Abstract
Cell-to-cell movement of beet yellows closterovirus requires four structural proteins and a 6-kDa protein (p6) that is a conventional, nonstructural movement protein. Here we demonstrate that either virus infection or p6 overexpression results in association of p6 with the rough endoplasmic reticulum. The p6 protein possesses a single-span, transmembrane, N-terminal domain and a hydrophilic, C-terminal domain that is localized on the cytoplasmic face of the endoplasmic reticulum. In the infected cells, p6 forms a disulfide bridge via a cysteine residue located near the protein's N terminus. Mutagenic analyses indicated that each of the p6 domains, as well as protein dimerization, is essential for p6 function in virus movement.
Transport of plant viruses within and between cells is an active process that requires the function of virus-coded movement proteins (MPs). By definition, MPs are specialized proteins that are essential for the translocation of viral genomes or virions, but they are not required for virus genome replication or encapsidation. Viral MPs belong to several distinct protein families, each of which seems to exhibit a unique functional profile (10, 26). Many virus genera possess not one but two or three MPs. In addition, cell-to-cell movement of some viruses requires proteins whose primary functions are in genome replication or encapsidation (8, 9, 21).
Among several present models of virus movement, two have approached canonic status (10, 26). One is a Tobacco mosaic virus (TMV) model (4). The only TMV MP, the 30-kDa protein p30, is able to bind viral RNA and guide it through the plasmodesmata (13). Its additional activities include modification of plasmodesmatal gating properties and interactions with microtubules, actin microfilaments, endoplasmic reticulum (ER) (28, 31, 38), and a cell wall-specific host enzyme (12).However, the exact mechanistic contributions of these MP associations to intracellular movement are a matter of debate (6, 17, 41). Likewise, the transport mechanism of the RNA-MP complex through plasmodesmata largely remains a mystery. The leading model proposes that TMV-type MPs recruit a preexisting host machinery for intercellular trafficking (19, 27). Interestingly, both rod-shaped RNA viruses related to TMV and several icosahedral RNA and single-stranded DNA viruses appear to follow this movement paradigm (16, 26).
The second well-recognized model applies to several families of the icosahedral RNA viruses and pararetroviruses (35, 42). The MPs of these viruses reorganize plasmodesmata by inducing formation of the tubules through which mature virions translocate from cell to cell. The MP and endomembrane secretion system appear to be sufficient for tubule formation, whereas intact cytoskeleton is required for proper positioning of the tubules relative to plasmodesmata (24).
Mounting evidence indicates that the filamentous potexviruses do not fit in any of the abovementioned models. The 25-kDa MP (p25) of Potato virus X (PVX) possesses nucleoside triphosphatase and RNA helicase activities and is able to disassemble virions in a polar manner (29). p25 was the first viral MP for which a role in suppression of the host RNA silencing defense response was demonstrated (44). In addition to p25, the quadripartite PVX movement machinery includes two membrane-bound MPs and a capsid protein (CP), each of which is essential, but not sufficient, for virus translocation (11, 23).
The family Closteroviridae in general and the Beet yellows virus (BYV) in particular occupy a special niche among models of plant virology due to their large RNA genomes, exceptionally long filamentous virions, and a five-component machinery for cell-to-cell movement (14). Four of the BYV movement-associated proteins are the virion components. One is a major CP which encapsidates most of the virion RNA. The three others are the minor CP (CPm), a 64-kDa protein (p64), and a homolog of the ∼70-kDa heat shock proteins (Hsp70h). Remarkably, CPm, p64, and Hsp70h assemble virion tails that were proposed to function as a specialized movement device (3, 30). The only “conventional” BYV MP is a 6-kDa hydrophobic protein (p6). Although p6 is not required for assembly of the movement-competent, tailed virions, it is essential for BYV movement from cell to cell (2, 3, 33).
In this work, we demonstrate that BYV p6 is inserted into ER membranes with its C-terminal hydrophilic domain facing the cytosol. The Cys-3 residue of p6 is present within the ER lumen and is involved in the formation of the disulfide bond. Mutational analysis of p6 revealed that the short luminal, transmembrane, and cytosolic regions of this protein are each essential for p6 function in BYV cell-to-cell movement.
MATERIALS AND METHODS
Isolation and chemical treatments of the microsomal fractions.
Symptomatic leaves from BYV-infected Nicotiana benthamiana plants were ground in lysis buffer (20 mM HEPES [pH 6.8], 150 mM potassium acetate, 250 mM mannitol, 1 mM MgCl2), and the homogenate was clarified by centrifugation at 3,000 × g for 10 min at 4°C. The supernatant was centrifuged at 30,000 × g for 1 h at 4°C to yield the soluble (S30) and the crude (P30) microsomal fractions. Microsomal pellets were resuspended in 10 vol of 100 mM Na2CO3 (pH 11), 4 M urea, or original lysis buffer and incubated for 30 min on ice (39).
Membranes were collected by centrifugation at 30,000 × g for 30 min and resuspended in the original volume of lysis buffer. The S30 and P30 fractions were prepared and boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer with or without dithiothreitol (DTT) as indicated in Results.
Alternatively, P30 was resuspended in lysis buffer containing 1% Triton X-100, incubated on ice for 30 min, and centrifuged at 30,000 × g for 1 h. The resulting pellet was resuspended in lysis buffer in a volume equal to that of the original sample.
Treatment with Triton X-114 was performed by resuspending P30 in lysis buffer containing 1% Triton X-114, clarifying by centrifugation at 0°C, and incubating the lysate at 37°C for 10 min with subsequent centrifugation at 10,000 × g at room temperature to allow separation of aqueous and organic phases (5, 39). The lower phase, corresponding to the detergent-rich fraction, was washed by addition of fresh buffer lacking Triton X-114 and vortexing. The tube was placed on ice for 10 min, and phase separation was repeated.
Sucrose gradient fractionation, immunoblotting, and proteinase treatments.
Plant material was harvested and ground in lysis buffer containing either 0.1 or 5 mM MgCl2. P30 prepared as described above was loaded on top of 20 to 60% linear sucrose gradients containing lysis buffer with corresponding concentrations of MgCl2 (39, 46). Gradients were centrifuged for 16 h at 100,000 × g in a Beckman SW40 rotor at 4°C, and 15 fractions were collected starting from the top. Aliquots from each fraction were separated using SDS-PAGE. Immunoblot analyses were done using rabbit polyclonal antibodies to BYV p6 (33), BiP (a gift from Maarten Chrispeels, University of California, San Diego), or green fluorescent protein (GFP) (Living Color antibodies; Clontech, San Jose, Calif.).
For proteinase K treatments, microsomal pellets were resuspended in lysis buffer and loaded on top of discontinuous sucrose gradients consisting of 20 and 60% sucrose. Gradients were centrifuged for 2 h at 100,000 × g. A fraction containing closed-cell vesicles originating from disrupted ER (top of 60% sucrose phase) was collected. Aliquots of this fraction were treated with 100 μg of proteinase K/ml at 0°C in the presence or absence of 1% Triton X-100 (37). After 20 min, the reactions were quenched by the addition of 2 mM phenylmethylsulfonyl fluoride and the products were used for immunoblotting.
Molecular cloning and analyses of the mutant viruses and p6 variants.
The alanine-scanning and premature stop codon mutations listed in Table 1 were introduced into the p6 open reading frame (ORF) by using p65 M plasmid as described previously (2). The SnaBI-NdeI fragments from the resulting mutant plasmids were engineered to the pBYV-GFP plasmid that harbored a full-length cDNA clone of BYV tagged by the insertion of the GFP expression cassette (34). The corresponding in vitro RNA transcripts were used to inoculate Claytonia perfoliata plants, and the cell-to-cell movement of the resulting viruses was assayed as described previously (34).
TABLE 1.
Mutation analysis of p6 function in BYV cell-to-cell movement
| BYV variant | p6 mutation | Avg diam of infection foci (no. of cells ± SD)a | Maximal focus diam (no. of cells) |
|---|---|---|---|
| BYV-GFP | NAb | 6.7 ± 2.8 | 20 |
| A1 | D2A | 1.1 ± 0.05 | 3 |
| A2 | C3A | 1.0 | 1 |
| A3 | R6A | 1.1 ± 0.08 | 2 |
| A4 | G14A | 2.2 ± 0.11 | 9 |
| A5 | C18A | 1.2 ± 0.13 | 7 |
| A6 | C23A | 1.2 ± 0.02 | 2 |
| A7 | W29A | 2.2 ± 0.3 | 7 |
| A8 | K33A | 1.1 ± 0.07 | 3 |
| A9 | R38A | 2.3 ± 0.34 | 10 |
| A10 | E45A | 1.3 ± 0.37 | 4 |
| A11 | R47A | 1.9 ± 0.07 | 7 |
| A12 | R50A | 1.9 ± 0.04 | 6 |
| Stop33 | K33Stop | 1.0 | 1 |
| Stop34 | Q34Stop | 1.0 | 1 |
| Stop38 | R38Stop | 1.0 | 1 |
| Stop45 | E45Stop | 1.0 | 1 |
| Stop48 | F48Stop | 1.2 ± 0.5 | 3 |
| Stop51 | S51Stop | 1.8 ± 0.9 | 5 |
SD, standard deviation.
NA, not applicable.
Alternatively, wild-type or mutant p6 ORFs were inserted into a pCB-derived minibinary vector for Agrobacterium-mediated overexpression in the N. benthamiana leaves (32, 36). To generate fusion of the p6 and GFP ORFs (p6/GFP), enhanced GFP ORF(Clontech) was inserted in-frame downstream from the coding sequence of p6. Analogously, enhanced GFP ORF was inserted in-frame upstream from the sequence encoding the C-terminal, hydrophilic domain of p6 (codons 32 to 54) to generate a fusion ORF, designated GFP/C-p6.
To transiently express p6 variants, the minibinary plasmids were introduced into Agrobacterium tumefaciens strain C58 GV2260 by electroporation. Leaves of N. benthamiana plants were infiltrated with the resulting agrobacteria and used either for immunoblot analyses or for microscopic detection of the p6/GFP fusion products (32). Confocal laser scanning microscopy was done 2 days after leaf infiltration by using an inverted Leica DMIRBE microscope equipped with a TCS4D laser and a band-pass fluorescein isothiocyanate filter.
RESULTS
BYV p6 is an integral membrane protein.
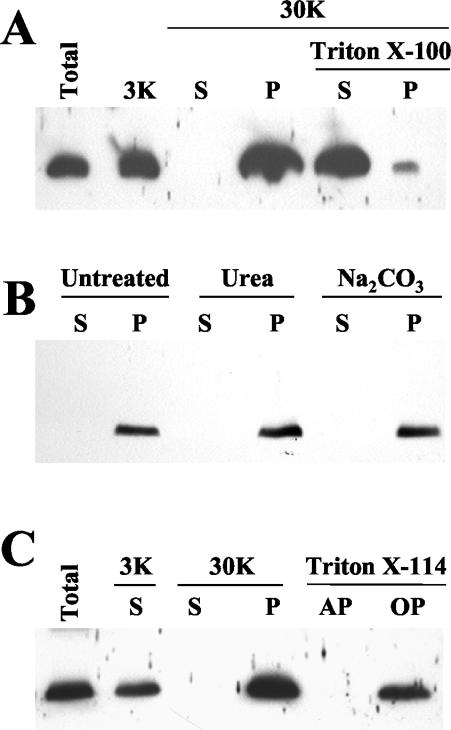
Computer analysis of the p6 amino acid sequence consistently predicted that p6 is a membrane protein with a single, α-helical transmembrane domain spanning residues 8 to 32 (Dense Alignment Surface [DAS] transmembrane prediction server), 9 to 32 (Kyte-Doolittle hydrophobicity plot), or 12 to 31 (PHD program). Subcellular fractionation of the extracts derived from the BYV-infected plant tissue was used to test computer predictions. The immunoblot analysis revealed that virtually all p6 was present in a pellet fraction (P30) following centrifugation at 30,000 × g (Fig. 1A). Because this fraction contains membrane-derived microsomes, we assumed that p6 is indeed a membrane-associated protein. This assumption was supported by solubilization of p6 by the treatment of P30 with the nonionic detergent Triton X-100 (Fig. 1A) (37). To distinguish between the luminal and membrane-associated localizations of p6, P30 was treated with Na2CO3 (pH 11). Such treatment renders microsomes to open membranous sheets, thus releasing the soluble luminal proteins (39). Because p6 remained in the pellet (Fig. 1B), we concluded that it is tightly associated with the membranes. To determine whether p6 is a peripheral membrane protein, the P30 fraction was treated with urea (37, 39). This treatment did not release p6 (Fig. 1B), suggesting that p6 is anchored within the membrane. To confirm this conclusion, P30 was also treated with Triton X-114, a detergent that forms a separate phase to which the membrane lipids and hydrophobic proteins are partitioned (5). Because p6 was detected in the hydrophobic, but not the aqueous, fraction (Fig. 1C), we concluded that p6 is an integral membrane protein.
FIG. 1.
Immunoblot analyses, using anti-p6 serum, of the extracts from BYV-infected plants. Lanes: Total, protein extract prior to fractionation; 3K, supernatant following extract centrifugation at 3,000 × g; S and P, supernatant and pellet, respectively, following centrifugation at 30,000 × g; AP and OP, aqueous and organic phases, respectively, following extraction with Triton X-114. Samples treated with Triton X-100, urea, or Na2CO3 buffer are marked accordingly.
p6 resides in the rough ER.
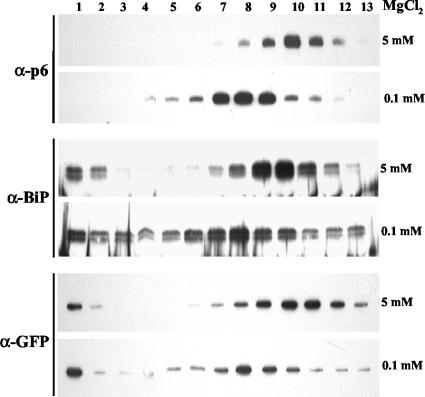
Sucrose density gradient centrifugation in the presence of 5 mM MgCl2 was employed to fractionate p6-containing membranes derived from the BYV-infected plants. Immunoblot analysis of the gradient fractions using p6-specific antibody revealed the peak of p6 in fractions 9 to 11 (Fig. 2, top), a pattern suggestive of p6 association with the ER membranes. Indeed, probing the same gradient fractions with the antibody to a residential ER protein, BiP, demonstrated cofractionation of BiP and p6 (Fig. 2, middle). Furthermore, in the presence of 0.1 mM MgCl2, both p6 and BiP peaked in fractions 7 to 9 (Fig. 2). Similar results were obtained with the ER-targeted GFP (Fig. 2, bottom) (18). Because such mobility shift due to release of the ER-associated ribosomes is characteristic of the rough ER (37, 39, 46), we concluded that p6 is likely present in the rough ER. It is worth noting that both BiP and ER-targeted GFP, but not p6, were also observed at the top of the gradient (Fig. 2). This could be due to release of ER-luminal BiP and GFP from the popped ER vesicles, whereas no such release could be expected for p6 if it were an integral membrane protein.
FIG. 2.
Immunoblot analyses of the protein extracts following separation in the sucrose gradients, with the fraction numbers shown at the top. The types of antisera used for analysis are indicated at left, and MgCl2 concentrations are shown at right. BiP, ER-resident marker protein.
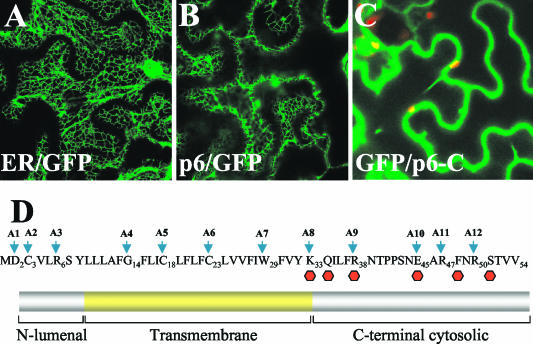
To visualize p6 within the living cells, we fused GFP to the C terminus of p6 and overexpressed the resulting p6/GFP product in leaf tissue. Confocal laser scanning microscopy revealed that p6/GFP was localized primarily to a reticulate network, which was indistinguishable from that observed in transgenic 16c plants engineered to express the ER-targeted GFP (Fig. 3A and B). Interestingly, fusion of the C-terminal, hydrophilic domain of p6 to the GFP C terminus (GFP/C-p6) resulted in the uniform distribution of this product in cytoplasm (Fig. 3C). This result is compatible with the notion that p6 is targeted to the ER via its N-terminal, hydrophobic domain rather than via interaction of the C-terminal, hydrophilic domain with the peripheral ER protein. Taken together, the results presented above establish p6 as an integral membrane protein that is targeted to the rough ER in the BYV-infected cells or upon transient expression.
FIG. 3.
(A to C) Confocal laser scanning microscopy analysis of the 16c transgenic plants that express ER-targeted GFP (A), the p6/GFP fusion (B), or the GFP fusion to the C-terminal, hydrophilic domain of p6 (C). The green corresponds to the GFP fluorescence, and the occasional red spots represent the autofluorescent chloroplasts. (D) Amino acid sequence (top) and membrane topology (bottom) of BYV p6. A1 to A12 and arrows indicate the alanine-scanning mutations introduced into indicated positions of p6. Red hexagons indicate premature stop codon mutations replacing the residues shown above.
Membrane topology of p6.
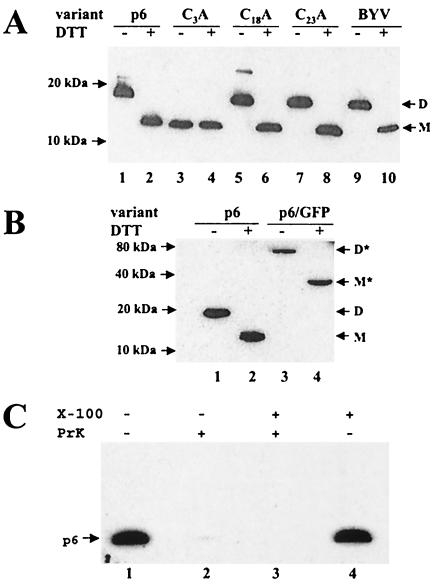
A single topogenic domain of p6 functions as both signal and anchor for insertion into membrane. Therefore, p6 could be either a type II membrane protein with its C terminus located within the ER lumen or a type III membrane protein with a luminal N terminus (40). Whatever is the case, the luminal segment of p6 must be exposed to an oxidizing environment that promotes disulfide bond formation via available Cys residues (15). Because p6 possesses a Cys residue at the third position from the N terminus (Fig. 3D), whereas the other two cysteines are buried within the membrane, the ability of p6 to dimerize would suggest that it is a type III membrane protein. To investigate the possibility of p6 dimerization, we performed an electrophoretic analysis of the protein extracts from BYV-infected plants (Fig. 4A, lanes 1 and 2) or plants that transiently expressed p6 (Fig. 4A, lanes 9 and 10) under nonreducing versus reducing conditions. Strikingly, for both extracts, these analyses revealed dramatically different p6 mobilities depending on the presence of DTT in the protein dissociation buffer. Under reducing conditions, p6 migrated as an ∼12-kDa protein, whereas its mobility under nonreducing conditions corresponded to an ∼18-kDa protein. This result is compatible with the ability of p6 to dimerize. We assume that anomalously slow migration of the p6 monomers and dimers is due to p6 hydrophobicity.
FIG. 4.
(A) Analysis of dimerization of wild-type p6 and three alanine mutants targeting each of the cysteine residues present in p6 (see also Fig. 3D and Table 1). Lanes: p6, agrobacterium-mediated expression of p6; BYV, virus-infected plants. The presence or absence of DTT in the protein dissociation buffer is indicated above the lanes with a plus or minus sign, respectively. D, p6 dimer; M, p6 monomer. Positions of the protein markers are shown at left. In both panels A and B, p6 was detected using immunoblotting and p6 antiserum raised against the C-terminal hydrophilic domain of p6. (B) Dimerization of the p6/GFP fusion product. The designations are the same as in panel A, except for M* and D*, which correspond to a monomer and a dimer formed by the p6/GFP fusion product, respectively. (C) Treatment of the resuspended P30 fraction of the p6-containing protein extracts with proteinase K (PrK) in the presence or absence of Triton X-100 as indicated at the top.
To determine whether, as expected, Cys-3 is required for p6 dimerization, we replaced this residue with Ala and expressed the resulting mutant protein in plants. For comparison, Ala was also substituted for Cys-18 and Cys-23, each of which is located within the transmembrane domain (Fig. 3D) and not expected to form disulfide bridges. In complete agreement with these predictions, mutation of Cys-3, but not of Cys-18 or Cys-23, resulted in loss of p6 dimerization (Fig. 4A, lanes 3 to 8). Because formation of disulfide bonds catalyzed by protein disulfide isomerase is feasible only within the ER lumen (15), we concluded that the N-terminal segment of p6 including Cys-3 is indeed present in this compartment and that p6 is likely a type III membrane protein.
The ∼18-kDa p6 dimer could be either a homodimer or a heterodimer formed by p6 with an apparent molecular mass of 12 kDa and by another protein of ∼6 kDa. To distinguish between homo- and heterodimerization of p6, we compared dimerization of p6 to that of the p6/GFP fusion product (Fig. 4B). The p6/GFP molecular mass of ∼38 kDa estimated by SDS-PAGE under reducing conditions (Fig. 4B, lane 4) was reasonably close to the sum of the molecular mass of GFP (∼27 kDa) and the apparent molecular mass of the p6 monomer (∼12 kDa). If this protein were to form a homodimer, the expected molecular mass should be ∼76 kDa. Alternatively, if p6/GFP were to form a heterodimer with the same hypothetical protein as in the case of p6, its estimated molecular mass should be ∼44 kDa [molecular mass of p6/GFP plus the difference between the molecular masses of the p6 dimer and monomer: 38 kDa + (18 kDa − 12 kDa) = 44 kDa]. Because the estimated molecular mass of the p6/GFP product analyzed under nonreducing conditions was ∼76 kDa (Fig. 4B, lane 4), it could be assumed that p6/GFP and, likely wild-type p6, form homodimers.
To confirm a type III membrane topology of p6, a sucrose gradient-purified microsomal fraction derived from plants transiently expressing p6 was treated with proteinase K in the absence or presence of Triton X-100 (37). It was expected that if the hydrophilic C-terminal domain of p6 is present in the cytoplasm, it would be proteolytically degraded with or without membrane solubilization by Triton X-100. Conversely, if the C-terminal domain is present in the ER lumen, it would be protected from digestion in the absence, but not in the presence, of a detergent. Because the antibody used to detect p6 was raised against synthetic peptide corresponding to the C-terminal 23 residues of p6 (33), digestion of the C-terminal domain should abolish p6 immunogenicity. As shown in Fig. 4C, the obtained results are clearly compatible with the former, but not the latter, scenario. That is, p6 lost its C-terminal immunogenic determinants following proteinase K treatment in both the absence and presence of the detergent. The same results were obtained when trypsin was used for the treatment (data not shown). Taken together with the ability of p6 to dimerize via the Cys-3 residue, these data establish p6 as a type III membrane protein with its hydrophilic, C-terminal region facing the cytosol.
Mutational analysis of p6 function in BYV cell-to-cell movement.
Alanine-scanning mutagenesis was used to map structure-to-function relations within the p6 molecule. Twelve Ala replacement mutations were introduced along the entire p6 sequence (Fig. 3D and Table 1). The mutant p6 variants were engineered into a cDNA clone of BYV tagged via insertion of the GFP reporter to visualize replication and cell-to-cell movement of the virus in indicator plants (34). Approximately 60 to ∼250 of the individual infection foci were analyzed for each of the BYV variants.
Each of the three mutations introduced into the short, N-terminal, luminal segment of p6 (A1 to A3) resulted in the dramatic reduction or complete loss of virus ability to move from cell to cell (Table 1). Likewise, all four mutations that targeted the transmembrane domain (A4 to A7) also affected virus movement. Finally, five Ala replacements of the positively or negatively charged residues in the C-terminal hydrophilic domain of p6 (A8 to A12) all resulted in reduced movement. These data indicate that p6 function is unusually prone to structural changes within each of the three topological regions of this membrane protein. It should be emphasized that the A2 mutation that replaced a Cys-3 residue involved in p6 dimerization stands alone in that all of the detected 93 infection foci were unicellular. In contrast, each of the other alanine-scanning mutants was able to generate at least a few multicellular foci (Table 1). This result underscores the critical nature of Cys-3 and suggests that dimerization is essential to p6 function in BYV cell-to-cell movement.
To further probe the cytosolic domain of p6, six premature stop codons were introduced along its length (Fig. 3D). As shown in Table 1, progressive truncation of p6 resulted in gradual loss of its function. Only two mutants that truncated p6, by three and six residues, were able to move, albeit inefficiently. Even though the length and amino acid sequence of the C-terminal domain are not conserved among p6 orthologs encoded by the members of the genus Closterovirus (data not shown), both alanine-scanning and truncation analyses point to very rigid structure-to-function relations within this domain.
DISCUSSION
The endomembranes in general and ER in particular play conspicuous yet poorly understood roles in virus-cell interactions. Many, but not all, positive-strand RNA viruses recruit ER for the formation of the membrane-enveloped spherules to which replication complexes are sequestered (1). In addition, several plant virus MPs that function in a TMV-like, comovirus-like, or PVX-like manner were found in tight association with the ER (4, 22, 23, 25, 29, 45). In most of these cases, the mechanistic significance of the MP-ER association is unclear. The apparent connection of the plasmodesmata with ER elements and the presence of the appressed ER in the desmotubule prompted the suggestion that viruses need to modify and/or recruit plasmodesma-associated ER for their movement. It has also been demonstrated that pharmacological inactivation of membrane trafficking from the ER to the Golgi apparatus and beyond affects localization of the TMV MP to the cell periphery (20). Analogously, it has been demonstrated that membrane trafficking is required either for proper targeting of the comoviral MPs or for the ability of MPs to form movement-associated tubules (24, 35).
The present model of the ER-associated form of the 30-kDa TMV MP features two transmembrane domains, with the termini of both proteins facing the cytosol (7). A similar model was developed for the unrelated, 9-kDa MP of a carmovirus (43). So far, the 6-kDa BYV MP is unique among other MPs due to its small size and a single-span, type III membrane topology (Fig. 3D). However, because p6 forms disulfide bonds via its Cys-3 residue, the resulting homodimer also contains two cytosolic domains.
How does such a small protein molecule provide a critical contribution to the movement of unusually large BYV virions? Although the mechanism of p6 action remains a matter of speculation, two possibilities are compatible with the existing data. The first assumes that the modification of the ER membranes by p6 is required to promote BYV movement. Such a modification could be needed to release nascent virions from ER-derived vesicular aggregates, where BYV RNA is synthesized and likely assembled. p6 could also promote virion transport to the cell periphery in association with the ER-derived vesicles. A second possibility is that p6 modifies the secretion pattern of the infected cell. Such a modification could be negative, i.e., suppression of the secretion of the ER-synthesized antiviral factors. It also could be positive, i.e., stimulation of the secretion of the factors required for virus transport. Interestingly, each of the 18 point mutations introduced into ER-luminal, transmembrane, or cytosolic segments of p6 resulted in partial or complete loss of function. These unusually stringent structural requirements for this minute MP suggest its involvement in critical interactions with the host and/or viral factors.
Is p6 distributed uniformly within the ER network of the infected cell, or is it confined to a subset of ER elements? Is p6 capable of association with the Golgi apparatus or plasma membranes? These and other related questions will be addressed by using p6 fused to a monomeric red fluorescent protein in combination with GFP-tagged organelles. In addition, p6 provides an excellent model for probing the ER structure and function. In particular, the small size and simple membrane topology of p6 are useful for probing the mechanisms of targeting and retention of transmembrane proteins in the ER as well as the structural requirements for proper membrane orientation and disulfide bond formation.
Acknowledgments
We thank Jim Carrington and Christophe Ritzenthaler for critical reading of the manuscript, David Baulcombe for 16c transgenic plants, Maarten Chrispeels for the BiP-specific antibody, and Jim Haseloff for the ER-targeted GFP.
The research was supported by grants from the National Institutes of Health (R1GM53190) and the U.S. Department of Agriculture (CSREES 2001-35319-10875) to V.V.D.
REFERENCES
- 1.Ahlquist, P., A. O. Noueiry, W. M. Lee, D. B. Kushner, and B. T. Dye. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 77:8181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alzhanova, D. V., Y. Hagiwara, V. V. Peremyslov, and V. V. Dolja. 2000. Genetic analysis of the cell-to-cell movement of beet yellows closterovirus. Virology 268:192-200. [DOI] [PubMed] [Google Scholar]
- 3.Alzhanova, D. V., A. Napuli, R. Creamer, and V. V. Dolja. 2001. Cell-to-cell movement and assembly of a plant closterovirus: roles for the capsid proteins and Hsp70 homolog. EMBO J. 20:6997-7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beachy, R. N., and M. Heinlein. 2000. Role of p30 in replication and spread of TMV. Traffic 1:540-544. [DOI] [PubMed] [Google Scholar]
- 5.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 6.Boyko, V., J. Ferralli, J. Ashby, P. Schellenbaum, and M. Heinlein. 2000. Function of microtubules in intercellular transport of virus RNA. Nat. Cell Biol. 2:826-832. [DOI] [PubMed] [Google Scholar]
- 7.Brill, L. M., R. S. Nunn, T. W. Kahn, M. Yeager, and R. N. Beachy. 2000. Recombinant tobacco mosaic virus movement protein is an RNA-binding, alpha-helical membrane protein. Proc. Natl. Acad. Sci. USA 97:7112-7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callaway, A., D. Giesman-Cookmeyer, E. T. Gillock, T. L. Sit, and S. A. Lommel. 2001. The multifunctional capsid proteins of plant RNA viruses. Annu. Rev. Phytopathol. 39:419-460. [DOI] [PubMed] [Google Scholar]
- 9.Carrington, J. C., P. E. Jensen, and M. C. Schaad. 1998. Genetic evidence for an essential role for potyvirus CI protein in cell-to-cell movement. Plant J. 14:393-400. [DOI] [PubMed] [Google Scholar]
- 10.Carrington, J. C., K. D. Kasschau, S. K. Mahajan, and M. C. Schaad. 1996. Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8:1669-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman, S., G. Hills, J. Watts, and D. Baulcombe. 1992. Mutational analysis of the coat protein gene of Potato virus X: effects on virion morphology and viral pathogenicity. Virology 191:223-230. [DOI] [PubMed] [Google Scholar]
- 12.Chen, M.-H., J. Sheng, G. Hind, A. K. Handa, and V. Citovsky. 2000. Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J. 19:913-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Citovsky, V., M. L. Wong, A. L. Shaw, B. V. Prasad, and P. Zambryski. 1992. Visualization and characterization of tobacco mosaic virus movement protein binding to single-stranded nucleic acids. Plant Cell 4:397-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolja, V. V. 2003. Beet yellows virus: the importance of being different. Mol. Plant Pathol. 4:91-98. [DOI] [PubMed] [Google Scholar]
- 15.Frand, A. R., J. W. Cuozzo, and C. A. Kaiser. 2000. Pathways for protein disulphide bond formation. Trends Cell Biol. 10:203-209. [DOI] [PubMed] [Google Scholar]
- 16.Fujiwara, T., D. Giesman-Cookmeyer, B. Ding, S. A. Lommel, and W. Lucas. 1993. Cell-to-cell trafficking of macromolecules through plasmodesmata potentiated by the red clover necrotic mosaic virus movement protein. Plant Cell 5:1783-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillespie, T., P. Boevink, S. Haupt, A. G. Roberts, R. Toth, T. Valentine, S. Chapman, and K. J. Oparka. 2002. Functional analysis of a DNA-shuffled movement protein reveals that microtubules are dispensable for the cell-to-cell movement of tobacco mosaic virus. Plant Cell 14:1207-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haseloff, J., K. R. Siemering, D. C. Prasher, and S. Hodge. 1997. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94:2122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haywood, V., F. Kragler, and W. J. Lucas. 2002. Plasmodesmata: pathways for protein and ribonucleoprotein signalling. Plant Cell 14(Suppl.):S303-S325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinlein, M., H. S. Padgett, J. S. Gens, B. G. Pickard, S. J. Casper, B. L. Epel, and R. N. Beachy. 1998. Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10:1107-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirashima, K., and Y. Watanabe. 2001. Tobamovirus replicase coding region is involved in cell-to-cell movement. J. Virol. 75:8831-8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, M., L. Jongejan, H. Zheng, L. Zhang, and J. F. Bol. 2001. Intracellular localization and movement phenotypes of alfalfa mosaic virus movement protein mutants. Mol. Plant-Microbe Interact. 14:1063-1074. [DOI] [PubMed] [Google Scholar]
- 23.Krishnamurthy, K., M. Heppler, R. Mitra, E. Blancaflor, M. Payton, R. S. Nelson, and J. Verchot-Lubicz. 2003. The Potato virus X TGBp3 protein associates with the ER network for virus cell-to-cell movement. Virology 309:135-151. [DOI] [PubMed] [Google Scholar]
- 24.Laporte, C., G. Vetter, A.-M. Loudes, D. G. Robinson, S. Hillmer, C. Stussi-Garaud, and C. Ritzenthaler. 2003. Involvement of the secretory pathway and the cytoskeleton in intracellular targeting and tubule assembly of grapevine fanleaf virus movement in tobacco BY-2 cells. Plant Cell 15:1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence, D. M., and A. O. Jackson. 2001. Interactions of the TGB1 protein during cell-to-cell movement of Barley stripe mosaic virus. J. Virol. 75:8712-8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazarowitz, S. G., and R. N. Beachy. 1999. Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11:535-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, J. Y., B. C. Yoo, M. R. Rojas, N. Gomez-Ospina, L. A. Staehelin, and W. J. Lucas. 2003. Selective trafficking of non-cell-autonomous proteins mediated by NtNCAPP1. Science 299:392-396. [DOI] [PubMed] [Google Scholar]
- 28.McLean, B. G., J. Zupan, and P. C. Zambryski. 1995. Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell 7:2101-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morozov, S. Y., and A. G. Solovyev. 2003. Triple gene block: modular design of a multifunctional machine for plant virus movement. J. Gen. Virol. 84:1351-1366. [DOI] [PubMed] [Google Scholar]
- 30.Napuli, A. J., D. V. Alzhanova, C. E. Doneanu, D. F. Barofsky, E. V. Koonin, and V. V. Dolja. 2003. The 64-kDa capsid protein homolog of beet yellows virus is required for assembly of virion tails. J. Virol. 77:2377-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oparka, K. J., and A. G. Roberts. 2001. Plasmodesmata. A not so open-and-shut case. Plant Physiol. 125:123-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng, C.-W., V. V. Peremyslov, A. R. Mushegian, W. O. Dawson, and V. V. Dolja. 2001. Functional specialization and evolution of leader proteinases in the family Closteroviridae. J. Virol. 75:12153-12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peremyslov, V. V., and V. V. Dolja. 2002. Identification of the subgenomic mRNAs that encode 6-kDa movement protein and Hsp70 homolog of beet yellows virus. Virology 295:299-306. [DOI] [PubMed] [Google Scholar]
- 34.Peremyslov, V. V., Y. Hagiwara, and V. V. Dolja. 1999. HSP70 homolog functions in cell-to-cell movement of a plant virus. Proc. Natl. Acad. Sci. USA 96:14771-14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pouwels, J., J. E. Carette, J. W. van Lent, and J. Wellink. 2002. Cowpea mosaic virus: effects on host cell processes. Mol. Plant Pathol. 3:411-418. [DOI] [PubMed] [Google Scholar]
- 36.Prokhnevsky, A. I., V. V. Peremyslov, A. J. Napuli, and V. V. Dolja. 2002. Interaction between long-distance transport factor and Hsp70-related movement protein of beet yellows virus. J. Virol. 76:11003-11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reichel, C., and R. N. Beachy. 1998. Tobacco mosaic virus infection induces severe morphological changes of the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 95:11169-11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reichel, C., P. Mas, and R. N. Beachy. 1999. The role of the ER and cytoskeleton in plant viral trafficking. Trends Plant Sci. 4:458-462. [DOI] [PubMed] [Google Scholar]
- 39.Schaad, M. C., P. E. Jensen, and J. C. Carrington. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16:4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spiess, M. 1995. Heads or tails—what determines the orientation of proteins in the membrane. FEBS Lett. 369:76-79. [DOI] [PubMed] [Google Scholar]
- 41.Storms, M. M. H., C. van der Schoot, M. Prins, R. Kormelink, J. W. M. van Lent, and R. W. Goldbach. 1998. A comparison of two methods of microinjection for assessing altered plasmodesmatal gating in tissues expressing viral movement proteins. Plant J. 13:131-140. [Google Scholar]
- 42.Thomas, C. L., and A. J. Maule. 1999. Identification of inhibitory mutants of Cauliflower mosaic virus movement protein function after expression in insect cells. J. Virol. 73:7886-7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vilar, M., A. Sauri, M. Monne, J. F. Marcos, G. von Heijne, E. Perez-Paya, and I. Mingarro. 2002. Insertion and topology of a plant viral movement protein in the endoplasmic reticulum membrane. J. Biol. Chem. 277:23447-23452. [DOI] [PubMed] [Google Scholar]
- 44.Voinnet, O., C. Lederer, and D. C. Baulcombe. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103:157-167. [DOI] [PubMed] [Google Scholar]
- 45.Ward, B. M., R. Medville, S. G. Lazarowitz, and R. Turgeon. 1997. The geminivirus BL1 movement protein is associated with endoplasmic reticulum-derived tubules in developing phloem cells. J. Virol. 71:3726-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wienecke, K., R. Glas, and D. G. Robinson. 1982. Organelles involved in the synthesis and transport of hydroxyproline-containing glycoproteins in carrot root discs. Planta 155:58-63. [DOI] [PubMed] [Google Scholar]