Abstract
Leukemia and lymphoma account for more than 60% of deaths in captive koalas (Phascolarctos cinereus) in northeastern Australia. Although the endogenizing gammaretrovirus koala endogenous retrovirus (KoRV) was isolated from these koalas, KoRV has not been definitively associated with leukemogenesis. We performed KoRV screening in koalas from the San Diego Zoo, maintained for more than 45 y with very limited outbreeding, and the Los Angeles Zoo, maintained by continuously assimilating captive-born Australian koalas. San Diego Zoo koalas are currently free of malignant neoplasias and were infected with only endogenous KoRV, which we now term subtype “KoRV-A,” whereas Los Angeles Zoo koalas with lymphomas/leukemias are infected in addition to KoRV-A by a unique KoRV we term subtype “KoRV-B.” KoRV-B is most divergent in the envelope protein and uses a host receptor distinct from KoRV-A. KoRV-B also has duplicated enhancer regions in the LTR associated with increased pathology in gammaretroviruses. Whereas KoRV-A uses the sodium-dependent phosphate transporter 1 (PiT1) as a receptor, KoRV-B employs a different receptor, the thiamine transporter 1 (THTR1), to infect cells. KoRV-B is transmitted from dam to offspring through de novo infection, rather than via genetic inheritance like KoRV-A. Detection of KoRV-B in native Australian koalas should provide a history, and a mode for remediation, of leukemia/lymphoma currently endemic in this population.
Mammals have been infected with retroviruses for millions of years, and sometimes these viruses remain in an exogenous state following integration into the host genome, replicating to high titers to increase the likelihood of onward transmission and, in some cases, pathologic conditions. Retroviruses can also exist in an endogenous state after integrating into the host germ line. These endogenous retroviruses (ERVs) are genetically inherited and are present in all cells of the offspring. Most ERVs degenerate as a consequence of recombination or mutations that disrupt genes encoding retroviral proteins (1). Before the discovery of the koala ERV (KoRV), mammalian ERVs were thought to be millions of years old and not, for the most part, to produce infectious particles. KoRV has been estimated to be ∼100 y old (2) and retains its ability to form infectious virus capable of transpecies transmission (3). As a result of the recency of the endogenization event, the koala genome contains inherited and newly integrated forms of KoRV. The latter represents infectious KoRV and may represent a combination of unknown variations. However, KoRV isolates so far detected in Australia, and in zoos in Germany and Japan, have shown very little genetic diversity (>99% sequence identity). KoRV, like its closely related virus originally obtained from a gibbon with lymphoma, gibbon ape leukemia virus (GALV) (4), has been etiologically linked to leukemias and lymphomas resulting in increased mortality. Although elevated plasma viremia has been linked to increased GALV-associated pathologic processes (5), the invariable genetic identity does not resemble the role that other leukogenic gammaretroviruses play in disease development and secondary transmission such as murine leukemia virus (MLV) or feline leukemia virus (FeLV). Specifically, FeLV and MLV show significant differences in sequence variability especially in their envelope (env) genes that frequently results in different FeLV or MLV isolates that use distinct receptors to infect cells.
To investigate diversity among KoRV isolates, we assessed the infection status of 13 koalas in the United States from a cohort of animals housed at the Los Angeles Zoo (LAZ) and 28 from the San Diego Zoo (SDZ). All contain the previously characterized endogenous KoRV (4). Because three LAZ koalas from one family group died from neoplastic malignancies previously associated with KoRV, we searched for evidence that KoRV variants might explain the pathologic condition observed in this family.
Results
PCR Amplification of Unique KoRV Envelope.
Initial PCR amplification of viral sequences from specimens obtained from the LAZ was performed by using genomic DNA prepared from blood or tissue and from viral RNA present in plasma, with primers specific to the KoRV env gene and LTR (Figs. 1 and 2). Analysis of env sequences demonstrated the existence in all assessed koalas of a KoRV envelope gene almost identical with those previously obtained (GenBank accession nos. AF151794 and JQ244835–JQ244839) (2, 4) and from cultured peripheral blood mononuclear cells (PBMCs) from koalas in a zoo in Germany (3) (accession no. DQ174772) and a zoo in Japan (6) (GenBank accession no. AF151794). Similar KoRV env sequences were also detected in the plasma and blood of all 28 koalas from the SDZ. Notably, in addition to the exclusively homologous endogenous KoRV detected in previous studies, a unique KoRV env sequence was identified in blood or tissue samples from 6 of 13 koalas from the LAZ, including three koalas that died of lymphoma (Table 1). The death of these three koalas (koalas 2, 4, and 5; Table 1) was attributed to lymphoid leukemias. Postmortem examination of the three LAZ KoRV-B–positive koalas revealed disseminated lymphoid malignancy involving various organs, including bone marrow, spleen, thymus, lymph nodes, brain, adipose tissue, lung, liver, skin, eye, and mammary gland. Seventy-five percent of koala 2’s neoplastic cells stained positive with CD3, and 80% were positive labeled with CD3E antibody. Two other deaths (koalas 1 and 13) were not associated with a malignant neoplasia, and these koalas were KoRV-A–positive but KoRV-B–negative (Table 1). A sixth animal (koala 12) was a KoRV-B–positive joey ejected from pouch of a KoRV-B–positive dam at approximately 1 mo. As a result of the age of the joey, definitive necropsy results related to cause of death could not be obtained.
Fig. 1.
Organization of KoRV proviral genome shows locations of primers used to clone KoRV-B. Gray triangles represent primers specific to KoRV-B. Asterisks indicate primers used in previous studies (5).
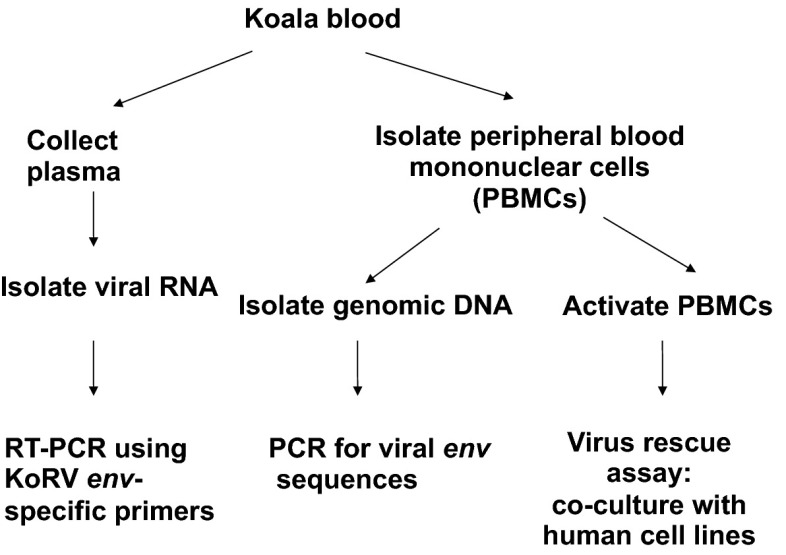
Fig. 2.
Experimental design used to detect KoRV in koala blood samples.
Table 1.
Detection of KoRV-B by PCR in plasma, genomic DNA and using virus rescue assays
| Koala no. | Age, y* | Plasma vRNA | Genomic DNA | Live virus rescue | Necropsy results |
| 1 | 7.6 | NA | —† | NA | Unknown |
| 2‡ | 4.1 | NA | +† | NA | Lymphoma |
| 3‡ | 6.2 | + | +§ | + | — |
| 4‡ | 5.4 | + | +§ | +¶ | Lymphoma |
| 5‡ | 5.5 | + | +† | + | Lymphoma |
| 6 | 11.1 | — | —§ | — | — |
| 7 | 3.3 | — | —§ | — | — |
| 8‡ | 1.8 | + | +§ | + | — |
| 9 | 2.6 | — | —† | — | — |
| 10 | 11.1 | — | —§ | — | — |
| 11 | 7.2 | — | —§ | — | — |
| 12‡ | 0.1 | NA | +† | NA | NA |
| 13 | 0.5 | NA | —† | NA | Pneumonia |
NA, specimens not available.
Live koala ages are calculated as of January 31, 2013. The ages of deceased koalas reflect the age at death.
Tissue sample (koalas 1 and 13, liver; koalas 2 and 5, spleen; koala 12, joey dermis).
KoRV-B–positive animal.
Whole blood sample.
KoRV-A/B propagated in MDTF cells expressing appropriate receptors.
As the present KoRV isolate unique to LAZ koalas is highly related to, but clearly distinct from, the KoRV isolate identified earlier in Australia, Germany, and Japan, and presently in both LAZ and SDZ, we provisionally refer to the KoRV isolate as KoRV-B, and the original isolate as KoRV-A. Detection of KoRV-B env sequences was independently confirmed at the Centers for Disease Control laboratory by using freshly collected blood from multiple time points from one koala (koala 4; Table 1) using a real-time PCR assay with primers designed from the KoRV-B env sequence.
Isolation of KoRV-B Virus.
To isolate KoRV-B, a viral marker rescue assay was developed by using 293T-GFP cells that contain an integrated replication incompetent retroviral vector expressing GFP (Fig. S1). Proteins encoded by an infectious retrovirus can mobilize the GFP-expressing retroviral genome. HT1080 target cells exposed to supernatant collected after coculturing koala PBMCs with 293T-GFP cells expressed GFP (turning green under a fluorescent microscope), indicating GFP vector mobilization by infectious retrovirus from koala PBMCs (Figs. S1 and S2). PCR of infected HT1080 cells by using KoRV-specific primers confirmed the existence of KoRV-A from all nine LAZ resident animals with blood samples available and KoRV-B from four of them. By using primers specific for this unique env sequence and primers flanking 3′ and 5′ LTR (Fig. 1), we obtained the complete genome of KoRV-B (GenBank accession no. KC779547) from genomic DNA obtained from blood samples. Sequence analysis of the KoRV-B genome shows that it differs from KoRV-A mainly in the U3 region of the LTR and env sequences. The U3 region is important in regulating gene expression, as it contains the transcriptional promoter and regulatory sequences (enhancer elements) that can modulate transcription activity. The U3 region present in the 5′ and 3′ LTR direct viral gene expression and can also direct expression of adjacent host genes. When such host genes have oncogenic potential, LTR-mediated activation plays a key role in pathogenicity (7). The U3 region of the KoRV-B LTR has four tandem repeats each 18 nt in length (5′-ACGGAATATCTGTGGTCA-3′); these repeats, by analogy with other retroviruses, contain core enhancer elements (Fig. 3). This element is present only as a single copy in the KoRV-A U3 region and contains a G in place of the A at the fifth position of the motif (Fig. 3).
Fig. 3.
KoRV-B differs from KoRV-A in the LTR U3. An alignment of the nucleotides comprising the LTR regions of KoRV-A and KoRV-B. Nucleotide insertions in KoRV-B LTR are highlighted in gray. U3, R, and U5 boundaries are defined according to those described for the LTR of the KoRV-A genome (GenBank accession no. AF151794). Transcription regulatory signals such as the CAAT sequence, the promoter (TATAAAA), and polyadenylation signals AATAAA are shown within boxes.
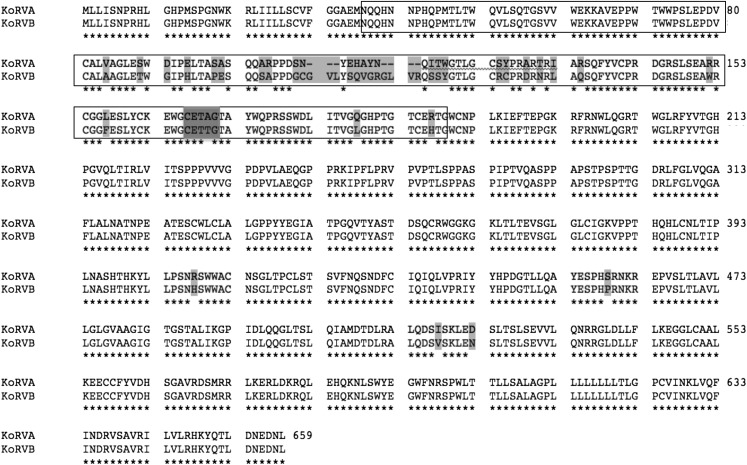
The Env protein encoded by KoRV-B has several distinguishing features. KoRV-B contains a CETTG motif, which is present in the envelope protein receptor-binding domain (RBD) of all infectious gammaretroviruses except KoRV-A isolates as well as all other uninducible endogenous gammaretroviruses (8) (Fig. 4). CETTG is a motif present in all exogenous and inducible endogenous gammaretroviruses but absent in ERVs. Attenuating this motif by introducing CETAG or CGTAG (in KoRVA isolates) in place of CETTG causes a reduction in cytopathic affects induced by fusion from without (8).
Fig. 4.
KoRV-B and KoRV-A differ in their envelope regions: alignment of amino acid residues encoded by the KoRV-A and KoRV-B env genes. Consensus amino acid sequences appear with an asterisk below the residue alignment. Residues that differ between KoRV-A and KoRV-B are highlighted in light gray. The boundary between the surface unit (SU) and transmembrane domain (TM) is shown. Proposed receptor-binding domain (RBD) region is indicated in boxes. The CETAG sequence present in KoRV-A and the canonical motif CETTG found in KoRV-B are highlighted in dark gray.
KoRV-A and KoRV-B Use Distinct Receptors.
Another important feature of KoRV-B is the difference between KoRV-A and KoRV-B RBDs (Fig. 4). As 35 of the 40 amino acid residues that differ between KoRV-A and KoRV-B envelope proteins are in the RBD region, we constructed retroviral vectors pseudotyped with KoRV-A, KoRV-B, or GALV envelopes (Fig. 5B) to assess the functional impact of these differences. It was previously established that KoRV-A and GALV use the same sodium-dependent phosphate transporter membrane protein (SLC20A1, also called PiT1) to infect human cells (9). To assess whether KoRV-B also uses this receptor, KoRV-B pseudotyped vectors were used in interference assays. Interference assays are based on the observation that cells productively infected with a retrovirus are resistant to challenge infection by a second retrovirus that uses the same membrane receptor. As shown in Fig. S3, Top, human HT1080 cells are susceptible to KoRV-A, GALV, and KoRV-B enveloped retroviral vectors (Fig. 5B). HT1080 cells productively infected with a chimeric replication competent virus containing a KoRV-A or GALV envelope (Fig. 5A) are resistant to challenge infection by vectors bearing KoRV-A or GALV envelopes but remain susceptible to vectors with KoRV-B envelopes (Fig. S3, Bottom, and Table 2). Thus, KoRV-B does not use the same receptor as GALV and KoRV-A to infect human cells. Expression of the human cDNA for PiT1 in murine cells rendered these cells susceptible to KoRV-A and GALV but did not confer susceptibility to KoRV-B pseudotyped vectors (Table 2), providing additional proof that KoRV-B does not use human PiT1 as a receptor.
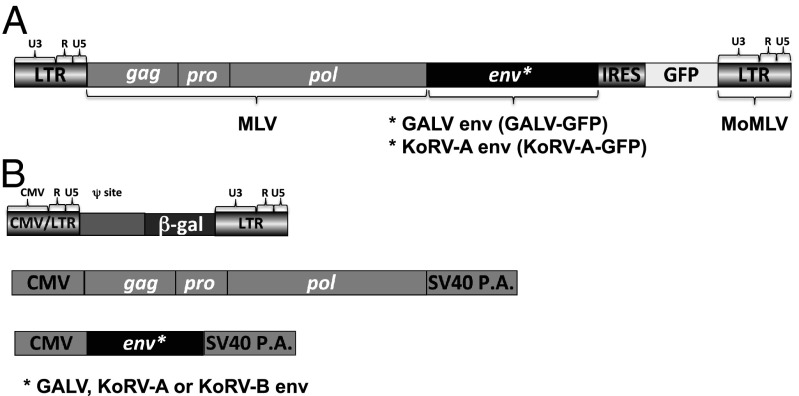
Fig. 5.
(A) Schematic representation of the replication competent retrovirus containing a KoRV-A or GALV as an env gene. IRES, internal ribosome entry site. A replication-competent MoMLV with an IRES-GFP cassette between the env and 3′LTR was used as a template to construct GALV-GFP replacing the MLV env with the GALV env (18). An insertion of TCC just upstream of the splice acceptor (SA) provides the GALV-GFP with improved infection and replication properties (18). Replacement of the GALV env with the KoRV-A env gives rise to replication competent MLV using KoRV-A as an envelope. (B) Plasmids used in transient transfection in 293T cells to produce GALV, KoRV-A, or KoRV-B pseudotyped retroviral vectors referenced in Table 2. After transfection CMV promoter drives the transient expression of MoMLV gagpol, GALV, KoRV-A, or KoRV-B env, and MoMLV LTR-based viral vector genomic RNA encoding nuclear localized β-gal gene.
Table 2.
KoRV-B uses THTR1 (SLC19A2) as a receptor
| Species/cell line | Receptor* | KoRV-A | KoRV-B |
| Human | |||
| HT1080 | — | (3.2 ± 1.0) × 104† | (1.3 ± 0.6) × 105 |
| HT1080/KoRV-A | — | <5 | (7.7 ± 1.8) × 104 |
| HT1080/GALV | — | <5 | (1.0 ± 0.4) × 105 |
| Murine | |||
| MDTF | — | <5 | <5 |
| MDTFPiT1 | hPiT1 | (5.6 ± 3.7) × 104 | <5 |
| MDTFTHTR1 | hTHTR1 | <5 | (3.1 ± 1.0) × 106 |
| MDTFTHTR1/D93H | hTHTR1/D93H | <5 | (2.3 ± 1.0) × 103 |
The letter “h” represents “human.”
β-Gal blue focus forming units per milliliter ± SD obtained from at least three independent experiments.
Gammaretroviruses exhibit a propensity to use as receptors carrier facilitator transporter proteins found on the surface of a wide variety of cell types (10). To investigate further the possible host cell receptor used by KoRV-B, we expressed a panel of known human transporters/gammaretroviral receptors in KoRV-B–resistant murine cells. These included the sodium-dependent phosphate transporter 2 (PiT2 or SLC20A2), the riboflavin receptor SLC52A1 (or PAR2), the sodium-dependent neutral amino acid transporter type 2 (or ASCT2), THTR1 (or SLC19A2), and the functionally uncharacterized gammaretroviral receptor Xpr1. Only THTR1 functioned as a receptor for KoRV-B, as demonstrated by its ability to render previously resistant mus dunni tail fibroblast (MDTF) cells susceptible to KoRV-B infection (Table 2). In contrast, KoRV-A was not able to use THTR1 but retained the ability to use PiT1. We were also able to propagate KoRV-B by exposing MDTF-THTR1 cells to supernatant from the coculture of 293T-GFP cells and PBMCs (Fig. S2) of koala 4 (Table 1).
In general, gammaretroviral receptor and transporter function are not coupled within the gammaretroviral receptor/transporter protein family (11). To investigate whether THTR1 requires retention of its ability to transport thiamine to facilitate KoRV-B entry, we used a point mutant of THTR1 (D93H) that lacks the thiamine transporter function but traffics to the cell membrane like the native protein (12). This mutant transporter retains KoRV-B receptor properties, albeit at a reduced efficiency, when expressed in KoRV-B–resistant murine cells (Table 2). Thus, KoRV-B infection of cells expressing THTR1 is independent of the thiamine transporter function of THTR1.
KoRV-B Is Transmitted from Infected Dam to Offspring.
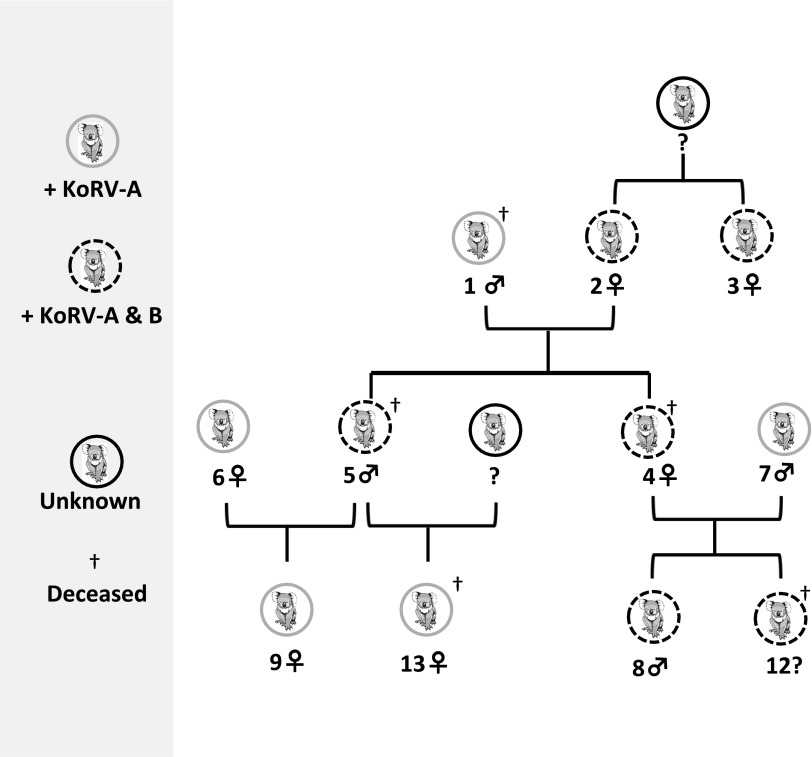
GALV transmission has been reported to occur in utero, postnatally and from infected gibbons to uninfected gibbons through contact transmission (13), and via exposure to feces (14). That KoRV transmission occurs in a similar manner, and does not appear to be vertically transferred in the germ line, is suggested by the observation that KoRV-B–positive offspring (joeys) within the LAZ study group all arose from KoRV-B–positive dams, whereas joeys with KoRV-B–positive sire and KoRV-B–negative dams were not infected with KoRV-B. As shown in Fig. 6, all KoRV-B–positive koalas are in one genetically linked family group, in which two KoRV-B–positive joeys (koalas 4 and 5) had a KoRV-B–positive dam (koala 2) and a KoRV-B–negative sire (koala 1). The KoRV-B–positive sire (koala 5) had two KoRV-B–negative joeys (koalas 9 and 13) with a KoRV-B–negative dam (koala 6), and the KoRV infection status of the dam of offspring koala 13 is unknown (Fig. 6). Furthermore, two KoRV-B–positive joeys (koalas 8 and 12) were born to a KoRV-B–positive dam 4 and a KoRV-B–negative sire koala 7 (Fig. 6). Two of six KoRV-B–positive koalas were Australian-born (koalas 2 and 3), and the rest were born in US zoos. For the KoRV-B–negative koalas from the LAZ population, four were captive-born Australia koalas (koalas 1, 6, 10, and 11) and one was born at LAZ. Therefore, Australian-born koalas at the LAZ contain a mixture of KoRV-B–positive and KoRV-B–negative animals. Necropsy tissue from a 6-wk-old joey (koala 12) that died in pouch and was ejected from dam 4 was KoRV-B–positive, suggesting that KoRV-B may also be transmitted in utero or in milk ingested in pouch.
Fig. 6.
Dam to offspring transmission of KoRV-B, but not sire to offspring transmission. Standard sex symbols denoting male and female are used in the family tree.
Discussion
KoRV-A isolates are unusual in their high degree of genetic conformity. Of the 38 koalas analyzed from SDZ and LAZ, all contain KoRV-A sequences, most of the envelope sequences are closely related to, or, in many cases, identical to the previously reported KoRV-A envelope sequences (2–4). KoRV-A envelope variants have been reported (2, 9) (GenBank accession nos. ABH05084 and ABH05085); however, none of these variants are as divergent as the envelope sequence variation observed between the five exogenous GALV isolates (15, 16). KoRV-B (GenBank accession no. KC779547) represents a degree of genetic diversity not previously observed among all reported KoRV-A isolates. This diversity results in distinct phenotypic features. KoRV-B employs the thiamine transporter THTR1 as a receptor. Interestingly, FeLV subtype A, a strain of FeLV that is transmitted horizontally from cat to cat, also uses THTR1 as a receptor (17). Even though KoRV-B and FeLV-A use the same transporter THTR1 to infect cells, FeLV-A has a host range restricted to feline cells (7). By contrast, KoRV-B can infect a wide range of cells, derived from different species, including human cell lines (Fig. S3), in culture, consistent with the ability of KoRV-B to use divergent THTR1 orthologues. Replication-competent KoRV-B and KoRV-A can infect and be passaged in MDTF-THTR1 or MDTF-PiT1 cells, respectively, as evident by the ability of mobilized RT43.2GFP to be passaged multiple rounds in MDTF-THTR1 or MDTF-PiT1 cells (Fig. S2). In addition to differences in receptor employment, three potential BRCA1 binding sites (18) distinguish KoRVB from KoRVA, and are present in the U3 tandem repeat regions of KoRV-B and two other potential BRCA1 binding sites in the KoRV-B U3 not found in the corresponding nucleotide stretch from 64 to 95 of KoRV-A are contained in KoRVB (Fig. 3). Additional potential transcription factor binding sites present in the U3 of KoRV-B but not KoRV include those for transcription factors TCF3, GATA-1, MafG, Myb, and Spi-B binding (18).
All koalas examined at the SDZ and the LAZ were born in the 21st century. Seven of the 28 SDZ koalas died or were euthanized as a result of complications of nonmalignant disease including degenerative age-related conditions, anemia and associated bone marrow hypoplasia, intussusception, or degenerative joint disease. Historically, at least nine koalas at SDZ not in our study population died from leukemia/lymphoma, and testing of archived tissues, if available, for KoRV-B is needed to determine the strength of an association of this virus with disease. It is also unclear if anemia or bone marrow hypoplasia is associated with KoRV infection. Examination of the koala pedigrees at LAZ showed that the first generation of KoRV-B–positive koalas was born to sires and dams bred and born in Australia. When combined with the negative results for koalas currently at the SDZ, these findings suggest that KoRV-B may have originated in Australian koalas and then spread within this single zoo, but this hypothesis will require expanded testing of captive and wild animals for confirmation.
Discovering the origin of KoRV-B and how it acquired its distinct genotypic and phenotypic changes will require additional studies. Given the prevalence of KoRV-A in koalas, it is possible that KoRV-B is a recombinant between infectious KoRV-A and KoRV sequences present in the koala genome. The rise of a new retroviral subtype from such recombination has occurred in the FeLV family of retroviruses. FeLV subgroup B is a recombinant of exogenous FeLV-A and endogenous genomic feline sequences. Whether KoRV-A serves as a founder virus in a manner analogous to FeLV-A giving rise to different KoRV subgroups or variants in addition to KoRV-B will need further investigation. Sequencing the koala genome will help resolve the composition of endogenous retroviral fragments that may have contributed to the generation of the KoRV-B subgroup and other KoRV subgroups or variants.
The correlation between the presence of KoRV-B infectious virus and malignant disease in koalas is strong even though the assessed sample size is small and we cannot exclude participation of KoRV-A in the observed pathologic conditions. Nonetheless, the ability to assess KoRV-B status, and therefore the likelihood of susceptibility to neoplastic malignancy, could be of tremendous importance in sustaining and managing the koala population in captivity and understanding better the epidemiology of KoRV infection. Furthermore, assessment of KoRV-B infection in the Australian native koala population may provide important insights into the prevalence and spread of KoRV-B that potentially threatens the existence of the koala species as a whole. Preventing KoRV-B–positive dams from breeding, sequestering KoRV-B–positive koalas from the rest of the koala population, and treating KoRV-B–positive animals with antiretroviral agents may all be sensible approaches to reducing the impact of KoRV-B infection on the koala population. It will also be important to reassess the role of KoRV-A alone, or in concert with KoRV-B, in the pathologic processes associated with these viruses.
Materials and Methods
Sample Collection.
Most SDZ and LAZ samples were collected during 2010 to 2012; tissue samples of animals 1, 2, and 13 were collected at necropsy in 2008 and 2009. EDTA-treated whole blood samples were collected opportunistically from nine koalas housed at the LAZ and 25 koalas at the SDZ in accordance with their Institutional Animal Care and Use Committee protocols. Various tissues from four koalas and an aborted joey were collected at necropsy and stored at −80 °C.
Plasmids.
The human THTR1 (huTHTR1) expressing plasmid pLSN-huTHTR1, the feline THTR1 plasmid pFB-neo-FeTHTR1, and pGALV-GFP were provided by Julie Overbaugh (Fred Hutchinson Cancer Research Center, Seattle, WA), Chet Tailor (University of Toronto, Toronto, Canada), and Christopher Logg (University of California, Los Angeles, CA), respectively. pKoRV-A-GFP was constructed by replacing GALV env in pGALV-GFP (19) with the KoRV-A env at the HpaI and MluI restriction sites. To create phuTHTR1D93H the aspartic acid (residue 93) codon THTR1 was mutated to a histidine codon with complementary primers 5′-CCT GTG TTC CTT GCC ACA CAC TAC CTC CGT TAT AAA CC-3′ and 5′-GGT TTA TAA CGG AGG TAG TGT GTG GCA AGG AAC ACA GG-3′ by using the QuikChange mutagenesis kit (Stratagene) according to the manufacturer’s instructions. KoRV-A and KoRV-B expression plasmids were constructed by cloning KoRV-A and KoRV-B envelope into pCIneo expression vector. All constructs were verified by sequencing.
Cell Lines.
293T human embryonic kidney cells (CCL 11268; American Type Culture Collection), Mus dunni tail fibroblast MDTF cells (20), and human HT1080 cells (CCL-121; American Type Culture Collection) were maintained in DMEM supplied with 10% FBS, penicillin 100 U/mL , and streptomycin 100 µg/mL.
Transfection and Transduction.
A Profection calcium phosphate transfection kit (Promega) was used to transfect 293T cells by using plasmids encoding viral envelope (KoRV-A, KoRV-B, or GALV), Moloney MLV (MoMLV) gagpol, and a retroviral genome encoding β-gal as an indicator. Viral supernatants were then passed through a 0.45-µm syringe filter and stored at −80 °C. To determine the host range of KoRVs, plasmids encoding KoRV-A or B env were used to produce viral vectors. To establish a 293T cell line expressing a replication-incompetent retroviral genome encoding GFP (RT43.2GFP; Fig. S1), a plasmid encoding RT43.2GFP was transfected into 293T cells together with two plasmids encoding VSV-G env and MoMLV gagpol (Fig. 5B). 293T cells were exposed to supernatants containing vector particles for 72 h to establish the 293T-GFP cell line. The expression of GFP in more than 90% of 293T-GFP cells was confirmed by flow cytometry. MDTF cells expressing huTHTR1 or huTHTR1D91H were made by transduction of MDTF with VSV-G enveloped retroviral vectors expressing individual plasmid followed with G418 selection. Transduction of HT1080 cells with GALV-GFP and KoRV-A-GFP replication-competent virus (Fig. 5A) generated HT1080 productively infected with KoRV-A or GALV enveloped viruses. Productive infection was confirmed by flow cytometry showing >90% green fluorescent cells.
Infection.
Target cells (4 × 104 cells per well) were seeded in a 24-well plate. The next day, cells were transduced with viral vectors pseudotyped with KoRV-A, KoRV-B, or GALV env genes in the presence of 10 µg/mL Polybrene. The β-gal (LacZ) gene in the genome of the retroviral vector was used as an indicator of infection. X-Gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) staining was carried out 48 h after exposing cells to supernatant. Vector titer was determined by quantifying the number of blue colonies obtained per milliliter of virus supernatant.
Specimen Preparation and Virus Isolation.
Genomic DNA was extracted from the blood and necropsy tissues by using a Wizard Genomic DNA Purification Kit (Promega). Plasma for RT-PCR testing was prepared by centrifugation of 1 to 2 mL of each blood sample at 1800 × g for 5 min. Plasma was passed through a 0.45-µm filter (Millipore), and 2 to 4 mL of PBS solution was mixed gently with the remaining blood pellet to obtain PBMCs by Ficoll–Hypaque (GE Healthcare) separation. PBMCs were then stimulated with 5 µg/mL phytohemagglutinin in RPMI medium containing 20% (vol/vol) FBS (inactivated at 65 °C for 30 min), penicillin 100 U/mL, and streptomycin 100 µg/mL for 1 wk. Stimulated PBMCs were then cocultured with 293T-GFP cells for 4 to 8 wk, and cell supernatants were collected, filtered through a 0.45-µm syringe, and used to infect HT1080, MDTFTHTR1, or MDTFPiT1 cells. The expression of GFP in KoRV-infected cells was viewed by fluorescent microscopy. KoRV infection was confirmed by PCR of genomic DNA prepared from infected cells.
PCR and RT-PCR.
Primers sequences used for amplification of KoRV-B are listed in Fig. 1. The complete KoRV-B provirus was PCR-amplified by using two primer sets to generate two overlapping halves of the KoRV-B genome. Initial PCR amplification of a 3.2-kb KoRV-B sequence from genomic DNA was accomplished with primers P7 located in polymerase (pol) region and P3 complementary to the end of 3′ U3 region (Fig. 1). The KoRVB genomic sequence accession number is KC779547 in GenBank. PCR was performed by using the Takara PrimeSTAR HS DNA polymerase. KoRV-A env was also detected by using primer pair P3/P7. To produce the 5′ KoRV-B specific provirus fragment, LA Taq DNA polymerase (Takara) was used according to the instructions for large-fragment PCR. By using primer pair P5 (complementary to KoRV-B–specific envelope sequences) and P6 (complementary to 5′ end of LTR; Fig. 1), a 6.4-bp fragment containing 5′ LTR, gagpol, and partial KoRV-B env sequences was obtained by gel purification and cloned by using a TOPO XL PCR Cloning Kit (Invitrogen).
For RT-PCR, viral RNA was isolated from koala plasma using the QIAamp Viral RNA mini kit (Qiagen) in accordance with the manufacturer’s recommendations. Contaminating DNA was removed by using Turbo DNase (Ambion). A SuperScript First-Strand synthesis kit (Invitrogen) was used for first-strand cDNA synthesis of RNA prepared from plasma. For detection of KoRV-B, nested PCR was performed by using primer P1/P5 as the outer primer pair and P2/P4 as the inner primer pair. For each of the analyzed sample, reverse transcriptase-negative and RNA template-negative controls were performed in parallel.
Generic and type-specific primers were also designed for the detection of KoRV and differentiation of each subtype by using real-time PCR (Table S1) at the Centers for Disease Control. The KoRV PCR assays used the following cycling conditions: 95 °C for 9 min, 95 °C for 30 s, and 62 °C for 30 s for 55 cycles using ampliTaqGold (ABI) on a BioRad CFX96 Touch Real-Time instrument. A total of 200 ng of PBMC DNA or 50 μL equivalents of plasma RNA were used in real-time PCR or real-time RT-PCR, respectively. The integrity of the koala PBMC DNA was confirmed by B-actin PCR by using SYBR Green I incorporation during PCR according to Tarlinton et al. (21).
Supplementary Material
Acknowledgments
We thank Dr. Robin Weiss and Dr. Laura Levy for helpful comments; James Nagle and Deborah Kauffman (National Institute of Neurological Disorders and Stroke sequencing facility, National Institutes of Health) for sequencing; Jill Russ, Mickeyas Alemayehu, Joseph Tran, and Laura Li for expert technical assistance; and Dr. Michael M. Garner (Northwest ZooPath) for providing tissue samples. This work was supported by National Institute of Mental Health intramural funding.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. KC779547).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304704110/-/DCSupplemental.
References
- 1.Dewannieux M, et al. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 2006;16(12):1548–1556. doi: 10.1101/gr.5565706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ávila-Arcos MC, et al. One hundred twenty years of koala retrovirus evolution determined from museum skins. Mol Biol Evol. 2013;30(2):299–304. doi: 10.1093/molbev/mss223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fiebig U, Hartmann MG, Bannert N, Kurth R, Denner J. Transspecies transmission of the endogenous koala retrovirus. J Virol. 2006;80(11):5651–5654. doi: 10.1128/JVI.02597-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanger JJ, Bromham LD, McKee JJ, O’Brien TM, Robinson WF. The nucleotide sequence of koala (Phascolarctos cinereus) retrovirus: A novel type C endogenous virus related to Gibbon ape leukemia virus. J Virol. 2000;74(9):4264–4272. doi: 10.1128/jvi.74.9.4264-4272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tarlinton R, Meers J, Hanger J, Young P. Real-time reverse transcriptase PCR for the endogenous koala retrovirus reveals an association between plasma viral load and neoplastic disease in koalas. J Gen Virol. 2005;86(pt 3):783–787. doi: 10.1099/vir.0.80547-0. [DOI] [PubMed] [Google Scholar]
- 6.Miyazawa T, Shojima T, Yoshikawa R, Ohata T. Isolation of koala retroviruses from koalas in Japan. J Vet Med Sci. 2011;73(1):65–70. doi: 10.1292/jvms.10-0250. [DOI] [PubMed] [Google Scholar]
- 7.Bolin LL, Levy LS. Viral determinants of FeLV infection and pathogenesis: Lessons learned from analysis of a natural cohort. Viruses. 2011;3(9):1681–1698. doi: 10.3390/v3091681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oliveira NM, Satija H, Kouwenhoven IA, Eiden MV. Changes in viral protein function that accompany retroviral endogenization. Proc Natl Acad Sci USA. 2007;104(44):17506–17511. doi: 10.1073/pnas.0704313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliveira NM, Farrell KB, Eiden MV. In vitro characterization of a koala retrovirus. J Virol. 2006;80(6):3104–3107. doi: 10.1128/JVI.80.6.3104-3107.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prassolov V, et al. Mus cervicolor murine leukemia virus isolate M813 belongs to a unique receptor interference group. J Virol. 2001;75(10):4490–4498. doi: 10.1128/JVI.75.10.4490-4498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bottger P, Pedersen L. Two highly conserved glutamate residues critical for type III sodium-dependent phosphate transport revealed by uncoupling transport function from retroviral receptor function. J Biol Chem. 2002;277(45):42741–42747. doi: 10.1074/jbc.M207096200. [DOI] [PubMed] [Google Scholar]
- 12.Baron D, Assaraf YG, Drori S, Aronheim A. Disruption of transport activity in a D93H mutant thiamine transporter 1, from a Rogers Syndrome family. Eur J Biochem. 2003;270(22):4469–4477. doi: 10.1046/j.1432-1033.2003.03839.x. [DOI] [PubMed] [Google Scholar]
- 13.Kawakami TG, Sun L, McDowell TS. Natural transmission of gibbon leukemia virus. J Natl Cancer Inst. 1978;61(4):1113–1115. [PubMed] [Google Scholar]
- 14.Kawakami TG, Sun L, McDowell TS. Infectious primate type-C virus shed by healthy gibbons. Nature. 1977;268(5619):448–450. doi: 10.1038/268448a0. [DOI] [PubMed] [Google Scholar]
- 15.Ting YT, Wilson CA, Farrell KB, Chaudry GJ, Eiden MV. Simian sarcoma-associated virus fails to infect Chinese hamster cells despite the presence of functional gibbon ape leukemia virus receptors. J Virol. 1998;72(12):9453–9458. doi: 10.1128/jvi.72.12.9453-9458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parent I, et al. Characterization of a C-type retrovirus isolated from an HIV infected cell line: Complete nucleotide sequence. Arch Virol. 1998;143(6):1077–1092. doi: 10.1007/s007050050357. [DOI] [PubMed] [Google Scholar]
- 17.Mendoza R, Anderson MM, Overbaugh J. A putative thiamine transport protein is a receptor for feline leukemia virus subgroup A. J Virol. 2006;80(7):3378–3385. doi: 10.1128/JVI.80.7.3378-3385.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5(4):276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 19.Logg CR, Baranick BT, Lemp NA, Kasahara N. Adaptive evolution of a tagged chimeric gammaretrovirus: Identification of novel cis-acting elements that modulate splicing. J Mol Biol. 2007;369(5):1214–1229. doi: 10.1016/j.jmb.2007.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lander MR, Chattopadhyay SK. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ectropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J Virol. 1984;52(2):695–698. doi: 10.1128/jvi.52.2.695-698.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarlinton RE, Meers J, Young PR. Retroviral invasion of the koala genome. Nature. 2006;442(7098):79–81. doi: 10.1038/nature04841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.