE analysis and Coomassie blue staining of purified IBDV and of the material present in the lower and upper bands. (C) Western blot analyses of purified virus and of the material present in the lower and in the upper bands with an anti-pVP2/VP2 antibody or with an anti-VP3 antibody. The relative molecular weights (in thousands), determined by reference to marker proteins, are indicated on the left.
The [−10, −6] C terminus of VP3 is a VP1 binding domain.
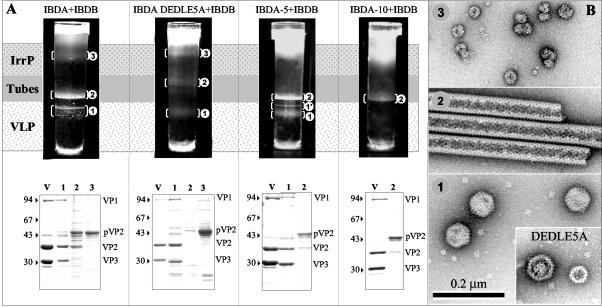
VP3 has been shown to interact with VP1 through at least its last 10 carboxy-terminal residues (12, 18). Coexpression of VP1 with the polyprotein led to the formation of VLPs containing VP1 (13). It could thus be postulated that the VP3-VP1 interactions controlled the assembly of the viral particles. To test this hypothesis and better characterize the interactions between VP1 and the C terminus of VP3, a recombinant baculovirus driving expression of VP1 (BacIBDB) was constructed. First, coinfection of Sf9 cells with BacIBDB and BacΔIBDA was carried out. Cell lysates were extracted, and purified assemblies were submitted to density gradient centrifugation (Fig. 7A). Three bands were observed in the gradient. Electron microscopy revealed that the lower band mainly contains VLPs, the middle band contains tubes, and the upper band contains irregular particles (Fig. 7B). SDS-PAGE and MALDI-TOF analysis showed that VLPs contained the capsid proteins VP2 and VP3 as well as the polymerase VP1. While the major component of VLPs was VP2, tubes and irregular particles were mainly constituted by pVP2. The assembly process was associated with maturation of pVP2 to VP2. VP1 thus promoted the assembly of VP2 and VP3 into VLPs containing VP1. Second, VP1 and IBDA polyproteins mutated at their C termini were coexpressed. Coinfections of Sf9 cells with BacIBDB and BacΔIBDA − 5, or BacΔIBDA-DEDLE5A were carried out, cell lysates were extracted, and the assemblies were further purified by density centrifugation (Fig. 7A). When VP1 was coexpressed with IBDA − 5 or IBDA-DEDLE5A, three bands were identified in the gradients. Electron microscopy of negatively stained preparations showed that the lower bands (band 1) contained VLPs (Fig. 7B). In the case of IBDA-DEDLE5A, small VLPs having a diameter of about 40 nm were also detected in this band. For IBDA − 5, a minor band (1′) was visualized and contained VLPs like the major VLP band (band 1). The middle band (band 2) contained tubes, and the upper band (band 3) contained irregular particles. For IBDA-DEDLE5A, the tube band also contained disk-like structures. These structures, occasionally present in different preparations, were not further characterized. Coexpression of VP1 and IBDA − 10 did not give rise to VLPs, but instead gave rise to tubes. Importantly, SDS-PAGE and MALDI-TOF analysis showed that the VLPs contained VP2, the mutated VP3 and VP1 (Fig. 7A). The tubes and the irregular particles were mainly constituted of pVP2. Due to the lack of material, the protein composition of band 1′ was not ascertained. Since it has been shown that only the last 10 C-terminal residues of VP3 were involved in VP1 interaction (18), the fact that IBDA-DEDELE5A and IBDA − 5 VLPs contained VP1 showed that the [−10, −6] C-terminal domain of VP3 was sufficient to allow VP1 interaction. The fact that small VLPs were obtained with IBDA-DEDLE5A suggested that the last five residues were also implicated in the control of the capsid geometry.
FIG. 7.
Analysis of the structures produced by coexpression of the VP1 polymerase with the IBDA polyprotein and the mutated forms IBDA-DEDLE5A, IBDA − 5, and IBDA − 10. (A, upper panel) After 18 h of centrifugation in a CsCl density of 1.30 at 35,000 rpm, the gradients were illuminated and photographed. The visualized bands were numbered (1 to 3) according to the assemblies they were shown to contain (see panel B). (A, lower panel) SDS-PAGE and Coomassie blue staining of IBDV virions (V) and of the material present in the different bands. Protein assignment was carried out by trypsin digestion and MALDI-TOF analysis. The presence of VP1 was thus ascertained in the different types of VLPs. The relative molecular weights (in thousands), determined by reference to marker proteins, are indicated on the left. IrrP, irregular proteins. (B) Material collected from the band present in the gradients was negatively stained with 2% uranyl acetate. Large numbers of VLPs in band 1, tubes in band 2, and irregular particles in band 3 were observed. For IBDA-DEDLE5A, disk-like structures were also observed. Due to the small amount of material in band 1′, it could not be differentiated from band 1. Note that with the DEDLE5A mutant, small capsids with a different geometry were identified.
DISCUSSION
When expressed in recombinant expression systems, the capsid proteins of many viruses self-assemble, producing VLPs. In the case of IBDV, although the IBDA polyprotein contains the two capsid proteins VP2 and VP3, its expression in insect cells did not result in efficient assembly of VLPs. The polyprotein led to the formation of tubes mainly constituted of pVP2, the precursor of the external capsid protein VP2. These tubes, which had a diameter of about 55 nm, were morphologically indistinguishable from the type I tubes identified in IBDV-infected cells (4, 14). We previously showed that when an exogenous sequence (the GFP sequence) was fused to the C terminus of VP3, the capsid proteins were able to self-assemble into VLPs. These particles had the same diameter and apparent geometry as the virions, and, remarkably, full processing of VP2 occurred during the formation of these structures (4). Here, we showed that replacement of the GFP sequence with an unrelated sequence (a truncated ovalbumin) at the C terminus of VP3 also allowed efficient capsid assembly and VP2 processing. These results suggested that the capsid assembly signal was not sequence specific and censored by a domain close to the fusion site, the C terminus of VP3.
The C terminus of VP3 controls capsid assembly.
We showed that the last C-terminal residues [−5, −1] of VP3 negatively controlled the capsid assembly process when the IBDA polyprotein was expressed without any other viral components. While the expression of the wild-type polyprotein gave rise to tubes, expression of a polyprotein in which the last five C-terminal residues of VP3 were deleted resulted in VLP formation. These particles had the same diameter and apparent geometry as the virions and were constituted by deleted VP3s and VP2, demonstrating a full processing of pVP2 during the formation of these structures. When a larger truncation was made at the C terminus of VP3, VLP assembly did not occur, suggesting that the [−10, −6] C-terminal domain of VP3 was essential to the pVP2/VP2-VP3 interaction. In addition, our data suggested that the last 5 aa constituted a negative control to capsid assembly. Deletion, substitution, or shielding of the C terminus of VP3 through association with exogenous proteins or peptides suppressed the negative constraint and allowed VLP assembly.
We confirmed that the VP3 C terminus is highly susceptible to proteases (13), even in the presence of a fusion protein. However, we demonstrated that the VP3 cleavage was abrogated when protease inhibitors were added during infection and extraction of the expressed assemblies. All experiments were thus carried out with protease inhibitors. Under these conditions, VP3 cleavage was inhibited and did not thus interfere with the assembly processes. Our data with recombinant polyproteins showed that capsid assembly was indeed regulated through interactions between VP3 and an exogenous protein. During the virus life cycle, it may be postulated that VP1 plays the role of the exogenous sequence, regulating the capsid assembly process. Indeed, VP1-VP3 interactions have been shown to extend over the last 10 aa residues of VP3 (18) and to promote capsid assembly.
The C terminus of VP3 presents two functional domains: VP1 (and possibly pVP2/VP2) binding and assembly control.
We confirmed that VP1 leads to VLP assembly and is encapsidated during this process (13). Having determined that the last five C-terminal residue of VP3 controlled VLP formation, we studied the encapsidation of VP1 into VLPs containing VP3 that had been truncated or had substitutions on its last five residues. We showed that VLPs composed of mutated VP3s did indeed contain VP1, demonstrating that the C terminus of VP3 presented two domains: one involved in the VP1 binding and a second involved in the control of capsid assembly. We furthermore observed that the last domain played a role in determining the capsid geometry. While the assembly control domain extended over the last 5 aa of VP3, the VP1 binding domain extended over the upstream five residues. The fact that the expression of IBDA − 10 gave rise to pVP2 tubes while expression of IBDA − 5 resulted in VLP formation suggested that the [−10, −6] VP1 binding domain of VP3 could also interact with pVP2/VP2. In VLPs and virions, VP1 molecules were present in a much smaller amount than VP3. This observation might suggest that the VP3 C-terminus shielding by VP1 was required on a limited number of VP3 molecules to promote capsid assembly. Alternatively, it may be thought that VP1 might bind to several VP3 molecules. In all cases, due to its critical role in capsid morphogenesis, VP1 could be considered an integral capsid protein, suggesting that the capsid may play a role in replication, as well as in genome packaging. Reverse genetics studies have shown that the last residues of VP3 are necessary to the virus rescue (18). This observation suggested that the assembly control domain is involved in another crucial function such as polymerase regulation or genomic packaging or release.
pVP2 processing and VLP formation.
VLP formation and pVP2 processing were two correlated processes, showing that the viral protease VP4, which is associated with VP2 maturation, was also important for virus assembly.
In conclusion, we have shown that IBDV capsid assembly is a complex process regulated by a single amino acid located at the C terminus of VP3 and by interaction with the viral polymerase VP1. Finally, while the last 5 aa of VP3 appeared to determine the capsid architecture, the five residues immediately upstream [−10, −6] were shown to be involved in VP1 binding and possibly in interactions with pVP2/VP2.
Acknowledgments
We thank J.-F. Bouquet and F.-X. Legros (Mérial) for providing monoclonal antibody 1C6 specific for pVP2/VP2 and IBDV, N. Eterradossi (AFSSA) for monoclonal antibody 20 specific for VP3, C. Henry (INRA) for performing the MALDI-TOF analyses, and Wendy Brandt-Williams (INRA) and Félix Rey (CNRS) for critical reading of the manuscript.
REFERENCES
- 1.Birghan, C., E. Mundt, and A. E. Gorbalenya. 2000. A non-canonical lon proteinase lacking the ATPase domain employs the Ser-Lys catalytic dyad to exercise broad control over the life cycle of a double-stranded RNA virus. EMBO J. 19:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Böttcher, B., N. A. Kiselev, V. Y. Stel'Mashchuk, N. A. Perevozchikova, A. V. Borisov, and R. A. Crowther. 1997. Three-dimensional structure of infectious bursal disease virus determined by electron cryomicroscopy. J. Virol. 71:325-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle, J. S., C. Koniaras, and A. M. Lew. 1997. Influence of cellular location of expressed antigen on the efficacy of DNA vaccination: cytotoxic T lymphocyte and antibody responses are suboptimal when antigen is cytoplasmic after intramuscular DNA immunization. Int. Immunol. 9:1897-1906. [DOI] [PubMed] [Google Scholar]
- 4.Chevalier, C., J. Lepault, I. Erk, B. Da Costa, and B. Delmas. 2002. The maturation process of pVP2 requires assembly of infectious bursal disease virus capsids. J. Virol. 76:2384-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cosgrove, A. S. 1962. An apparently new disease of chicken—avian nephrosis. Avian Dis. 6:385-389. [Google Scholar]
- 6.Da Costa, B., C. Chevalier, C. Henry, J.-C. Huet, S. Petit, J. Lepault, H. Boot, and B. Delmas. 2002. The capsid of infectious bursal disease virus contains several small peptides arising from the maturation process of pVP2. J. Virol. 76:2393-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Granzow, H., C. Birghan, T. C. Mettenleiter, J. Beyer, B. Köllner, and E. Mundt. 1997. A second form of infectious bursal disease virus-associated tubule contains VP4. J. Virol. 71:8879-8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harkness, J. W., D. J. Alexander, M. Pattison, and A. C. Scott. 1975. Infectious bursal disease agent: morphology by negative stain electron microscopy. Arch. Virol. 48:63-73. [DOI] [PubMed] [Google Scholar]
- 9.Lejal, N., B. Da Costa, J.-C. Huet, and B. Delmas. 2000. Role of Ser-652 and Lys-692 in the protease activity of infectious bursal disease virus VP4 and identification of its substrate cleavage sites. J. Gen. Virol. 81:983-992. [DOI] [PubMed] [Google Scholar]
- 10.Leong, J. C., D. Brown, P. Dobos, F. S. B. Kibenge, J. E. Ludert, H. Müller, E. Mundt, and B. Nicholson. 2000. Family Birnaviridae, p. 481-490. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, D. J. McGeoch, J. Maniloff, M. A. Mayo, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the International Committee on the Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 11.Lombardo, E., A. Maraver, J. R. Castón, J. Rivera, A. Fernández-Arias, A. Serrano, J. L. Carrascosa, and J. F. Rodriguez. 1999. VP1, the putative RNA-dependent RNA polymerase of infectious bursal disease virus, forms complexes with the capsid protein VP3, leading to efficient encapsidation into virus-like particles. J. Virol. 73:6973-6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maraver, A., R. Clemente, J. F. Rodriguez, and E. Lombardo. 2003. Identification and molecular characterization of the RNA polymerase-binding motif of infectious bursal disease virus inner capsid protein VP3. J. Virol. 77:2459-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maraver, A., A. Oña, F. Abaitua, D. González, R. Clemente, J. A. Ruiz-Diaz, J. R. Castón, F. Pazos, and J. F. Rodriguez. 2003. The oligomerization domain of VP3, the scaffolding protein of infectious bursal disease virus, plays a critical role in capsid assembly. J. Virol. 77:6438-6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez-Torrecuadrada, J. L., J. R. Caston, M. Castro, J. L. Carrascosa, J. F. Rodriguez, and J. I. Casal. 2000. Different architectures in the assembly of infectious bursal disease virus capsid proteins expressed in insect cells. Virology 278:322-331. [DOI] [PubMed] [Google Scholar]
- 15.Özel, M., and H. Gelderblom. 1985. Capsid symmetry of viruses of the proposed Birnavirus group. Arch. Virol. 84:149-161. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez, A. B., and J. F. Rodriguez. 1999. Proteolytic processing in infectious bursal disease virus: identification of the polyprotein cleavage sites by site-directed mutagenesis. Virology 262:190-199. [DOI] [PubMed] [Google Scholar]
- 17.Tacken, M. G., P. J. Rottier, A. L. Gielkens, and B. P. Peeters. 2000. Interactions in vivo between the proteins of infectious bursal disease virus: capsid protein VP3 interacts with the RNA-dependent RNA polymerase, VP1. J. Gen. Virol. 81:209-218. [DOI] [PubMed] [Google Scholar]
- 18.Tacken, M. G., B. P. Peeters, A. A. M. Thomas, P. J. Rottier, and H. J. Boot. 2002. Infectious bursal disease virus capsid protein VP3 interacts both with VP1, the RNA-dependent RNA polymerase, and with viral double-stranded RNA. J. Virol. 76:11301-11311. [DOI] [PMC free article] [PubMed] [Google Scholar]