Abstract
The motor neuron (MN)–hexamer complex consisting of LIM homeobox 3, Islet-1, and nuclear LIM interactor is a key determinant of motor neuron specification and differentiation. To gain insights into the transcriptional network in motor neuron development, we performed a genome-wide ChIP-sequencing analysis and found that the MN–hexamer directly regulates a wide array of motor neuron genes by binding to the HxRE (hexamer response element) shared among the target genes. Interestingly, STAT3-binding motif is highly enriched in the MN–hexamer–bound peaks in addition to the HxRE. We also found that a transcriptionally active form of STAT3 is expressed in embryonic motor neurons and that STAT3 associates with the MN–hexamer, enhancing the transcriptional activity of the MN–hexamer in an upstream signal-dependent manner. Correspondingly, STAT3 was needed for motor neuron differentiation in the developing spinal cord. Together, our studies uncover crucial gene regulatory mechanisms that couple MN–hexamer and STAT-activating extracellular signals to promote motor neuron differentiation in vertebrate spinal cord.
Keywords: LIM homeodomain factor, LIF, Lhx3, Isl1, NLI
The combinatorial action of transcription factors is a prevalent strategy for achieving cellular complexity in the CNS. However, how the combinatorial action of transcription factors leads to the expression of distinct batteries of terminal differentiation genes, which together establish a specific cellular identity; how the cell fate-specifying transcription factors interact with extracellular cues remain unclear. To address these questions, it is essential to identify both the cis-regulatory elements in the genome, which recruit a specific combination of transcription factors, and the target genes associated with those cis-regulatory elements.
One of the best examples of combinatorial transcription codes has emerged from studies of spinal motor neuron (MN) development (1). Two LIM-homeodomain (LIM-HD) factors, LIM homeobox 3 (Lhx3) and Islet-1 (Isl1), are vital for directing MN fate specification in the developing spinal cord (2–5). During this process, two Isl1:Lhx3 dimers bind to nuclear LIM interactor (NLI, also known as LDB for LIM domain binding) that has a self-dimerization domain, thereby forming the MN–hexamer complex (Fig. 1A and Fig. S1A) (2, 6). The combinatorial expression of Lhx3 and Isl1 is capable of triggering MN specification in chick spinal cord, ES cells (ESCs), and induced pluripotent stem cells (2, 6–8). In contrast to MNs, during the specification of V2 interneurons, two Lhx3s and two NLIs form a tetrameric complex, which directs the V2-interneuron fate (Fig. S1A) (2, 9). Thus, the combinatorial action of Lhx3 and Isl1, via the formation of the MN–hexamer, is critical to determine MN identity over V2-interneuron fate. However, key questions remain unanswered. First, does the MN–hexamer directly control terminal differentiation genes that are required for consolidating the functional identity of MNs? Second, does the MN–hexamer collaborate with other transcription factors and/or extracellular signals to control its downstream target genes for MN differentiation? Identification of transcriptional regulators and extrinsic cues cooperating with the MN–hexamer would provide important insights into MN development. In this regard, it is noteworthy that leukemia inhibitory factor (LIF) signaling and its downstream transcription factor STAT3 have been implicated in MN survival and regeneration following nerve injury (10–13). However, little is known about the role of STAT3 and LIF signaling in MN differentiation. LIF family cytokines bind receptor complexes, including Interleukin 6 Signal Transducer (IL6ST, also known as gp130) and LIF receptor (LIFR) subunits, and activate JAKs, which subsequently phosphorylate STAT3 transcription factors and enable STAT3 to activate its target genes (14).
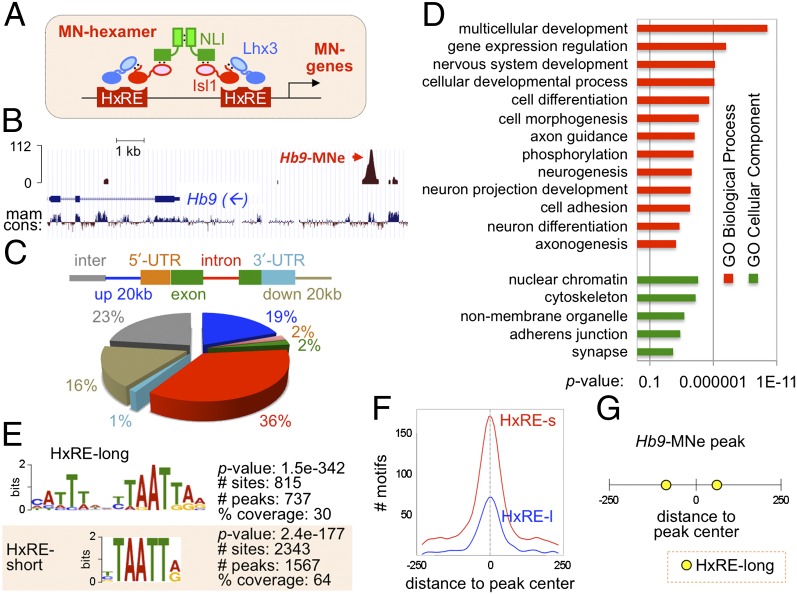
Fig. 1.
Genome-wide mapping of the MN–hexamer targets. (A) Schematic representation of the MN–hexamer consisting of two Lhx3, two Isl1, and two NLI molecules. We hypothesize that, in differentiating MNs, the MN–hexamer binds to the HxRE in MN genes and induces their expression. (B) The ChIP-seq analysis identified a strong Isl1-Lhx3–bound peak in the MN–specific enhancer of Hb9 gene (Hb9-MNe), the only known MN–hexamer–binding site. mam cons, mammalian conservation index. The arrow indicates transcription direction of the Hb9 gene. (C) Peak locations with respect to RefSeq gene models. The gene body includes all exons and introns. Percent of peaks in each category is shown in the pie chart. Inter, intergenic (more than 20 kb from the gene body). (D) GO term analysis of the genes associated with Isl1-Lhx3–bound peaks. X-axis shows P value. (E) HxRE-long and HxRE-short motifs are enriched in 2,455 Isl1-Lhx3–bound peaks. P value, the enrichment of each binding motif in comparison with the local background; # sites, total number of each motif found in 2,455 peaks; # peaks, number of peaks that contain at least one given motif; % coverage, the proportion of peaks that contain at least one motif among the total 2,455 peaks. (F) Graphical representation of the frequency of each motif in relation to the central position (0 in x axis) of all Isl1-Lhx3–bound peaks. The y axis shows the number of motifs in each position. HxRE-long (HxRE-l) and HxRE-short (HxRE-s) motifs are highly concentrated toward the center of the peaks. (G) Schematic representation of the location of HxRE motifs in the peak covering the Hb9-MNe region. The number shows the relative position within the peak (0, the central position). Two HxRE-long motifs are found in ±70 positions.
In this study, we uncovered a genome-wide map of MN–hexamer–binding sites via ChIP-sequencing (seq) methods. This led to the finding that the MN–hexamer directly activates terminal differentiation genes. Furthermore, the ChIP-seq analyses also revealed STAT3, which is activated by LIF-signaling, as a major collaborating factor of the MN–hexamer in MN development. Together, our findings provide critical insights into the gene regulatory networks for MN fate specification in which the MN–hexamer forms a feed-forward regulatory network with STAT3.
Results
Genome-Wide Mapping of the MN–Hexamer–Binding Regions.
To identify direct targets of the MN–hexamer and transcription factors collaborating with the MN–hexamer in an unbiased and comprehensive manner, we performed genome-wide ChIP-seq analysis of MN–hexamer target genes. To immunopurify only MN–hexamer targets that recruit Isl1 and Lhx3 simultaneously, we adopted mouse ESC lines that express a FLAG-tagged Isl1-Lhx3 fusion protein upon doxycycline (Dox) treatment, named inducible MN-ESCs (iMN-ESCs) (8). The Isl1-Lhx3 construct consists of full-length Isl1 and Lhx3 connected by a short linker and therefore keeps an optimal equi-molar ratio of Isl1 and Lhx3 in forming the MN–hexamer complex with endogenous NLI in cells (Fig. S1A). Consequently, Isl1-Lhx3 is highly efficient and specific in inducing MN differentiation in chick neural tube and mouse ESCs (8), suggesting that Isl1-Lhx3 forms a functional MN–hexamer complex that activates the expression of target MN genes. The inclusion of a Flag epitope in the Isl1-Lhx3 construct makes it possible to use readily available high quality α-FLAG antibodies for ChIP. Thus, iMN-ESCs provide excellent tools to identify MN–hexamer targets using ChIP-seq.
We purified the MN–hexamer–bound chromatin fragments by a single ChIP with α-Flag antibody from Dox-induced iMN-ESCs, in which FLAG-Isl1-Lhx3 was highly expressed (Fig. S1B). As a negative control, we generated a ChIP-seq library from FLAG-immunoprecipitated chromatin from non-Dox treated iMN-ESCs that did not express FLAG-Isl1-Lhx3. Comparison of Isl1-Lhx3–bound and control ChIP-seq data sets with the QuEST peak calling algorithm (15) identified a final set of 2,455 unique genomic regions, called “peaks,” which were significantly enriched in the Dox-treated ChIP-seq library compared with the Dox-untreated negative control library, at a false discovery rate <0.0051 (Dataset S1). As a validation of our approach, a strong peak was detected in MN-specific enhancer of the Hb9 gene (Hb9-MNe) that we have previously identified as an MN–hexamer target (6, 16, 17) (Fig. 1B). Sequences of Isl1-Lhx3–bound peaks displayed a significantly higher degree of evolutionary conservation than randomly chosen sequences (P value <2.2e−16), implying functional significance of the peaks.
The Isl1-Lhx3–bound peaks are distributed throughout the gene bodies, in the vicinity of the gene-coding regions (within 20 kb from a RefSeq transcription unit), and in intergenic regions (Fig. 1C). To discover the potential targets of the MN–hexamer, we identified genes that have the Isl1-Lhx3–bound peaks in the gene bodies or within 20 kb from a RefSeq transcription unit (Dataset S2). If no gene was found within the selected range from a peak, the nearest genes to the peak were chosen as potential targets. The analysis of gene ontology (GO) terms for the target genes within 20-kb range or nearest genes from the ChIP-seq peaks revealed that the MN–hexamer targets are highly enriched for the genes involved in nervous system development, cell differentiation, cell morphogenesis, neurogenesis, axon guidance, cell adhesion, and axonogenesis (Fig. 1D). These data suggest that the MN–hexamer directly controls a wide battery of genes involved in essential aspects of MN differentiation.
Next, we performed the analyses to combine the ChIP-seq data with the RNA-seq data, which identified transcriptome changes upon expression of Isl1-Lhx3 in iMN-ESC–derived spinal neurons (8). Among 975 differentially expressed genes with cutoff of at least 1.5-fold changes, 281 genes have Isl1-Lhx3–bound peaks within 20 kb from their transcription units. Seventy-seven percent of these genes (167 genes) were up-regulated by Isl1-Lhx3 (Dataset S3), consistent with the idea that the MN–hexamer functions mainly as a transcriptional activator complex (6, 16). Thus, our analyses identified highly likely direct target genes of the MN–hexamer, which recruit Isl1-Lhx3 and, at the same time, are induced by Isl1-Lhx3 in their expression. Notably, the ChIP score was not directly correlated with fold induction of associated genes (Fig. S2A), suggesting that stronger binding of Isl1-Lhx3 does not necessarily result in a higher induction of the target gene.
Identification of the in Vivo MN–Hexamer–Binding Motifs.
To identify in vivo motifs recruiting the MN–hexamer, we used two complementary motif discovery algorithms, Multiple Expectation Maximization for Motif Elicitation (MEME) (18) and Discriminative Regular Expression Motif Elicitation (DREME) (19), on a total of 2,455 peaks. The MEME algorithm uses expectation maximization to discover probabilistic models of DNA-binding by single transcription factors or transcription factor complexes, increasing the chance to identify de novo sequence patterns in ChIP-seq peaks. The DREME algorithm uses a nonprobabilistic model, thus complementing MEME with its ability to find very short motifs that MEME often fails to detect. These analyses discovered a 15-nucleotide-long motif, termed HxRE (hexamer response element)-long in ∼30% of the Isl1-Lhx3–bound peaks and a shortened HxRE sequences, named HxRE-short (Fig. 1E and Dataset S4). A 2,343 HxRE-short motif was detected in 1,567 Isl1-Lhx3–bound peaks (∼64% of total peaks), indicating that a substantial fraction of the peaks have multiple HxRE-short motifs. When we combined the counting of HxRE-long and HxRE-short, 2,648 HxRE sites were found in ∼69% of the Isl1-Lhx3–bound regions (1,687 peaks). Around 41% HxRE-containing peaks have at least two HxRE motifs. Both HxRE-long and HxRE-short motifs are strongly enriched around the center of the 500-bp-long peaks (position 0 in Fig. 1F), indicating that these HxREs serve as a motif directly recruiting the MN–hexamer to these genomic regions. The MEME analysis identified two HxRE-long motifs in 500-bp Hb9-MNe, which we have previously mapped as the binding sites for the MN–hexamer using mutational analyses (16) (Fig. 1G). These two HxREs are ∼150 bp apart and evenly spaced from the summit of the peak. Each of HxREs in Hb9-MNe is critical for the MN-specific enhancer activity in the developing embryos (16), implying that the HxRE motifs that we identified are functional in vivo. Together, our motif analyses uncovered in vivo response elements of the MN–hexamer, providing important insights into the mechanisms to control MN gene transcription.
Enrichment of Binding Sites for STAT3 in Isl1-Lhx3–Bound Peaks.
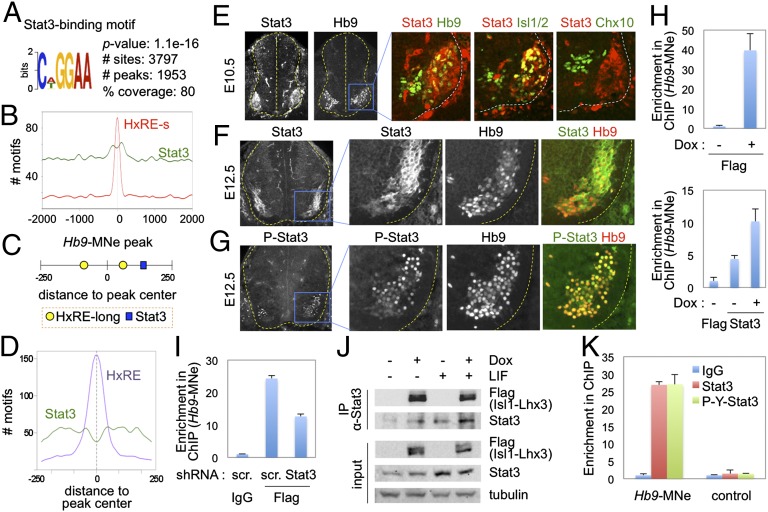
To systematically identify the transcription factors that cooperate with the MN–hexamer, we compared the motifs, which were identified to be significantly enriched in the Isl1-Lhx3–bound peaks by MEME and DREME algorithms, to a database of known transcription factor–binding motifs using the TOMTOM algorithm (20). One of the most abundant non-HxRE motifs in the peaks was STAT3 binding site (Fig. 2A and Fig. S2B). A total of 3,797 STAT3 sites were found in ∼80% of total peaks, many of which have multiple STAT3 sites (Dataset S4). Unlike HxRE motifs that are enriched in −100 bp to +100 bp positions relative to peak summit, STAT3 motif is more broadly distributed, being significantly enriched within 300 bp from the center of the peaks, compared with the genomic regions outside of these areas (Fig. 2B). Approximately 79% of HxRE-containing peaks also have at least one STAT3 motif, including the Isl1-Lhx3–bound peak in Hb9 (Fig. 2C). In these peaks that have both HxRE and STAT3 sites, HxRE motifs are enriched around the summit of the peaks, whereas STAT3 site is broadly found with a relatively low frequency in the center (Fig. 2D). These results suggest that HxRE serves as a direct binding site for the MN–hexamer and that STAT3 is corecruited with the MN–hexamer to the MN–hexamer–target genes.
Fig. 2.
ChIP-seq and expression studies suggest roles of STAT3 in MN development. (A) The significant enrichment of STAT3-binding motif in 2,455 Isl1-Lhx3–bound peaks. P value, the enrichment of STAT3 motif in comparison with the local background; # sites, total number of the STAT3 motif found in 2,455 peaks; # peaks, number of peaks that contain at least one STAT3 motif; % coverage, the proportion of peaks that contain at least one STAT3 motif among the total 2,455 peaks. (B) The frequency of HxRE- and STAT3-binding motifs in all Isl1-Lhx3–bound peaks from −2 kb to +2 kb regions from the central position (0 in x axis). The y axis shows the number of motifs in each position. STAT3 site is significantly enriched in −300- to +300-bp regions of the peaks compared with other genomic areas. (C) Schematic representation of the location of the motifs in the Hb9-MNe region. The number shows the relative position within the peak (0, the central position). (D) The frequency of each motif in 1,330 peaks, which contain both HxRE and STAT3 motifs, in relation to the central position (0 in x axis) of the peaks. HxRE motifs are enriched around the summit of the peaks, whereas STAT3 site is broadly found with a relatively low frequency in the center. (E–G) Immunohistochemical analyses in E10.5 (E) and E12.5 (F and G) mouse developing spinal cord. STAT3 is specifically expressed and activated in embryonic MNs. Spinal cords are outlined by dotted lines. (H) ChIP assays in iMN-ESCs with or without induction of FLAG-tagged Isl1-Lhx3. The cells were treated with LIF in all conditions. STAT3 is recruited to Hb9-MNe, and Isl1-Lhx3 expression facilitates STAT3-binding to Hb9-MNe. (I) ChIP assays in P19 cells transfected FLAG-Isl1-Lhx3 and shRNA constructs. The cells were treated with LIF in all conditions. Isl1-Lhx3–binding to Hb9-MNe is reduced when STAT3 is knocked down. scr., scrambled sequences. (J) Coimmunoprecipitation (CoIP) analyses in iMN-ESC–derived MNs, treated with or without Dox and/or LIF. STAT3 is coimmunopurified with Isl1-Lhx3 independently of LIF, which triggers phosphorylation of STAT3. (K) In vivo ChIP assays in E12.5 spinal cords to monitor the recruitment of STAT3 and Y705-phosphorylated STAT3 to the MN–hexamer–bound MN-specific enhancer. The phosphorylated STAT3 was recruited specifically to Hb9-MNe in embryonic spinal cords. Control indicates the genomic area that does not have the MN–hexamer bound peak. Error bars (H, I, and K) represent the SD.
STAT3 Is Highly and Specifically Expressed in Developing MNs.
Our results predict that STAT3 is expressed in embryonic MNs. Indeed, STAT3 was strongly induced at ∼E10 in MNs soon after Isl1+Lhx3+ MNs were born, and was continuously expressed in MNs in high levels at later stage of development (Fig. 2 E and F). To test whether STAT3 is transcriptionally active in MNs, we performed immunohistochemical analyses with an anti–phospho-STAT3 antibody that detects only Tyr705-phosphorylated STAT3, a transcriptionally active form of STAT3. Phosphorylated STAT3 was detected specifically in the nuclei of MNs (Fig. 2G), indicating that STAT3 in embryonic MNs is activated by upstream signals and is capable of enhancing the transcription of its target genes.
STAT3 Associates with the MN–Hexamer and Is Recruited to the MN–Hexamer Target Genes.
The coenrichment of HxRE and STAT3 sites in the MN–hexamer–bound genomic regions and the MN-specific expression of STAT3 suggest that STAT3 could be a partner transcription factor collaborating with the MN–hexamer to regulate at least a subset of MN genes. To test whether STAT3 binds to the MN–hexamer target genomic regions, we monitored the recruitment of STAT3 to a group of Isl1-Lhx3–bound peaks that have both HxRE and STAT3 sites (Fig. S3A) in iMN-ESCs cultured with or without Dox. As expected, Isl1-Lhx3 bound to each peak in Dox-treated iMN-ESCs (Fig. 2H and Fig. S3B). Interestingly, STAT3 was recruited to the STAT3 site-bearing peaks in the absence of Isl1-Lhx3, while STAT3 binding was weakly enhanced upon the expression of Isl1-Lhx3 (Fig. 2H and Fig. S3C), suggesting that the MN–hexamer is not required for STAT3 recruitment to the hexamer target genes that have STAT3 sites but it is capable of promoting STAT3 binding to the targets. Next, we tested whether STAT3 also promotes the MN–hexamer-binding to its targets by monitoring the recruitment of Isl1-Lhx3 to the HxRE+STAT3 site+ peaks in P19 cells transfected with or without STAT3 knockdown vector. Knocking down STAT3 significantly attenuated Isl1-Lhx3 binding to the peak regions (Fig. 2I and Fig. S4), indicating that STAT3 facilitates the recruitment of the MN–hexamer to its target genes. These results prompted us to test the association between the MN–hexamer and STAT3. Interestingly, Isl1-Lhx3 associated with endogenous STAT3 independently of LIF signaling, which triggers phosphorylation of STAT3 at Tyr705 residue and elicits the transcriptional activity of STAT3 in iMN-ESC–derived MNs (Fig. 2J). Together, our data suggest that although the MN–hexamer and STAT3 alone can bind to their respective binding motifs, the association of MN–hexamer and STAT3 facilitates their recruitment to the target MN genes.
To examine the binding of endogenous STAT3 to the MN–hexamer target genes in vivo, we performed ChIP assays using E12.5 mouse embryonic spinal cords using α-STAT3 and α-Tyr705-phosphorylated STAT3 antibodies and found that the phosphorylated form of STAT3 is recruited to the Hb9-MNe (Fig. 2K). These results indicate that STAT3 is recruited to the MN–hexamer target genes in the developing spinal cord.
STAT3 Collaborates with the MN–Hexamer to Activate the MN Gene Enhancer.
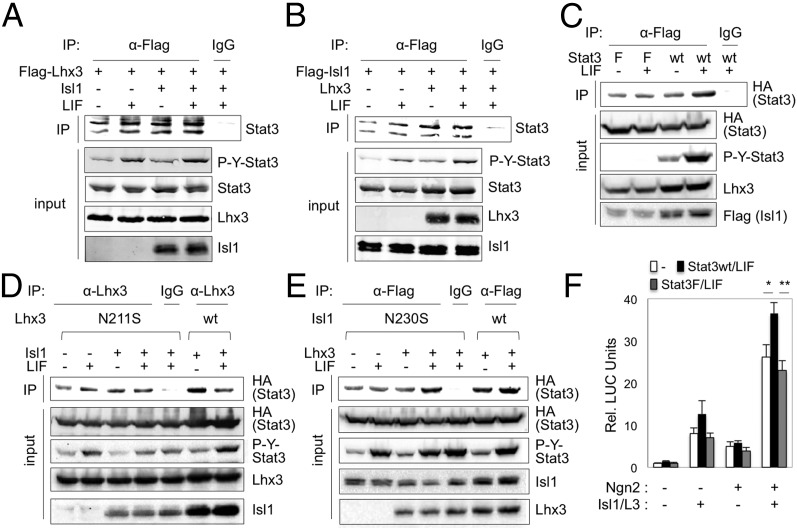
To test whether STAT3 phosphorylation is needed for the association between STAT3 and the MN–hexamer, we expressed HA-tagged STAT3 with Isl1 and/or Lhx3 in HEK293T cells that express NLI endogenously, treated cells with or without LIF, and immunopurified Isl1/Lhx3-associating proteins. Both Isl1 and Lhx3 associated with STAT3 independently of LIF signaling (Fig. 3 A and B). Consistently, STAT3F mutant, in which Tyr705 residue is mutated to phenylalanine and thus is refractory to the signal-dependent STAT3 phosphorylation, was also coimmunopurified with Isl1, similarly to STAT3 WT (Fig. 3C). To test whether DNA binding of the MN–hexamer is required for STAT3/MN–hexamer association, we used DNA-binding defective point mutants of Isl1 and Lhx3, Isl1-N230S, and Lhx3-N211S, respectively (2), for CoIP assays and found that Lhx3-N211S and Isl1-N230S associate with STAT3 (Fig. 3 D and E). Together, our data indicate that the association of STAT3 and the MN–hexamer is independent of STAT3 phosphorylation and the DNA-binding activity of the MN–hexamer.
Fig. 3.
STAT3 associates with the MN–hexamer in a STAT3-Tyr705-phosporylation– and DNA-binding–independent manner, but enhances the MN–hexamer–directed transcription in a STAT3-Tyr705-phosporylation–dependent manner. (A–E) CoIP analyses in HEK293T cells, transfected with vectors as indicated at the top of each panel. The cells were treated with or without LIF 16 h before CoIP assays. Isl1 and Lhx3 associate with STAT3 in cells independently of LIF (A and B) and STAT3-Y705 phosphorylation (C). DNA-binding defective point mutants of Lhx3 and Isl1, Lhx3-N211S and Isl1-N230S, respectively, also associate with STAT3 (D and E). (F) Luciferase reporter assays in P19 cells using Hb9-MNe:LUC reporter with expression vectors as indicated below the graph. STAT3 WT, but not phosphorylation-defective STAT3F mutant, enhanced the activation of Hb9-MNe:LUC by coexpression of Isl1, Lhx3, and Ngn2 in the presence of LIF. Error bars represent the SD. *P < 0.05, **P < 0.005 in two-tailed Student t test.
Although neither STAT3-acativating signals nor STAT3 phosphorylation is required for the association between STAT3 and MN–hexamer, it is possible that they are needed for STAT3-directed transcriptional activation of MN genes. To monitor the effect of STAT3 and STAT3-activating signals on the MN–hexamer–mediated transcriptional activation, we performed luciferase (LUC) reporter assays using Hb9-MNe:LUC in P19 cells. STAT3 activation via expression of STAT3 WT and LIF treatment enhanced the transcriptional activation of Hb9-MNe by Isl1, Lhx3, and Neurogenin2 (Ngn2), whereas nonphosphorylatable STAT3F mutant failed to enhance the MN–hexamer–directed transcriptional activation (Fig. 3F). These data indicate that STAT3 cooperates with the MN–hexamer to activate the MN gene enhancers and that this transcriptional cooperation requires STAT3-Tyr705 phosphorylation by upstream signals.
STAT3 Plays Important Roles for MN Differentiation.
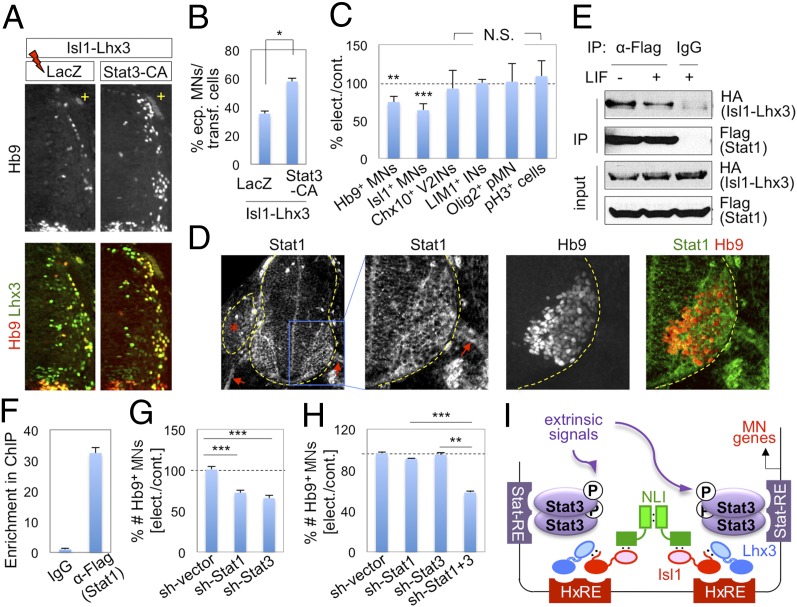
To test whether STAT3 affects the MN–hexamer–mediated MN differentiation during spinal cord development, we examined ectopic MN formation triggered by expression of Isl1-Lhx3, with or without a constitutively active STAT3 (STAT3-CA) that is transcriptionally active independently of upstream signaling, in the developing chick spinal cord (Fig. 4 A and B). Isl1-Lhx3 triggered ectopic MN differentiation in ∼35% of electroporated cells in the dorsal spinal cord. STAT3-CA increased the efficiency of ectopic MN generation to ∼58% when coelectroporated with Isl1-Lhx3, whereas STAT3-CA alone did not induce ectopic MNs. These data indicate that STAT3 is capable of enhancing MN differentiation potential of the MN–hexamer.
Fig. 4.
STAT1 and STAT3 play important roles for MN differentiation in the developing spinal cord. (A and B) The immunohistochemical analyses of ectopic MN formation in chick spinal cord electroporated with Isl1-Lhx3 along with either LacZ control or STAT3-CA. Only the electroporated (+) side of the dorsal neural tube is shown (A). The expression of STAT3-CA increases the efficiency of Isl-Lhx3–triggered ectopic MN formation. (C) Analyses of chick spinal cord electroporated with STAT3-DNs. Inhibition of STAT3 activity using STAT3-DNs resulted in reduction of the number of Hb9+/Isl1+ MNs, but did not change the number of Chx10+ V2 interneurons, Lim1+ interneurons, Olig2+ MN progenitors, or pH3+ proliferation cells. (B and C) Error bars, SD; *P < 0.01, **P < 1.0 × 10−4, ***P < 1.0 × 10−5 in two-tailed Student t test; N.S., no significant change. (D) The immunohistochemical analyses of STAT1 in E10.5 mouse developing spinal cord. *Dorsal root ganglion; arrows indicate motor axons. (E) CoIP analyses in HEK293T cells transfected with HA-Isl1-Lhx3 and FLAG-STAT1. STAT1 associates with Isl1-Lhx3 independently of LIF. (F) ChIP assays in P19 cells transfected with Isl1-Lhx3 and FLAG-STAT1 and treated with LIF. STAT1 is recruited to Hb9-MNe. Error bars, SD. (G and H) Knockdown analyses in chicken embryos electroporated with shRNA constructs. Error bars, SEM; **P < 1.0 × 10−4, ***P < 1.0 × 10−5 in two-tailed Student t test. (I) The MN–hexamer and STAT3 collaborate to control MN gene transcription. STAT3 associates with the MN–hexamer and is recruited to the enhancers of MN genes, which have HxREs and STAT3-response elements (STAT-RE). This leads to the transcriptional cooperation to induce the expression of MN genes.
To further examine the role of STAT3 in MN development, we inhibited the action of endogenous STAT3 by expressing dominant negative forms of STAT3 (STAT3-DNs), STAT3F and STAT3D, which lack tyrosine phosphorylation and DNA-binding activity of STAT3, respectively (21), in the chick neural tube. Expression of STAT3-DNs reduced the number of Hb9+ MNs by ∼26%, compared with the unelectroporated control side (Fig. 4C and Fig. S5A). Similar results were also obtained using Isl1 as an MN marker (Fig. 4C), indicating that blockade of STAT3 activity led to a substantial suppression of MN formation. STAT3-DNs neither reduced the number of Olig2+ MN progenitors nor affected cell death or proliferation (Fig. 4C). In addition, STAT3-DNs did not inhibit generation of interneurons that do not express STAT3, such as Chx10+ V2 interneurons and Lim1+ interneurons (Fig. 4C and Fig. S5A).
Together, our data indicate that STAT3 plays important roles in MN specification during spinal cord development.
STAT1 and STAT3 Function Redundantly to Promote MN Differentiation.
Next, we asked whether other STAT proteins, which share DNA-binding motifs with STAT3 (22) and thus could bind to STAT3 site in MN genes, play redundant roles with STAT3 in MN differentiation. Similarly to STAT3, STAT1 expression was induced as MNs were born and was maintained in postmitotic MNs (Fig. 4D). Furthermore, like STAT3, STAT1 associated with Isl1-Lhx3 independently of LIF signaling, and was recruited to the MN–hexamer target Hb9-MNe (Fig. 4 E and F). Knocking down STAT1 or STAT3 individually using shRNA constructs resulted in reduction of Hb9+ MNs by ∼28 and ∼34%, respectively (Fig. 4G and Fig. S5B), suggesting that STAT1 as well as STAT3 are important for MN generation. Moreover, although low concentration of sh-STAT1 or sh-STAT3 construct alone did not cause reduction of MNs, simultaneous knockdown of STAT1 and STAT3 by coelectroporating the same concentration of both constructs decreased the number of Hb9+ MNs by ∼42% (Fig. 4H). Together, our data suggest that STAT1 and STAT3 play redundant roles in promoting MN differentiation.
Discussion
The Role of the MN–Hexamer as a Terminal Selector for the MN Fate.
Past studies have uncovered a network of inductive signaling cues and transcription factors that direct specification of spinal MNs, a critical component of motor circuits (1, 23). Sonic hedgehog (Shh), an inductive cue secreted from the floor plate, and Olig2, a basic helix-loop-helix transcription factor that specifies the MN progenitors, led to up-regulation of Lhx3 and Isl1, two DNA-binding components of the MN–hexamer (24–28). The gain- and loss-of-function studies suggest that the MN–hexamer functions as a terminal selector of the MN fate that is positioned downstream of Shh and progenitor transcription regulators (2, 6–8). However, it remained unknown whether the MN–hexamer directly controls the expression of a terminal differentiation gene battery that establishes unique MN properties or whether it functions as an intermediate activator that turns on a master transcription factor that specifies the MN fate. Moreover, it is unclear whether, and if so how, the MN–hexamer collaborates with other transcription factors and/or extracellular signals to manifest its critical biological actions. Aiming to answer these important questions, we identified comprehensive genome-wide occupancy map of the MN–hexamer using ChIP-seq technology. Our ChIP-seq results indicate that the MN–hexamer directly controls a wide range of terminal differentiation genes that are involved in neurogenesis, axonogenesis, axon guidance, and synaptic function via in vivo cis-regulatory elements that recruit the MN–hexamer. A significant fraction of the peaks has more than one HxRE, consistent with the idea that the MN–hexamer target genes would bear at least two HxREs, each of which binds to Isl1:Lhx3 dimer (Fig. 1A). Notably, the in vivo HxRE motif deduced from the ChIP-seq peaks shows higher degeneracy than the in vitro motif inferred from the SELEX (for systematic evolution of ligands by exponential enrichment) screening method (6), consistent with the notion that SELEX selects only high-affinity sites for the given transcription factors (29). The less optimal nature of the in vivo HxRE motifs could be beneficial to allow other transcription factors, such as Ngn2 (16) and STAT1/3, to cooperate with the MN–hexamer in facilitating the target gene transcription, providing a convergence point with signaling pathways and other transcription factors. In the future, it will be interesting to investigate how the potential targets of the MN–hexamer, identified in this study, control diverse MN properties, such as axon guidance and formation of neuromuscular junctions.
The Transcriptional Regulatory Networks for MN Fate Specification and Differentiation.
Our genome-wide occupancy analysis of the MN–hexamer revealed the cross-talk between the MN–hexamer and STAT proteins in MN gene regulatory networks (Fig. 4I). Our data support a model in which the complex of the MN–hexamer and STAT3 is recruited to the enhancer regions containing both HxREs and STAT-binding motifs in MN genes (Fig. 4I). Then, upon arrival of extrinsic signals, STAT3 enhances the transcriptional activity of the MN–hexamer, facilitating the expression of MN genes. Notably, STAT3 is a latent transcription factor that is activated in response to various extracellular ligands (30). Our studies revealed that STAT3-activating signaling pathway is activated in MNs and that STAT3-activating signaling, which triggers STAT3 Tyr705 phosphorylation, is required for STAT3 to enhance the MN–hexamer–mediated transcriptional activation. Together, the cross-talk between the MN–hexamer and STAT3 provides crucial insights into mechanisms by which extracellular signals influence specification and differentiation of MNs. Our data indicate that STAT1 and STAT3, which recognize the similar STAT-binding motif, play redundant roles in promoting MN differentiation. Consistently, conditional deletion of the STAT3 gene in postmitotic neurons was not sufficient to cause the loss of embryonic MNs, although it led to defects in MN survival after nerve lesion in postnatal life (13). Future analysis of mouse models deficient in multiple STAT genes or upstream signaling molecule common to all STATs will enhance our understanding of the role of LIF-STAT signaling pathway in MN development.
An important aspect of MN fate commitment is to suppress alternative cell fates, especially V2 interneurons that share developmental regulators with MNs along the lineage specification (1). The binding of the MN–hexamer to a MN-specific enhancer in the Hb9 gene, predicted from previous studies (6, 16, 17), was confirmed by ChIP-seq in this study. Combined with the reports that Hb9 inhibits aberrant V2-interneuron genes in MNs (6, 31, 32), these findings support a feed-forward loop model that, besides directly activating MN genes, the MN–hexamer also inhibits the expression of V2-interneuron genes by inducing Hb9 (6).
The MN–hexamer acts in a concerted manner with multiple transcription regulators, such as STAT1/3, Hb9, and Ngn2, to establish the MN development program in which these transcription factors are interconnected by feedback or feed-forward loops that stabilize developmental lineage decision. This intricately connected gene network would allow the MN–hexamer to induce terminal differentiation genes that dictate various functional aspects of MNs.
Materials and Methods
Details are provided in SI Materials and Methods. For ChIP-seq assays, iMN-ESCs were cultured on 0.1% gelatin-coated dishes in the ESC growth media lacking LIF in the presence or absence of doxycycline (2 μg/mL), which induces the expression of FLAG-tagged Isl1-Lhx3, for 1 d. Approximately 4 × 106 cells were subject to ChIP assays. ChIP DNA samples were prepared for sequencing according to the Illumina protocol and sequenced with the Illumina Genome Analyzer IIx. The sequence reads of 50 or 36 bp generated from Illumina GApipeline were mapped to mouse reference genome (NCBI37, mm9) using BWA software (v0.5.8a) with default parameters (33). The peak calling was conducted with QuEST v2.4 software (15) using the “transcription factor” option (bandwidth 30 bp, region size 300 bp) and the recommended peak calling parameters (30-fold Isl1-Lhx3-ChIP to control enrichment for seeding the regions, threefold Isl1-Lhx3-ChIP enrichment for extending the regions, and threefold Isl1-Lhx3-ChIP to control fold enrichment). For the analysis of peak location, we used mm9 RefSeq gene annotation. We assigned the peaks to the exons that exclude untranslated regions, 5′ untranslated regions, 3′ untranslated regions, introns, up to 20 kb upstream from the 5′ end of a transcription start site (TSS), up to 20 kb downstream from the 3′ transcription end site (TES), and intergenic regions (with a distance greater than 20 kb upstream from any TSS or 20 kb downstream from any TES). To associate each peak to genes, the potential target RefSeq genes were identified with the following criterion: at least one ChIP-seq peak located from 20 kb upstream or downstream of a gene. If no gene is found within the selected range from a peak, the nearest genes to the peak were chosen as potential targets. Analysis of GO terms was performed using DAVID (34). The MEME-ChIP Suite (19) was used for motif analysis with some modifications based on its standard protocol.
Supplementary Material
Acknowledgments
We thank Juhee Kim and Seongkyung Seo for excellent technical support, Richard Goodman and Gail Mandel for help with data analysis on the bioinformatics computation facilities, and Rui Chen for help with Chip-seq assays. This work was supported by the National Institutes of Health (NIH)/National Institute of Neurological Disorders and Stroke Grant R01 NS054941, the March of Dimes Foundation, and the Christopher and Dana Reeve Foundation (to S.-K.L.); NIH/NIDDK Grant R01 DK064678 and the World Class University program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology Grant R31-10105 (to J.W.L.); and Research Settlement Fund for the new faculty of Seoul National University, POSCO TJ Park Science Fellowship, Basic Science Research Program Grant 2012R1A1A1001749, and Bio and Medical Technology Development Program Grant 2012M3A9C6050508 of the National Research Foundation funded by the Korean government, and National Research and Development Program for Cancer Control, Ministry of Health and Welfare, Republic of Korea Grant 1220120 (to S.L.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302676110/-/DCSupplemental.
References
- 1.Lee SK, Pfaff SL. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat Neurosci. 2001;4(Suppl):1183–1191. doi: 10.1038/nn750. [DOI] [PubMed] [Google Scholar]
- 2.Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell. 2002;110(2):237–249. doi: 10.1016/s0092-8674(02)00823-1. [DOI] [PubMed] [Google Scholar]
- 3.Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84(2):309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- 4.Sharma K, et al. LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell. 1998;95(6):817–828. doi: 10.1016/s0092-8674(00)81704-3. [DOI] [PubMed] [Google Scholar]
- 5.Tanabe Y, William C, Jessell TM. Specification of motor neuron identity by the MNR2 homeodomain protein. Cell. 1998;95(1):67–80. doi: 10.1016/s0092-8674(00)81783-3. [DOI] [PubMed] [Google Scholar]
- 6.Lee S, et al. A regulatory network to segregate the identity of neuronal subtypes. Dev Cell. 2008;14(6):877–889. doi: 10.1016/j.devcel.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hester ME, et al. Rapid and efficient generation of functional motor neurons from human pluripotent stem cells using gene delivered transcription factor codes. Mol Ther. 2011;19(10):1905–1912. doi: 10.1038/mt.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S, et al. Fusion protein Isl1-Lhx3 specifies motor neuron fate by inducing motor neuron genes and concomitantly suppressing the interneuron programs. Proc Natl Acad Sci USA. 2012;109(9):3383–3388. doi: 10.1073/pnas.1114515109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joshi K, Lee S, Lee B, Lee JW, Lee SK. LMO4 controls the balance between excitatory and inhibitory spinal V2 interneurons. Neuron. 2009;61(6):839–851. doi: 10.1016/j.neuron.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M, Sendtner M, Smith A. Essential function of LIF receptor in motor neurons. Nature. 1995;378(6558):724–727. doi: 10.1038/378724a0. [DOI] [PubMed] [Google Scholar]
- 11.DeChiara TM, et al. Mice lacking the CNTF receptor, unlike mice lacking CNTF, exhibit profound motor neuron deficits at birth. Cell. 1995;83(2):313–322. doi: 10.1016/0092-8674(95)90172-8. [DOI] [PubMed] [Google Scholar]
- 12.Oppenheim RW, et al. Cardiotrophin-1, a muscle-derived cytokine, is required for the survival of subpopulations of developing motoneurons. J Neurosci. 2001;21(4):1283–1291. doi: 10.1523/JNEUROSCI.21-04-01283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schweizer U, et al. Conditional gene ablation of Stat3 reveals differential signaling requirements for survival of motoneurons during development and after nerve injury in the adult. J Cell Biol. 2002;156(2):287–297. doi: 10.1083/jcb.200107009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turnley AM, Bartlett PF. Cytokines that signal through the leukemia inhibitory factor receptor-beta complex in the nervous system. J Neurochem. 2000;74(3):889–899. doi: 10.1046/j.1471-4159.2000.0740889.x. [DOI] [PubMed] [Google Scholar]
- 15.Valouev A, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5(9):829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SK, Pfaff SL. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron. 2003;38(5):731–745. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- 17.Lee SK, Jurata LW, Funahashi J, Ruiz EC, Pfaff SL. Analysis of embryonic motoneuron gene regulation: Derepression of general activators function in concert with enhancer factors. Development. 2004;131(14):3295–3306. doi: 10.1242/dev.01179. [DOI] [PubMed] [Google Scholar]
- 18.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machanick P, Bailey TL. MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics. 2011;27(12):1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8(2):R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bousquet C, Melmed S. Critical role for STAT3 in murine pituitary adrenocorticotropin hormone leukemia inhibitory factor signaling. J Biol Chem. 1999;274(16):10723–10730. doi: 10.1074/jbc.274.16.10723. [DOI] [PubMed] [Google Scholar]
- 22.Horvath CM. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem Sci. 2000;25(10):496–502. doi: 10.1016/s0968-0004(00)01624-8. [DOI] [PubMed] [Google Scholar]
- 23.Jessell TM. Neuronal specification in the spinal cord: Inductive signals and transcriptional codes. Nat Rev Genet. 2000;1(1):20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 24.Martí E, Bumcrot DA, Takada R, McMahon AP. Requirement of 19K form of Sonic hedgehog for induction of distinct ventral cell types in CNS explants. Nature. 1995;375(6529):322–325. doi: 10.1038/375322a0. [DOI] [PubMed] [Google Scholar]
- 25.Roelink H, et al. Floor plate and motor neuron induction by different concentrations of the amino-terminal cleavage product of sonic hedgehog autoproteolysis. Cell. 1995;81(3):445–455. doi: 10.1016/0092-8674(95)90397-6. [DOI] [PubMed] [Google Scholar]
- 26.Ericson J, Morton S, Kawakami A, Roelink H, Jessell TM. Two critical periods of Sonic Hedgehog signaling required for the specification of motor neuron identity. Cell. 1996;87(4):661–673. doi: 10.1016/s0092-8674(00)81386-0. [DOI] [PubMed] [Google Scholar]
- 27.Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31(5):773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- 28.Mizuguchi R, et al. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron. 2001;31(5):757–771. doi: 10.1016/s0896-6273(01)00413-5. [DOI] [PubMed] [Google Scholar]
- 29.Klug SJ, Famulok M. All you wanted to know about SELEX. Mol Biol Rep. 1994;20(2):97–107. doi: 10.1007/BF00996358. [DOI] [PubMed] [Google Scholar]
- 30.Darnell JE., Jr STATs and gene regulation. Science. 1997;277(5332):1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 31.Thaler J, et al. Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron. 1999;23(4):675–687. doi: 10.1016/s0896-6273(01)80027-1. [DOI] [PubMed] [Google Scholar]
- 32.Arber S, et al. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron. 1999;23(4):659–674. doi: 10.1016/s0896-6273(01)80026-x. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.