Significance
Resistance to chemotherapy is one of the major challenges in oncology. Neuroblastoma is the most common extracranial solid tumor in childhood, and the successful response of high-risk patients to chemotherapy remains poor. Our work showed that the so far poorly studied histone deacetylase (HDAC)10 promotes autophagy-mediated cell survival and signals poor outcome in independent high-risk patient cohorts. Inhibition of HDAC10 sensitized tumor cells for cytotoxic drug treatment. These results offer HDAC10 as a potential biomarker for treatment response of high-risk tumors and open new avenues for developing selective treatment strategies to bypass drug resistance of these tumors.
Keywords: drug resistance, HDAC inhibitor, childhood tumors
Abstract
Tumor cells activate autophagy in response to chemotherapy-induced DNA damage as a survival program to cope with metabolic stress. Here, we provide in vitro and in vivo evidence that histone deacetylase (HDAC)10 promotes autophagy-mediated survival in neuroblastoma cells. We show that both knockdown and inhibition of HDAC10 effectively disrupted autophagy associated with sensitization to cytotoxic drug treatment in a panel of highly malignant V-MYC myelocytomatosis viral-related oncogene, neuroblastoma derived-amplified neuroblastoma cell lines, in contrast to nontransformed cells. HDAC10 depletion in neuroblastoma cells interrupted autophagic flux and induced accumulation of autophagosomes, lysosomes, and a prominent substrate of the autophagic degradation pathway, p62/sequestosome 1. Enforced HDAC10 expression protected neuroblastoma cells against doxorubicin treatment through interaction with heat shock protein 70 family proteins, causing their deacetylation. Conversely, heat shock protein 70/heat shock cognate 70 was acetylated in HDAC10-depleted cells. HDAC10 expression levels in high-risk neuroblastomas correlated with autophagy in gene-set analysis and predicted treatment success in patients with advanced stage 4 neuroblastomas. Our results demonstrate that HDAC10 protects cancer cells from cytotoxic agents by mediating autophagy and identify this HDAC isozyme as a druggable regulator of advanced-stage tumor cell survival. Moreover, these results propose a promising way to considerably improve treatment response in the neuroblastoma patient subgroup with the poorest outcome.
Autophagy is an evolutionarily highly conserved process that can be induced by metabolic or therapeutic stress, such as DNA damage-inducing drugs (1). The two dominant types of autophagy are macroautophagy and chaperone-mediated autophagy (CMA) (2). Macroautophagy is regulated by autophagy-related genes (ATGs), including beclin-1 (ATG6) and microtubule-associated protein 1 light chain 3 (LC3) (ATG8), and involves the sequestration of cytoplasmic components within a double-membrane structure called the autophagosome and successive delivery to lysosomes for degradation (reviewed in ref. 3). CMA targets specific cytosolic proteins to the lysosomes for protein degradation (4). During CMA, the cytosolic chaperone heat shock cognate (Hsc)70 binds proteins targeted for degradation and translocates them to the lysosomes (5), where they bind to the substrate protein receptor lysosome-associated membrane protein type 2A (LAMP-2A) (6).
Inhibition of histone deacetylases (HDACs) by HDAC inhibitors (HDACis) have been shown to cause significant anti-tumor effects, including cell-cycle arrest, differentiation, and apoptosis, in a broad spectrum of hematologic and solid tumors (reviewed in ref. 7). The efficacy of HDACis are currently being evaluated for treating various cancers in clinical trials (7–9). Recent research carried out in several tumor cell lines has shown that apoptosis induced by HDACis is accompanied by autophagy (10–17). The functional role of autophagy during HDACi-induced cell death is controversial, and interpretations range from assisting the cell death to assisting cell survival. Numerous cytosolic and nuclear proteins are modified by posttranscriptional acetylation (18), including the proapoptotic protein, p53 (19); the macroautophagy regulator, α-tubulin (20); and the CMA chaperone protein, heat shock protein (Hsp)90 (21). The human HDAC family consists of four classes: class I (HDAC1, -2, -3, and -8); class II, which is subdivided into class IIa (HDAC4, -5, -7, and 9) and class IIb (HDAC6 and -10); class III (sirtuin 1 to -7); and class IV (HDAC11) (7, 22, 23). The class IIb family member HDAC6 is well investigated and has been shown to participate in stress responses via α-tubulin deacetylation and Hsp90 interaction (20, 21, 24). Much less is known about HDAC10, the second member of class IIb histone deacetylases (25).
Neuroblastoma, a common pediatric tumor of the sympathetic nervous system, encompasses a wide spectrum of clinical courses ranging from localized to highly aggressive disease (26). The majority of neuroblastoma patients with advanced stage, high-risk tumors categorized as stage 4 by the International Staging System (INSS) continue to have a poor prognosis despite the continuous intensification of chemotherapy over the last 10 y (26, 27). Approximately 30% of patients in this group, however, respond well to chemotherapy for as yet unknown reasons. Here, we investigated whether expression of any of the classical HDAC family members in advanced neuroblastomas were capable of predicting poor and good treatment response in this high-risk neuroblastoma subgroup commonly treated with intense multimodal chemotherapy. We then set out to unravel the HDAC10-mediated mechanism of cell survival of advanced stage neuroblastomas.
Results
HDAC10 Expression in Neuroblastomas Predicts Treatment Outcome.
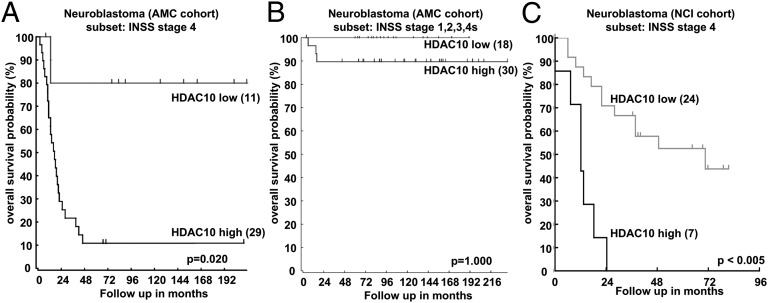
Expression levels of genes encoding single HDAC enzymes have prognostic value in pediatric tumors of the nervous system (28–30). Here, we examined whether HDAC expression levels might serve as a biomarker for treatment success in the high-risk subgroup of neuroblastoma patients. We reanalyzed publicly available expression data [Academic Medical Center (AMC) cohort; Gene Expression Omnibus (GEO) database accession no. GSE16476] from 40 advanced stage primary neuroblastomas (INSS stage 4) from patients before treatment with multimodal chemotherapy using the Web-based R2 microarray database (http://r2.amc.nl) (31) to determine HDAC1 to -11 expression levels. From all 11 classical HDACs, only HDAC10 expression significantly correlated with poor overall survival in this patient cohort (Table S1). Low HDAC10 expression in the tumor correlated with excellent long-term patient survival, with an overall survival probability of 80%, whereas high expression reduced overall survival probability to 11% (Fig. 1A). HDAC10 expression could not significantly separate patients with low-risk tumor stages (1–3 and 4s), into different prognostic groups (Fig. 1B). To confirm this observation, we assessed HDAC10 expression in an independent patient cohort from the National Cancer Institute (NCI) Neuroblastoma Prognosis Database (32), which is publicly available at the Oncogenomics Data Center (http://home.CCR.cancer.gov/oncology/oncogenomics). Elevated HDAC10 expression in advanced INSS stage 4 tumors also significantly correlated with poor overall patient survival in this cohort (Fig. 1C). HDAC10 expression in other highly malignant pediatric tumors of the nervous system, such as medulloblastoma [Heidelberg cohort (33); GEO accession no. GSE28245 (http://r2.amc.nl)] patients significantly separated recurrence and survival (Fig. S1 A and B). Thus, HDAC10 expression in neuroblastomas and medulloblastomas before patient treatment separates the survival probability of patients from independent cohorts and may, therefore, serve as useful biomarker to predict treatment outcome in pediatric patients with high-risk pediatric tumors of the nervous system.
Fig. 1.
HDAC10 tumor expression separates treatment outcome of high-risk neuroblastoma patients. (A) Kaplan–Meier curves are shown for overall survival in high-risk neuroblastoma patients (INSS stage 4; AMC cohort) whose tumors expressed low (n = 11) or high (n = 29) levels of HDAC10. Scan modus was used for cutoff determination, and P values were corrected for multiple testing (Bonferroni). The R2 microarray analysis and visualization platform (http://r2.amc.nl) was used for calculations and is the source of the data. (B) Kaplan–Meier curves are shown for overall survival in low-risk neuroblastoma patients (INSS stages 1, 2, 3, and 4s; AMC cohort) whose tumors expressed low (n = 18) or high (n = 30) levels of HDAC10. Tests were run in the same way and using the same tool as in A. (C) Kaplan–Meier curves are shown for overall survival in high-risk neuroblastoma patients (INSS stage 4; NCI cohort; n = 31) whose tumors expressed low (n = 24) or high (n = 7) levels of HDAC10. Expression data were obtained from the Neuroblastoma Prognosis Database of the NCI Oncogenomics Data Center (http://home.CCR.cancer.gov/oncology/oncogenomics). Cutoff was determined using a P value minimization check, and P values were corrected for multiple testing using the Bonferroni method.
HDAC10 Promotes Autophagic Processing.
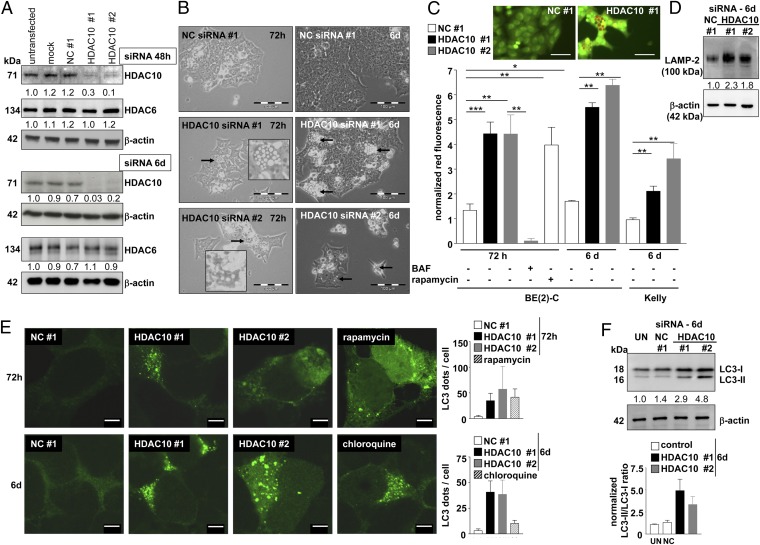
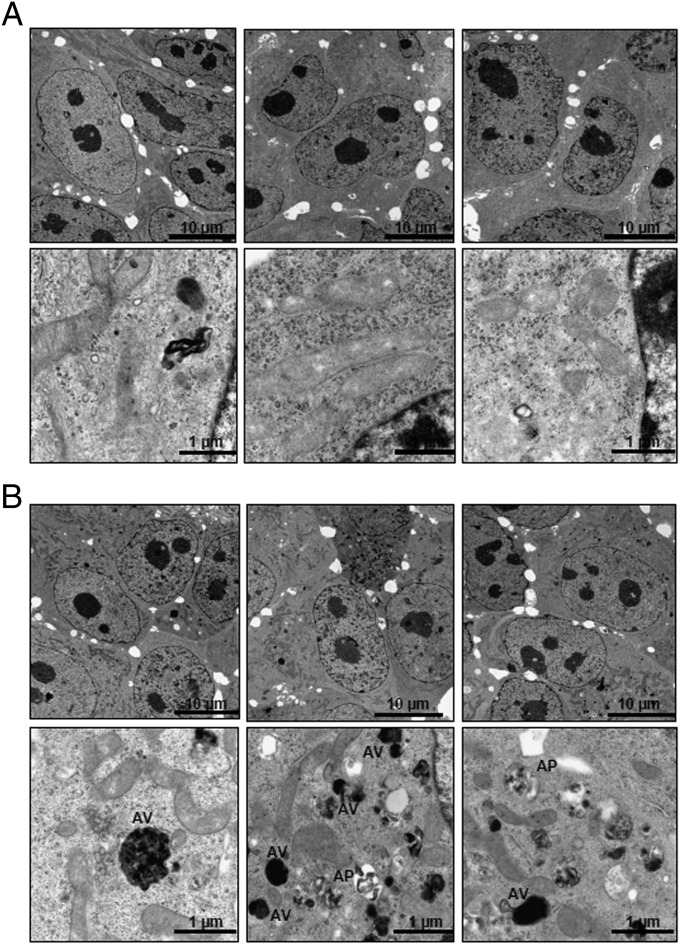
Correlation and gene-set analysis on the AMC neuroblastoma cohort using the R2 microarray database revealed that ATG4D expression, which is necessary for autophagosome formation during the induction of autophagy, positively correlated with HDAC10 expression (Table S2 and Fig. S1C) and that the term “regulation of autophagy” was overrepresented (Table S2). Because we also identified HDAC10 expression in pretreatment tumors as a biomarker for response to polychemotherapy in several patient cohorts, we hypothesized that HDAC10 might have a key function in regulating autophagy and cell survival after exposure to cytotoxic agents. We performed a series of experiments to detect the involvement of HDAC10 in autophagic pathways. HDAC10 was depleted by specific siRNAs against different regions of the HDAC10 mRNA sequence but not of the closely related HDAC6 sequence (Fig. 2A). HDAC10 depletion increased the number of large cytoplasmic vesicles in a time-dependent manner (Fig. 2B). Staining with the acidotropic dye acridine orange and detection of the shift to red fluorescence 72 h after HDAC10 siRNA transfection of BE(2)-C cells revealed the accumulation of acidic vesicular organelles (AVOs), including lysosomes and late endosomes (Fig. 2C and Fig. S2). This shift was fully inhibited by treating the cells with bafilomycin A1, a lysosomal proton pump inhibitor (Fig. 2C). The increase in AVOs after HDAC10 knockdown was comparable to the AVO accumulation in rapamycin-treated cells (Fig. 2C). Rapamycin is known to induce autolysosome formation and was used as a positive control. The number of AVOs remained elevated in viable neuroblastoma cells 6 d after siRNA transfection (Fig. 2C). HDAC10 knockdown also enhanced LAMP-2 expression, which is an AVO marker protein (Fig. 2D). To investigate whether AVO accumulation upon HDAC10 knockdown is part of macroautophagic processes, expression of the LC3-phosphatidylethanolamine conjugate (LC3-II), an autophagy marker localized to the autophagosome membrane, was assessed (2). EGFP-LC3–positive autophagic organelles (green dots) increased over time following siRNA transfection (Fig. 2E), and HDAC10 knockdown increased the expression of endogenous LC3-II (Fig. 2F). Electron microscopy confirmed the formation of autophagosomes and electron-dense lysosomes 72 h after HDAC10 knockdown (Fig. 3). These results indicate a blockade of autophagic flux.
Fig. 2.
Depletion of HDAC10 promotes accumulation of acidic vesicular organelles. (A) Western blot of whole-cell lysates of BE(2)-C cells transfected with siRNAs #1 and #2 targeting HDAC10 showing HDAC10 and HDAC6 protein expression 48 h and 6 d after transfection. β-Actin expression served as a loading control. The ratio of HDAC10:β-actin or HDAC6:β-actin expression relative to the untransfected control was inserted below the Western blots for HDAC10 and HDAC6 expression, respectively. Expression in the three control situations are also shown: mock, mock-transfected cells; NC #1, cells transfected with negative control siRNA; untransfected, cells not undergoing transfection and cultured in normal medium. (B) Phase-contrast micrographs of cells 72 h and 6 d after transfection with siRNAs targeting HDAC10 (siRNA #1 and #2) or negative control NC #1 siRNA. Morphological alterations suggestive of autophagy, such as massive cytosolic vacuolization of the cells, appeared only in cells after HDAC10 knockdown and are indicated by arrows. (C) Accumulation of acidic organelles 72 h and 6 d after transfection of BE(2)-C and Kelly cells with two siRNAs specific for HDAC10 (HDAC10 #1 and #2) or the negative control siRNA (NC #1) detected after acridine orange staining (red fluorescence) by FACS for quantification and on a microscope for visualization microscopically (inlay). (Scale bar: 50 µm.) Cells were also treated with 10 nM bafilomycin A1 (BAF) or 100 nM rapamycin (positive control). Means from at least three independent experiments are shown, and error bars represent SEM. Asterisks indicate the level of significance between testing groups from an unpaired two-tailed t test. *P < 0.05; **P < 0.01; ***P < 0.001. (D) Expression of the LAMP-2 lysosomal marker protein is shown in Western blots of whole-cell lysates 6 d after transfection with HDAC10 siRNAs #1 and #2 or negative control siRNA (NC #1). β-Actin was used as a loading control. Numbers indicate LAMP-2 expression relative to the negative control, normalized to β-actin expression. (E) Autophagosome formation was visualized after HDAC10 knockdown by transient transfection with the EGFP-LC3 expression construct. Punctate staining is indicative for the redistribution of EGFP-LC3 to autophagosomes. Pictures were taken 72 h and 6 d after transfection with siRNAs. Cells treated for 24 h with 100 nM rapamycin or for 5 h with 25 µM chloroquine are shown as positive controls. ImageJ quantification of three independent experiments, each including at least 25 cells per treatment is shown to the right of the representative pictures for the respective time points. HDAC10 #1, HDAC10 siRNA #1; HDAC10 #2, HDAC10 siRNA #2; NC #1, negative control siRNA #1. (F) LC3-I and LC3-II expression in whole-cell lysates of BE(2)-C cells 6 d after transfection with HDAC10 siRNAs #1 or #2 or negative control siRNA (NC #1) detected by Western blotting. β-Actin was used as a loading control. The ratios of LC3-II to LC3-I expression normalized to untransfected cells (UN) are included below the Western blot, and the normalized ratios from three independent experiments are shown in the bar graph below.
Fig. 3.
Depletion of HDAC10 increases the presence of autophagic structures. Representative transmission electron micrographs of BE(2)-C cells 72 h after transfection with negative control (NC) siRNA #1 (A) or siRNA #1 against HDAC10 (B). In B, autophagosomes (AP) and acidic vesicles (AV) with electron-dense material were apparent under the higher magnification.
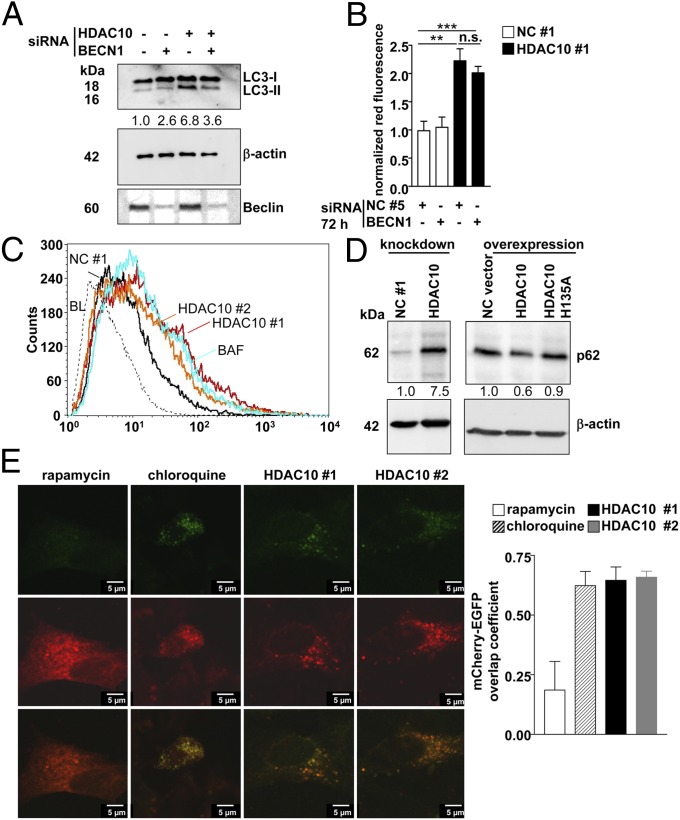
Knockdown of BECN1 or ATG7, both necessary for de novo autophagosome formation, did not affect AVO accumulation after HDAC10 depletion (Fig. 4 A and B and Fig. S3 A and B). These results further indicate that HDAC10 depletion impairs the progression of autophagy rather than inducing it. This observation was validated quantitatively by flow-cytometric detection of EGFP-LC3–expressing BE(2)-C cells. HDAC10 knockdown caused an accumulation of EGFP-LC3 to a similar extent as treatment with the lysosomal inhibitor bafilomycin A1 (Fig. 4C). We monitored accumulation of the autophagy substrate p62/sequestosome 1 (34) to directly assess autophagic flux inhibition (35). Rapamycin treatment of BE(2)-C cells induced p62 degradation, whereas bafilomycin A1 and chloroquine treatment resulted in p62 accumulation (Fig. S3C). HDAC10 knockdown also increased p62 levels (Fig. 4D). Conversely, ectopic expression of wild-type HDAC10, but not the catalytically inactive mutant H135A (25), reduced p62 levels (Fig. 4D). These results demonstrate that HDAC10 regulates autophagic flux and that the intact catalytic domain of HDAC10 is required for this regulation.
Fig. 4.
Autophagic flux requires HDAC10. (A) Western blot showing LC3-I and LC3-II expression in whole-cell lysates of BE(2)-C cells 6 d after transfection with HDAC10 siRNA #1 or BECN1 siRNA. Anti-Beclin antibody was used to control for BECN1 knockdown, and β-actin was used as a loading control. Ratios of LC3-II to LC3-I are shown below the Western blot. (B) Accumulation of acidic organelles (red fluorescence) detected by acridine orange staining in FACS analysis after knockdown of HDAC10 with and without BECN1 knockdown compared with the negative controls, as indicated. Means of at least three independent experiments are shown. Error bars represent SEM. Significant differences between groups were detected by unpaired two-tailed t test. **P < 0.01; ***P < 0.001; n.s., not significant. (C) EGFP-LC3 accumulation in BE(2)-C cells 6 d after transfection with the siRNA indicated was detected in FACS analysis and is shown as log10 fluorescence. Untransfected BE(2)-C cells were treated with 100 nM bafilomycin A1 (BAF), a lysosomal inhibitor that served as a positive control for the inhibition of autophagic flux. One representative result from at least three independent experiments is shown. (D) Expression of p62 protein levels upon knockdown or forced HDAC10 overexpression in BE(2)-C cells. β-Actin served as a loading control. Numbers indicate p62 expression normalized to β-actin expression. HDAC10 H135A, expression construct for HDAC10 with the H135A mutation in the deacetylase domain; NC #1, negative control siRNA; NC vector, empty expression construct. (E) Fluorescence microscopic detection of autophagosome-lysosome fusion in BE(2)-C cells transiently transfected with the mCherry-EGFP-LC3B expression construct 6 d after HDAC10 knockdown (HDAC10 #1 and HDAC10 #2). Untransfected BE(2)-C cells were treated with 100 nM rapamycin as a control for effective fusion and treated with 25 µM chloroquine as a control for defective fusion. Green signals for EGFP-LC3B expression indicate autophagosomes. Red signals for mCherry-LC3B expression indicate autophagosomes and autophagolysosomes. Yellow overlay indicates autophagosomes only, whereas red-only signals in the overlay indicate autophagolysosomes and the overlap coefficient, determined from at least five cells per condition, is shown in the bar graph (means ± SEM) to the right. (Scale bar: 5 µm.)
HDAC10 depletion combined with chloroquine treatment, a lysosomal inhibitor, did not increase LC3-II levels above levels produced by chloroquine treatment alone (Fig. S3D). Because accumulation of mitochondria is indicative for an insufficient autophagic degradation of organelles, we observed mitochondria following HDAC10 knockdown in BE(2)-C cells with MitoTracker Green staining. Significant accumulation of mitochondria (Fig. S3E) accompanied by increased reactive oxygen species (ROS) formation (Fig. S3F) occurred after HDAC10 depletion. To investigate whether HDAC10 depletion disables the efficient fusion of autophagosomes to lysosomes, BE(2)-C cells were transfected with the mCherry-EGFP-LC3B plasmid. The acid-sensitive green fluorescence is lost upon the efficient fusion between autophagosomes and lysosomes because of the low pH of the autolysosome, shifting the fluorescence to red. Nonacidic autophagosomes fluoresce yellow because of the overlapping of red mCherry tag and the green fluorescing EGFP tag on the LC3 protein. The overlap coefficient was determined for quantification (Fig. 4E). HDAC10 depletion disabled efficient autophagosome-lysosome fusion.
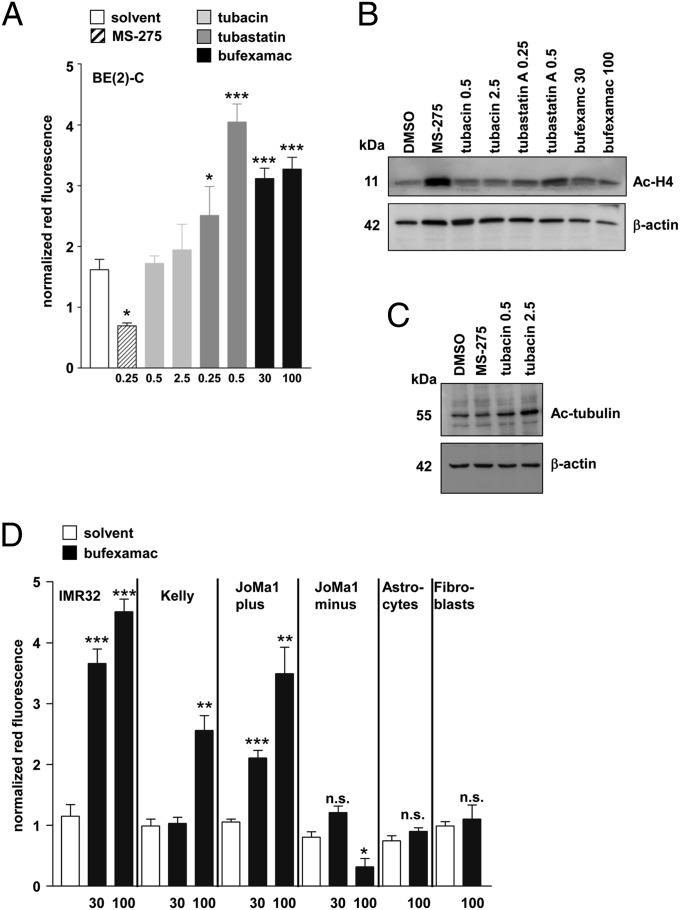
BE(2)-C cells were treated with different class IIb HDAC inhibitors, as well as the class I selective HDAC inhibitor MS-275, and AVO development was followed by acridine orange staining. Bufexamac selectively inhibits HDAC10 and -6 (36), whereas tubastatin inhibits HDAC10 with higher affinity than HDAC6 (Fig. S4A), and tubacin selectively inhibits only HDAC6 (37). Treatment with bufexamac and tubastatin, but neither tubacin nor MS-275, significantly increased the number of AVOs (Fig. 5A). MS-275 class I inhibitory action was confirmed by demonstration of histone 4 acetylation (Fig. 5B). We also determined tubulin acetylation in tubacin-treated cells, demonstrating on-target activity of the compound (Fig. 5C). The results are in line with HDAC6 knockdown experiments, which also did not increase the number or AVOs (Fig. S4B). Bufexamac treatment caused AVO accumulation in several neuroblastoma cell lines and MYC-transformed JoMa1 cells but not in untransformed astrocytes or fibroblasts or normal JoMa1 cells (Fig. 5D). The neural crest-derived JoMa1 cells represent a neuroblastoma cell of origin isogenic model that can be retained in a transformed, immortalized, and undifferentiated state via (Z)-4-hydroxytamoxifen (4-OHT) supplementation to activate MYC expression and in a differentiated state upon 4-OHT removal (38). Thus, small-molecule inhibition of HDAC10 phenocopies the effect of HDAC10 depletion. Taken together, our data demonstrate a critical function for HDAC10 in regulating autophagic processing in tumor cells.
Fig. 5.
HDAC10 inhibition induces accumulation of acidic organelles. (A) BE(2)-C cells treated with class I HDAC inhibitor MS-275 and class IIb HDAC inhibitors (tubacin, tubastatin, or bufexamac) as indicated (in µM). The accumulation of acidic organelles (normalized red fluorescence) was monitored by FACS after staining with acridine orange. (B) Acetylation of histone 4 (Ac-H4) upon treatment of BE(2)-C cells with HDAC inhibitors as indicated. β-Actin served as a loading control. (C) Acetylation of tubulin upon treatment of BE(2)-C cells with MS-275 and tubacin as indicated. β-Actin served as a loading control. (D) The BE(2)-C, Kelly, and IMR32 neuroblastoma cell lines; the neural crest-derived JoMa1 cells, which can be retained in an immortalized and undifferentiated state via 4-OHT supplementation to activate MYC expression (JoMa1 plus) and in an differentiated state upon 4-OHT removal (JoMa1 minus); and untransformed astrocytes and fibroblasts were treated with 30 or 100 µM bufexamac for 24 h and then stained with acridine orange. The accumulation of acidic organelles was quantified by FACS analysis (normalized red fluorescence). Red fluorescence in treated cells was normalized to red fluorescence in untreated cells in all experiments. Bars represent means (±SEM) of at least three independent experiments. Significant differences between groups were tested using an unpaired, two-tailed t test. **P < 0.01; ***P < 0.001.
HDAC10 Inhibition Sensitizes Drug-Resistant Neuroblastoma Cells to Doxorubicin Treatment.
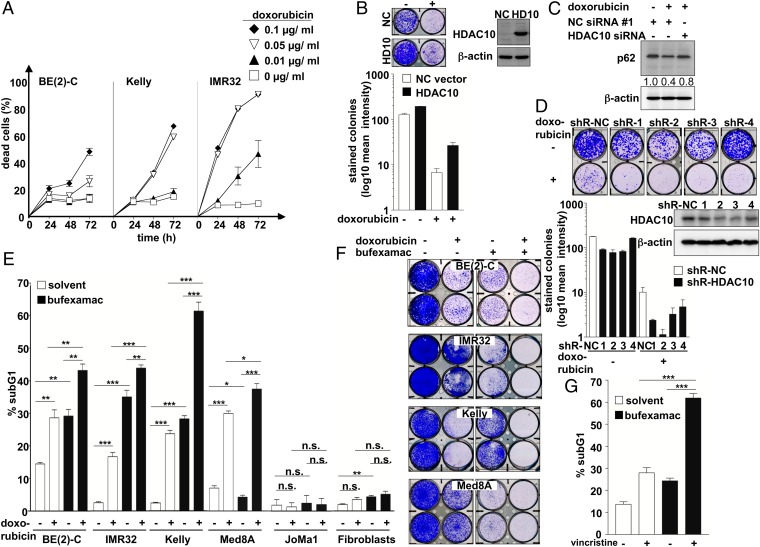
It has been proposed that tumor cells use autophagy as a survival mechanism to cope with stress induced by cytotoxic drugs, such as doxorubicin (1, 39, 40). Because HDAC10 depletion or inhibition blocks autophagy in our neuroblastoma model, we hypothesized that HDAC10 inhibition would sensitize neuroblastoma cells to drug-induced cell death. The V-MYC myelocytomatosis viral-related oncogene, neuroblastoma derived (MYCN)-amplified neuroblastoma cell lines BE(2)-C, Kelly, and IMR32 have been described as relatively insensitive to doxorubicin treatment (41). Among these, BE(2)-C cells were most resistant, and IMR32 cells were most sensitive (Fig. 6A). Interestingly, endogenous HDAC10 expression was lower in IMR32 cells than in BE(2)-C and Kelly cells (Fig. S5A). Stable HDAC10 overexpression protected IMR32 cells against doxorubicin-induced toxicity in longer-term colony assays (Fig. 6B). The treatment of the relatively resistant BE(2)-C cells with high-dose doxorubicin enhanced autophagic flux, measurable by p62 degradation, and this doxorubicin-induced p62 degradation was abolished by HDAC10 depletion (Fig. 6C). HDAC10-depleted BE(2)-C cells were more sensitive to doxorubicin (Fig. 6D and Fig. S5 B–D). In contrast, untransformed fibroblasts were not sensitized to doxorubicin by HDAC10 depletion (Fig. S5D). We treated neuroblastoma, medulloblastoma, and untransformed cells with the class IIb HDAC inhibitor bufexamac in combination with doxorubicin and then measured cell death via DNA fragmentation assay and poly(ADP-ribose) polymerase (PARP) cleavage. Whereas bufexamac and doxorubicin cotreatment significantly enhanced cell death of tumor cells, an isogenic, normal neural crest-derived cell model (JoMa1) exhibited no increase in cell death, which indicated tumor selectivity of this treatment combination (Fig. 6E and Fig. S6). Of note, in longer-term colony assays, cotreatment of cells with bufexamac and doxorubicin resulted in effective killing of neuroblastoma and medulloblastoma cells (Fig. 6F). Similar results were obtained for bufexamac and vincristine cotreatment (Fig. 6G and Fig. S5F). In summary, blocking autophagic flux through HDAC10 inhibition enhanced the tumor-specific toxicity to DNA damage-inducing drugs currently used in patient treatment. Our results suggest a promising tumor-selective approach for advanced-stage, chemotherapy-resistant tumors.
Fig. 6.
HDAC10 expression supports neuroblastoma cell survival. (A) BE(2)-C, Kelly, and IMR32 cell lines were treated with increasing concentrations of doxorubicin (0.01, 0.05, 0.1 µg/mL) and then monitored at 24, 48, and 72 h after treatment for cell death using trypan blue staining (dead cells, trypan blue-staining cells). (B) Stable hygromycin-resistant HDAC10-overexpressing IMR32 cells (filled bars) or hygromycin-resistant empty vector IMR32 control cells (NC vector; open bars) were treated with doxorubicin (0.0025 µg/mL). Colonies were stained (inlay picture) and quantified (bar diagram). Western Blot shows HDAC10 overexpression of the stably transfected cells; β-actin was used as a loading control. (C) BE(2)-C cells were treated with high-dose doxorubicin (0.5 µg/mL) after HDAC10 knockdown. Western Blot shows p62 expression after 48 h of doxorubicin treatment. Expression in control-transfected (NC siRNA #1) and untreated controls are shown as indicated. Numbers indicate p62 expression normalized to β-actin expression. (D) BE(2)-C cells were stably transfected with four different shRNAs targeting HDAC10 (shR-1, -2, -3, -4) or the negative control (shR-NC). Cells were treated with 0.05 µg/mL doxorubicin or normal culture medium, colonies were stained after 10 d, and results were quantified (bar diagram; shR-NC, open bars; shR-HDAC10, filled bars). Western Blot shows HDAC10 expression of the stably transfected cells; β-actin was used as a loading control. (E) Embryonal tumor-derived cell lines and normal murine neural crest-derived cells (JoMa1), as well as human fibroblasts, were treated with the HDAC6/10 inhibitor bufexamac alone or in combination with the cytotoxic agent doxorubicin. Where treatment is indicated, all cell types were treated with 30 µM bufexamac for 6 d. Doxorubicin treatment, where indicated, started 72 h before the detection of apoptotic cells by PI staining of ethanol-fixed cells and was applied at varied concentrations: neuroblastoma cell lines, BE(2)-C (0.1 µg/mL), Kelly (0.05 µg/mL), and IMR32 (0.01 µg/mL), the Med8A medulloblastoma cell line (0.1 µg/mL), and JoMa1 (0.01 µg/mL) and fibroblasts (0.1 µg/mL). Bars reflect the amount of cells in the sub-G1 area of the cell cycle profile. (F) BE(2)-C cells were treated with bufexamac alone or in combination with doxorubicin, and colonies were stained after 10 d. Where treatment is indicated, all cell types were treated with 30 µM bufexamac. Doxorubicin treatment, where indicated, varied in concentration: BE(2)-C (0.1 µg/mL), Kelly (0.05 µg/mL), IMR32 (0.01 µg/mL), and Med8A (0.1 µg/mL). (G) BE(2)-C cells were treated with 30 µM bufexamac for 6 d, where indicated, and cotreated with 10 ng/mL vincristine for the last 72 h, where indicated. Bars reflect the amount of cells in the sub-G1 area of the cell cycle profile of PI-stained ethanol-fixed cells. All bars represent means (±SEM) of at least three independent experiments. Significant differences between groups were tested using an unpaired, two-tailed t test. *P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant.
HDAC10 Controls Lysosomal Activity and Resistance to Cytotoxic Drugs via Interaction with Hsp70 Family Proteins.
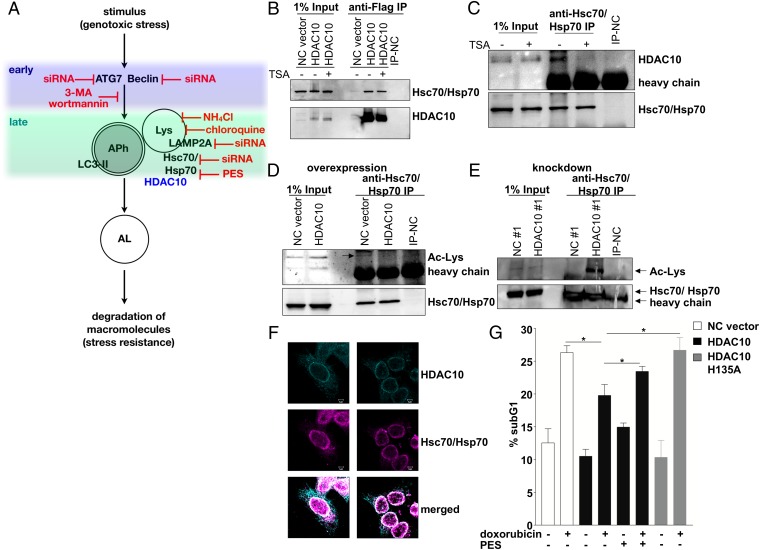
We next set out to narrow down the critical point of interference during the process of autophagy. BECN1, ATG7, LAMP-2A, and the Hsp70 family proteins are required at different points during autophagy or for different types of autophagy. BECN1 and ATG7 are required for autophagosome formation, LAMP-2A is required for CMA, HDAC6 plays a role in the clearance of misfolded proteins by autophagy, and the Hsp70 protein family with the two major family members, Hsc70 and Hsp70, is an essential mediator of lysosomal degradation. Constitutively expressed Hsc70 enables the transport of proteins marked for degradation via LAMP-2–driven translocation to lysosomes (6), whereas Hsp70 expression induced by stress functions as a guardian of lysosomal membrane integrity (42–44). We blocked autophagy during cytotoxic treatment in BE(2)-C cells at the different points shown in Fig. 7A using siRNA-mediated knockdown or inhibitor treatment. ATG7, LAMP2A, or HDAC6 knockdown protected the neuroblastoma cells from doxorubicin-mediated cytotoxicity, whereas BECN1 knockdown had a slight sensitizing effect. HSC70 knockdown and doxorubicin treatment resulted in complete lethality (Tables 1 and 2), which could be confirmed by longer-term colony assays (Table 1 and Fig. S7A). Cotreatment of the BE(2)-C cell line with doxorubicin and one of the lysosomal inhibitors [NH4Cl, chloroquine, or 2-phenylethynesulfonamide (PES)] also significantly decreased cell viability (Table 3). Longer-term colony assays confirmed complete loss of cell viability in chloroquine/doxorubicin and PES/doxorubicin cotreated cells, respectively (Table 3 and Fig. S7B). PES interacts with Hsp70 family proteins to disrupt the association with interacting proteins and impair lysosomal function and inhibit autophagy (45, 46). Treatment with wortmannin or 3-MA, which inhibit the early stage of autophagosome formation, did not enhance neuroblastoma cell cytotoxicity to doxorubicin (Table 3 and Fig. S7B). Thus, failed autophagy because of impaired lysosomal activity enhanced neuroblastoma cell toxicity to doxorubicin.
Fig. 7.
HDAC10 physically interacts with Hsc70/Hsp70 and affects its acetylation. (A) Graphical representation of the macroautophagy process, with indications of where interventions occur via siRNA knockdown and drug treatment. Early inhibitors interfere with the formation of autophagosomes (APh). Late inhibitors block the autophagic process via interference with lysosomal (Lys) activity and prevention of autolysosome (AL) formation. (B) Whole-cell extracts (input) of BE(2)-C cells transfected with either a HDAC10 expression construct (HDAC10) or empty vector (NC vector) and treated with 75 nM TSA or solvent control, as indicated, were immunoprecipitated with an antibody directed against the FLAG tag. HDAC10 and Hsc70/Hsp70 proteins in the extracts and immunoprecipitated complexes were detected on Western blots. IP-NC indicates the immunoprecipitation negative control. (C) Whole-cell extracts (input) of BE(2)-C cells treated with 75 nM TSA or solvent control, as indicated, were immunoprecipitated with an antibody directed against the Hsc70/Hsp70 proteins. HDAC10 and Hsc70/Hsp70 proteins in the extracts and immunoprecipitated complexes were detected on Western blots. IP-NC indicates the immunoprecipitation negative control. (D and E) Detection of lysine acetylation (Ac-Lyc) in Hsc70/Hsp70 on Western blots of whole-cell extracts (input) and proteins immunoprecipitated with an antibody against the Hsc70/Hsp70 proteins. BE(2)-C cells were transfected with either a HDAC10 expression construct (HDAC10) or empty vector (NC vector) (D) or with siRNA against HDAC10 (HDAC10 #1) or a negative control siRNA (NC#1) (E) before extraction and immunoprecipitation. (F) Immunofluorescent microscopy of BE(2)-C cells transfected with FLAG-tagged HDAC10. Cells were immunostained for the FLAG tag (cyan) and for Hsc70/Hsp70 (magenta). Cytoplasmic colocalization is shown by overlay in white. (G) IMR32 cells overexpressing HDAC10 or HDAC10-H135A or transfected with empty vector (NC vector) were treated 48 h with 0.01 µg/mL doxorubicin and the Hsc70/Hsp70 inhibitor PES (10 µM), where indicated. Apoptotic cells in the sub-G1 area of the cell cycle profile of PI-stained and ethanol-fixed cells were measured and are displayed as means (±SEM) of at least three independent experiments. Significant differences between groups were tested using a paired, two-tailed t test. *P < 0.05.
Table 1.
Failed autophagy enhances antineuroblastoma activity of doxorubicin treatment (knockdown studies)
| Target mRNA | siRNA-mediated knockdown | Doxorubicin treatment (0.1 μg/mL) | siRNA-mediated knockdown and doxorubicin treatment |
| Metabolic activity,* % | |||
| HDAC10 | 35 ± 3 | 52 ± 3 | 17 ± 1 |
| HSC70/HSPA8 | 13 ± 2 | 52 ± 3 | 8 ± 2 |
| BECN1 | 98 ± 7 | 55 ± 8 | 40 ± 5 |
| ATG7 | 136 ± 12 | 47 ± 6 | 76 ± 16 |
| LAMP2 | 132 ± 12 | 35 ± 8 | 104 ± 19 |
| HDAC6 | 146 ± 9 | 71 ± 4 | 108 ± 7 |
| Sub-G1,† % | |||
| HDAC10 #1 | 28 ± 2 | 28 ± 1 | 49 ± 2 |
| HDAC10 #2 | 19 ± 3 | 47 ± 4 | |
| HSC70/HSPA8 | 60 ± 3 | 28 ± 1 | 78 ± 1 |
| BECN1 | 10 ± 2 | 28 ± 1 | 37 ± 3 |
| ATG7 | 12 ± 0.2 | 28 ± 1 | 22 ± 1 |
| LAMP2 | 12 ± 1 | 28 ± 1 | 34 ± 2 |
| HDAC6 | 15 ± 2 | 28 ± 1 | 27 ± 2 |
| Mean intensity of stained colonies (10 d)‡ | |||
| HDAC10 #1 | 46 ± 5 | 8 ± 1 | 0.8 ± 0.7 |
| HDAC10 #2 | 16 ± 1 | 0.1 ± 0.1 | |
| HSC70/HSPA8 | 43 ± 9 | 8 ± 1 | 0.1 ± 0.1 |
| BECN1 | 121 ± 1 | 8 ± 1 | 5 ± 1 |
| ATG7 | 76 ± 1 | 8 ± 1 | 24 ± 7 |
| LAMP2 | 100 ± 3 | 8 ± 1 | 3 ± 1 |
| HDAC6 | 122 ± 6 | 8 ± 1 | 18 ± 4 |
Relative metabolic activity measured with WST-1 assay, normalized to NC siRNA and displayed as means ± SEM. 6 d after knockdown and 72 h after drug treatment.
Percentage of PI-stained cells in the sub-G1 area of the cell cycle profile ± SEM. 6 d after knockdown and 72 h after drug treatment.
Mean intensity of crystal violet-stained colonies quantified via ImageJ software ± SEM.
Table 2.
Failed autophagy enhances antineuroblastoma activity of doxorubicin treatment (knockdown studies)
| Target mRNA | Knockdown of RNA expression, % |
| HDAC10 | 67 ± 2 |
| HSC70/HSPA8 | 92 ± 5 |
| BECN1 | 84 ± 3 |
| ATG7 | 71 ± 2 |
| LAMP2 | 84 ± 2 |
| HDAC6 | 69 ± 5 |
Table 3.
Failed autophagy enhances anti-neuroblastoma activity of doxorubicin treatment (inhibitor studies)
| Inhibitor of autophagic flux (applied concentration) | Inhibitor treatment | Doxorubicin treatment (0.1 µg/mL) | Combined inhibitor and doxorubicin treatment |
| Metabolic activity,* % | |||
| 3-MA (10 mM) | 67 ± 8 | 73 ± 8 | 61 ± 5 |
| Wortmannin (100 nM) | 78 ± 5 | 80 ± 1 | 70 ± 2 |
| Chloroquine (15 µM) | 36 ± 7 | 83 ± 7 | 24 ± 3 |
| NH4Cl (5 mM) | 94 ± 20 | 82 ± 7 | 54 ± 8 |
| PES (10 µM) | 28 ± 17 | 84 ± 18 | 8 ± 4 |
| Sub-G1,† % | |||
| 3-MA (10 mM) | 1.6 ± 0.2 | 21 ± 2 | 12 ± 1 |
| Wortmannin (100 nM) | 3.8 ± 0.7 | 21 ± 2 | 22 ± 2 |
| Chloroquine (15 µM) | 19 ± 1 | 21 ± 2 | 41 ± 4 |
| NH4Cl (5 mM) | 5 ± 0.2 | 21 ± 2 | 36 ± 1 |
| PES (10 µM) | 9 ± 1 | 21± 2 | 34 ± 1 |
| Mean intensity of stained colonies (10 d)‡ | |||
| 3-MA (10 mM) | ND | ND | ND |
| Wortmannin (100 nM) | 157 ± 7 | 16 ± 1 | 14 ± 4 |
| Chloroquine (15 µM) | 19 ± 5 | 16 ± 1 | 1 ± 0.2 |
| NH4Cl (5 mM) | 107 ± 4 | 16 ± 1 | 2 ± 0.5 |
| PES (10 µM) | 37 ± 5 | 16 ± 1 | 0.5 ± 0.1 |
ND, not determined.
Relative metabolic activity measured with WST-1 assay, normalized to NC siRNA and displayed as means ± SEM. 6 d after inhibitor treatment and 72 h after drug treatment.
Percentage of PI-stained cells in the sub-G1 area of the cell cycle profile ± SEM. 6 d after inhibitor treatment and 72 h after drug treatment.
Mean intensity of crystal violet stained colonies quantified via ImageJ software ± SEM.
Because we determined that sensitization of neuroblastoma cells toward doxorubicin takes place at the level of the lysosomes and involves Hsc70/Hsp70 (scheme in Fig. 7A), we hypothesized that HDAC10 is involved in controlling this step of autophagic processing to create drug resistance. Recently, Lai et al. reported that HDAC10 was complexed together with deacetylated Hsc70 in HEK293 cells (47). To test whether HDAC10 interacts with Hsp70 family proteins in neuroblastoma cells, we transfected BE(2)-C cells with FLAG-tagged HDAC10 and immunoprecipitated HDAC10-containing complexes. Interestingly, Hsc70/Hsp70 coprecipitated with FLAG-tagged HDAC10 (Fig. 7B and Fig. S7C). Treatment with the HDAC10-selective inhibitors bufexamac and tubastatin A (Fig. S7D), as well as broad-spectrum inhibitor trichostatin A (TSA) (Fig. 7B), reduced the amount of Hsc70/Hsp70 interacting with FLAG-tagged HDAC10. Immunoprecipitation using an antibody recognizing both Hsc70 and Hsp70 demonstrated that endogenous HDAC10 also interacts with Hsc70/Hsp70 (Fig. 7C). TSA treatment eliminated the interaction between endogenous Hsc70/Hsp70 and HDAC10 proteins, which is consistent with data from Lai et al. that HDAC10 binds deacetylated Hsc70 (47). HDAC10 overexpression resulted in the loss of detectable Hsc70/Hsp70 acetylation (Fig. 7D), whereas HDAC10 knockdown induced Hsc70/Hsp70 acetylation (Fig. 7E). Colocalization studies confirmed that both proteins colocalize in the cytoplasm of the cells and at perinuclear regions, typically harboring late endosomes (Fig. 7F). To confirm whether the cytotoxic protective function of HDAC10 depends on functional Hsc70/Hsp70, HDAC10 expression was enforced in IMR32 cells, which were then cotreated with the Hsc70/Hsp70 inhibitor PES and doxorubicin. In control experiments, catalytically inactive HDAC10-H135A variant was overexpressed. PES treatment inhibited the interaction of HDAC10 with Hsc70/Hsp70 (Fig. S7E) and abolished the protective function of HDAC10 against doxorubicin (Fig. 7G). These data imply that HDAC10 promotes resistance to doxorubicin via direct interaction with Hsc70/Hsp70, resulting in its deacetylation.
Discussion
Histone deacetylases regulate the processes of differentiation, cell cycle, and apoptosis through the deacetylation of epigenetic, indirect epigenetic and nonepigenetic substrates. However, the cellular and molecular mechanisms controlled by individual HDAC isozymes are poorly understood (8, 30, 48). Here, we report a critical function of HDAC10 in regulating autophagic flux and cell survival. The HDAC class IIb family includes HDAC6 and HDAC10. Whereas numerous publications on HDAC6 function as a microtubule-associated deacetylase involved in the response to misfolded protein stress via degradation by autophagy have been published (49–52), little is known about the molecular and cellular function of HDAC10. HDAC10 has been reported to be involved in melanogenesis via repressor deacetylation (47) and in thioredoxin up-regulation in gastric cancer cells (53). A HDAC10 knockout phenotype has yet to be described. Thus, the identification of HDAC10 as an important regulator of autophagy and lysosomal fusion is an important and previously undescribed cellular function of HDAC10. Knockdown of HDAC10 expression did not affect HDAC6 expression, ruling out a compensatory mechanism. Neither treatment with the HDAC6-selective inhibitor tubacin nor HDAC6 knockdown induced AVO accumulation, indicating that these two class IIb HDAC family members have different functions in regulating protein degradation in neuroblastoma cells. We have reported previously that HDAC8 controls differentiation and that HDAC2 controls apoptosis in neuroblastoma cells (29), underscoring the selective cellular function of individual HDAC isozymes in this tumor type. Intriguingly, of all 11 classical HDACs, only the expression of HDAC10 was a significant biomarker separating the clinical outcome of advanced (stage 4) neuroblastoma patients.
Autophagy is a complicated process that can have opposing effects on tumorigenic and tumor progression processes. In advanced-stage tumors, autophagy very likely represents a prosurvival mechanism helping cells to cope with cellular stress and resist cytotoxic treatments. DNA damage-inducing drugs, such as doxorubicin, have been described to induce autophagy in tumor cells as a mechanism for survival and resistance (1, 40). We show that productive autophagy with efficient autophagosome-lysosome fusion depends on HDAC10 and that HDAC10 depletion enhances the susceptibility of tumor cells to chemotherapeutic drugs, such as doxorubicin. Of note, HDAC10 depletion itself impaired viability of the tumor cells, very likely through the inefficient clearing of mitochondria and increased ROS production. In line with these results, enforced HDAC10 expression mediated doxorubicin resistance. HDAC6 knockdown did not enhance susceptibility but enhanced resistance of neuroblastoma cells to doxorubicin. We also demonstrated that HDAC10 interacts with Hsps in neuroblastoma cells. The major members of the Hsp70 family are Hsc70 and Hsp70, both controlling many important cellular functions, including protein folding, ubiquitin-mediated protein degradation, lysosomal protein degradation, and lysosomal membrane integrity (reviewed in ref. 54). It is tempting to speculate that the interaction of HDAC10 with Hsp70 family proteins, which results in their deacetylation, stabilizes lysosomal function and membrane integrity (44). However, further experiments are necessary to unravel the direct molecular link.
We showed that HDAC10 expression in primary tumors signaled poor survival of advanced stage neuroblastoma patients. HDAC10 expression also correlated with autophagy in gene-set analysis, primarily with ATG4D, which regulates autophagosome biogenesis through LC3 processing (55). Thus, our results identify HDAC10 as a regulator of neuroblastoma cell survival and a potential biomarker for autophagy dependency and the prediction of chemotherapy response in patients with high-risk neuroblastomas, as well as being potentially useful for patients with other highly malignant pediatric tumors of the nervous system. Interruption of autophagic flux by selective inhibition of HDAC10 functionality for the lysosomes through an Hsc70/Hsp70-dependent mechanism (see also Fig. 7A) could represent a promising approach to sensitize aggressive neuroblastomas to chemotherapy. Taken together, these results identify HDAC10 as a druggable regulator of advanced-stage tumor cell survival and a potential marker for predicting therapy response in the neuroblastoma subgroup with the poorest outcome of all. Implementation of this approach in clinical trials may dramatically improve the survival of children who currently have almost no chance of cure.
Materials and Methods
Cell Culture.
Cell culture and transient transfections were performed as described previously (29). Details are supplied in SI Materials and Methods.
Reagents.
The class IIb inhibitors bufexamac (Sigma) (36), tubastatin (Biozol; 5 mM stock), and tubacin (Enzo Life Sciences; 1 mM stock) were dissolved in DMSO. Bafilomycin A1 (10 µM stock), rapamycin (100 µM stock), and wortmannin (1 mM stock) were all obtained from Sigma and dissolved in DMSO. Chloroquine (Sigma; 100 mM stock), 3-methyladenine (3-MA) (Sigma; 200 mM stock), NH4Cl (Carl Roth; 1 M stock), PES (Sigma; 10 mM stock), doxorubicin (Biozol; 1 mg/mL stock), and vincristine (Enzo Life Sciences, 5 mM stock) were dissolved in H2O. Determination of Kd values for tubastatin were performed as described in ref. 56. The details of Kd value calculation is supplied in SI Materials and Methods.
Real-Time Reverse Transcription–PCR.
Real-time reverse transcription–PCR was performed as described previously (29). Primer pairs are listed in SI Materials and Methods.
Coimmunoprecipitation.
Cells were lysed in 50 mM Tris⋅HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100 buffer containing a protease inhibitor mixture (Roche). Total cell lysates were precleared for 1 h with anti-rabbit IgG TrueBlot beads (eBioscience) and then incubated for 2.5 h with antibodies at 4 °C. Immunocomplexes were captured with anti-rabbit IgG TrueBlot beads (eBioscience) or with anti-FLAG M2-conjugated agarose (Sigma) by incubation at 4 °C for 2 h on a rotatory mixer. Coprecipitated proteins were detected using Western blotting as described previously (29). Antibodies are listed in SI Materials and Methods.
Cell-Viability and Cell-Death Assays.
Cell viability was measured by automated trypan blue staining on the Vi-Cell XR Cell Viability Analyzer (Beckman Coulter). Apoptosis quantification of propidium iodide (PI) stained and ethanol-fixed cells by flow cytometry was performed as described previously (57).
For detection of PARP cleavage, anti-PARP (4C10-5; BD Pharmingen) was used. For all cell death assays, trypsinized cells were pooled with corresponding supernatant, centrifuged, and resuspended in cell culture media. Metabolic activity was colorimetrically determined using the WST-1 assay (Roche) according to the manufacturer’s instructions.
Colony Assay.
In six-well plates, 1,000–30,000 cells, depending on the cell line, were seeded and treated as indicated. Viable colonies were stained after 10 d with crystal violet. For quantification, the mean intensity of each well of the 8-bit binary picture was measured with ImageJ software.
Acridine Orange, EGFP-LC3, and ROS Detection.
The acidotropic dye acridine orange was used to detect and quantify AVOs. Acridine orange undergoes a shift from green to red fluorescence in functional (auto)lysosomes (58). Cells were stained with 1 μg of acridine orange (Sigma) per milliliter of phenol red-free medium for 30 min at 37 °C and then collected and resuspended in 300 µL of phenol red-free medium for FACS analysis. Green (510–530 nm) and red (>650 nm) fluorescent emission from living cells stimulated with blue laser (488 nm) was detected using the FACSCalibur flow cytometer (Becton Dickinson). Autophagic flux was determined based on pH quenching and lysosomal degradation of EGFP-LC3 (59). EGFP fluorescence of living cells was determined with FACSCalibur flow cytometer (Fl-1 channel: 510–530 nm) following transfection and indicated drug treatments. ROS generation was quantified using 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (Molecular Probes) according to the manufacturer’s instructions. Cells were incubated for 20 min with 5 µM dye, and fluorescence intensity was measured using FACSCalibur flow cytometer. Mitochondria were stained and quantified using MitoTracker Green (Invitrogen) according to the manufacturer’s instructions. Cells were incubated for 20 min with 50 nM dye, and fluorescence intensity was measured using FACSCalibur flow cytometer. All FACSCalibur data were analyzed using the CellQuest Pro software.
Electron and Fluorescent Microscopy.
For electron microscopy, 6 × 104 cells were seeded onto glass slides in six-well plates. Cells were fixed in Karnovsky’s solution [4% (wt/vol) paraformaldehyde and 2% (wt/vol) glutaraldehyde] 72 h after transfection and then postfixed in 2% (wt/vol) osmium tetroxide in 0.2 M cacodylate buffer, before step-wise dehydration in ethanol and embedding in araldite. Ultrathin sections (60–70 nm) were cut on an ultracut UCT microtome (Leica). Sections were contrasted with uranyl acetate and lead citrate and analyzed on an EM 900 electron microscope (Zeiss). For immunostaining, cells were fixed onto glass slides with 2% paraformaldehyde in PBS and then permeabilized with 0.1% (vol/vol) Triton X-100 in PBS before incubation with primary antibodies for 2 h and secondary antibodies for 1 h at room temperature. Primary antibodies were as follows: rabbit anti-Hsp70/Hsc70 (H-300; Santa Cruz Biotechnology) and mouse anti-FLAG-M2 (Sigma). Secondary antibodies were as follows: Cy3-labeled anti-rabbit IgG (Molecular Probes) and Alexa488-labeled anti-mouse IgG (Molecular Probes). Immunostained cells and cells transfected with EGFP-LC3 or mCherry-EGFP-LC3B were viewed using the 63× objective on an LSM700 laser-scanning confocal microscope (Zeiss).
Web-Based Gene-Expression Analysis.
R2 (http://r2.amc.nl) and the online oncogenomics database (http://pob.abcc.ncifcrf.gov/cgi-bin/JK) were used to investigate HDAC10 mRNA expression in previously analyzed cohorts of primary neuroblastomas (AMC und NCI) and medulloblastomas (Heidelberg), with the respective GEO accession nos. GSE16476 (neuroblastoma AMC) and GSE28245 (medulloblastoma Heidelberg). The following probe sets were used to detect HDAC10 expression: neuroblastoma (AMC), 226672_s_at; neuroblastoma (NCI), 811025; and medulloblastoma (Heidelberg), A_23_P211673. Patient characteristics for the neuroblastoma (AMC), neuroblastoma (NCI), and medulloblastoma (Heidelberg) cohorts were previously published (31–33).
Statistical Analysis.
All cell culture experiments were performed in duplicate or triplicate, and each experiment was repeated at least three times. A two-tailed t test was performed using GraphPad Prism Version 3.00 (GraphPad Software) to compare treatment groups. P values of less than 0.05 were considered as significant. The Bonferroni-corrected P value, which denotes the significance in survival potential when performing a scan, was calculated to compare Kaplan–Meier curves.
Supplementary Material
Acknowledgments
We thank C. Rütz, R. Straub, and A. Bittmann for excellent technical assistance, and K. Astrahantseff for manuscript editing. We acknowledge the support of the German Cancer Research Center (DKFZ) Light Microscopy Facility. Dr. T.-P. Yao kindly provided the HDAC10 constructs. This work was supported by German Federal Ministry of Education and Research NGFNplus Grant 01GS0896, a Heinrich F. C. Behr stipend from the DKFZ (to J.-P.L.), the FRONTIER and OLYMPIA MORATA programs of the University of Heidelberg (H.E.D.), a B. Braun Foundation grant (to T.M.), and a Wilhelm Sander Foundation grant (to T.M. and I.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.A.J. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300113110/-/DCSupplemental.
References
- 1.Pan Y, et al. Targeting autophagy augments in vitro and in vivo antimyeloma activity of DNA-damaging chemotherapy. Clin Cancer Res. 2011;17(10):3248–3258. doi: 10.1158/1078-0432.CCR-10-0890. [DOI] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3(3):181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 3.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290(5497):1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dice JF. Chaperone-mediated autophagy. Autophagy. 2007;3(4):295–299. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- 5.Agarraberes FA, Dice JF. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J Cell Sci. 2001;114(Pt 13):2491–2499. doi: 10.1242/jcs.114.13.2491. [DOI] [PubMed] [Google Scholar]
- 6.Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28(18):5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5(9):769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 8.Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: What are the cancer relevant targets? Cancer Lett. 2009;277(1):8–21. doi: 10.1016/j.canlet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 9.Glaser KB. HDAC inhibitors: Clinical update and mechanism-based potential. Biochem Pharmacol. 2007;74(5):659–671. doi: 10.1016/j.bcp.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Shao Y, Gao Z, Marks PA, Jiang X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2004;101(52):18030–18035. doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Q, et al. Autophagy induced by suberoylanilide hydroxamic acid in Hela S3 cells involves inhibition of protein kinase B and up-regulation of Beclin 1. Int J Biochem Cell Biol. 2008;40(2):272–283. doi: 10.1016/j.biocel.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 12.Thomas S, Thurn KT, Biçaku E, Marchion DC, Münster PN. Addition of a histone deacetylase inhibitor redirects tamoxifen-treated breast cancer cells into apoptosis, which is opposed by the induction of autophagy. Breast Cancer Res Treat. 2011;130(2):437–447. doi: 10.1007/s10549-011-1364-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez G, Torres K, Lev D. Autophagy blockade enhances HDAC inhibitors’ pro-apoptotic effects: Potential implications for the treatment of a therapeutic-resistant malignancy. Autophagy. 2011;7(4):440–441. doi: 10.4161/auto.7.4.14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu YL, et al. Autophagy potentiates the anti-cancer effects of the histone deacetylase inhibitors in hepatocellular carcinoma. Autophagy. 2010;6(8):1057–1065. doi: 10.4161/auto.6.8.13365. [DOI] [PubMed] [Google Scholar]
- 15.Carew JS, et al. Autophagy inhibition enhances vorinostat-induced apoptosis via ubiquitinated protein accumulation. J Cell Mol Med. 2010;14(10):2448–2459. doi: 10.1111/j.1582-4934.2009.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto S, et al. Suberoylanilide hydroxamic acid (SAHA) induces apoptosis or autophagy-associated cell death in chondrosarcoma cell lines. Anticancer Res. 2008;28(3A):1585–1591. [PubMed] [Google Scholar]
- 17.Carew JS, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110(1):313–322. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 19.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90(4):595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 20.Hubbert C, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417(6887):455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 21.Bali P, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: A novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280(29):26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 22.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem J. 2003;370(Pt 3):737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem. 2002;277(28):25748–25755. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez-Gonzalez A, et al. Role of the aggresome pathway in cancer: Targeting histone deacetylase 6-dependent protein degradation. Cancer Res. 2008;68(8):2557–2560. doi: 10.1158/0008-5472.CAN-07-5989. [DOI] [PubMed] [Google Scholar]
- 25.Guardiola AR, Yao TP. Molecular cloning and characterization of a novel histone deacetylase HDAC10. J Biol Chem. 2002;277(5):3350–3356. doi: 10.1074/jbc.M109861200. [DOI] [PubMed] [Google Scholar]
- 26.Brodeur GM. Neuroblastoma: Biological insights into a clinical enigma. Nat Rev Cancer. 2003;3(3):203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 27.Berthold F, et al. Myeloablative megatherapy with autologous stem-cell rescue versus oral maintenance chemotherapy as consolidation treatment in patients with high-risk neuroblastoma: A randomised controlled trial. Lancet Oncol. 2005;6(9):649–658. doi: 10.1016/S1470-2045(05)70291-6. [DOI] [PubMed] [Google Scholar]
- 28.Milde T, et al. HDAC5 and HDAC9 in medulloblastoma: Novel markers for risk stratification and role in tumor cell growth. Clin Cancer Res. 2010;16(12):3240–3252. doi: 10.1158/1078-0432.CCR-10-0395. [DOI] [PubMed] [Google Scholar]
- 29.Oehme I, et al. Histone deacetylase 8 in neuroblastoma tumorigenesis. Clin Cancer Res. 2009;15(1):91–99. doi: 10.1158/1078-0432.CCR-08-0684. [DOI] [PubMed] [Google Scholar]
- 30.Oehme I, Deubzer HE, Lodrini M, Milde T, Witt O. Targeting of HDAC8 and investigational inhibitors in neuroblastoma. Expert Opin Investig Drugs. 2009;18(11):1605–1617. doi: 10.1517/14728220903241658. [DOI] [PubMed] [Google Scholar]
- 31.Molenaar JJ, et al. Copy number defects of G1-cell cycle genes in neuroblastoma are frequent and correlate with high expression of E2F target genes and a poor prognosis. Genes Chromosomes Cancer. 2012;51(1):10–19. doi: 10.1002/gcc.20926. [DOI] [PubMed] [Google Scholar]
- 32.Chen QR, et al. An integrated cross-platform prognosis study on neuroblastoma patients. Genomics. 2008;92(4):195–203. doi: 10.1016/j.ygeno.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Remke M, et al. FSTL5 is a marker of poor prognosis in non-WNT/non-SHH medulloblastoma. J Clin Oncol. 2011;29(29):3852–3861. doi: 10.1200/JCO.2011.36.2798. [DOI] [PubMed] [Google Scholar]
- 34.Bjørkøy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171(4):603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bantscheff M, et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat Biotechnol. 2011;29(3):255–265. doi: 10.1038/nbt.1759. [DOI] [PubMed] [Google Scholar]
- 37.Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci USA. 2003;100(8):4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molenaar JJ, et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat Genet. 2012;44(11):1199–1206. doi: 10.1038/ng.2436. [DOI] [PubMed] [Google Scholar]
- 39.Chen N, Karantza V. Autophagy as a therapeutic target in cancer. Cancer Biol Ther. 2011;11(2):157–168. doi: 10.4161/cbt.11.2.14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hundeshagen P, Hamacher-Brady A, Eils R, Brady NR. Concurrent detection of autolysosome formation and lysosomal degradation by flow cytometry in a high-content screen for inducers of autophagy. BMC Biol. 2011;9:38. doi: 10.1186/1741-7007-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gogolin S, et al. MYCN/MYC-mediated drug resistance mechanisms in neuroblastoma. Int J Clin Pharmacol Ther. 2010;48(7):489–491. doi: 10.5414/cpp48489. [DOI] [PubMed] [Google Scholar]
- 42.Nylandsted J, et al. Heat shock protein 70 promotes cell survival by inhibiting lysosomal membrane permeabilization. J Exp Med. 2004;200(4):425–435. doi: 10.1084/jem.20040531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doulias PT, et al. Involvement of heat shock protein-70 in the mechanism of hydrogen peroxide-induced DNA damage: The role of lysosomes and iron. Free Radic Biol Med. 2007;42(4):567–577. doi: 10.1016/j.freeradbiomed.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 44.Kirkegaard T, et al. Hsp70 stabilizes lysosomes and reverts Niemann-Pick disease-associated lysosomal pathology. Nature. 2010;463(7280):549–553. doi: 10.1038/nature08710. [DOI] [PubMed] [Google Scholar]
- 45.Leu JI, Pimkina J, Pandey P, Murphy ME, George DL. HSP70 inhibition by the small-molecule 2-phenylethynesulfonamide impairs protein clearance pathways in tumor cells. Mol Cancer Res. 2011;9(7):936–947. doi: 10.1158/1541-7786.MCR-11-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leu JI, Pimkina J, Frank A, Murphy ME, George DL. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell. 2009;36(1):15–27. doi: 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai IL, et al. Histone deacetylase 10 relieves repression on the melanogenic program by maintaining the deacetylation status of repressors. J Biol Chem. 2010;285(10):7187–7196. doi: 10.1074/jbc.M109.061861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Witt O, Deubzer HE, Lodrini M, Milde T, Oehme I. Targeting histone deacetylases in neuroblastoma. Curr Pharm Des. 2009;15(4):436–447. doi: 10.2174/138161209787315774. [DOI] [PubMed] [Google Scholar]
- 49.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280(48):40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 50.Kawaguchi Y, et al. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115(6):727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 51.Pandey UB, Batlevi Y, Baehrecke EH, Taylor JP. HDAC6 at the intersection of autophagy, the ubiquitin-proteasome system and neurodegeneration. Autophagy. 2007;3(6):643–645. doi: 10.4161/auto.5050. [DOI] [PubMed] [Google Scholar]
- 52.Lee JY, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29(5):969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee JH, et al. Inhibition of histone deacetylase 10 induces thioredoxin-interacting protein and causes accumulation of reactive oxygen species in SNU-620 human gastric cancer cells. Mol Cells. 2010;30(2):107–112. doi: 10.1007/s10059-010-0094-z. [DOI] [PubMed] [Google Scholar]
- 54.Daugaard M, Rohde M, Jäättelä M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007;581(19):3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 55.Mariño G, et al. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem. 2003;278(6):3671–3678. doi: 10.1074/jbc.M208247200. [DOI] [PubMed] [Google Scholar]
- 56.Sharma K, et al. Proteomics strategy for quantitative protein interaction profiling in cell extracts. Nat Methods. 2009;6(10):741–744. doi: 10.1038/nmeth.1373. [DOI] [PubMed] [Google Scholar]
- 57.Oehme I, Bösser S, Zörnig M. Agonists of an ecdysone-inducible mammalian expression system inhibit Fas Ligand- and TRAIL-induced apoptosis in the human colon carcinoma cell line RKO. Cell Death Differ. 2006;13(2):189–201. doi: 10.1038/sj.cdd.4401730. [DOI] [PubMed] [Google Scholar]
- 58.Moriyama Y, Takano T, Ohkuma S. Acridine orange as a fluorescent probe for lysosomal proton pump. J Biochem. 1982;92(4):1333–1336. doi: 10.1093/oxfordjournals.jbchem.a134053. [DOI] [PubMed] [Google Scholar]
- 59.Shvets E, Fass E, Elazar Z. Utilizing flow cytometry to monitor autophagy in living mammalian cells. Autophagy. 2008;4(5):621–628. doi: 10.4161/auto.5939. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.